Abstract
Previously we have shown that interferon (IFN)-α induced apoptosis is predominantly mediated by the upregulation of tumor necrosis factor related apoptosis-inducing ligand (TRAIL) via the caspase-8 pathway. It was also shown that recruitment of mitochondria in IFN-α induced apoptosis involves the cleavage of BH3 interacting domain death agonist (Bid) to truncated Bid (tBid). In the present study, we demonstrate that tBid induced by IFN-α2a activates mitochondrial Bak to trigger the loss of mitochondrial membrane integrity, consequently causing release of apoptosis-inducing factor (AIF) in ovarian cancer cells, OVCAR3. AIF translocates from the mitochondria to the nucleus and induces nuclear fragmentation and cell death. Both a small molecule Bid inhibitor (BI-6C9) or Bid-RNA interference (RNAi) preserved mitochondrial membrane potential, prevented nuclear translocation of AIF, and abrogated IFN-α2a-induced cell death. Cell death induced by tBid was inhibited by AIF-RNAi, indicating that caspase-independent AIF signaling is the main pathway through which Bid mediates cell death. This was further supported by experiments showing that BI-6C9 did not prevent the release of cytochrome c from mitochondria to cytosol, while the release of AIF was prevented. In conclusion, IFN-α2a-induced apoptosis is mediated via the mitochondria-associated pathway involving the cleavage of Bid followed by AIF release that involves Bak activation and translocation of AIF from the mitochondria to the nucleus in OVCAR3 cells.
Keywords: IFN-α, apoptosis, mitochondria, AIF, Bid, Bak
1. Introduction
Interferons (IFNs) are members of a family of cytokines that have antiviral, antiproliferative, and immunomodulatory properties.[1] There are several types of IFNs, each of which interacts with a type-specific receptor complex. Type I IFNs, which include IFN-α, IFN-β, and IFN-ϖ, are ubiquitously induced in mammals by viruses and other factors, (eg. dsRNA) and interact with the IFN-α receptor (IFNAR) subunits 1 and 2.[2] Activated T lymphocytes and NK cells produce the single species of type II IFN (IFN-γ), which interacts with the IFN-γ receptor (IFNGR) subunits 1 and 2. Nearly every cell type expresses receptors for type I IFNs and IFN-γ.[3–5] The recently characterized type III IFNs include IFN-λ1 (IL-29), IFN-λ2 (IL-28A), and IFN-λ3 (IL-28B), which bind to the IFN-λ receptor (IFNLR1) and the IL-10Rβ subunit (IL-10Rβ). All IFNs exhibit species specificity.[2, 6]
The clinical applications of IFN-α are treatment of viral infections such as hepatitis B and C, and treatment of certain malignancies such as malignant melanoma, hairy cell leukemia, Kaposi sarcoma, chronic myelogenous leukemia and renal cell carcinoma (in combination with Avastin). IFN-β has been used in the treatment of relapsing-remitting multiple sclerosis.[3] In contrast, IFN-γ is the sole type 2 IFN, and is predominantly produced by T helper cell type 1 lymphocytes and natural killer cells. IFN-γ is used clinically for the treatment of chronic granulomatous disease and congenital osteopetrosis, but it has been generally unsuccessful as an antitumor agent.[3]
Apoptosis, a form of programmed cell death (PCD), is an essential process for both development and maintenance of tissue homeostasis and was first recognized by Kerr et al.[7] It has been well established that apoptosis in most cells is induced through the activation of either mitochondrial (intrinsic) pathway or death receptor (extrinsic) pathway.[8, 9] Both ways lead to cell shrinkage, membrane blebbing, chromatin condensation and formation of apoptotic bodies.[10] Apoptotic cell death largely proceeds in either a caspase-dependent or caspase-independent manner.[11] Several studies provide evidence that, in response to apoptotic stimuli, mitochondria can release caspase-independent cell death effectors such as cytochrome c, Apoptosis-Inducing Factor (AIF) and endonuclease G (EndoG).[11, 12]
BH3 interacting domain death agonist (Bid), a substrate of caspase-8, is activated in the Fas/Apo-1 (CD95), tumor necrosis factor (TNF)-α, and tumor necrosis factor related apoptosis-inducing ligand (TRAIL) receptor-mediated cell death.[13, 14] Once Bid is cleaved and activated, it is translocated to the mitochondria as tBid and induces the release of proapoptotic proteins such as cytochrome c, AIF and EndoG.[13, 15] The ability of Bid to induce release of cytochrome c, AIF or EndoG is mediated by the proapoptotic Bcl-2 family proteins Bak or Bax, because Bid can facilitate the insertion of Bak or Bax into the mitochondrial membrane to form functional oligomers.[15–17] Double-knockout mouse embryonic fibroblasts (MEFs) (Bak−/− and Bax−/−) are resistant to apoptosis by various agents, and mice deficient in both Bak and Bax survived anti-Fas antibody treatment.[17] There is increasing evidence to suggest the involvement of Bak and Bax in the release of cytochrome c, and it was reported that mutations in the Bax or Bak gene render cells resistant to apoptosis.[18]
Type I IFNs have potent pro-apoptotic activities in cell lines representing various histologic origins, suggesting that the induction of apoptosis is important to their overall physiological functions. IFNs induce apoptosis indirectly by upregulating the expression of death ligands, such as TRAIL and Fas ligand, which activate the extrinsic Fas-associated death domain (FADD)/Caspase-8 signaling pathway.[19–21] IFNs can also modulate mitochondrial outer membrane stability, in part, by activating pro-apoptotic Bcl-2 members Bak and Bax.[22] This leads to release of pro-apoptotic proteins such as cytochrome c, AIF and EndoG and finally results in depolarization of the inner-mitochondrial membrane.[23] Pretreatment with type I IFNs can also sensitize cells to the pro-apoptotic effects of other therapeutics.[24] Although IFN-induced apoptosis is caspase-dependent and involves the mitochondria, the identities of all of the mediators responsible for disruption of the mitochondria during this process are still unknown.[25]
In the present study, we investigated IFN-α2a induced apoptotic signaling in ovarian cancer cells (OVCAR3). We found that IFN-α2a induced apoptosis is mediated via the mitochondria-associated pathway involving the cleavage of Bid followed by AIF release that involves Bak activation and translocation of AIF from the mitochondria to the nucleus in OVCAR3 cells.
