Abstract
The assembly of a highly-parallel force spectroscopy tool requires careful placement of single-molecule targets on the substrate and the deliberate manipulation of a multitude of force probes. Since the probe must approach the target biomolecule for covalent attachment, while avoiding irreversible adhesion to the substrate, the use of the polymer microsphere as force probes to create the tethered bead array poses a problem. Therefore, the interactions between the force probe and the surface must be repulsive at very short distances (< 5 nm) and attractive at long distances. To achieve this balance, the chemistry of the substrate, force probe, and solution must be tailored to control the probe-surface interactions. In addition to an appropriately designed chemistry, it is necessary to control the surface density of the target molecule in order to ensure that only one molecule is interrogated by a single force probe. We used gold-thiol chemistry to control both the substrate’s surface chemistry and the spacing of the studied molecules, through a competitive binding of the thiol-terminated DNA and an inert thiol forming a blocking layer. For our single molecule array, we modeled the forces between the probe and the substrate using DLVO theory and measured their magnitude and direction with colloidal probe microscopy. The practicality of each system was tested using a probe binding assay to evaluate the proportion of the beads remaining adhered to the surface after application of force. We have translated the results specific for our system to general guiding principles for preparation of tethered bead arrays and demonstrated the ability of this system to produce a high yield of active force spectroscopy probes in a microwell substrate. This study outlines the characteristics of the chemistry needed to create such a force spectroscopy array.
Keywords: DLVO theory, single molecule, force spectroscopy, non-specific binding, surface attachment, DNA array
Introduction
In the push towards personalized medicine, low cost DNA sequencing is essential.1,2 To lower the final cost of sequencing, we have proposed a sequencing strategy that uses force spectroscopy to detect the conformational changes of DNA in the course of a stepwise ligation. This approach to sequencing requires a controlled ligation of short DNA strands of known composition to the DNA strand in question followed by mechanical stretching of individual molecules to determine the success or failure of the ligation (as in sequencing by synthesis).3 Since the detection step is non-optical (i.e. does not use fluorescent labels), natural enzymes and DNA oligos can be used, thus, simplifying the process and ultimately lowering the cost.2 The principle component of such sequencing device is an array (random or organized) of single molecules to be sequenced.4 The array is constructed by attaching DNA strands to microscopic force probes (e.g. magnetic-fluorescent microspheres) and, at the opposite termini, to the surface of a fluid cell, which is capable of exchanging reagents. A force field (either magnetic or dielectrophoretic) is then applied to pull on the probe and generate force versus extension curves for multiple DNA molecules in parallel.5–8 Wide field microscopy enables simultaneous observation of multiple force probes.3,9 The observation of the stretching behavior on such a large sampling scale can give insight into sequence effects in DNA, polyelectrolyte stretching behavior,10 as well as DNA-protein interactions.11
The interpretation of the recorded force-extension curves of the DNA, by determining the proportion of hybridized DNA, gives a direct indication to the outcome of each ligation step. This technique would have practical value only if thousands of the DNA strands can be manipulated at once.12 Implementation of such a force-spectroscopy-on-a-chip device, working on numerous single molecules simultaneously, presents several challenges. To create a highly parallel, high throughput force spectroscopy method, our design must include an array of probes that can withstand the shear force of multiple exchanges of reagent solutions.13–17 The chosen conjugation procedure should result in a high yield of active probes, i.e. much greater number of beads bound to a single DNA molecule than number of beads with multiple tethers or bound non-specifically to the surface of the chip.5 While biomolecular conjugation (e.g. using biotin-avidin,18,19 dig-antidig pairs,20 or both11) is the most common approach used with magnetic (or optical) tweezers, we investigated use of covalent chemistry to ensure robust attachment that can withstand the strong shear, allow the system remain stable for days, and resist disintegration under high pulling forces. The study of such forces and systems can lead to a better understanding of dispersion and adhesion of particles, as well as bacterial adhesion to surfaces.21 Here, we report on tuning the surface chemistry and reaction conditions for bead attachment to fabricate a high efficiency, high density single-molecule-bead array suitable for use in experiments on single-molecule manipulation with tethered beads.
Approach
In developing procedures to create a surface with properties suitable for magnetic tweezers3,6,22–25 or dielectrophoretic (DEP) tweezers,9,26 one must (i) control the surface density of DNA oligomers, (ii) enable robust attachment of oligos to both the surface of the solid support and the force probe (i.e. microscopic bead); and (iii) ensure that bead attachment occurs only via a terminal group of the DNA, whereas the surrounding area resists non-specific adhesion of the bead. To implement these features, we have developed a series of chemical modifications to the probe, DNA, and substrate (Figure 1 and Figure S1, Supporting Information). To attach the DNA oligomers as well as add an organic blocking layer, we have chosen to use self-assembled monolayers (SAMs) of thiols on gold due to easy, reproducible chemistry and flexibility in the choice of the ω-functional group.
Figure 1.
