SUMMARY
Amino acids stimulate cell growth and suppress autophagy through activation of mTORC1. The activation of mTORC1 by amino acids is mediated by Rag guanosine triphosphatase (GTPase) heterodimers on the lysosome. The molecular mechanism by which amino acids regulate the Rag GTPase heterodimers remains to be elucidated. Here, we identify SH3BP4 (SH3 domain-binding protein 4) as a binding protein and a negative regulator of Rag GTPase complex. SH3BP4 binds to the inactive Rag GTPase complex through its Src homology 3 (SH3) domain under conditions of amino acid starvation and inhibits the formation of active Rag GTPase complex. As a consequence, the binding abrogates the interaction of mTORC1 with Rag GTPase complex and the recruitment of mTORC1 to the lysosome, thus inhibiting amino acid-induced mTORC1 activation and cell growth and promoting autophagy. These results demonstrate that SH3BP4 is a negative regulator of the Rag GTPase complex and amino acid-dependent mTORC1 signaling.
INTRODUCTION
In eukaryotes, amino acids act not only as building blocks of proteins but also as mediators of signal transduction for cell growth. The signaling function of amino acids, especially branched-chain amino acids such as leucine, is mainly mediated through mammalian target of rapamycin (mTOR), a Ser/Thr kinase conserved from yeast to mammals. mTOR interacts with raptor to form mTOR complex 1 (mTORC1) that regulates protein synthesis, cell growth and autophagy in response to the availability of amino acids, glucose and growth factors (Hara et al., 2002; Jung et al., 2010; Kim et al., 2002; Loewith et al., 2002). Hyperactivation of mTORC1 has been identified in a number of human cancers including prostate cancer, multiple myeloma and hamartoma syndromes, and impairment of mTOR regulation has also been linked to diabetes, obesity and aging (Goberdhan and Boyd, 2009; Harrison et al., 2009; Selman et al., 2009; Um et al., 2004; Wullschleger et al., 2006). Given the broad function of mTOR and its implication in many human diseases and physiological states, it is important to understand the mechanism underlying amino acid-dependent mTORC1 signaling.
Amino acids activate mTORC1 via Rag GTPases that are evolutionarily conserved in eukaryotes from yeast to mammals (Binda et al., 2009; Kim et al., 2008; Sancak et al., 2008). Mammalian cells have four Rag GTPases: RagA, RagB, RagC and RagD (Sekiguchi et al., 2001). Rag GTPases are distinct from other small GTPases as they form heterodimeric complexes consisting of RagA or RagB and RagC or RagD (Dubouloz et al., 2005; Hirose et al., 1998; Kim et al., 2008). In yeast, Gtr1p and Gtr2p, which are orthologues of RagA and RagC respectively, form a heterodimeric complex playing similar roles as Rag GTPase complexes (Binda et al., 2009; Dubouloz et al., 2005). The activity of Rag GTPase heterodimers depends on whether RagA and RagB are bound to GTP or GDP. The Rag GTPase complex containing GTP-bound RagA or RagB is active in stimulating mTORC1 in response to amino acids, whereas the complex containing GDP-bound RagA or RagB is inactive (Kim et al., 2008; Sancak et al., 2008).
Several binding proteins of Rag GTPases were identified in recent studies. Ragulator is a lysosomal protein complex that binds and enables the Rag GTPase complex to recruit mTORC1 to the lysosomal membrane where mTORC1 is activated by Rheb GTPases (Sancak et al., 2010). Ragulator was also shown to bind to vacuolar H(+)-adenosine triphosphatase (v-ATPase) that regulates Rag GTPases in response to amino acids in the lysosomal lumen (Zoncu et al., 2011). In Drosophila, Rag GTPases interact with mitogen-activated protein 4 kinase 3 (MAP4K3) (Bryk et al., 2010). In mammalian cells, MAP4K3 regulates mTORC1 in response to amino acids (Findlay et al., 2007; Yan et al., 2010). Rag GTPases interact with p62 (SQSTM1), a ubiquitin-binding protein degraded by autophagy (Duran et al., 2011). They also regulate mTORC1 localization to the TOR-autophagy spatial coupling compartment (TASCC) (Narita et al., 2011), indicating an intimate crosstalk between the Rag-mTORC1 pathway and autophagy. Most recently, leucyl-tRNA synthetase (LRS) was found to interact with RagD in an amino acid-dependent manner and function as a GTPase-activating protein (GAP) for RagD (Han et al., 2012). In yeast, Vam6p was identified as a binding protein of Gtr1p and as a guanine nucleotide exchange factor (GEF) for Gtr1p (Binda et al., 2009). Although these studies have provided important insight into the regulation of Rag GTPases, the molecular mechanisms by which amino acids regulate the activity of Rag GTPases remains not clearly understood.
In this study, we describe the discovery of a Rag GTPase-binding protein, SH3BP4, that negatively regulates Rag GTPase-mTORC1 signaling. We determined that SH3BP4 binds to the inactive Rag GTPase complex under leucine-deprived conditions and suppresses the formation of active Rag GTPase complex thereby inhibiting mTORC1 signaling. We found that the inhibitory effect of SH3BP4 on mTORC1 signaling depends on the interaction between SH3BP4 and Rag GTPases that is mediated by the SH3 domain of SH3BP4. Based on these findings, we define SH3BP4 as a negative regulator of Rag GTPase complex and amino acid-dependent Rag GTPase-mTORC1 signaling.
RESULTS
Identification of SH3BP4 as a Rag GTPase-binding protein
To identify proteins that bind to Rag GTPases, we stably expressed flag-tagged RagB in HEK293T cells and isolated RagB-binding proteins by flag-affinity chromatography. Following our previously described procedure to prepare samples for mass spectrometry (Vander Haar et al., 2007) (Figure S1A), we identified proteins that were specifically eluted with flag-tagged RagB. The pool of identified proteins contained RagC and RagD, confirming that our purification isolated the physiological binding partners of RagB. The pool also contained a protein called SH3BP4 or TTP (transferrin receptor-trafficking protein), a vertebrate-specific 963 amino acid protein that was previously shown to bind dynamin and Eps15 (epidermal growth factor receptor pathway substrate 15) and regulate endocytosis of transferrin receptor (TfR) (Tosoni et al., 2005). This protein possesses one SH3 domain, three NPF motifs for binding the Eps15 homology domain and putative binding sites for proteins involved in clathrin-mediated endocytosis (Figure 1A) (Dunlevy et al., 1999). TTP is an acronym for a zinc finger protein tristetraprolin, and it has a function independent of TfR as it becomes obvious from our study. Thus, the protein was named SH3BP4 throughout this manuscript following the nomenclature by the HUGO Gene Nomenclature Committee.
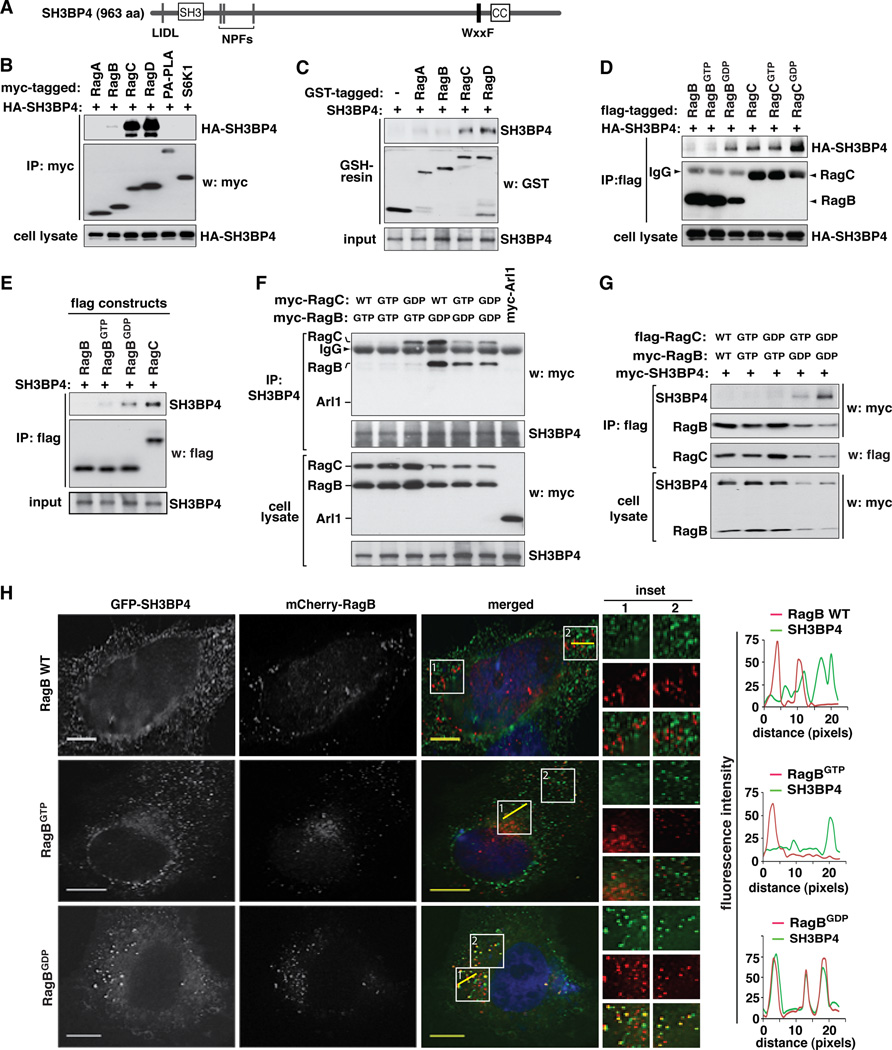
Figure 1. SH3BP4 preferentially binds to the GDP-bound state of Rag GTPases.