2. Material and Methods
2.1. Cell and reagents
OVCAR3 cells were cultured in RPMI 1640 complete medium (Invitrogen, Carlsbad, CA) supplemented with 10% (v/v) heat-inactivated fetal bovine serum, 2mM L-glutamine, 100 units/ml penicillin, and 100mg/ml streptomycin at 37 °C in a humidified 5% CO2 incubator. IFN-α2a was obtained from Hoffman La Roche (Nutley, NJ). The Bid inhibitor BI-6C9 and the inhibitor of apoptosome formation NS3694 were obtained from (Sigma-Aldrich, St Louis, MO). Antibodies purchased were as follows: rabbit polyclonal anti-Bid, caspase-3, caspase-9, poly ADP-ribose polymerase (PARP). apoptotic protease-activating factor-1 (Apaf-1), EndoG, AIF and β-actin (Cell Signaling, Danvers, MA); mouse anti- caspase-8, cytochrome c and Heat shock protein 60 (Hsp60) (BD Biosciences, San Jose, CA); rabbit polyclonal anti-Bax (Millipore, Temecula, CA) and rabbit polyclonal anti-Bak (Epitomics, Inc., Burlingame, CA); rabbit polyclonal anti-Lamin B1(Abcam, Cambridge, MA).
2.2. Antiproliferative assay
6×103 OVCAR3 cells pre well were seeded in 96-well plate and incubated for 24hrs. IFN-α2a was then added for an additional 72 hrs and cell viability was evaluated using 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl- 2H-tetrazolium bromide (MTT) as described previously.[26] Briefly, after treatment as indicated, followed by incubation with MTT (Sigma-Aldrich) for 4 hrs, the stained living cells were solubilized with acidified isopropanol (0.04 N HCI in isopropanol) and the absorbance was measured at 570 nm. Experiments were carried out in triplicate in duplicate plates.
2.3. RNA interference
OVCAR3 cells (1.5×106 in 10cm dish or 6×103/well in 96-well plate) were seeded and incubated for 24hrs in antibiotic-free RPMI 1640 containing 10% FBS and 2 mM L-glutamine. After incubation for 24hrs, cells were transfected with 20 nM RNA interference (RNAi) oligos complexed in Lipofectamine 2000 (Invitrogen) in Opti-MEM media (Invitrogen) for 24 hrs. IFN was then added for an additional 24 hrs. The media was then removed and replaced with RPMI 1640 containing 2% FBS, 2 mM L-glutamine, 50 U/ml penicillin G and 50 mg/ml streptomycin. To determine efficiency of RNAi-mediated gene silencing, cells were harvested and lysates were prepared and analyzed, as described for Western blot analysis. RNAi oligo purchased were as follows: Bid (Oligo ID: 141377 and 141378); Bax (141354 and 141355); Bak (184085 and 184087); Apaf-1 (141235); Cytochrome c (147625); Caspase-9 (189012); AIF (113512 and 113513) from Invitrogen: Apaf-1 (J003456-13); Cytochrome c (J017355-06); Caspase-9 (J003309-05); ENDOG(J004426-08 and J004426-09) from Thermo Fisher Scientific Dharmacon (Lafayette, CO).
2.4. Plasmid and transfection
The plasmid pDsRed2-Bid for expression of DsRed fused to the C-terminus of Bid to allow translocation analyses of full-length Bid and tBid was purchased commercially (Clontech Laboratories, Inc., Mountain View, CA). Plasmid transfections of 3×105 OVCAR3 cells seeded in 6-well plates were performed in antibiotic-free growth medium using FuGENE HD (Roche Diagnostic Corporation; Indianapolis, IN). Controls were treated with 100 ul/ml OPTI-MEM only and vehicle controls with 3ul/ml FuGENE HD.
2.5. Western blot analysis
Cells were harvested as indicated, lysed in mammalian protein extraction (M-PER) reagent lysis buffer (Pierce Chemical, Rockford, IL) containing Halt protease inhibitor and Halt phosphatase inhibitor mixtures (Thermo Fisher Scientific, Waltham, MA) for 1 hour on ice. Protein concentrations were determined by measuring the absorbance at 280 nm, using a Nanodrop spectrophotometer (Nanodrop Technologies, Wilmington, DE), and equal amounts of protein were separated by SDS-PAGE using 10 to 20% Tris-glycine gels, (Invitrogen Corp., Gaithersburg, MD) under reducing conditions, and transferred to nitrocellulose membranes. Membranes were blocked with 5% fat-free milk in PBS containing 0.05% Tween 20 for 1hr and incubated with each antibody overnight at 4°C. Secondary goat anti-mouse and goat anti-rabbit Abs were purchased from Southern Biotechnology Associates (Birmingham, AL). Membranes were developed using the SuperSignal West Femto ECL kit (Thermo Fisher Scientific) and visualized with an LAS-3000 charge-coupled device camera system (Fujifilm Medical System, Stamford, CT).
2.6. Immunostaining and confocal laser scanning microscopy
For immunofluorescence analysis, OVCAR3 cells were seeded in 6-well plates at a density of 2×104 cells/well. Mitochondria were stained with MitoTracker (Invitrogen) according to the manufacturer’s protocol or visualized by CellLight Reagents BacMam 2.0 (Invitrogen). After treatment, cells were fixed with 4% paraformaldehyde (PFA). The cells were permeabilized by exposure for 30 min to 0.1% Triton X-100, and were then placed in blocking solution (3% horse serum) for 30 min. Cells were then exposed to each antibody overnight at 4°C, followed by incubation for 1 hr with Alexa Fluor 488 or 594-labeled goat anti-rabbit IgG for 30 min at RT in the dark. The specificity immunoreactivity was controlled by omission of the primary antibody. Nuclei were counterstained with DAPI (Invitrogen). Images of stained cells were obtained by confocal microscopy (Leica SP5, Leica Microsystems, Exton, PA). All data were processed with Leica LAS AF software (Version 2.2.1, Leica Microsystems).
2.7. Flow cytometry for cell death and mitochondrial membrane potential (ΔΨm) measurements
Phosphatidylserine exposure on the outer layer of the plasma membrane was detected using the MEHBCYTO Apoptosis Kit (MBL, Nagoya, Japan) according to the manufacturer’s instructions. In brief, 2×105 cells were pelleted following treatment and washed in PBS (pH 7.4). Next, the cells were resuspended in 100 ul of binding buffer containing annexin V-FITC and propidium iodide (PI) for 15 min at RT in the dark. Prior to flow cytometry analysis, 400 ul of binding buffer were added to the cells.
For ΔΨm determination, Mitochondrial Membrane Potential Detection Kit (Stratagene, La Jolla, CA, USA) was used according to the manufacturer’s instructions. Briefly, 1×106 cells were pelleted following treatment, resuspended in 500ul of 1× JC-1 reagent and incubated in a humidified 5% CO2 incubator at 37 °C for 15 min. Next, cells were pelleted and washed in 2 ml of 1× assay buffer. This step was repeated. Finally, cells were pelleted, resuspended in 1×assay buffer, and analyzed by flow cytometry. Analysis was performed using FlowJo software (Tree Star, Ashland, OR).