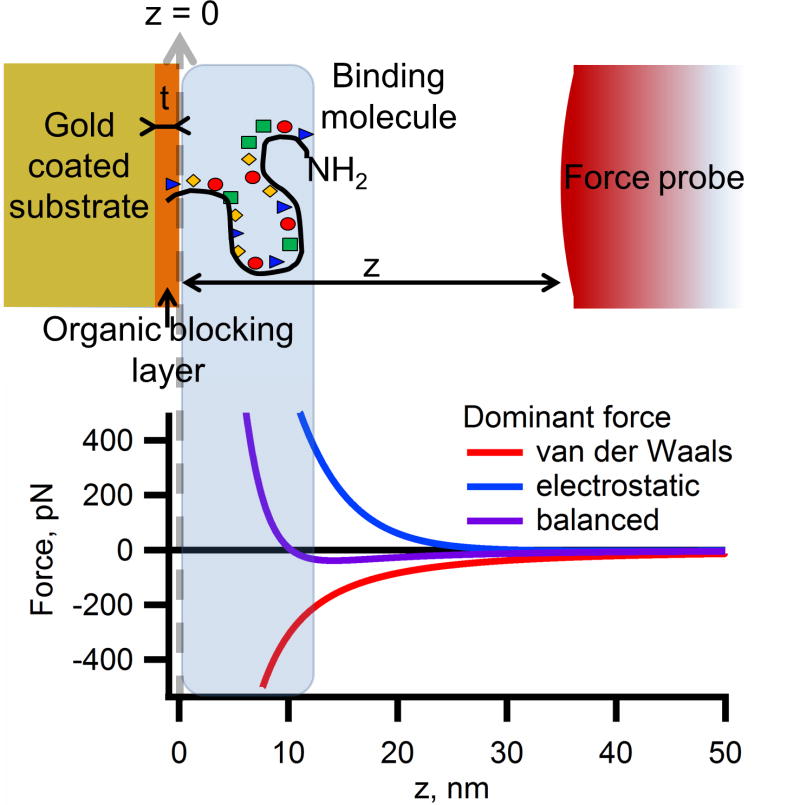
Representation of a force profile for a probe (microsphere) approaching a surface. To bind to the DNA molecule, the probe must interact with the amine terminal group by passing into the area represented by the blue-gray shaded box. If the probe comes in contact with the surface, it will most likely adhere strongly, so it must not pass completely through the region indicated by a shaded box. The graph shows three representative force-distance profiles as the probe approaches the surface. The repulsive electrostatic force can be too strong and prevent the probe from approaching the target molecule. Conversely, the van der Waals attractive forces can be too strong and cause the probe to stick irreversibly to the surface. If the forces become balanced, the probe will be attracted to the point where it may bind to the DNA, but is repelled at a very close proximity to the surface.
The most critical aspect of assembling the fluid cell for magnetic or DEP tweezers is designing appropriate surface and solution chemistry that allows the bead to bind to the DNA, but not the substrate. 27–29 Typical force-distance profile of a probe approaching a surface can be described by an extended Derjaguin and Landau, Verwey and Overbeek (DLVO) theory that treats overall interaction of a particle and a surface as the sum of the electrostatic, van der Waals, and steric forces.30–33 The combination of the repulsive forces (steric and electrostatic), which decay exponentially with separation, and attractive forces (van der Waals), which decreases as an inverse square function of separation, creates a distinct set of force distance profiles (Figure 1). These forces compete with each other and either push the probe away from the surface or pull the probe into contact with the surface. If the van der Waals force is too strong then the force will be attractive at all distances, however, if the repulsive electrostatic force dominates, then the probe will repel from the surface at all separations. By tuning the magnitude of the component forces in the total interaction, the probe-surface separation where the forces become repulsive can be optimized so that the probe is allowed to approach the surface and bind covalently to the DNA molecule without nonspecifically binding to the surface.
To determine the conditions for such a discerning binding, we tuned the magnitude and range of both the attractive and repulsive interactions between the probe and the surface by changing the pH or ionic strength of the solution, introducing surfactant, as well as by varying the makeup, functionality, and the thickness of the blocking layer. In this paper, we outline a strategy to optimize the binding efficiency of microscopic probes to immobilized DNA. This strategy is a result of modeling of forces in this system, direct measurements of bead-surface forces with atomic force microscopy, and, ultimately, empirical optimization to create a selective binding scheme with probes that bind to the DNA and do not interact with the substrate.
Experimental methods
Expanded details for procedures outlined below are given in section S2 of the supporting information.
Substrate preparation
Gold coated glass slides were reacted with a 10 mM solution of thiol blocking molecule [mercaptopropionic acid (MPA), mercaptohexanoic acid (MHA), mercaptoundecanoic acid (MUA), mercaptohexadecanoic acid (MHDA), or mercaptoundecyl tetraethylene glycol (MutEG)] prepared either in a pH 7.4 phosphate buffer with 1 M sodium chloride or in ethanol, depending on the solubility of the molecule.
End modification of DNA
The procedure to end modify genomic DNA is described elsewhere.29 We created a 142-mer strand of end modified DNA by starting with a 30-mer strand of DNA complement to the 30 bases in the middle of the desired sequence. We purchased 71-mers of the DNA each with half the desired sequence, one strand having an amine group on the 5′ end and the other strand having a thiol group on the 3′ end. The two ends were hybridized with the 30-mer and subsequently ligated together to form the full 142-mer with the proper functionality at the 3′ and 5′ termini.
Probe Preparation
Microspheres were prepared by emulsification of polymer solutions that contained ~10 nm diameter magnetite nanoparticles (20 mass %) and organic fluorescent dye.3 For DEP tweezers, the probes were fabricated without magnetite.
Probe mounting
The magnetite-doped fluorescent probes were attached to a tipless triangular silicon nitride cantilever (Nanoworld PNP-TR-TL, approximate spring constant of 30 pN/nm) using Asylum Research MFP-3D atomic force microscope (AFM) mounted on an inverted optical microscope.34 To do so, the suspension of probes in water was first dried on a clean glass microscope slide alongside microdroplets of epoxy. The cantilever was lowered onto a microdroplet by manually adjusting the micropositioner, then lifted and lowered onto a colloidal probe. The probe was lifted off the substrate and the epoxy cured leaving a cantilever with a spherical probe.
AFM measurements
Force spectroscopy was conducted using an open fluid cell setup of the Asylum Research MFP-3D Bio AFM. Buffer solutions were pipetted directly onto the sample.
Probe binding assays
The probe binding assays were performed in samples comprising a 4 mm thick polydimethylsiloxane mask with 4 mm diameter wells on top of a gold substrate. In each well the desired blocking layer was deposited. The probes were allowed to settle on the substrate in solution for 15 min and a fluorescent image of the probes was taken. A permanent magnet was placed on top of the wells, producing an approximate force of 200 pN to detach the probes from the surface, and a fluorescent image was taken after five minutes. The amount of probes before and after the applied magnet was counted.
Mapping of fluorescent labeled DNA
A short DNA oligo modified with a fluorescent dye at the 3′-end and a thiol at the 5′-end was used as a test molecule to determine DNA surface binding. The DNA was added to a gold substrate with MutEG in both (i) a competitive fashion followed by a second addition of MutEG and (ii) a two-step fashion where the DNA was added alone followed by a subsequent addition of MutEG. Fluorescent images of the individual DNA molecules were taken and quantified.