(A) Schematic representation of structural features of SH3BP4. SH3BP4 possesses one LIDL motif (a putative clathrin binding motif), one SH3 domain, three NPF repeats (putative Eps15 homology domain binding sites), one WxxF motif (a putative AP2 binding site), and one coiled-coil (CC) region. (B) SH3BP4 interacts with Rag GTPases. HA-tagged SH3BP4 was co-expressed with myc-tagged constructs in HEK293T cells. The amount of HA-SH3BP4 co-immunopurified with myc-tagged constructs was analyzed by Western blotting (WB). PA-PLA and S6K1 are control proteins. (C) SH3BP4 directly binds to Rag GTPases. The amount of SH3BP4 bound to GST (control) or GST-tagged Rag GTPases immobilized on glutathione resin was analyzed by WB. The proteins were prepared from bacteria. (D) SH3BP4 preferentially binds to RagBGDP and RagCGDP. Flag-tagged RagB and RagC WT or mutants were expressed with HA-tagged SH3BP4 in HEK293T cells. The amount of HA-SH3BP4 bound to flag-tagged Rag GTPases was analyzed by WB. The arrow head points to IgG band. (E) SH3BP4 directly binds to RagBGDP. The amount of SH3BP4 bound to flag-tagged Rag GTPases was analyzed by WB. The proteins were purified from bacteria. (F) SH3BP4 preferentially binds to the inactive Rag GTPase complex. Myc-tagged Rag WT and mutants were expressed in HEK293T cells. The amount of Rag GTPases co-immunopurified with endogenous SH3BP4 was analyzed by WB. Arl1 was used as a control. (G) SH3BP4 binds to the inactive Rag complex but not the active Rag complex. Rag GTPases and SH3BP4 were expressed in HEK293T cells. The amounts of SH3BP4 and RagB co-immunopurified with RagC were analyzed by WB. (H) SH3BP4 colocalizes with RagBGDP but barely with WT or RagBGTP. GFP-SH3BP4 (green) and mCherry-RagB (red) transiently expressed in HeLa cells were visualized by fluorescence microscope. (Inset) Higher magnification demonstrates colocalization of SH3BP4 and RagBGDP (yellow). Nuclei were stained with DAPI (blue). Scale bar, 10 µm. Line scans on the right hand side indicate the degree of colocalization between SH3BP4 and RagB, which correlate to the lines drawn in the inset boxes of the merged images.
SH3BP4 preferentially binds to GDP-bound forms of Rag GTPases
SH3BP4 was co-immunopurified with either RagC or RagD but not with cytosolic S6K1 or membrane-bound PA-PLA used as negative controls (Figure 1B). In a reciprocal manner, RagC was co-immunopurified with SH3BP4 but not with the control proteins (Figure S1B). To determine whether SH3BP4 can directly interact with Rag GTPases, we performed glutathione S-transferase (GST) pull-down assay using SH3BP4 and Rag GTPases purely prepared from bacteria. SH3BP4 was pulled down with GST-tagged RagC and RagD but barely with GST-tagged RagA and RagB (Figure 1C). This result suggests that SH3BP4 has a higher binding affinity toward the monomeric forms of RagC and RagD than those of RagA and RagB.
To assess the effects of the nucleotide-bound states of Rag GTPases on the interaction between SH3BP4 and Rag GTPases, we analyzed the binding affinity of SH3BP4 toward Rag GTPase mutants mimicking GTP- or GDP-bound states (Dubouloz et al., 2005; Gao and Kaiser, 2006; Hirose et al., 1998; Kim et al., 2008; Sancak et al., 2008; Sekiguchi et al., 2001). RagB wild type (WT) and its mutant mimicking the GTP-bound state (RagBGTP) did not bind to SH3BP4, whereas its mutant mimicking the GDP-bound state (RagBGDP) was able to bind SH3BP4 (Figure 1D and Figure S1C). Like RagBGDP, RagAGDP was also able to bind to SH3BP4 (Figure S1C). This result suggests that SH3BP4 might bind only to the GDP-bound state of RagA and RagB, but not their GTP-bound states or uncharged states. By contrast, RagC interacted with SH3BP4 regardless of its nucleotide-bound status (WT, RagCGTP, and RagCGDP) (Figure 1D). SH3BP4 showed a stronger binding affinity toward RagCGDP than either WT or RagCGTP, suggesting that GDP-bound states of both RagB and RagC are likely favored for binding by SH3BP4. Through in vitro analysis of the interaction between recombinant proteins purified from bacteria, we confirmed that RagBGDP can directly interact with SH3BP4 with an affinity comparable to that of the SH3BP4-RagC interaction (Figure 1E).
SH3BP4 preferentially binds to the inactive Rag GTPase complex
The preferential binding of SH3BP4 to RagBGDP over RagBGTP prompted us to test whether SH3BP4 has a higher binding affinity toward the inactive Rag complex than the active Rag complex. Although RagC and RagCGTP as monomeric forms could bind to SH3BP4 (Figure 1D), they did not bind to SH3BP4 when they were co-expressed with RagBGTP (Figure 1F and Figure S1D). This result suggests that RagBGTP might have a negative effect on the binding of RagC or RagCGTP to SH3BP4. By contrast, RagCGDP interacted with SH3BP4 in RagBGTP-expressing cells (Figure 1F and Figure S1D). It was noteworthy however that the immune complex containing RagCGDP and SH3BP4 did not contain RagBGTP, suggesting that RagCGDP in association with SH3BP4 is free of RagBGTP. This suggests that SH3BP4 would not form a stable complex with RagBGTP and RagCGDP simultaneously. To confirm that SH3BP4 does not form a stable interaction with the RagBGTP-containing active complexes, we immunopurified RagBGTP-RagCGTP and RagBGTP-RagCGDP complexes from HEK293T cells and analyzed the presence of SH3BP4. SH3BP4 was not co-immunopurified with the RagBGTP-containing complexes (Figure 1G). This result demonstrates that SH3BP4 may not form a stable interaction with the active Rag GTPase complexes.
On the other hand, all three forms of RagC (WT, RagCGTP and RagCGDP) were able to interact with SH3BP4 when they were co-expressed with RagBGDP (Figure 1F and Figure S1D). Since RagBGDP forms inactive Rag complexes, we hypothesized that SH3BP4 might preferentially bind to inactive Rag complexes. Supporting the hypothesis, SH3BP4 was co-immunopurified with RagBGDP-containing inactive complexes, such as RagBGDP-RagCGTP and RagBGDP-RagCGDP complexes, from HEK293T cells (Figure 1G). To further validate the interaction, we monitored the colocalization of SH3BP4 to RagB, RagBGTP or RagBGDP in HeLa cells. Confirming the preferential binding of SH3BP4 to inactive Rag GTPase complex in cells, SH3BP4 colocalized to RagBGDP but barely to RagB WT or RagBGTP (Figure 1H and Figure S1E – S1G). Taken together, these results suggest that SH3BP4 preferentially binds to inactive Rag GTPase complex.