2.8. Determination of Bax and Bak oligomerization and activation
For detection of Bax and Bak oligomerization, 1×106 cells were harvested, washed, and resuspended in 80 ul of 100 mM EDTA/PBS (pH 7.4), incubated with 1mM cross-linking agent bismaleimidohexane (BMH) for 30 min at room temperature, quenched with 100 mM dithiothreitol (Sigma-Aldrich) for 15 min, pelleted, and processed for Western blotting.
For detection of activated Bax and Bak by flow cytometry, 1×106 cells were washed in PBS (pH 7.4) and fixed in 400 ul of 0.25% paraformaldehyde for 5 min, subsequently washed two times with 2% FBS in PBS (pH 7.4), and incubated in 50 ul of staining buffer (2% FBS and 100 ug/ml digitonin in PBS (pH 7.4)) with a conformation-specific mouse monoclonal antibody against Bax and Bak for 30 min at room temperature. Then, cells were washed and resuspended in 50 ul of staining buffer containing 0.25ug Alexa Fluor 488-labeled goat anti-rabbit IgG for 30 min in the dark. Cells were washed again and analyzed by flow cytometry. Analysis and histogram overlays were performed using FlowJo software.
2.9. Mitochondria and nuclei isolation
Mitochondria were isolated using the Qproteome Mitochondrial Isolation Kit (Qiagen, Valencia, CA, USA) following the manufacturer’s instructions. Briefly, 2 ×107 cells were washed in ice-cold PBS (pH 7.4) and incubated in lysis buffer with protease inhibitor (supplied with the kit) for 10 min on an orbital shaker at 4°C. Samples were centrifuged for 10 min at 1000 g at 4°C, and the pellet was resuspended in a disruption buffer with protease inhibitor (supplied with the kit). Cells were disrupted on ice by 10 rounds of aspiration and ejection through a 21-gauge needle. The lysate was centrifuged for 10 min at 1000 g at 4°C, and the resulting supernatant was centrifuged at 6000 g at 4°C to pellet mitochondria. Mitochondria were washed with ice-cold storage buffer (supplied with the kit) and centrifuged for 20 min at 6000 g at 4°C. Cytoplasmic and nuclear proteins were isolated using Qproteome nuclear protein kit (Qiagen) following the manufacturer’s instructions. Briefly, 1×107cells Cells were harvested and lysed in lysis buffer. Twenty-five microliters of detergent solution was added to the cell suspension and then centrifuged. The supernatant was retained as the cytosolic fraction. The remaining pellet was incubated with extraction buffer by gentle agitation at 4°C for 30 min and was then centrifuged. The supernatant was retained as the nuclear fraction. Samples were snap-frozen on dry ice and stored at −80°C.
2.10. Statistical analysis
Statistical comparisons of mean values were done by one-way ANOVA. Statistical analysis was performed using JMP version 8.0 (SAS Institute, Cary, NC, USA). A P value of less than 0.05 was considered to be statistically significant.
3. Results
3.1. Bid inhibition attenuates growth-inhibiting effect of IFN-α2a and inhibits IFN-α2a-induced apoptosis
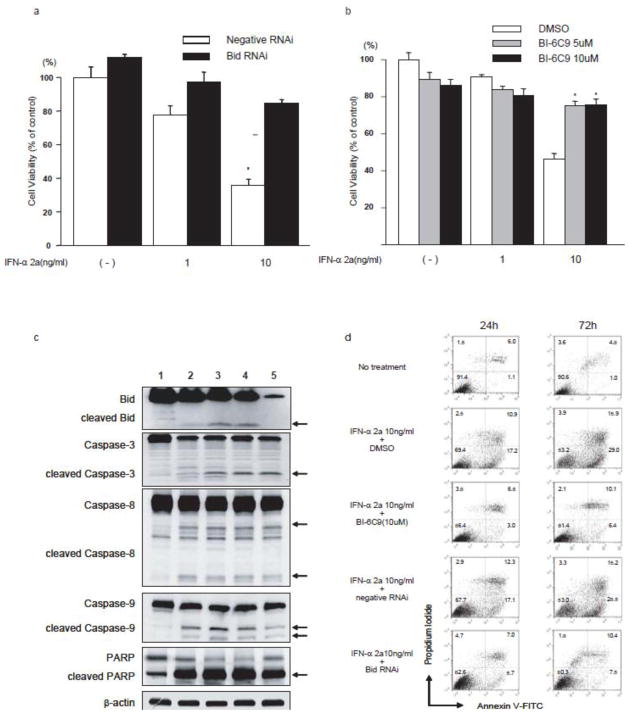
To examine whether Bid inhibition attenuates the growth-inhibiting effect of IFN-α2a in OVCAR3 cells, an antiproliferative assay was performed. As was previously shown, Bid-RNAi significantly reduced growth-inhibiting effect of IFN-α2a.[14] In addition to the RNAi approach, the specific Bid inhibitor BI-6C9 was applied to confirm the essential role of Bid in the growth-inhibiting effect by IFN-α2a. After cells were incubated with Bid-RNAi or specific Bid inhibitor BI-6C9 at 37°C for 24hrs, IFN-α2a was added at the concentration of 1, or 10 ng/ml, followed by incubation for 72hrs. As shown in Fig. 1a and 1b, Bid-RNAi or BI-6C9 significantly reduced the growth-inhibiting effect of IFN-α2a. (Supplementary Figure 1, BID-RNAi in the absence of IFN-α2a).
Fig. 1.
Bid inhibition protects OVCAR3 cells against IFN-α2a-induced apoptosis (a–b)Bid-RNAi or Bid inhibitor, BI-6C9 significantly attenuated IFN-α2a-induced cell death as determined by antiproliferative assay. (c)Cleavage of caspases, PARP and Bid were assessed by immunoblot analysis after treatment for 24 hrs. (d)Flow cytometry analysis of the apoptotic population after treatment of IFN-α2a with BI-6C9 or Bid-RNAi for 24 and 72 hrs. Cells were stained with Annexin V and PI. Quadrants are defined as live (lower left), early apoptotic (lower right), late apoptotic (upper right), and necrotic (upper left). Numbers refer to the percentage of cells in each quadrant. (a–b)* p < 0.05 in comparison with no treatment. Lanes: (c)1, No treatment; 2, IFN-α2a: 1ng/ml; 3, IFN-α2a: 10ng/ml; 4, IFN-α2a: 10ng/ml + BI-6C9 (10uM); 5, IFN-α2a: 10ng/ml+ Bid RNAi.