Force spectroscopy demonstration
To conduct the force spectroscopy experiments, we fabricated a microwell array as described elsewhere.9,35,36 A glass substrate was coated with a thick gold layer, followed by a layer of 4 μm deep, 7 μm diameter wells, with the gold on their floors etched away and finally topped with a thin gold layer. DNA was added to the surface in a 1 HM solution with 10 mM phosphate buffer (pH 8) with 100 mM NaCl followed by a surface modification with MutEG (chosen as the best blocker from the probe binding assays) followed by the conjugation of the probes. The two step binding process was chosen to maximize the probe-DNA binding efficiency. The probes were reacted to the amine groups at the free end of the DNA. The whole chip was integrated into a fluid cell and an AC electric field was applied to conduct stretching of single DNA oligomers with the DEP tweezers.
Results and discussion
Control of DNA surface density
To conduct DEP tweezers it is important to bind the probe to the surface using only one DNA molecule. We have determined (see summary given in the section S3 of the SI) that it is best to use a competitive binding process (where the DNA and MutEG are reacted at the same time) when it is important to space the DNA and a two-step binding process when it is important to ensure a high yield of the reaction with the probes.
Theoretical modeling of force-distance profiles
To gain insight into the behavior of the forces between a surface and a probe, we used extended DLVO theory to model how various parameters affect the magnitude and character of respective interactions.30,31,34,37–42 This theory states that the total force acting upon the system is the sum to the electrostatic (Fel), steric (Fst), and van der Waals (FvdW) forces,
| (1) |
The total force is now easily broken down into the three components that can all be independently manipulated. In practice, one can adjust system variables (e.g. solution or surface composition) to obtain the desired force-distance profile (Figure 1).
By comparing the electrostatic, van der Waals, and steric forces we can begin to build a profile and qualitatively determine the effects of changing the parameters. The forces can be written as
| (2) |
| (3) |
and
| (4) |
We define all the parameters involved as the separation between the probe and substrate (z), probe radius (R), the surface potential of both the substrate and the probe (Ψs and Ψp), and the inverse Debye length of the medium (κ), ε0 is the permittivity of free space and εr is the relative permittivity of the medium, Hopm is the Hamaker constant between the organic blocking layer and the probe across water, Hspm is the Hamaker constant between the probe and the gold metal substrate across the blocking layer and water, γ is the grafting density of the polymer chains protruding from the surface (in 1/m2), and Rg is the radius of gyration of the polymer.
This set of equations makes it clear that all three types of forces have identical dependence on R, thus differences in the probe size we will only be manifested as a change in the magnitude of the forces acting on the probe and not the character of the force-distance profile. 43,44 Therefore, small probes will be easier to remove from the surface and less likely to adhere permanently. On the other hand, the magnetic (or DEP)45 force acting on a super-paramagnetic (or dielectric) particle scales as R3, thus resulting in a substantial loss in magnitude of the pulling force for small R, making a reduction of the radius of the particles impractical.
In our modeling we are able to ignore the effects of gravity, since the force of gravity of polymer probes of this size is under 1 pN, which is well below the magnitude of the electrostatic and van der Waals forces. Additionally from these equations it becomes clear that the behavior of the probe substrate interactions become hard to predict at distances shorter than 5 nm due to the steric interactions. The radius of gyration of the solvated polymers that may hang off the probe are on the order of 1–2 nm making this steric interaction very short range.34,40,46,47 It is also very difficult to characterize the grafting density of these molecules on the probe making the magnitude of these forces unknown. Furthermore, this analysis does not take into account the roughness of the substrate and the probes, which can cause deviations in the forces at short ranges (see Figure S3). For these reasons, we choose to study the probe-substrate interaction forces only at distances greater than 5 nm.
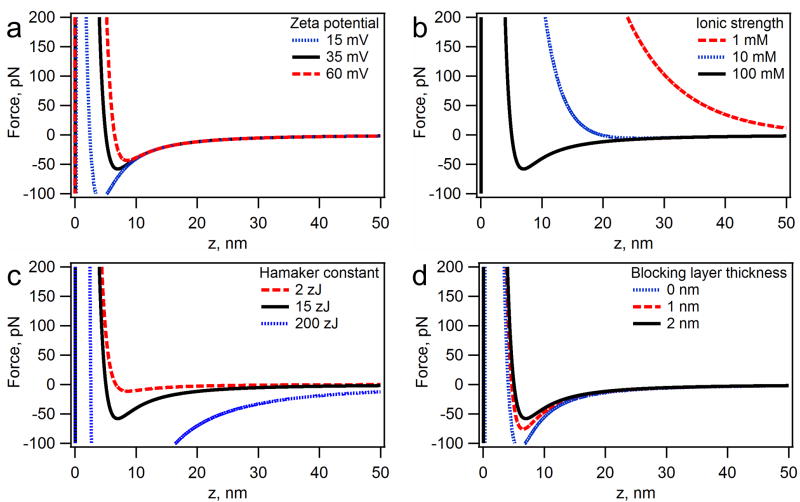
In our system, the thickness of the blocking layer, the Debye length (by way of the solution ionic strength), the probe and substrate potentials,30,31,48 and the Hamaker constants49–59 can be targeted to change the system properties the easiest. The theoretical responses of tuning these parameters can be seen in Figure 2. Each of the systems is relatively easy to tune. The ionic strength can be changed by altering the concentration of sodium chloride, the blocking layer thickness is tuned by changing the number of carbons in the chain of the thiol blocking moiety, the surface potential are a function of the pH or the nature of the surface functional groups, and the Hamaker constants depend on the materials used. Of the four properties, the Hamaker constant is the most difficult to change since significant alterations in its value would require drastic changes in the substrate and/or probe, thus requiring a different scheme for surface chemistry.
Figure 2.
The effects of tuning the parameters of the system based on DLVO interactions between a polymer bead (4.5 μm diameter) and an organic layer (thiol monolayer) on gold in water. The zero distance is set at the organic layer/water interface. The black curves in each graph represent a standard set of conditions, where ionic strength = 100 mM, zeta potential = 35 mV, blocking layer thickness = 2 nm, the blocking layer-water-probe Hamaker constant = 2 zJ, and the substrate-water-probe Hamaker constant of 15 zJ. The effects of zeta potential (a), ionic strength (b), substrate-water-probe Hamaker constant (c), and the blocking layer thickness (d) were evaluated.