SH3BP4 binding to Rag GTPases is enhanced under leucine deprived condition
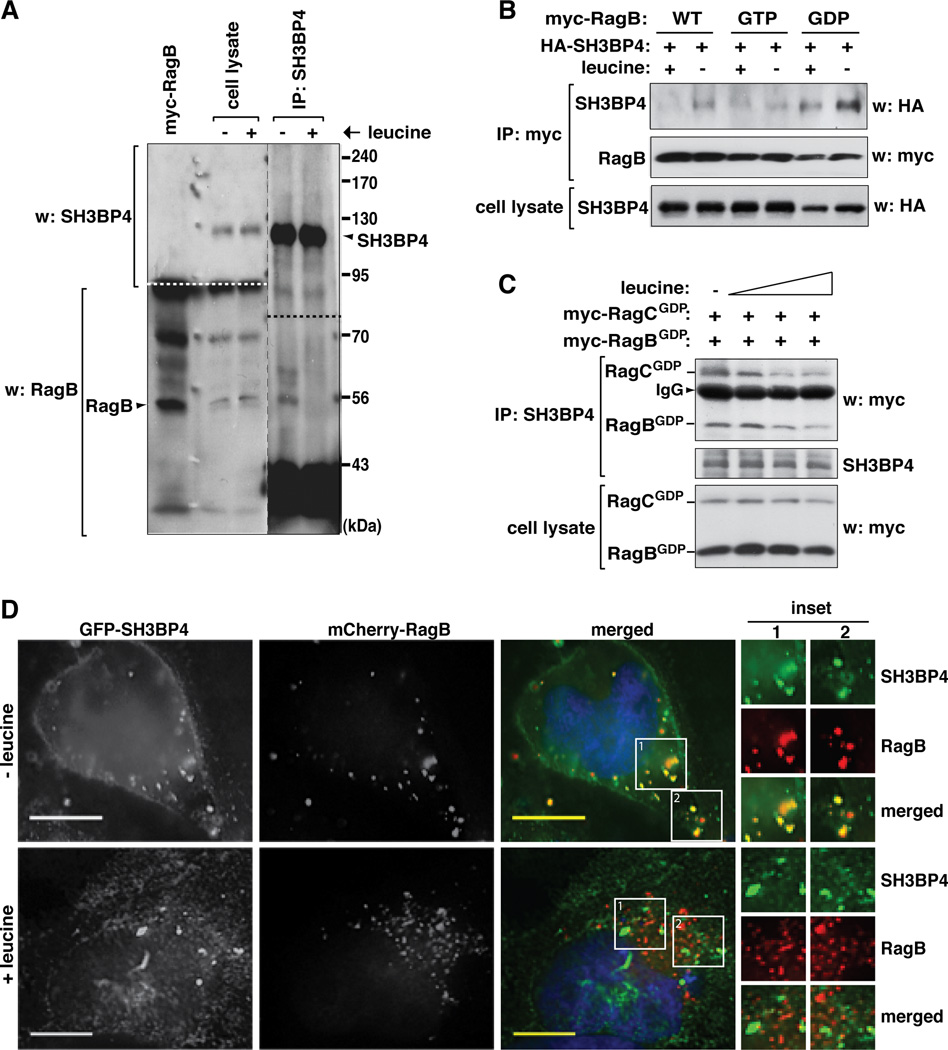
If SH3BP4 preferentially binds to the inactive Rag GTPase complexes, we hypothesized that SH3BP4 binding to Rag GTPases might increase under amino acid starvation. Since leucine is the most potent stimulator of mTORC1 signaling among amino acids (Kim et al., 2002), we analyzed the interaction in the presence or absence of leucine in the medium. Supporting the hypothesis, leucine deprivation enhanced the amount of RagB recovered with SH3BP4 at endogenous levels (Figure 2A). The leucine deprivation-induced up-regulation of interaction could be recapitulated by SH3BP4 and Rag GTPases overexpressed in HEK293T cells (Figure 2B). Leucine deprivation enhanced the interaction of SH3BP4 with RagB WT and RagBGDP but barely with RagBGTP. The amount of SH3BP4 bound to RagBGDP was much greater than that bound to RagB WT, as we would expect from the stronger binding affinity of SH3BP4 toward RagBGDP than RagB WT (Figure 1D and E). We further confirmed that the association of SH3BP4 with co-expressed RagBGDP and RagCGDP is gradually reduced by leucine at increasing concentrations in the medium (Figure 2C). These results suggest that the interaction of SH3BP4 with RagBGDP, but not with RagBGTP, is enhanced by leucine deprivation. The leucine-dependent interaction between SH3BP4 and RagB could be confirmed by fluorescence image analysis of GFP-SH3BP4 and mCherry-RagB in HeLa cells (Figure 2D and Figure S2). Combined, these results consistently show that SH3BP4 binding to Rag GTPases is negatively regulated by leucine.
Figure 2. The SH3BP4-Rag interaction is regulated by amino acids.
(A) SH3BP4 binding to RagB is negatively regulated by amino acids. HeLa cells were starved of leucine for 40 min then supplemented with leucine at 104 µg/ml (+) or cultured further without leucine for 10 min (−). All other amino acids were present in the medium as described previously (Kim et al., 2002). Endogenous SH3BP4 was isolated using anti-SH3BP4 antibody. The amount of endogenous RagB recovered with SH3BP4 was analyzed by WB. (B) SH3BP4 binding to RagBGDP is negatively regulated by amino acids. Myc-tagged RagB WT, RagBGTP or RagBGDP was expressed with HA-SH3BP4 in HEK293T cells. Cells were treated with leucine as described in Fig. 2A. The amount of HA-SH3BP4 recovered with myc-RagB was analyzed by WB. (C) SH3BP4 binding to the RagBGDP-RagCGDP complex was gradually reduced by increasing levels of leucine in the medium. HEK293T cells expressing myc-tagged RagBGDP and RagCGDP were treated with leucine (0, 1.04, 10.4, or 104 µg/ml) as described in Fig. 2A. The amounts of Rag GTPases co-immunopurified with endogenous SH3BP4 were analyzed by WB. (D) Leucine deprivation induces the colocalization of SH3BP4 with RagB. GFP-tagged SH3BP4 was coexpressed with mCherry-RagB in HeLa cells. Cells were treated with leucine as described in Fig. 2A. The colocalization of SH3BP4 with RagB was analyzed by fluorescence microscope. Scale bar, 10 µm.
SH3BP4 negatively regulates amino acid-mTORC1 signaling
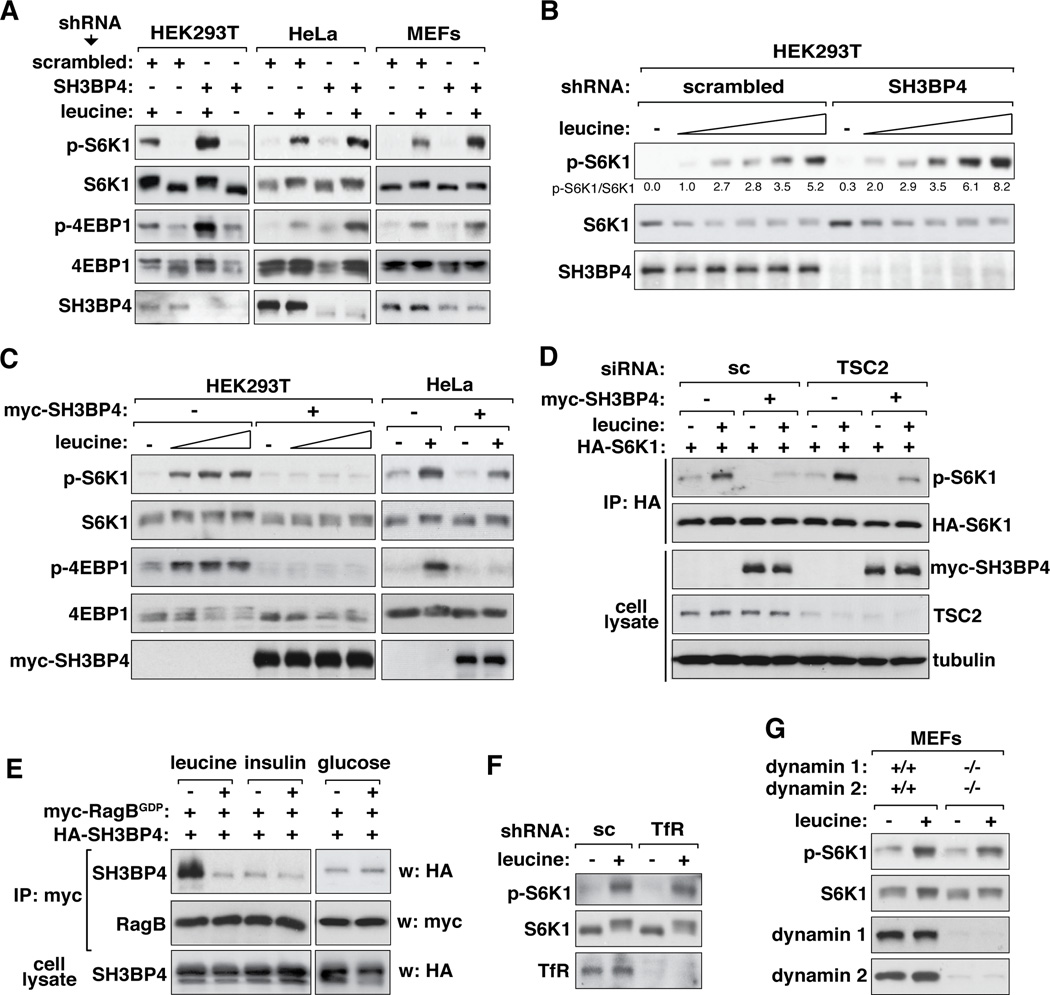
Knowing that SH3BP4 preferentially binds to inactive Rag GTPase complex, we wondered if SH3BP4 regulates amino acid-mTORC1 signaling. We first investigated the effects of SH3BP4 knockdown on mTORC1 signaling in response to leucine in the medium. Knocking down SH3BP4 in HEK293T, HeLa or MEFs induced a drastic increase in leucine-stimulated S6K1 phosphorylation at Thr389, the mTORC1-dependent phosphorylation site (Figure 3A and B and Figure S3A). The increase in the S6K1 phosphorylation occurred in a broad range of leucine concentrations in the medium (Figure 3B). The S6K1 phosphorylation was detected even in the absence of leucine in the medium when SH3BP4 was knocked down (Figure S3A, lanes 1, 7). The up-regulation of S6K1 phosphorylation by SH3BP4 knockdown was corroborated by an increase in phosphorylation of 4E-BP1 at Ser65, another mTORC1 target site (Figure 3A). Consistent with the negative effect of SH3BP4 on mTORC1 signaling, either stable or transient overexpression of SH3BP4 almost completely suppressed the leucine-induced S6K1 and 4E-BP1 phosphorylation in HEK293T and HeLa cells (Figure 3C and Figure S3B and C).