Western blot analyses for Bid, caspase-3, -8, -9, and PARP were carried out to determine whether IFN-α2a-induced apoptotsis through cleavage of Bid, caspase-3, -8, -9, and PARP. As shown in Fig. 1c, western blot analysis revealed that Bid had been cleaved to tBid after incubation with IFN-α2a, confirming our previous results.[14] Similarly, cleavage of caspase-3, -8, -9, and PARP, which are mediators of apoptosis, were observed after treatment with IFN-α2a. Bid-RNAi inhibited cleavage of Bid, however BI-6C9 did not, which is consistent with the mechanism of BI-6C9.[27]
Flow cytometry analysis by annexin V-FITC and PI staining showed that Bid inhibition drastically reduced IFN-α2a induced apoptosis at both 24 and 72hr. As shown in Fig. 1d, apoptosis was seen in 28.1% of the cells at 24hr, 44.9% at 72hr with IFN-α2a and DMSO, 29.4% at 24hr, 43.7% at 72hr with IFN-α2a and negative-RNAi, 9.8% at 24hr, 16.5% at 72hr with IFN-α2a and BI-6C9 and 12.7% at 24hr, 17.9% at 72hr with IFN-α2a and Bid-RNAi.
3.2. IFN-α2a induces Bid-dependent ΔΨm depolarization
We next examined whether IFN-α2a induces Bid-dependent ΔΨm depolarization. As shown in Fig. 2a, IFN-α2a treatment caused a significant loss of mitochondrial membrane potential. The Bid inhibiton with BI-6C9 or Bid-RNAi (shown previously[14]) significantly prevented IFN-α2a-induced breakdown of the mitochondrial membrane potential, suggesting that Bid inhibiton significantly prevented Bid translocation to the mitochondria and consequently prevented IFN-α2a-induced collapse of the mitochondrial membrane potential in OVCAR3 cells.[14]
Fig. 2.
The Bid inhibition prevents IFN-α2a-induced mitochondrial depolarization. (a)OVCAR3 cells were treated with IFN-α2a for 24 and 72 hrs after pre-treatment with BI-6C9 or Bid-RNAi. Cells were collected and JC-1 was used to measure ΔΨm by flow cytometry as described in Materials and methods. The ratios (%) for upper right and lower right quadrants in the dot plots indicate the ratios of live cells and apoptotic cells, respectively (Live cells whose ΔΨm is intact; apoptotic cells whose ΔΨm is collapsed). Experiments were done three times with essentially similar results. (b)BI-6C9 or Bid-RNAi prevented the accumulation of Bid at the mitochondria and retained the homogeneous distribution of Bid in the cytosol. Cells expressing a Bid-DsRed fusion protein and Mito-GFP show colocalization of mitochondrial staining imaged by confocal microscopy exposed to IFN-α2a for 24hrs after pretreatment of BI-6C9 or Bid-RNAi for 24hrs. Bid (red), mitochondria (green), nuclear staining (DAPI, blue), and merge of the three patterns are shown The yellow/orange color represents the merge of red and green.
In addition, prevention of Bid translocation to the mitochondria was confirmed by immunostaining with confocal laser scanning microscopy. Bid inhibition-related reduction of IFN-α2a cytotoxicity was associated with a lack of Bid translocation to the mitochondria in experiments using pDsRed2-Bid vector and Mitotracker Green staining. Bid translocation to the mitochondria cause the merge of DsRed2-Bid and Mitotracker Green color, resulting in appearance of yellow/orange. As shown in Fig. 2b, Bid translocation to the mitochondria was prevented with BI-6C9 or Bid-RNAi, indicating that Bid translocation is essential for IFN-α2a induced apoptosis.
3.3. IFN-α2a-induced apoptosis is accompanied by Bak oligomerization
We then examined the extent to which Bax and Bak had undergone oligomerization in response to IFN-α2a, using two different techniques. We first investigated whether Bax and Bak underwent oligomerization using the cross-linking agent BMH. As shown in Fig. 3a, cross-linked Bak protein complexes were observed in OVCAR3 cells treated with 10 ng/ml of IFN-α2a for 24hrs, however, cross-linked Bax protein complexes were not. Bak oligomerization was prevented with BI-6C9. This result was confirmed with an active conformation-specific monoclonal Bak antibody and flow cytometry analysis in which activation causes a shift to the right of the resulting histogram and bar graph valued histogram, as shown in Fig. 3b. These findings were consistent with results obtained using immunostaining with confocal laser scanning microscopy. As shown in Fig. 3c, IFN-α2a treatment induced cross-linked Bak protein complexes, which was prevented with BI-6C9 or Bid-RNAi.
Fig. 3.
IFN-α2a-induced cell death is associated with Bak activation, mitochondrial translocation, and membrane insertion. OVCAR3 cells were cultured with IFN-α2a with or without BI-6C9 for 24hrs and (a)processed for determination of Bak oligomerization by immunoblot analysis, Lanes:1, No treatment; 2, IFN-α2a: 10ng/ml; 3, IFN-α2a: 10ng/ml + BI-6C9 (10uM). or (b)Bak activation by flow cytometry analysis. Histograms show Bak-associated fluorescence in vehicle-treated (dotted line), IFN-α2a treated (black closed) IFN-α2a with BI-6C9 treated (gray closed), and numbers refer to the increase in mean fluorescence intensity.
Right bar graphs represent the value of histograms for Bax and Bak activation. (c)Bax and Bak translocation to Mitochondria imaged by confocal microscopy in OVCAR3 cells exposed to IFN-α2a for 24hrs after pretreatment with BI-6C9 or Bid-RNAi for 24hrs. Bax or Bak (both green), mitochondria (red), nuclear staining (DAPI, blue), and merge of the three patterns are shown. IFN-α2a treated cells show translocation of Bak to mitochondria. BI-6C9 or Bid-RNAi prevented the translocation of Bak to mitochondria. (d)RNAi targeted for Bax and Bak were transfected into OVCAR3 cells. After 24 hrs incubation, the cells were exposed to IFN-α2a of indicated concentration and incubated for another 72 hrs. The knockdown of Bak reduced growth-inhibiting effect of IFN-α2a, whereas knockdown of Bax did not.*= p < 0.05 in comparison (b)without BI-6C9 treatment or (d)with negative RNAi treatment.
Furthermore, as shown in Fig. 3d, even though Bax RNAi knocked down Bax and Bak RNAi knocked down Bak, only Bak-RNAi significantly attenuated IFN-α2a induced cytotoxicity in OVCAR3 cells (Supplementary Fig. 2). Collectively, these findings suggest that IFN-α2a-induced cytotoxicity is predominantly mediated by Bid translocation to the mitochondria, which promote Bak controlled ΔΨm.
3.4. Apoptosome formation is not required for IFN-α2a-induced Bid-mediated cell death
As shown in Fig. 4a–c, there were no significant differences in cell viability with IFN-α2a and the described gene-specific RNAi, although each RNAi reduced its respective protein level. In the experiment with the diarylurea compound, NS3694 which modulates apoptosome formation, there was no significant difference in cell viability as well.[28] In addition, as shown in Fig. 4d, Flow cytometry analysis by annexin V-FITC and PI staining showed that there were no significant difference in IFN-α2a-induced apoptosis with each apoptosome related RNAi (Caspase-9, cytochrome c and Apaf-1) or NS3694.