Using this model, we can predict the trends that will emerge by varying experimental parameters for the system (Figure 2).60,61 Each individual parameter changes both the magnitude of the force-distance curve and its shape. The repulsive electrostatic forces acting on the probe will grow with the increase in the surface charge (e.g. the zeta potential), while the attractive van der Waals forces will remain constant. Therefore, the probe can be detached from the surface using weaker pulling forces (Figure 2a). In addition, the increased potential pushes the stable position of the probe (zero force) further away from the surface. Of all the system parameters at our disposal, the tuning of the ionic strength is by far the most dramatic. With a low ionic strength, the probes are unable to approach the surface, whereas a high ionic strength will completely screen the electrostatic forces giving the system minimum repulsion (Figure 2b). If we alter the substrate component of the Hamaker constant, we can decrease or increase the depth of the attractive well without changing the distance of the force minimum (maximum adhesive force) from the surface (Figure 2c). The blocking layer thickness has a similar effect, however, the task of tuning the layer thickness is much easier than changing the substrate material (Figure 2d).
The theoretical force-distance profiles indicate that in order to bind a microscopic force probe to the ssDNA strand with Rg=12 nm (200 base long) the ionic strength must be greater than 100 mM to allow the probe to approach the surface.53,62 The remaining parameters control mainly the magnitude of the force minimum (maximum adhesive force). Since our magnetic/DEP tweezers setup is capable of applying forces of up to 20–40 pN,3,9 it is important that the maximum adhesion force is below that value under most experimental conditions. Convenient surface chemistry dictated our choice of gold as a substrate for magnetic/DEP tweezers and effectively fixed the value of the Hamaker constant. Therefore, we must use a system with a thick blocking layer as well as a mid-range surface potential (~ −40 mV) to allow the probe to approach the DNA without adhering to the surface. Using this theoretical description, we systematically tested multiple conditions to determine a range of the above parameters where the specific binding of the probes is optimal, thus creating a general scheme for our dielectrophoretic/magnetic tweezers platform.
Tailoring the forces on the probes as they approach the surface
To verify the predictions of the theory and to obtain a complete quantitative description of the binding of the probes to the surface, we used two independent experimental approaches: (i) force spectroscopy with a probe mounted to an AFM cantilever and (ii) wide-area multi-probe binding assays. We conducted these experiments under various conditions (altering ionic strength, pH, and blocking layer thickness) and measured the force-distance profiles as well as the probability of binding to the surface. The purpose of conducting both sets of experiments is to directly measure the forces at long range (>5 nm) with high accuracy using AFM and to quantify the practical effects of the short range forces using the adhesion assays. The wide-area binding assays consist of two steps. First, the probes settle by gravity on the surface and an image of their spatial distribution and population is recorded. A magnet is then applied and a second image is taken to determine the fraction of probes remaining on the surface after application of force (Figure S4).
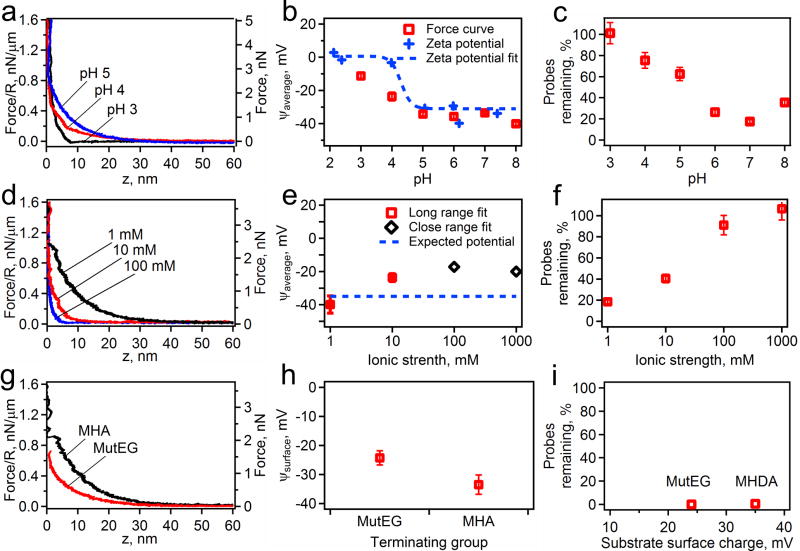
Figure 3 shows a summary of all the results analyzing the effects of altering surface and solution chemistry on the binding of probes to a substrate. The effects on the pH are summarized in Figure 3a–c. It is obvious from all the force curves that as the pH increases the system becomes increasingly repulsive. The fits of these force curves to equation for electrostatic force (equation 2 and 3) results in the trend for surface potential that mimics that of the zeta potential measurements of the probes. The probe binding assays corroborate the results that the probes become repulsive only at high pH (pH>5). Variations in the ionic strength of the solution achieve a similar effect. The force-distance curves and their fits (Figure 3d, e) show that the system becomes highly repulsive with a decreasing ionic strength. The binding assays (Figure 3f) show that to prevent significant binding the probes need to be placed into solutions of an ionic strength less than 100 mM. By altering the surface chemistry we can alter the binding properties of the system (Figure 3g–i). The force curves show MutEG to be less repulsive than MHA coated surfaces, with the surface potentials fits giving 25 mV for MutEG and 35 mV for MHA SAMs. At low ionic strength, however, neither configuration has many probes remaining. From this set of data we can determine that to bring the probe within 12 nm of the surface without it adhering would require a pH greater than 6, and ionic strength of around 100 mM, and a surface potential above 25 mV.
Figure 3.
The effects of changing the pH with a constant ionic strength of 1 mM on an MHA surface, (a, b, c), the solutions ionic strength with a constant pH of 7 on an MHA surface, (d, e, f), and the surface chemistry with constant pH of 7 and ionic strength of 1 mM (g, h, i) on the electrostatic forces. The measured force profiles are shown as an AFM cantilever lowers a probe towards a surface under various conditions (a, d, g). The pH (a), ionic strength (d), and the surface chemistry (g) are labeled on each plot. From these force profiles, electrostatic properties, such as Debye length and the surface potential of the probe and substrate, are measured by fitting to DLVO theory (b, e, h). In b, the data is plotted with the measured zeta potential of the probes only (blue crosses) and shown with the fit of the zeta potential (blue dotted line) to expected pH titration curve. Finally the fraction of remaining probes on the surface after a magnet is applied is shown for all the conditions measured (c, f, i).