Figure 3. SH3BP4 negatively regulates mTORC1 signaling.
(A) SH3BP4 knockdown enhanced amino acid-dependent mTORC1 signaling. HEK293T, HeLa and MEFs stably transduced by SH3BP4 shRNA or scrambled shRNA were treated with leucine as described in Fig. 2A. The levels of p-S6K1 (Thr389) and p-4E-BP1 (Ser65) in cell lysate were monitored by WB. (B) HEK293T cells stably transduced by shRNAs were treated with leucine at 0, 1.04, 5.2, 10.4, 52 or 104 µg/ml as described in Fig. 2A. The level of p-S6K1 was analyzed by WB. (C) Overexpression of SH3BP4 inhibited mTORC1 signaling. HEK293T and HeLa cells stably expressing myc-SH3BP4 were treated with leucine (0, 1.04, 10.4, or 104 µg/ml for HEK293T and 104 µg/ml for HeLa) as described in Fig. 2A. The levels of p-S6K1 (Thr389) and p-4E-BP1 (Ser65) in cell lysate were analyzed by WB. (D) The negative effect of SH3BP4 on mTORC1 signaling is independent of TSC2. Myc-SH3BP4 and HA-S6K1 were expressed in TSC2-silenced or scrambled siRNA-transduced HEK293T cells. Cells were treated with leucine as described in Fig. 2A. The level of p-S6K1 in HA immunoprecipitate was analyzed by WB. (E) The SH3BP4-Rag interaction is regulated by leucine but neither by insulin nor glucose in the medium. HEK293T cells expressing myc-RagBGDP and HA-SH3BP4 were starved for leucine or glucose for 40 min and supplemented with leucine (104 µg/ml) or glucose (11 mM) for 10 min. Insulin (150 nM) was treated to cells and incubated for 1 hr after overnight serum starvation. The amount of HASH3BP4 recovered with myc-RagBGDP was analyzed by WB. (F) TfR knockdown does not inhibit mTORC1 signaling in HEK293T cells. TfR-silenced cells were treated with leucine as described in Fig. 2A. The level of p-S6K1 was monitored by WB. (G) Deficiency of both dynamin 1 and dynamin 2 does not affect S6K1 phosphorylation in MEF cells. Dynamin 1- and 2-deficient MEFs were treated with leucine as described in Fig. 2A. The level of p-S6K1 was monitored by WB.
Since mTORC1 is activated not only by amino acids but also by insulin and glucose, we investigated a possibility that SH3BP4 might regulate mTORC1 through insulin- or glucose-dependent signaling. Because mTORC1 cannot be fully activated if any signal derived from amino acids, glucose or insulin is lacking (Fingar and Blenis, 2004; Sancak et al., 2008; Shamji et al., 2003), we anticipated that SH3BP4 would also have inhibitory effects on insulin- and glucose-dependent mTORC1 signaling. As anticipated, insulin-stimulated S6K1 phosphorylation was enhanced by SH3BP4 knockdown and suppressed by SH3BP4 overexpression (Figure S3D). Neither knockdown nor overexpression of SH3BP4 affected the phosphorylation state of Akt at Ser473 (Figure S3D). SH3BP4 overexpression drastically inhibited S6K1 phosphorylation even in tuberous sclerosis complex 2 (TSC2)-silenced cells where mTORC1 is highly active (Figure 3D). Given that TSC2 functions in the Akt-mTORC1 pathway, this result suggests that SH3BP4 regulates mTORC1 in a manner independent of the Akt-TSC2 axis. We also found that glucose-stimulated S6K1 phosphorylation was enhanced by SH3BP4 knockdown and suppressed by SH3BP4 overexpression (Figure S3E). Neither knockdown nor overexpression of SH3BP4 affected the phosphorylation state of AMPK at Thr172, the indicator of AMPK activity (Figure S3E). The SH3BP4-RagBGDP interaction was regulated by leucine but neither by insulin nor glucose in the medium (Figure 3E), confirming that SH3BP4 has a specific effect on amino acid-dependent mTORC1 signaling.
We also considered a possibility that SH3BP4 might regulate mTORC1 through dynamin-dependent endocytosis of TfR based on the previously-reported function of SH3BP4 (Tosoni et al., 2005). Neither knockdown (Figure 3F) nor overexpression (Figure S3F) of TfR in HEK293T cells affected the leucine-stimulated phosphorylation of S6K1. The leucine-stimulated phosphorylation of S6K1 was barely affected by deficiency of dynamin 1 and dynamin 2 in MEFs (Figure 3G) or only marginally by overexpression of dynamin 1 (Figure S3G). We also considered a possibility that SH3BP4 might regulate mTORC1 via altering amino acid uptake. Neither knockdown nor overexpression of SH3BP4 induced any significant change in amino acid uptake by HEK293T cells (Figure S3H and I). These results suggest that SH3BP4 negatively regulates amino acid-mTORC1 signaling in a manner independent of TfR, dynamin and amino acid uptake.