Fig. 4.
Apoptosome formation is not required in IFN-α2a-induced Bid mediated cell death. (a)Downregulation of Apaf-1, cytochrome c and caspase-9 protein levels by their respective RNAi was verified by immunoblot analysis. OVCAR3 cells transfected with Apaf-1, cytochrome c or caspase-9-RNAi were harvested and lysed for immunoblot analysis. (b)Apaf-1, cytochrome c or caspase-9 did not attenuate IFN-α2a-induced cell death as determined by antiproliferative assay. (c)NS3694, inhibitor of apoptosome formation, did not attenuate IFN-α2a-induced cell death as determined by antiproliferative assay (d) Flow cytometry analysis of the apoptotic population after treatment of IFN-α2a with or without gene-specific RNAi for 24 and 72 hrs. Cells were stained with Annexin V and PI. Quadrants are defined as live (lower left), early apoptotic (lower right), late apoptotic (upper right), and necrotic (upper left). Numbers refer to the percentage of cells in each quadrant. (e)Cytochrome c and casapase-9 release from mitochondria imaged by confocal microscopy in OVCAR3 cells exposed to IFN-α2a for 24hrs after pretreatment of BI-6C9 or NS3694 for 24hrs. Cytochrome c (green), caspase-9 (red), nuclear staining (DAPI, blue), and merge of the three patterns are shown. IFN-α2a treated cells show colocalization of cytochrome c and caspase-9. BI-6C9 decreased colocalization of cytochrome c and caspase-9, meaning inhibition of apoptosome formation, while there was no significant change of colocalization of cytochrome c and caspase-9 with NS3694. Lanes: (a)1, No treatment; 2, negative RNAi 20nM; 3, negative RNAi 100nM; 4, target RNAi 20nM; 5, target RNAi 100nM.
These findings were confirmed with immunostaining using confocal laser scanning microscopy. As shown in Fig. 4e, IFN-α2a-induced apoptosome formation, which was shown as areas of co-localization in the merge panels as yellow. Apoptosome formation was prevented with NS3694, while BI-6C9 did not inhibit apoptosome formation.
3.5. AIF functions as a major pro-apoptotic factor in the IFN-α2a-induced Bid-mediated cell death through Bak oligomerization and activation
The potential role of a pro-apoptotic inducing protein in the execution of cell death downstream of Bid-mediated mitochondrial depolarization through Bak oligomerization, was further addressed by a gene silencing approach. As shown in Fig. 5a–c, AIF-RNAi reduced protein levels and significantly attenuated IFN-α2a-induced cell death in OVCAR3 cells, while cytochrome c-RNAi or EndoG-RNAi did not (Supplementary Fig. 3). Flow cytometry analysis by annexin V-FITC and PI staining showed that AIF-RNAi drastically reduced IFN-α2a-induced apoptosis at both 24 and 72hr, while neither cytochrome c nor EndoG-RNAi did not. As shown in Fig. 5d, apoptosis was induced in 26.8% at 24hr, 48.4% at 72hr with IFN-α2a and negative RNAi, 12.6% at 24hr, 13.5% at 72hr with IFN-α2a and AIF-RNAi, 21.5% at 24hr, 42.5% at 72hr with IFN-α2a and cytochrome c-RNAi and 20.1% at 24hr, 41.9% at 72hr with IFN-α2a and EndoG-RNAi. These results suggested that AIF but not cytochrome c and EndoG is responsible for IFN-α2a-induced Bid-mediated caspase-independent execution of cell death.
Fig. 5.
AIF involves IFN-α2a-induced Bid-mediated cell death. The RNAi gene silencing of (a)AIF did reduced growth-inhibiting effect of IFN-α2a, whereas knockdown of (b)EndoG or (c)cytochrome c did not. (d) Flow cytometry analysis of the apoptotic population after treatment of IFN-α2a with or without gene-specific RNAi for 24 and 72 hrs. Cells were stained with Annexin V and PI. Quadrants are defined as live (lower left), early apoptotic (lower right), late apoptotic (upper right), and necrotic (upper left). Numbers refer to the percentage of cells in each quadrant. * p < 0.05 in comparison with negative RNAi treatment.
3.6. Inhibition of Bid prevents the release of AIF from mitochondria by IFN-α2a
Finally, we confirmed the locations of AIF, cytochrome c and EndoG, which are integral inner mitochondrial membrane proteins that are often affected by mitochondrial transmembrane potential. Cells were treated with IFN-α2a for 24hrs, followed by isolation using the Qproteome cell compartment kit. As shown in Fig. 6a, AIF transferred from mitochondria to the nucleus after exposure to 10 ng/ml IFN-α2a for 24 hrs and AIF release from mitochondria to cytosol was inhibited with BI-6C9. Meanwhile, cytochrome c release was not inhibited with BI-6C9. EndoG was not released from mitochondria with IFN-α2a treatment. Hsp60, a mitochondria-associated protein, and lamin B1, a nuclear specific protein, were used to show the purity of the cellular fractions.
Fig. 6.
AIF release is associated with IFN-α2a-induced Bid-mediated cell death. (a)IFN-α2a induces AIF translocation into the cytosol and nuclei from the mitochondria. Cells were cultured with IFN-α2a with or without BI-6C9 for 24hrs. Cellular fractions (cytosolic, mitochondrial and nuclear) were then isolated using the Qproteome cell compartment kit. As controls to confirm the fidelity of the fractionation procedure, the expression of Hsp60, lamin B1, and β-actin was also determined. Experiments were done three times with essentially similar results. (b–d)AIF, cytochromec and EndoG release from mitochondria imaged by confocal microscopy in OVCAR3 cells exposed to IFN-α2a for 24hrs AIF (green), cytochrome c (green) and EndoG (green) and nuclear staining (DAPI, blue), and merge of the two patterns are shown. IFN-α2a treated cells show release and extensive nuclear fragmentation (white arrows) of AIF and cytochrome c, whereas EndoG was not released. BI-6C9 or Bid-RNAi decreased release of AIF from mitochondria to cytosol, however unaffected release of cytochrome c. Lanes: (a)1, No treatment; 2, IFN-α2a: 10ng/ml; 3, IFN-α2a: 10ng/ml + BI-6C9 (10uM).
These findings were confirmed with immunostaining using confocal laser scanning microscopy. As shown in Fig. 6b–d, IFN-α2a-induced AIF release from mitochondria to the nucleus, which was prevented with BI-6C9 or Bid-RNAi. Unlike AIF, cytochrome c release was not inhibited with BI-6C9 or Bid-RNAi. Consistent with the protein expression assay, EndoG was not released.