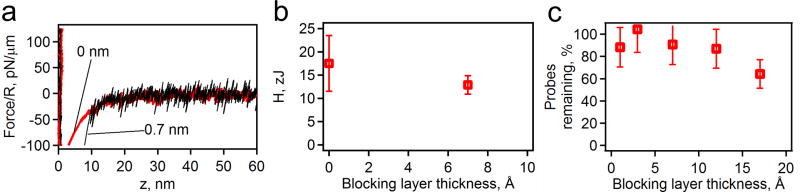
To understand effect of the attractive van der Waals interactions, we repeated the analysis of the approach curves from AFM force spectroscopy, as well as the probe binding assays (Figure 4). The solution had a pH of 3 to neutralize the acid groups on the probe and the surface, thus nearly eliminating all long-ranged repulsion in the system.54 This analysis has given three pieces of information. By taking the force curves on a system with and without a blocking layer (MHA), we were able to measure Hamaker constants of approximately 15 zJ for each set (Figure 4a and b), in close agreement with the theoretical calculations. The probe binding experiments conducted in a solution with a pH 3 and 100 mM ionic strength yielded close to 100 % binding for all blocking layers from zero to eleven-carbon chains (Figure 4c). It is not until the MHDA (a sixteen-carbon chain) is used as a blocking layer that a reduction from the almost complete adhesion is observed (consistent with our theoretical modeling). This consideration argues that, in the case of SAMs on Au surface chemistry, to produce a chip for use in massively parallel force spectroscopy, it is very important to have a long blocking layer to reduce the non-specific binding.
Figure 4.
AFM measurements of van der Waals interactions. (a) Representative force curves for the bead approaching substrates with blocking layers of varying thicknesses. The Hamaker constants for the probe attraction with the gold for each thickness (b) are calculated from the data in (a). The results of the probe binding assays (c) show the levels of adhesion for the different thicknesses in a series of carboxyl-terminated SAMs (formed by HS-(CH2)n-COOH, n=2, 5, 10, 15) at pH 3.
To study the effects of the surfactant (in this case Tween 20) on the system, we chose biological conditions close in value to the optimal conditions determined earlier: a MutEG surface and a solution with pH=7.4 and an ionic strength of 174 mM. To test the effects of the surfactant, we added Tween 20 up to 0.1 % v/v to these high ionic strength solutions and conducted force spectroscopy. The approach curves were essentially identical for all concentrations of Tween 20 (Figure S5a), including solutions containing no surfactant. The differences of the system with and without surfactant were found in the retraction curves, where we observed an average adhesion of 63±11 pN and 130±50 pN for curves recorded with and without surfactant. The presence of Tween 20 reduces the adhesion force of a polymer microsphere from a value well in excess of the maximum force attained with our electromagnet (F~ 40 pN) to one that is comparable to the force used to pull on these probes.
Using the same surfactant conditions we were able to see a decrease in the adhesion of probes to the surface in the binding assays. Increase in the concentration of Tween 20 show a dramatic drop (from 100 to 5–15 %) in the number of probes left on the surface after a magnet is applied and should greatly increase the number of active probes in our system (up to 95 % of the total population). These data indicate that to maximize the number of active probes in a bead-based platform (necessary for accurate and simultaneous force spectroscopy on a high number of molecules), it is highly advantageous to include non-ionic surfactant when one needs to work with solutions of high ionic strength (~100 mM).
Conducting highly parallel force spectroscopy
With the parameters governing bead-surface interactions fully quantified, we designed a procedure for binding force probes to single biomolecules in a force spectroscopy device3,59 with a reasonably high yield. Here, our strategy was to use DEP tweezers in a microwell format to conduct force spectroscopy on multiple DNA. The principles of this technique are described in details elsewhere.9 For an optimized device, one has to confine the probes to the microwell as well as center them. Overall, this setup requires a three-step approach: (i) introduction and positioning of the probes in the microwells, (ii) reaction of the probes with the DNA molecules, and (iii) the actual process of conducting a force spectroscopy experiment. Each of these steps requires slightly different solution conditions to achieve those goals.
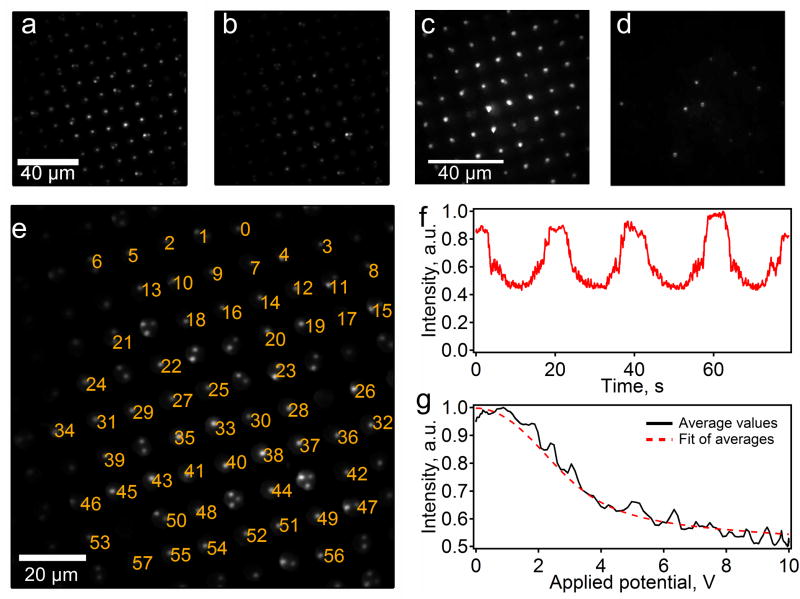
To attach the DNA to the surface and probe we determined the optimal conditions for immobilization of thiol-modified DNA (an exact procedure is described in the SI). The basic procedure includes blocking the surface with MutEG, reacting the probes under an ionic strength of 24 mM, and releasing the probes with a DEP force and low ionic strength conditions. We achieved the desired mode of binding of the beads to the DNA, as can be seen in Figure 5. We performed the force spectroscopy experiments on the DEP chips with and without DNA present. As expected, with no force present, we see the probes close to the surface in both samples (Figure 5a and c). When a DEP force is applied to the chip, regions with DNA show the force probes moving away from the surface without being pulled beyond the range of the evanescent field penetration (Figure 5b). The regions without DNA show the probes moving very far from the surface indicating that they are not tethered by DNA and, thus, become unobservable (Figure 5d).