SH3BP4 inhibits cell growth and promotes autophagy
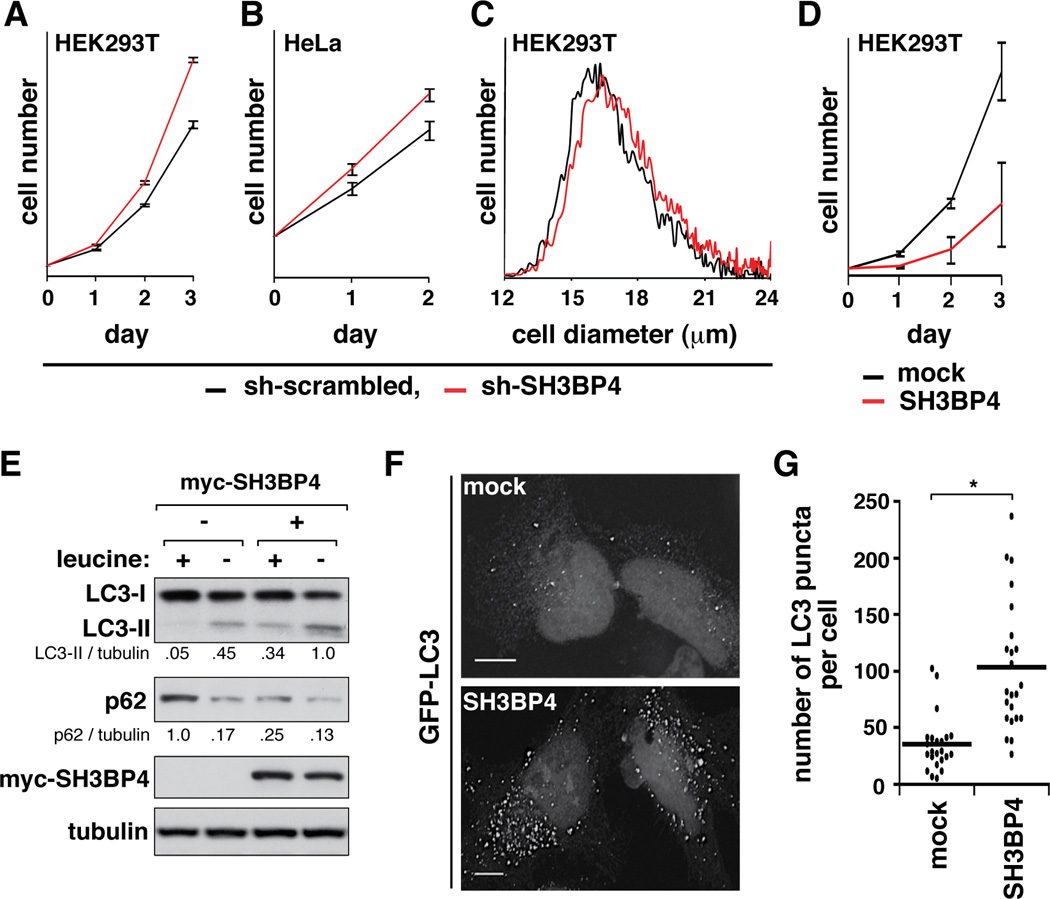
Knowing that SH3BP4 binds to the inactive Rag GTPase complex and inhibits mTORC1 signaling, we investigated if SH3BP4 has negative effects on cell growth and proliferation. Knockdown of SH3BP4 significantly enhanced proliferation rates of HEK293T and HeLa cells and increased mean size of HEK293T cells (Figure 4A–C and Figure S4). Consistently, overexpression of SH3BP4 in HEK293T cells dramatically reduced cell proliferation rate (Figure 4D). We also investigated whether SH3BP4 has effects on autophagy, a process downstream of mTORC1 (Hosokawa et al., 2009; Jung et al., 2009). SH3BP4 overexpression increased the level of endogenous microtubule-associated protein 1 light chain 3-II (LC3-II) and the formation of LC3 puncta and reduced the expression level of p62 (Figure 4E–G), all of which support an increase in autophagy (Mizushima and Yoshimori, 2007). These results support the inhibitory function of SH3BP4 in the amino acid-mTORC1 pathway.
Figure 4. SH3BP4 negatively regulates cell growth and promotes autophagy.
(A, B) SH3BP4 knockdown increased proliferation rates of HEK293T (A) and HeLa cells (B). Results are represented as means ± standard deviations (SD) (n = 3). (C) SH3BP4 knockdown increased mean size of HEK293T cells. The mean ± SD cell diameters (µm) are 16.7 ± 0.08 and 17.1 ± 0.01 for control and SH3BP4-silenced cells, respectively. (D) Stable overexpression of SH3BP4 in HEK293T cells significantly reduced cell proliferation rate. The error bars represent mean ± SD (n = 6). (E) SH3BP4 promotes autophagy. HEK293T cells stably expressing SH3BP4 were starved of leucine for 30 min (−) then supplemented with leucine (104 µg/ml) (+) for 2 hrs. The levels of LC3-I, LC3-II and p62 were analyzed by WB. (F) Overexpression of SH3BP4 induced the formation of LC3 puncta. GFP-tagged LC3 was expressed with HA-SH3BP4 in HeLa cells. Empty vector (mock) was used as a control for SH3BP4. GFP-LC3 puncta were visualized under a fluorescence microscope. Scale bar, 10 µm. (G) Quantitative analysis of GFP-LC3 puncta per cell (*, p < 0.001 from Student’s t-test, n = 23). Mean value is shown as a horizontal bar.
SH3BP4 inhibits Rag GTPase binding to mTORC1 thereby suppressing mTORC1 localization to lysosome
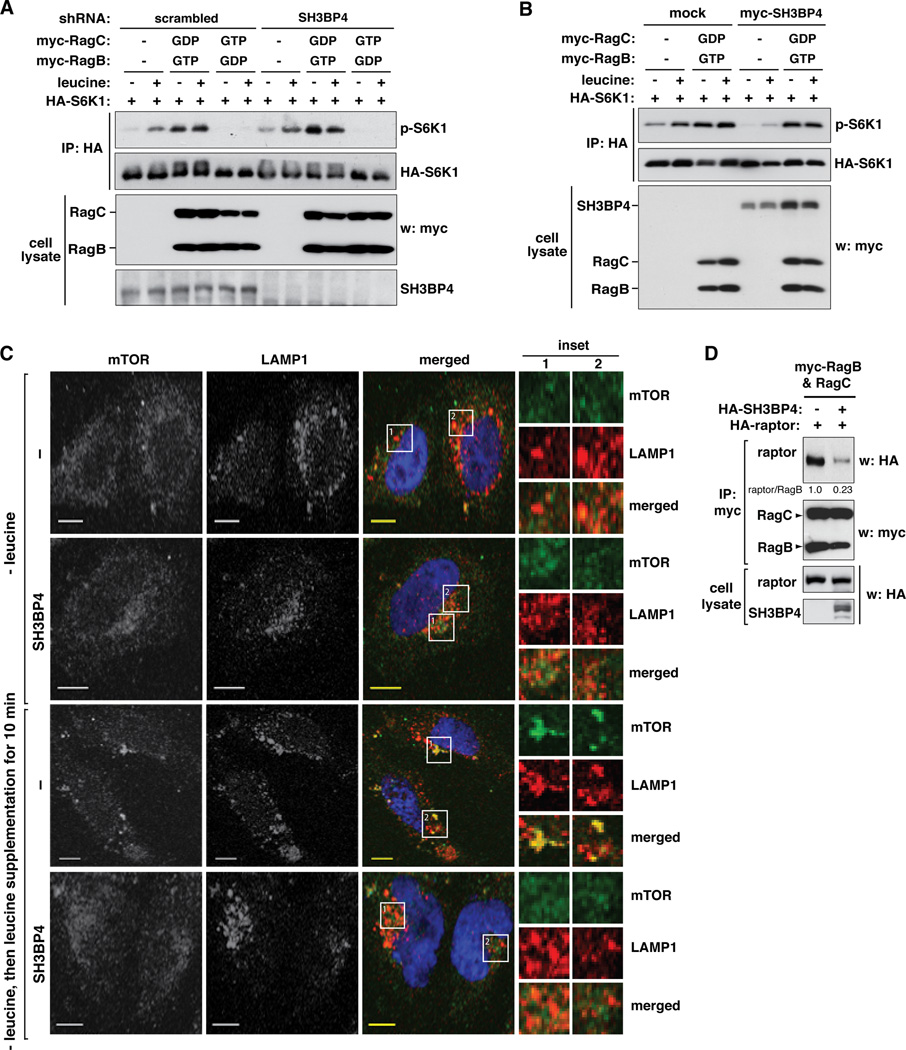
Since SH3BP4 binds to Rag GTPase complex, it is logical to reason that the inhibitory effect of SH3BP4 on mTORC1 signaling might be mediated through Rag GTPase complex. If SH3BP4 inhibits mTORC1 via Rag GTPases, SH3BP4 knockdown would not have any positive effect on mTORC1 signaling in cells expressing the dominant negative RagBGDP-RagCGTP complex. This was the case as SH3BP4 knockdown did not increase S6K1 phosphorylation in cells expressing the inactive complex (Figure 5A). In line with this result, SH3BP4 overexpression did not inhibit mTORC1 signaling in cells expressing the constitutively active RagBGTP-RagCGDP complex (Figure 5B). Combined, these results suggest that SH3BP4 negatively regulates mTORC1 signaling in a manner dependent upon the Rag GTPase complex.
Figure 5. SH3BP4 functions upstream of Rag GTPases to negatively regulate mTORC1.