4. Discussion
IFN-α is among the few cytokines to have an established role as an effective antitumor agent in some malignant diseases, notably several lymphoid malignancies.[29] The mechanism by which IFN exerts its antitumor activity is still ill-defined, but we and others have shown that IFN can be a potent inducer of apoptosis in some malignant cells, forming an attractive hypothesis for how it may act in cancer.[25, 30, 31] Favorable results have also been observed using treatment protocols where IFN has been combined with different cytostatic agents.[21, 32, 33] IFN may be predicted to have a different mode of activating the apoptosis machinery in comparison with chemotherapy; this may explain the positive effects observed in such combination protocols.
The present study demonstrates a key role for the proapoptotic Bcl-2 family protein Bid in mitochondrial membrane permeabilization and the subsequent nuclear translocation of AIF in OVCAR3 cells exposed to IFN-α2a. IFN-α2a-induced translocation of Bid to mitochondria, followed by mitochondrial membrane depolarization, Bak activation and AIF release to the nucleus. The pharmacological Bid inhibitor BI-6C9 and RNAi-mediated gene silencing, as shown previously both significantly preventing mitochondrial Bid translocation, migration of AIF to the nucleus, and, hence, IFN-α2a-induced cell death.
In order to optimize the use of IFN-α2a in the clinic, a better understanding of the molecular background to IFN-α2a-induced apoptosis is warranted. This prompted us to define molecular events leading to IFN-α-induced apoptosis. Our results show that IFN-α2a significantly disrupts the mitochondrial membrane potential (Fig. 2a), thus confirming previous work.[14] We thereby presume that mitochondria play an important role in IFN-α-induced apoptosis. Bcl-2 proteins are the main regulators of the mitochondrial-mediated death pathway by either suppressing or promoting mitochondrial changes in regulating the release of key proteins from mitochondria. The Bcl-2 family determines the cellular susceptibility of PCD.[34] How the Bcl-2 family proteins control the release of mitochondrial proteins is still unclear. However, it is clear that multidomain proapoptotic proteins such as Bax and Bak participate in this process.[35] Bax activation or Bak conformational changes lead to the formation of a mitochondrial pore that facilitates the release of mitochondrial proapoptotic proteins. Therefore, in apoptotic PCD, the combined genetic ablation of both Bax and Bak confer resistance to apoptosis.
In the present study, upon treatment of IFN-α2a, Bak protein was activated but not Bax (Fig. 3a–c), which suggested that Bak is necessary in the action of IFN-α2a on OVCAR3 cells. It is very interesting that these data well supported the hypothesis that Bax and Bak may perform different functions in different cell death models,[36] although Bax and Bak are similar in protein structure and have similar function in apoptosis.[37] However, the regulation of Bak and Bax activation is distinct, as is probably their role in IFN-α2a-induced apoptosis. Using immunostaining with confocal laser scanning microscopy, we also demonstrated that Bak activation occurred in the apoptotic response, prior to the AIF release and loss of ΔΨm, whereas Bax activation was not observed (Fig. 3c). Cells can probably die in response to IFN-α2a without activating Bax since there was no significant difference in the cell viability assay with Bax-RNAi. Furthermore, small molecule Bid inhibitor BI-6C9 or Bid-RNAi prevented the translocation of Bak to mitochondria, but did not influence the translocation of Bax in Western blotting assay (Fig. 3a) or flow cytometric analysis (Fig. 3b), implying that fluctuations in Bax levels are of less importance. In order to delineate the importance of Bak in IFN-α2a-induced apoptosis, it would have been helpful to use Bak−/− fibroblasts. However, in contrast to many established tumor cell lines and primary tumor cells, normal mouse embryo fibroblasts are not sensitive to mouse IFN-induced apoptosis and therefore these experiments cannot be preformed. [22]
Another important aspect of the present study addresses the role of AIF in mediating caspase-independent cell death downstream of Bid activation through Bak activation. The Bak/AIF pathway is a major caspase-independent apoptotic cascade.[11, 38] AIF is a flavoprotein with both oxidoreductase and DNA-binding domains but no intrinsic DNase activity.[11, 39, 40] AIF can promote cell survival or induce apoptosis. In mitochondria, AIF is involved in cellular respiration and is essential for cell survival.[39, 40]
Other pro-apoptotic factors such as cytochrome c, Smac/Diablo, Omi/HtrA2 and EndoG, which may execute cell death through caspase-dependent or caspase-independent mechanisms, are released with mitochondrial membrane permeabilization as well as AIF. For example, formation of the apoptosome by cytochrome c, Apaf-1, and procaspase-9 and downstream catalytic activation of caspase-3, -6, and -7 are widely established mechanisms of caspase-dependent cell death execution downstream of mitochondrial damage in many cells, including cancer cells. Here, we detected caspase-3 activation downstream of Bid, suggesting that this pathway may also play a role in ovarian cancer cell death in the present model (Fig. 1c). Caspase-3 inhibition alone, however, did not prevent IFN-α-induced cell death [14]. In addition, although our data strongly suggest a major role for caspase-independent cell death downstream of mitochondrial membrane permeabilization, apoptosome inhibition did not prevent IFN-α-induced cell death either (Fig. 4a–e). The release of AIF from mitochondria and its translocation to the nucleus is one of the events observed in OVCAR3 cells after exposure to IFN-α2a. Using Western blot analysis and immunofluoresence staining, we showed mitochondrial release of AIF and translocation to the nucleus, which occurred after the onset of IFN-α2a exposure. Although cytochrome c was released from mitochondria, Bid inhibitor BI-6C9 did not prevent the release of cytochrome c from mitochondria. On the other hand, EndoG was not released after IFN-α2a exposure (Fig. 6a–d). AIF translocation into the nucleus released from the mitochondria exposes the role of AIF downstream of mitochondrial demise to mediate cell death independent of other established execution mechanisms of intrinsic cell death pathways, such as apoptosome formation and activation of caspase-3. Furthermore, AIF siRNA significantly attenuated IFN-α2a induced cell death in OVCAR3 cells, whereas cytochrome c or EndoG-RNAi did not (Fig. 5a–d), clearly demonstrating that Bid cytoxicity is predominantly mediated by mitochondrial AIF release and hence caspase-independent cell death pathways.
In summary, our present study demonstrates that IFN–α2a induced apoptosis is mediated via the mitochondria-associated pathway which is Bid regulated, followed by AIF release that involves Bak activation and translocation to mitochondria in ovarian cancer cells, OVCAR3. Further studies using different types of cancer cells, will help to clarify the signaling events underlying the induction of apoptosis by IFN-α, and thereby provide insight for the development of improved ways to use IFN-α in the treatment of cancer and to identify new targets for cancer therapy.
Supplementary Material
Highlights.