Figure 5.
Force spectroscopy of ssDNA conducted using DEP tweezers on a system optimized using design principles based on results from this paper. Force probes tethered to DNA in wells before (a) and after (b) they were exposed to a DEP force. Free force probes settled in wells before (c) and after (d) the DEP force was applied. The brightness of the probes (a–d) varies due to Gaussian profile of the illumination laser beam and variations in the bead sizes. (e) A single frame from a movie of the probes, showing the indexing of the force probes for data analysis. (f) A representative intensity vs. voltage plot for one probe. (g) Fits of the data from the plot in part f.
To conduct the force spectroscopy using a positive DEP force, we applied 5 kHz electric field across the electrodes separated by a 100 μm gasket and modulated its amplitude between 0 and 10 V. This AC potential causes the probes to pull away from the substrate at a small force and we are able to track the movement and intensity of the individual probes using a custom bead tracking program. This program tracked 57 probes (out of total 85 occupied locations) labeled from 0 to 56 in Figure 5e and generated potential versus probe intensity curves for each bead (a representative curve is shown in Figure 5f and all the curves are compiled in the section S4 of supporting information). The majority of the non-indexed positions contain multiple probes, which appear to be bound and conducting some form of force spectroscopy, but not suitable for interpretation. Of the indexed force probes, we can extract usable force curves from 53 (or 93 %) of the single probes (see Figure S6). These 53 probes conducting force spectroscopy in one field of view is an order of magnitude improvement over the <5 active probes we have observed in our early experiments prior to optimization of surface and solution chemistry.3 We must note that some of the force curves do not comply with the shape expected for single molecule stretching. This aberration is most likely a result of multiple DNA molecules binding to a single probe and is the subject of ongoing experimentation to improve control of the spacing between DNA molecules on the surface of the substrate to avoid multiple tethering.
Since the stretching of single stranded DNA is well known, we can use these force curves both to calibrate forces acting on each probe and to determine if the force spectra are reasonable. To analyze the data, we averaged the normalized intensity-versus-applied-potential curves from one of the probes (Figure 5f) and fitted the result (Figure 5g) to the equation used in our previous work on magnetic tweezers:3,25
| (5) |
This equation is a statement of the force-extension relationship for the extensible freely jointed chain model,63–66 assuming exponentially decaying evanescent wave excitation and scaling of the DEP force with applied voltage as F=SfV2. The terms in the fitting equation include the total number of bases in the DNA strand (N=142), the contour length of a single base (lss=0.58 nm), the Kuhn length of the ssDNA (bss=1.4 nm,) and the segment elasticity of the ssDNA (Kss=905 pN,). The curve was fitted for the initial intensity of the probe, I0, the force sensitivity factor, Sf, and the penetration depth of the evanescent field, d. Upon fitting the equation to our data (Figure 5g), we determine that these parameters are equal to 0.953, 0.56 pN/V2, and 137 nm, respectively. The initial intensity is close to 1 since the curves were initially normalized to maximum intensity observed for a given probe. The penetration depth was found to be 137 nm, which is in the center of the typical range of values we get for our system.3 To determine the plausibility of the value obtained for the sensitivity factor, we determined a maximum force applied to the system by multiplying Sf by the maximum voltage squared. The estimate yields a maximum force of 56 pN, which is also a typical value for this setup.9 This analysis shows that one can conduct force spectroscopy in a parallel manner using finely tuned system to assemble high density tethered bead arrays.
Conclusions
We have successfully designed a force spectroscopy platform capable of performing parallel force spectroscopy on almost 80 % of probes bound to ssDNA. We used a general method for spacing thiol-modified DNA by means of a competitive binding with a thiol forming a blocking layer. The creation and tuning of a system for highly parallel force spectroscopy requires a non-trivial balance of many factors involved with the surface and solution chemistry of the system.
We were able to establish the guidelines for tuning the solution and surface chemistry of the flow cell by modeling the forces acting on the probes using DLVO theory, as well as AFM measurements and bead binding experiments. We have determined that the optimal conditions for specific binding include a long blocking layer (approximately 2 nm in thickness), a solution ionic strength in the range of 10–100 mM, and a surface potential of the surface and probe approaching 35 mV. The use of a non-ionic surfactant at low concentration (0.01–0.1 %) is critical to prevent adhesion of probes after they contact the surface. In our experiments, we used either addition of NaCl or change in the concentration of the phosphate buffer to control the ionic strength. In some biological systems divalent cations (such as Ca2+ and Mg2+) are important to certain biological structures or processes.67,68 For example, the melting point of dsDNAis raised in the presence of those divalent ions69 and Mg2+is required for the function of the DNA polymerase. Since one expects the effects of these divalent cations on ionic strength and the adhesion of the probes to the surface to be more pronounced than for monovalent cations, adjustments to the ionic strengths of the solutions should be made to optimize the experimental conditions.
By stretching ssDNA using DEP tweezers assembled with this method, we demonstrated that the proposed bead array can be used for force spectroscopy measurements in a highly parallel manner. The probes were allowed to approach the surface and bind weakly to it, hence to react with the immobilized single molecules and become specifically bound. Using deionized water, we were able to release the probes from intimate surface contact followed their attachment to DNA. By applying a DEP force to the beads in the microwells, we observed that almost all of the attached probes were active in force spectroscopy – we successfully demonstrated the ability to measure and fit the force spectra of individual DNA molecules. With further improvement to the yield in assembling active bead-biomolecule pairs in an organized bead array, this highly parallel technique can be used to conduct force spectroscopy on a variety of complex biological systems.