(A) Myc-tagged RagB and RagC constructs were expressed with HA-S6K1 in scrambled shRNA-transduced or SH3BP4-silenced HEK293T cells. Cells were treated with leucine as described in Fig. 2A. The level of p-S6K1 (Thr389) in HA immunoprecipitate was analyzed by WB. (B) Myc-tagged RagB and RagC constructs were expressed with myc-SH3BP4 and HA-S6K1 in HEK293T cells. Cells were treated with leucine as described in Fig. 2A. The level of p-S6K1 in HA immunoprecipitate was analyzed by WB. (C) SH3BP4 negatively regulates mTOR localization to the lysosome. GFP-tagged SH3BP4 or GFP alone (mock) was transiently expressed in HeLa cells. Two days after the transfection, cells were treated with leucine as described in Fig. 2A. Cells were stained using antibodies specific to mTOR (green) and LAMP1 (red) and visualized by fluorescence microscope. (Inset) Higher magnification demonstrates colocalization (yellow) of mTOR and LAMP1. Scale bar, 10 µm. (D) SH3BP4 suppresses the interaction between Rag GTPases and raptor. HA-tagged raptor and SH3BP4 were coexpressed with myc-tagged RagB and RagC in HEK293T cells. The amount of HA-raptor co-immunopurified with myc-Rag GTPases was analyzed by WB.
A previous study has shown that RagAGTP and RagBGTP, which form the active Rag GTPase complexes with RagC or RagD, bind to raptor and thereby recruit mTORC1 to the lysosome for mTORC1 activation (Sancak et al., 2010). Since SH3BP4 inhibits mTORC1 via Rag GTPases, we considered a possibility that SH3BP4 might suppress the localization of mTORC1 to the lysosome. mTOR and LAMP1, a lysosomal marker, became colocalized when leucine was supplemented to the medium (Figure 5C and Figure S5). The colocalization of mTOR and LAMP1 was suppressed in SH3BP4-overexpressing cells (Figure 5C and Figure S5). Through co-immunoprecipitation analysis, we confirmed that SH3BP4 suppresses the interaction between raptor and Rag GTPases (Figure 5D). This result led us to propose a model that SH3BP4 inhibits mTORC1 signaling through suppression of the Rag GTPase-mTORC1 interaction on the lysosome.
SH3BP4 binding to Rag GTPases through its SH3 domain is important for inhibition of mTORC1 signaling
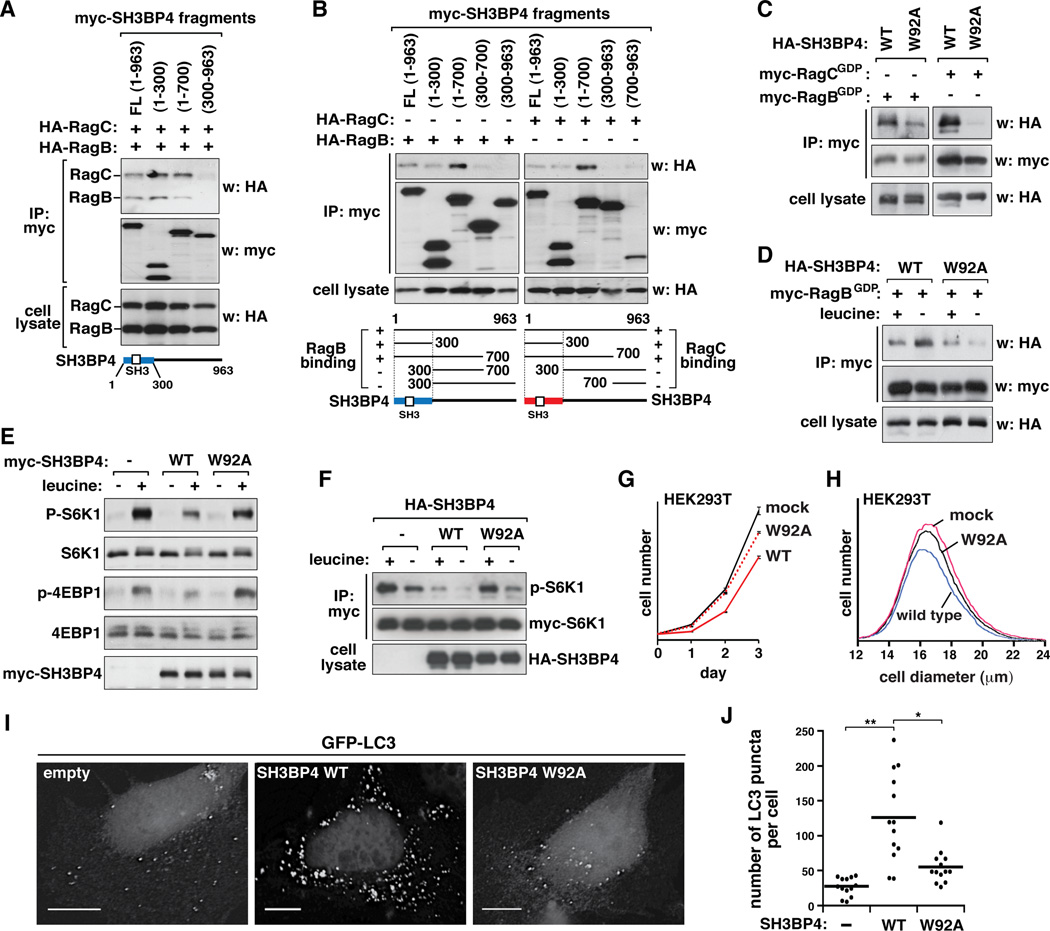
Although the results above suggest that SH3BP4 inhibits mTORC1 via Rag GTPases, it remains to be determined whether the inhibitory effect depends on the binding of SH3BP4 to Rag GTPases. To address this question, we attempted to identify a point mutation on SH3BP4 that prevents SH3BP4 binding to Rag GTPases. We first delimited the region of SH3BP4 that is involved in binding to Rag GTPases. We found that N-terminal fragments of SH3BP4 containing the SH3 domain bind to RagB and RagC that are in the heterodimeric state (Figure 6A) or in the monomeric states (Figure 6B). Because Rag GTPases contain a PxxP motif that is known to participate in binding SH3 domain-containing proteins (Figure S6A–S6D), we predicted that the SH3 domain of SH3BP4 near the N-terminus might be important for binding Rag GTPases. To test this possibility, we introduced a point mutation into the domain replacing Trp92 with alanine (W92A). Trp92 is located at a conserved position important for SH3 domain-specific interaction (Erpel et al., 1995; Musacchio et al., 1994; Panchamoorthy et al., 1994; Tosoni et al., 2005). The mutation suppressed SH3BP4 binding to Rag GTPases (Figure 6C and Figure S6E), suggesting that the interaction depends on the SH3 domain. The interaction disrupted by W92A mutation was not recovered by leucine deprivation in the medium, the condition that strengthens the SH3BP4-Rag GTPase interaction (Figure 6D). This result suggests that the SH3 domain-mediated interaction is important for amino acid-responsive interaction between SH3BP4 and Rag GTPases.
Figure 6. SH3BP4 binding to Rag GTPases via its SH3 domain is important for inhibition of mTORC1 signaling.
(A, B) N-terminal fragments of SH3BP4 bind to RagB and RagC in the dimeric form (A) or in the monomeric forms (B). Myc-tagged fragments of SH3BP4 were expressed with (A) both HA-tagged RagB and RagC or (B) RagB or RagC individually in HEK293T cells. Forty-eight hours post-transfection, cells were incubated in the medium without leucine for 40 min. The amounts of HA-tagged RagB and RagC bound to SH3BP4 fragments were analyzed by WB. (C) SH3BP4 W92A mutation suppresses the binding of SH3BP4 to RagB and RagC. HA-tagged SH3BP4 WT or W92A mutant was expressed with myc-tagged RagBGDP or RagCGDP in HEK293T cells. The amount of HA-SH3BP4 recovered with myc-RagBGDP or myc-RagCGDP was assessed by WB. (D) SH3BP4 W92A mutation impairs leucine-dependent regulation of SH3BP4-Rag GTPase interaction. HEK293T cells expressing SH3BP4 and RagBGDP as above in (C) were treated with leucine as described in Fig. 2A. The amount of HA-SH3BP4 bound to myc-RagBGDP was analyzed. (E) Stable or (F) transient overexpression of SH3BP4 W92A mutant abolished the inhibitory effect of SH3BP4 on mTORC1. HEK293T cells were treated with leucine as described in Fig. 2A and the phosphorylation states of endogenous S6K1 and 4EBP1 (E) or myc-tagged S6K1 (F) was analyzed by WB. (G) W92A mutation blunts the inhibitory effect of SH3BP4 on cell proliferation. HEK293T cells stably transduced by mock vector or SH3BP4 constructs were analyzed for cell proliferation. Results are represented as mean ± SD (n = 6). (H) W92A mutation suppresses the negative effect of SH3BP4 on cell growth. The mean ± SD cell diameters are 17.16 ± 0.08, 16.96 ± 0.09, and 17.10 ± 0.01 µm for mock, WT, and W92A cells, respectively. (I) W92A mutation suppresses the stimulatory effect of SH3BP4 on LC3 puncta formation in HeLa cells. GFP-LC3 puncta were analyzed as described in Figure 4F. Scale bar, 10 µm. (J) Quantitative analysis of GFP-LC3 puncta per cell (*, p < 0.05, **, p < 0.01, n = 13). Mean value is shown as a horizontal bar.