The growth-inhibiting effect of IFN-α2a is due to IFN-α2a induces Bid-dependent ΔΨm depolarization.
IFN-α2a-induced apoptosis is accompanied by Bak oligomerization
Apoptosome formation is not required for IFN-α2a-induced Bid-mediated cell death
AIF functions as a major pro-apoptotic factor in the IFN-α2a-induced Bid-mediated cell death through Bak oligomerization and activation
Inhibition of Bid prevents the release of AIF from mitochondria by IFN-α2a
Acknowledgments
We are grateful to members of Dr. Owen Schwartz’s laboratory, particularly S. Ganesan and S. Becker for their help with the confocal microscopy. We also thank the other members of the Kathryn C. Zoon’s laboratory, particularly Dr. Takaya Tsuno for many helpful discussions and suggestions. This work was supported by the Intramural Research Program of the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Abbreviations
- IFN
Interferon
- Bid
BH3 interacting domain death agonist
- AIF
Apoptosis-inducing factor
- RNAi
RNA interference
- PCD
programmed cell death
- EndoG
endonuclease G
- MEFs
mouse embryonic fibroblasts
- FADD
Fas-associated death domain
- PARP
Poly ADP-ribose polymerase
- Apaf-1
apoptotic protease-activating factor-1
- MTT
3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl- 2H-tetrazolium bromide)
- TNF
Tumor necrosis factor
- TRAIL
Tumor necrosis factor related apoptosis-inducing ligand
- PFA
Paraformaldehyde
- PI
Propidium iodide
- ΔΨm
Mitochondrial membrane potential
- BMH
Bismaleimidohexane
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pestka S, Langer JA, Zoon KC, Samuel CE. Interferons and their actions. Annual review of biochemistry. 1987;56:727–777. doi: 10.1146/annurev.bi.56.070187.003455. [DOI] [PubMed] [Google Scholar]
- 2.Takaoka A, Yanai H. Interferon signalling network in innate defence. Cellular microbiology. 2006;8:907–922. doi: 10.1111/j.1462-5822.2006.00716.x. [DOI] [PubMed] [Google Scholar]
- 3.Maher SG, Romero-Weaver AL, Scarzello AJ, Gamero AM. Interferon: cellular executioner or white knight? Current medicinal chemistry. 2007;14:1279–1289. doi: 10.2174/092986707780597907. [DOI] [PubMed] [Google Scholar]
- 4.Bach EA, Aguet M, Schreiber RD. The IFN gamma receptor: a paradigm for cytokine receptor signaling. Annual review of immunology. 1997;15:563–591. doi: 10.1146/annurev.immunol.15.1.563. [DOI] [PubMed] [Google Scholar]
- 5.Baron S, Finbloom J, Horowitz J, Bekisz J, Morrow A, Zhao T, Fey S, Schmeisser H, Balinsky C, Miyake K, Clark C, Zoon K. Near Eradication of Clinically Relevant Concentrations of Human Tumor Cells by Interferon-Activated Monocytes In Vitro. J Interferon Cytokine Res. doi: 10.1089/jir.2010.0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annual review of biochemistry. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 7.Kerr JF, Wyllie AH, Currie AR. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. British journal of cancer. 1972;26:239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Criollo A, Galluzzi L, Maiuri MC, Tasdemir E, Lavandero S, Kroemer G. Mitochondrial control of cell death induced by hyperosmotic stress. Apoptosis. 2007;12:3–18. doi: 10.1007/s10495-006-0328-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wajant H. The Fas signaling pathway: more than a paradigm. Science (New York, NY. 2002;296:1635–1636. doi: 10.1126/science.1071553. [DOI] [PubMed] [Google Scholar]
- 10.Zamzami N, Susin SA, Marchetti P, Hirsch T, Gomez-Monterrey I, Castedo M, Kroemer G. Mitochondrial control of nuclear apoptosis. The Journal of experimental medicine. 1996;183:1533–1544. doi: 10.1084/jem.183.4.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cregan SP, Dawson VL, Slack RS. Role of AIF in caspase-dependent and caspase-independent cell death. Oncogene. 2004;23:2785–2796. doi: 10.1038/sj.onc.1207517. [DOI] [PubMed] [Google Scholar]
- 12.Susin SA, Lorenzo HK, Zamzami N, Marzo I, Snow BE, Brothers GM, Mangion J, Jacotot E, Costantini P, Loeffler M, Larochette N, Goodlett DR, Aebersold R, Siderovski DP, Penninger JM, Kroemer G. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature. 1999;397:441–446. doi: 10.1038/17135. [DOI] [PubMed] [Google Scholar]
- 13.Li H, Zhu H, Xu CJ, Yuan J. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell. 1998;94:491–501. doi: 10.1016/s0092-8674(00)81590-1. [DOI] [PubMed] [Google Scholar]
- 14.Tsuno T, Mejido J, Zhao T, Phillips T, Zoon KC. Bid Is a Key Factor for Eliciting the Antiproliferative Activity of IFN-alpha Mediated by TRAIL. J Immunother. 2012;35:23–31. doi: 10.1097/CJI.0b013e3181ad4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim TH, Zhao Y, Barber MJ, Kuharsky DK, Yin XM. Bid-induced cytochrome c release is mediated by a pathway independent of mitochondrial permeability transition pore and Bax. The Journal of biological chemistry. 2000;275:39474–39481. doi: 10.1074/jbc.M003370200. [DOI] [PubMed] [Google Scholar]
- 16.Wei MC, Lindsten T, Mootha VK, Weiler S, Gross A, Ashiya M, Thompson CB, Korsmeyer SJ. tBID, a membrane-targeted death ligand, oligomerizes BAK to release cytochrome c. Genes & development. 2000;14:2060–2071. [PMC free article] [PubMed] [Google Scholar]
- 17.Wei MC, Zong WX, Cheng EH, Lindsten T, Panoutsakopoulou V, Ross AJ, Roth KA, MacGregor GR, Thompson CB, Korsmeyer SJ. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science (New York, NY. 2001;292:727–730. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang GQ, Gastman BR, Wieckowski E, Goldstein LA, Gambotto A, Kim TH, Fang B, Rabinovitz A, Yin XM, Rabinowich H. A role for mitochondrial Bak in apoptotic response to anticancer drugs. The Journal of biological chemistry. 2001;276:34307–34317. doi: 10.1074/jbc.M103526200. [DOI] [PubMed] [Google Scholar]