Supplementary Material
Acknowledgments
We acknowledge funding of this work by NIH grant R21 HG004141. We used instruments supported by NSF Major Research Instrumentation grant CHE-0923370.
Footnotes
Supporting Information Available: Scheme for approach to fabrication of single-molecule-bead arrays; detailed experimental methods; characterization of DNA surface density; probe and substrate roughness; representative result from the probe binding assay; experimental details on setup of the dielectrophoretic tweezers and the intensity-time curves for all probes identified in a single field of view.
References
- 1.Mukhopadhyay R. Analytical Chemistry. 2009;81:1736. doi: 10.1021/ac802712u. [DOI] [PubMed] [Google Scholar]
- 2.Fuller CW, Middendorf LR, Benner SA, Church GM, Harris T, Huang X, Jovanovich SB, Nelson JR, Schloss JA, Schwartz DC, Vezenov DV. Nat Biotechnol. 2009;27:1013. doi: 10.1038/nbt.1585. [DOI] [PubMed] [Google Scholar]
- 3.Oliver PM, Park JS, Vezenov D. Nanoscale. 2010;3:581. doi: 10.1039/c0nr00479k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song L, Ahn S, Walt DR. Analytical Chemistry. 2006;78:1023. doi: 10.1021/ac051417w. [DOI] [PubMed] [Google Scholar]
- 5.Nelson PC, Zurla C, Brogioli D, Beausang JF, Finzi L, Dunlap D. The Journal of Physical Chemistry B. 2006;110:17260. doi: 10.1021/jp0630673. [DOI] [PubMed] [Google Scholar]
- 6.De Vlaminck I, Henighan T, van Loenhout MTJ, Pfeiffer I, Huijts J, Kerssemakers JWJ, Katan AJ, van Langen-Suurling A, van der Drift E, Wyman C, Dekker C. Nano Letters. 2011;11:5489. doi: 10.1021/nl203299e. [DOI] [PubMed] [Google Scholar]
- 7.Plenat T, Tardin C, Rousseau P, Salome L. Nucleic Acids Res. 2012;40 doi: 10.1093/nar/gks250. in press, http://dx.doi.org/10.1093/nar/gks250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ribeck N, Saleh OA. Rev Sci Instrum. 2008;79:094301/1. doi: 10.1063/1.2981687. [DOI] [PubMed] [Google Scholar]
- 9.Cheng P, Barrett MJ, Oliver PM, Cetin D, Vezenov D. Lab Chip. 2011;11:4248. doi: 10.1039/c1lc20627c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McIntosh DB, Saleh OA. Macromolecules. 2011;44:2328. [Google Scholar]
- 11.Schlingman DJ, Mack AH, Mochrie SGJ, Regan L. Colloids and Surfaces B: Biointerfaces. 2011;83:91. doi: 10.1016/j.colsurfb.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan JB, Chee MS, Gunderson KL. Nat Rev Genet. 2006;7:632. doi: 10.1038/nrg1901. [DOI] [PubMed] [Google Scholar]
- 13.Burridge KA, Figa MA, Wong JY. Langmuir. 2004;20:10252. doi: 10.1021/la0489099. [DOI] [PubMed] [Google Scholar]
- 14.Moeller HC, Mian MK, Shrivastava S, Chung BG, Khademhosseini A. Biomaterials. 2008;29:752. doi: 10.1016/j.biomaterials.2007.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu J, Day D, Gu M. Opt Express. 2010;18:7611. doi: 10.1364/OE.18.007611. [DOI] [PubMed] [Google Scholar]
- 16.Danilowicz C, Greenfield D, Prentiss M. Analytical Chemistry. 2005;77:3023. doi: 10.1021/ac050057+. [DOI] [PubMed] [Google Scholar]
- 17.Cecchet F, Duwez AS, Gabriel S, Jérôme C, Jérôme R, Glinel K, Demoustier-Champagne S, Jonas AM, Nysten B. Analytical Chemistry. 2007;79:6488. doi: 10.1021/ac070155q. [DOI] [PubMed] [Google Scholar]
- 18.Wilchek M, Bayer EA. Analytical Biochemistry. 1988;171:1. doi: 10.1016/0003-2697(88)90120-0. [DOI] [PubMed] [Google Scholar]
- 19.Shumaker-Parry JS, Zareie MH, Aebersold R, Campbell CT. Analytical Chemistry. 2004;76:918. doi: 10.1021/ac034964v. [DOI] [PubMed] [Google Scholar]
- 20.Christoph K. Molecular and Cellular Probes. 1991;5:161. doi: 10.1016/0890-8508(91)90041-h. [DOI] [PubMed] [Google Scholar]
- 21.Fanga B, Gon S, Park M, Kumard KN, Rotello VM, Nussleind K, Santore MM. Colloids and Surfaces B: Biointerfaces. 2011;87:109. doi: 10.1016/j.colsurfb.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 22.Ribeck N, Saleh OA. Review of Scientific Instruments. 2008;79:094301. doi: 10.1063/1.2981687. [DOI] [PubMed] [Google Scholar]
- 23.Yang Y, Erb RM, Wiley BJ, Zauscher S, Yellen BB. Nano Letters. 2011;11:1681. doi: 10.1021/nl200189w. [DOI] [PubMed] [Google Scholar]
- 24.Erb RM, Son HS, Samanta B, Rotello VM, Yellen BB. Nature. 2009;457:999. doi: 10.1038/nature07766. [DOI] [PubMed] [Google Scholar]
- 25.Smith SB, Finzi L, Bustamante C. Science. 1992;258:1122. doi: 10.1126/science.1439819. [DOI] [PubMed] [Google Scholar]
- 26.Juárez JJ, Cui JQ, Liu BG, Bevan MA. Langmuir. 2011;27:9211. doi: 10.1021/la201478y. [DOI] [PubMed] [Google Scholar]
- 27.Kimura-Suda H, Petrovykh DY, Tarlov MJ, Whitman LJ. Journal of the American Chemical Society. 2003;125:9014. doi: 10.1021/ja035756n. [DOI] [PubMed] [Google Scholar]