In order to test if the SH3 domain-mediated interaction is important for inhibition of mTORC1 signaling, we expressed SH3BP4 WT or W92A mutant stably or transiently in HEK293T cells and analyzed the phosphorylation state of S6K1 and 4EBP1. The interaction-disrupting W92A mutation suppressed the inhibitory effect of SH3BP4 on leucine-stimulated phosphorylation of S6K1 and 4E-BP1 (Figure 6E and F). In line with this result, W92A mutation blunted the inhibitory effects of SH3BP4 on cell proliferation and growth (Figure 6G and H) and the stimulatory effect of SH3BP4 on the formation of LC3 puncta (Figure 6I and J and Figure S6F). Although we could not exclude a possibility that W92A mutation affects other unknown functions of SH3BP4, these results support our hypothesis that the SH3BP4-Rag GTPase interaction mediated by the SH3 domain is important for inhibition of mTORC1 signaling.
SH3BP4 binding to Rag GTPases inhibits the formation of active Rag GTPase complex
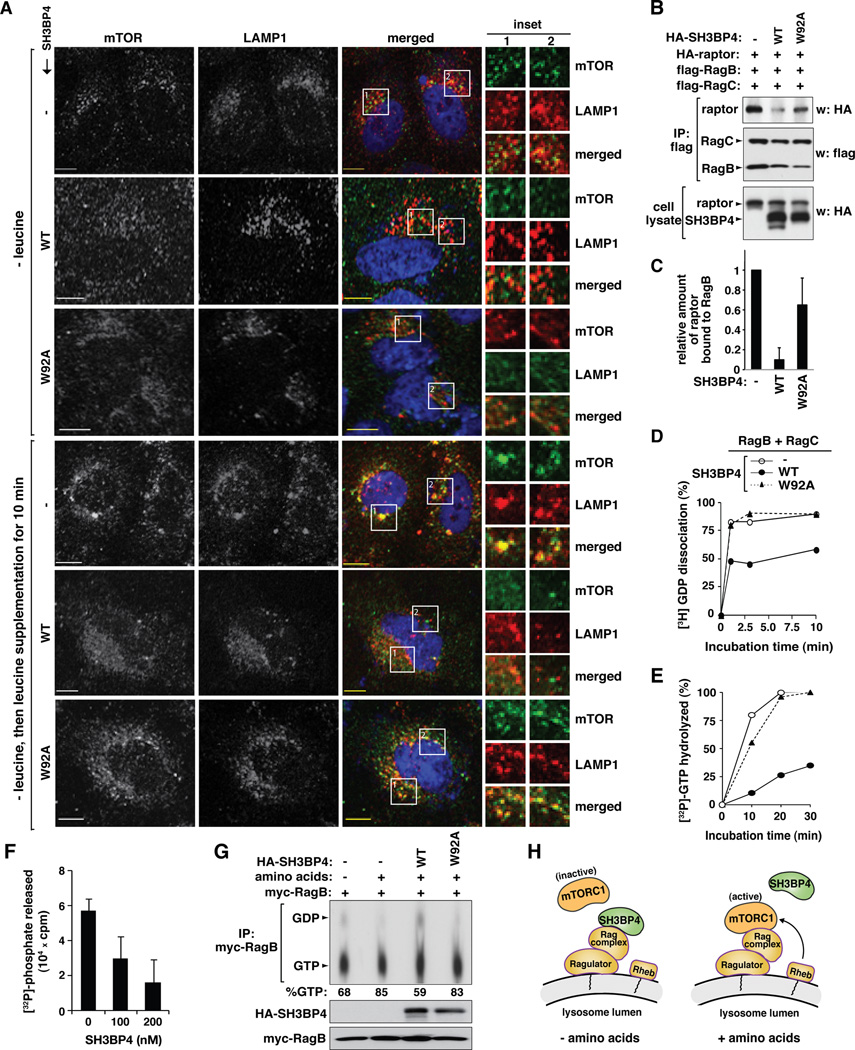
The results above support our hypothesis that SH3BP4 binds to the inactive Rag GTPase complex under conditions of amino acid starvation and inhibits the activation of Rag GTPase-mTORC1 signaling. A remaining question is how SH3BP4 binding inhibits the activity of the Rag GTPase complex. First, we wondered whether the inhibitory effect of SH3BP4 on mTOR localization to the lysosome can be attenuated by W92A mutation. SH3BP4 WT inhibited leucine-stimulated localization of mTOR to the lysosome, and this inhibitory effect was suppressed by W92A mutation (Figure 7A and Figure S7A). Consistent with this result, W92A mutation blunted the inhibitory effect of SH3BP4 on the interaction between raptor and Rag GTPases (Figure 7B and C). These results support our proposed model that the SH3 domain-dependent interaction between SH3BP4 and Rag GTPases is important to inhibit the raptor-Rag GTPase interaction and the recruitment of mTORC1 to the lysosome.
Figure 7. SH3BP4 inhibits the formation of active Rag GTPase complex.
(A) The SH3BP4-Rag interaction inhibits mTOR localization to the lysosome. GFP-tagged SH3BP4 WT or W92A was transiently expressed in HeLa cells. Control cells were transduced by GFP empty vector (−). The colocalization of endogenous mTOR (green) and LAMP1 (red) was analyzed as described in Fig. 5C. (B) The SH3BP4-Rag interaction is important for suppressing the raptor-Rag interaction. HA-tagged raptor and flag-tagged Rag GTPases were expressed with SH3BP4 WT or W92A in HEK293T cells. Control cells were transduced by empty vector (−) instead of SH3BP4 constructs. The amount of HA-raptor immunopurified with Rag GTPases was analyzed by WB. (C) Quantitation of raptor binding to RagB based on data from (B). Values are mean ± SD (n = 2). (D) SH3BP4 inhibits GDP dissociation from Rag GTPases, which is suppressed by W92A mutation. The y-axis represents the amount of 3H-GDP released from Rag GTPases as percent relative to the total amount of 3H-GDP bound to Rag GTPases prior to the addition of GTPγS. The used proteins were prepared from bacteria. The graph is a representative of three independent experiments. (E) SH3BP4 inhibits the GTP-hydrolysis activity of Rag GTPases, which is suppressed by W92A mutation. The amount of 32P released from Rag GTPases was analyzed over a time course and presented in the y-axis as percent relative to the GTP hydrolysis measured at thirty-minute incubation of Rag GTPases alone. The graph is a representative of three independent experiments. The same symbols were used for the graph as in (D). (F) The inhibitory effect of SH3BP4 on GTP hydrolysis by RagB-RagC complex was analyzed at 30 min of incubation with different concentrations of SH3BP4. The error bars represent mean ± SD (n= 4). (G) SH3BP4 inhibits amino acid-induced GTP loading onto RagB. HA-SH3BP4 WT or W92A was expressed with myc-RagB in HEK293T cells. Control cells were transduced by a mock vector (−) instead of SH3BP4 constructs. Cells were incubated with [32P] in the presence or absence of amino acids as described in Supplemental Experimental Procedures. The nucleotides bound on immunopurified myc-RagB were analyzed by thin layer chromatography followed by autoradiography. The number at the bottom indicates the % of GTP bound to RagB. (H) Model for the inhibitory function of SH3BP4 on Rag GTPase-mTORC1 signaling in response to amino acids.