- 19.Balachandran S, Kim CN, Yeh WC, Mak TW, Bhalla K, Barber GN. Activation of the dsRNA-dependent protein kinase, PKR, induces apoptosis through FADD-mediated death signaling. The EMBO journal. 1998;17:6888–6902. doi: 10.1093/emboj/17.23.6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balachandran S, Roberts PC, Kipperman T, Bhalla KN, Compans RW, Archer DR, Barber GN. Alpha/beta interferons potentiate virus-induced apoptosis through activation of the FADD/Caspase-8 death signaling pathway. Journal of virology. 2000;74:1513–1523. doi: 10.1128/jvi.74.3.1513-1523.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsuno T, Mejido J, Zhao T, Schmeisser H, Morrow A, Zoon KC. IRF9 is a key factor for eliciting the antiproliferative activity of IFN-alpha. J Immunother. 2009;32:803–816. doi: 10.1097/CJI.0b013e3181ad4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Panaretakis T, Pokrovskaja K, Shoshan MC, Grander D. Interferon-alpha-induced apoptosis in U266 cells is associated with activation of the proapoptotic Bcl-2 family members Bak and Bax. Oncogene. 2003;22:4543–4556. doi: 10.1038/sj.onc.1206503. [DOI] [PubMed] [Google Scholar]
- 23.Yanase N, Ohshima K, Ikegami H, Mizuguchi J. Cytochrome c release, mitochondrial membrane depolarization, caspase-3 activation, and Bax-alpha cleavage during IFN-alpha-induced apoptosis in Daudi B lymphoma cells. J Interferon Cytokine Res. 2000;20:1121–1129. doi: 10.1089/107999000750053799. [DOI] [PubMed] [Google Scholar]
- 24.Chawla-Sarkar M, Leaman DW, Jacobs BS, Borden EC. IFN-beta pretreatment sensitizes human melanoma cells to TRAIL/Apo2 ligand-induced apoptosis. J Immunol. 2002;169:847–855. doi: 10.4049/jimmunol.169.2.847. [DOI] [PubMed] [Google Scholar]
- 25.Thyrell L, Erickson S, Zhivotovsky B, Pokrovskaja K, Sangfelt O, Castro J, Einhorn S, Grander D. Mechanisms of Interferon-alpha induced apoptosis in malignant cells. Oncogene. 2002;21:1251–1262. doi: 10.1038/sj.onc.1205179. [DOI] [PubMed] [Google Scholar]
- 26.Miyake K, Shimada M, Nishioka M, Sugimoto K, Batmunkh E, Uto Y, Nagasawa H, Hori H. The novel hypoxic cell radiosensitizer, TX-1877 has antitumor activity through suppression of angiogenesis and inhibits liver metastasis on xenograft model of pancreatic cancer. Cancer letters. 2008;272:325–335. doi: 10.1016/j.canlet.2008.07.020. [DOI] [PubMed] [Google Scholar]
- 27.Becattini B, Sareth S, Zhai D, Crowell KJ, Leone M, Reed JC, Pellecchia M. Targeting apoptosis via chemical design: inhibition of bid-induced cell death by small organic molecules. Chemistry & biology. 2004;11:1107–1117. doi: 10.1016/j.chembiol.2004.05.022. [DOI] [PubMed] [Google Scholar]
- 28.Lademann U, Cain K, Gyrd-Hansen M, Brown D, Peters D, Jaattela M. Diarylurea compounds inhibit caspase activation by preventing the formation of the active 700-kilodalton apoptosome complex. Molecular and cellular biology. 2003;23:7829–7837. doi: 10.1128/MCB.23.21.7829-7837.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strander H, Einhorn S. Interferons and the tumor cell. Biotherapy (Dordrecht, Netherlands) 1996;8:213–218. doi: 10.1007/BF01877207. [DOI] [PubMed] [Google Scholar]
- 30.Sangfelt O, Erickson S, Castro J, Heiden T, Einhorn S, Grander D. Induction of apoptosis and inhibition of cell growth are independent responses to interferon-alpha in hematopoietic cell lines. Cell Growth Differ. 1997;8:343–352. [PubMed] [Google Scholar]
- 31.Miyake K, Imura S, Nishioka M, Batmunkh E, Sugimoto K, Ohmoto Y, Shimada M. Serum evaluation of soluble interferon-alpha/beta receptor and high-sensitivity C-reactive protein for diagnosis of the patients with gastrointestinal and hepatobiliary-pancreatic cancer. Cytokine. 49:251–255. doi: 10.1016/j.cyto.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 32.Smalley RV, Weller E, Hawkins MJ, Oken MM, O’Connell MJ, Haase-Statz S, Borden EC. Final analysis of the ECOG I-COPA trial (E6484) in patients with non-Hodgkin’s lymphoma treated with interferon alfa (IFN-alpha2a) plus an anthracycline-based induction regimen. Leukemia. 2001;15:1118–1122. doi: 10.1038/sj.leu.2402161. [DOI] [PubMed] [Google Scholar]
- 33.Miyake K, Tsuchida K, Sugino H, Imura S, Morine Y, Fujii M, Shimada M. Combination therapy of human pancreatic cancer implanted in nude mice by oral fluoropyrimidine anticancer agent (S-1) with interferon-alpha. Cancer chemotherapy and pharmacology. 2007;59:113–126. doi: 10.1007/s00280-006-0250-5. [DOI] [PubMed] [Google Scholar]
- 34.Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 35.Martinou JC, Green DR. Breaking the mitochondrial barrier. Nature reviews. 2001;2:63–67. doi: 10.1038/35048069. [DOI] [PubMed] [Google Scholar]
- 36.Cartron PF, Juin P, Oliver L, Martin S, Meflah K, Vallette FM. Nonredundant role of Bax and Bak in Bid-mediated apoptosis. Molecular and cellular biology. 2003;23:4701–4712. doi: 10.1128/MCB.23.13.4701-4712.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scorrano L, Korsmeyer SJ. Mechanisms of cytochrome c release by proapoptotic BCL-2 family members. Biochemical and biophysical research communications. 2003;304:437–444. doi: 10.1016/s0006-291x(03)00615-6. [DOI] [PubMed] [Google Scholar]
- 38.Daugas E, Susin SA, Zamzami N, Ferri KF, Irinopoulou T, Larochette N, Prevost MC, Leber B, Andrews D, Penninger J, Kroemer G. Mitochondrio-nuclear translocation of AIF in apoptosis and necrosis. Faseb J. 2000;14:729–739. [PubMed] [Google Scholar]
- 39.Daugas E, Nochy D, Ravagnan L, Loeffler M, Susin SA, Zamzami N, Kroemer G. Apoptosis-inducing factor (AIF): a ubiquitous mitochondrial oxidoreductase involved in apoptosis. FEBS letters. 2000;476:118–123. doi: 10.1016/s0014-5793(00)01731-2. [DOI] [PubMed] [Google Scholar]
- 40.Lipton SA, Bossy-Wetzel E. Dueling activities of AIF in cell death versus survival: DNA binding and redox activity. Cell. 2002;111:147–150. doi: 10.1016/s0092-8674(02)01046-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.