- 28.Opdahl A, Petrovykh DY, Kimura-Suda H, Tarlov MJ, Whitman LJ. Proceedings of the National Academy of Sciences. 2007;104:9. doi: 10.1073/pnas.0608568103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lim HI, Oliver PM, Marzillier J, Vezenov DV. Anal Bioanal Chem. 2010;397:1861. doi: 10.1007/s00216-010-3733-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andersson KM, Bergström L. Journal of Colloid and Interface Science. 2002;246:309. doi: 10.1006/jcis.2001.8021. [DOI] [PubMed] [Google Scholar]
- 31.Martines E, Csaderova L, Morgan H, Curtis ASG, Riehle MO. Colloids and Surfaces A: Physicochemical and Engineering Aspects. 2008;318:45. [Google Scholar]
- 32.Ono H, Jidai E. Colloid Polym Sci. 1976;254:17. [Google Scholar]
- 33.Dedkov GV, Dedkova EG, Tegaev RI, Khokonov KB. Tech Phys Lett. 2008;34:17. [Google Scholar]
- 34.Bower MJD, Bank TL, Giese RF, van Oss CJ. Colloids and Surfaces A: Physicochemical and Engineering Aspects. 2010;362:90. [Google Scholar]
- 35.Bijamov A, Shubitidze F, Oliver PM, Vezenov DV. J Appl Phys. 2010;108:104701/1. doi: 10.1063/1.3510481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bijamov A, Shubitidze F, Oliver PM, Vezenov DV. Langmuir. 2010;26:12003. doi: 10.1021/la1015252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chin CJ, Yiacoumi S, Tsouris C. Environmental Science & Technology. 2002;36:343. doi: 10.1021/es010905s. [DOI] [PubMed] [Google Scholar]
- 38.Wang J, Yoon RH. Langmuir. 2008;24:7889. doi: 10.1021/la800276r. [DOI] [PubMed] [Google Scholar]
- 39.Adler JJ, Rabinovich YI, Moudgil BM. J Colloid Interface Sci. 2001;237:249. doi: 10.1006/jcis.2001.7466. [DOI] [PubMed] [Google Scholar]
- 40.Grasso D, Subramaniam K, Butkus M, Strevett K, Bergendahl J. Reviews in Environmental Science and Biotechnology. 2002;1:17. [Google Scholar]
- 41.Vezenov DV, Noy A, Rozsnyai LF, Lieber CM. Journal of the American Chemical Society. 1997;119:2006. [Google Scholar]
- 42.Zheng J, Yeung ES. Analytical Chemistry. 2003;75:818. doi: 10.1021/ac026146t. [DOI] [PubMed] [Google Scholar]
- 43.Bevan MA, Prieve DC. Langmuir. 2000;16:9274. [Google Scholar]
- 44.Bevan MA, Prieve DC. The Journal of Chemical Physics. 2000;113:1228. [Google Scholar]
- 45.Baek SH, Chang WJ, Baek JY, Yoon DS, Bashir R, Lee SW. Analytical Chemistry. 2009;81:7737. doi: 10.1021/ac901211b. [DOI] [PubMed] [Google Scholar]
- 46.Israelachvili JN. Intermolecular and Surface Forces. 3. Elsevier; Burlington, Ma: 2011. [Google Scholar]
- 47.Dolan AK, Edwards SF. Proc R Soc London, Ser A. 1974;337:509. [Google Scholar]
- 48.Kar G. Journal of Colloid and Interface Science. 1973;44:347. [Google Scholar]
- 49.Bevan MA, Prieve DC. Langmuir. 1999;15:7925. [Google Scholar]
- 50.Israelachvili JN. Proc Roy Soc London, Ser A. 1972;331:39. [Google Scholar]
- 51.Israelachvili JN, Tabor D. Proceedings of the Royal Society of London A Mathematical and Physical Sciences. 1972;331:19. [Google Scholar]
- 52.Biggs S, Mulvaney P. J Chem Phys. 1994;100:8501. [Google Scholar]
- 53.Czarnecki J, Ichenskii VI. J Colloid Interface Sci. 1984;98:590. [Google Scholar]
- 54.Das S, Sreeram PA, Raychaudhuri AK. Nanotechnology. 2007;18:035501. doi: 10.1088/0957-4484/18/3/035501. [DOI] [PubMed] [Google Scholar]
- 55.French RH. J Am Ceram Soc. 2000;83:2117. [Google Scholar]
- 56.Herman MC, Papadopoulos KD. J Colloid Interface Sci. 1991;142:331. [Google Scholar]
- 57.Hutter JL, Bechhoefer J. J Vac Sci Technol, B. 1994;12:2251. [Google Scholar]
- 58.Roth CM, Lenhoff AM. J Colloid Interface Sci. 1996;179:637. [Google Scholar]
- 59.Parsegian VA. Van der Waals Forces. Cambridge University Press; Cambridge: 2006. [Google Scholar]
- 60.Bahukudumbi P, Bevan MA. The Journal of Chemical Physics. 2007;126:244702. doi: 10.1063/1.2739548. [DOI] [PubMed] [Google Scholar]
- 61.Wu HJ, Bevan MA. Langmuir. 2005;21:1244. doi: 10.1021/la047892r. [DOI] [PubMed] [Google Scholar]
- 62.Walz JY, Suresh L, Piech M. Journal of Nanoparticle Research. 1999;1:99. [Google Scholar]
- 63.Cui S, Yu Y, Lin Z. Polymer. 2009;50:930. [Google Scholar]
- 64.Towles KB, Garcia JFBHG, Phillips R, Nelson PC. Phys Biol. 2009;6:025001. doi: 10.1088/1478-3975/6/2/025001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rouzina I, Bloomfield VA. Biophysical Journal. 2001;80:882. doi: 10.1016/S0006-3495(01)76067-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smith SB, Cui Y, Bustamante C. Science. 1996;271:795. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 67.Lengyel A, Uhríková D, Klacsová M, Balgavý P. Colloids and Surfaces B: Biointerfaces. 2011;86:212. doi: 10.1016/j.colsurfb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 68.Lomakina EB, Waugh RE. Biophysical Journal. 2009;96:276. doi: 10.1016/j.bpj.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Duguid JG, Bloomfield VA. Biophysical Journal. 1995;69:2642. doi: 10.1016/S0006-3495(95)80134-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.