A possible mechanism for SH3BP4 to inhibit the raptor-Rag GTPase interaction might be through suppression of the formation of the active Rag GTPase complex. Given that SH3BP4 preferentially binds to the inactive Rag GTPase complex, we considered a possibility that SH3BP4 might inhibit the conversion of GDP-bound inactive forms of RagA/B to the GTP-bound active forms. Incubation of Rag GTPases with SH3BP4 WT in vitro resulted in suppression of GDP dissociation from Rag GTPases (Figure 7D and Figure S7B and C). The suppressive effect by SH3BP4 WT was almost completely abolished by SH3BP4 W92A mutation (Figure 7D). We also tested whether a fragment containing the N-terminal 300 amino acids of SH3BP4 that encompasses the SH3 domain can suppress GDP dissociation from Rag GTPases. We found no significant effect with this fragment (Figure S7D and E), suggesting that a region of SH3BP4 other than the SH3 domain is required for the inhibitory effect. We predicted that the suppression of GDP dissociation should be accompanied by reduction in the GTP hydrolysis activity of Rag GTPases. As predicted, the incubation of RagB and RagC with SH3BP4 WT, but not W92A mutant, inhibited the GTP hydrolysis activity of Rag GTPases (Figure 7E and F and Figure S7F and G). Consistent with this result, GTP loading onto RagB in response to amino acids was suppressed by SH3BP4 WT but not by SH3BP4 W92A (Figure 7G). In line with this, GTP loading onto RagB was increased by SH3BP4 knockdown (Figure S7H). Combined, these results support our proposed model that SH3BP4 binding to the inactive Rag GTPase complex impairs the formation of the active Rag GTPase complex (Figure 7H).
DISCUSSION
In this study, we define SH3BP4 as a binding protein of Rag GTPases and a negative regulator of amino acid-mTORC1 signaling. SH3BP4 directly binds to the inactive Rag GTPase complex under amino acid starved conditions and inhibits the formation of the active complex. Consistent with this proposed mechanism, mTORC1 activity in SH3BP4-silenced cells was not completely inhibited in leucine-deprived conditions and was maintained at higher levels over a broad range of leucine concentrations in the medium compared to that in control cells (Figure 3B and Figure S3A). Such a function of SH3BP4 might contribute to reduction in the basal activity of mTORC1 or prevention of hyper-activation of mTORC1, and might be linked to cancer or other human diseases associated with hyperactive mTORC1 signaling. Consistent with this notion, analysis of human cancer genome databases revealed that deletions of the SH3BP4 locus are found in 10 of 14 cancer subtypes across 2594 tumors (Table S1). Among the 10 subtypes, three subtypes (breast, renal and non-small cell lung cancers) showed the highest frequency of deletions in the SH3BP4 locus with a statistical significance. Thus, SH3BP4 might be a potential candidate of tumor suppressor that functions in amino acid-Rag GTPase-mTORC1 signaling.
Although our study provides strong evidence for the inhibitory effects of SH3BP4 on Rag GTPases, how it regulates Rag GTPases needs further investigation. According to our study (Figure 2B and C), SH3BP4 binding to Rag GTPases requires not only GDP charging of RagB but also an unknown process induced by amino acid starvation. It is noteworthy that RagC binding to SH3BP4 depends on the nucleotide-loaded states of RagB that is in a dimeric complex with RagC (Figure 1F and Figure S1D). This suggests that the dimeric interaction somehow affects the binding of SH3BP4 to Rag GTPases. Because the RagBGDP-RagCGDP complex could bind to SH3BP4 more strongly than the RagBGDP-RagCGTP complex (Figure 1G), SH3BP4 might bind to RagCGDP as well as RagBGDP in the complex. A detailed analysis of the stoichiometry of the interaction between Rag GTPases and SH3BP4 is necessary to better understand how they interact in the complex and how SH3BP4 inhibits Rag GTPases.
We found that a high amount of Rag GTPases is isolated as a GTP-bound form under amino acid starved conditions (Figure 7G). Recent reports have also shown a similar result (Duran et al., 2011; Sancak et al., 2008). A possible explanation for such a high level of GTP-bound forms under amino acid starvation might be that Rag GTPases bound to GDP are unstable or immunopurified less efficiently compared to GTP-bound forms. Alternatively, the affinity of GDP towards Rag GTPases might be lower than that of GTP in vitro, thus resulting in loss of the GDP-bound forms during in vitro isolation of Rag GTPases. Supporting this possibility, RagC was shown to bind GDP with a significantly lower affinity than GTP (Sekiguchi et al., 2001). It is also possible that there might exist a population of Rag GTPases that are not regulated by amino acids.
Our in vitro assay of Rag GTPase activities revealed that SH3BP4 might not act as a GEF or a GAP for Rag GTPases (Figure 7D and E). The in vitro assay also suggested that SH3BP4 inhibits Rag GTPases without requiring other molecules. The inhibitory function of SH3BP4 is similar to its previously identified function toward dynamin (Tosoni et al., 2005). This implies that SH3BP4 might regulate multiple GTPases, at least including dynamin and Rag GTPases. Given that SH3BP4 functions are implicated in endocytosis and endosomal trafficking, it is tempting to speculate that the regulation of multiple GTPases by SH3BP4 might be important for a spatial and temporal coordination of endocytosis, vesicle trafficking and mTOR localization in response to amino acids and other cellular signals.
How SH3BP4 regulates Rag GTPases in relation to Ragulator, v-ATPase and LRS remains to be determined. Although SH3BP4 would not be a direct sensor of amino acids, conformational changes or post-translational modifications may occur in either SH3BP4 or Rag GTPases to alter the interaction between SH3BP4 and Rag GTPases in response to amino acids. Supporting this notion, multiple phosphorylation sites have been reported for SH3BP4 (Chen et al., 2009; Chiba et al., 2009; Pan et al., 2008). Those phosphorylations of SH3BP4 could potentially be an important mechanism for mTORC1 regulation by diverse intracellular signals as well as amino acids. Although the presence of SH3BP4 homologues in lower eukaryotes is not clear, it remains possible that a protein with a divergent sequence might exist in lower eukaryotes and play a similar role as SH3BP4. Alternatively, SH3BP4 might have evolved to meet the greater regulatory complexity of higher eukaryotes. Further studies on the function of SH3BP4 may provide important insight into the regulatory mechanism of amino acid-mTORC1 signaling.
EXPERIMENTAL PROCEDURES
Identification of RagB-binding Proteins
Cell extract from HEK293T cells that stably express flag-tagged RagB was run through flag affinity chromatography. The fraction that was bound to the column was trypsinized and analyzed by mass spectrometry. A detailed procedure is described in the Supplemental Experimental Procedures.
Plasmids
Human and mouse SH3BP4 cDNAs obtained from the ATCC (Image# 2988648) and Open Biosystems (Image# 4947387) were cloned into pRK5 expression vector by PCR amplification. Rag GTPase mutant constructs were generated by site-directed mutagenesis. pLKO.1 shRNA vector was used to knockdown human or mouse SH3BP4. The target sequences are listed in the Supplemental Experimental Procedures.
Cell image analysis
The colocalization of proteins was analyzed by an Olympus FluoView 1000 IX2 inverted confocal microscope and a Deltavision Personal DV microscope (Applied Precision). A detailed procedure is described in the Supplemental Experimental Procedures.
In vitro GDP Dissociation and GTPase Assay
In vitro GDP dissociation and GTP hydrolysis assay were performed using Rag GTPases and SH3BP4 purified from bacteria and using [3H]-GDP and [γ32P]-GTP, respectively, as described in the Supplemental Experimental Procedures.
Other experimental procedures
Other experimental procedures, including materials, mass spectrometry, mutagenesis, co-immunoprecipitation, Western blotting, lentiviral transduction, recombinant protein production, cell number and size measurement, autophagy assay, and analysis of genome from tumor samples, are described in Supplemental Experimental Procedures.
Supplementary Material
ACKNOWLEDGEMENTS
We thank K. Martemyanov for GTPase assay; W. J. Hong for Arl1 clones; D. Billadeau for TfR and dynamin antibodies; P. De Camilli for dynamin KO MEFs; H. Towle, A. Lange, and Kim lab members for comments; Mass Spectrometry and Proteomics Center and Supercomputing Institute at the U of Minnesota for instrumentation and bioinformatic tools. This study was supported by the NIH (DK050456, DK083474, and GM097057) and the ADA (7-07-CD-08).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Information
Supplemental Information includes Supplemental Experimental Procedures, seven figures and one table.
There is no financial conflict of interest with this study.
REFERENCES
- Binda M, Peli-Gulli MP, Bonfils G, Panchaud N, Urban J, Sturgill TW, Loewith R, De Virgilio C. The Vam6 GEF controls TORC1 by activating the EGO complex. Mol Cell. 2009;35:563–573. doi: 10.1016/j.molcel.2009.06.033. [DOI] [PubMed] [Google Scholar]
- Bryk B, Hahn K, Cohen SM, Teleman AA. MAP4K3 regulates body size and metabolism in Drosophila. Dev Biol. 2010;344:150–157. doi: 10.1016/j.ydbio.2010.04.027. [DOI] [PubMed] [Google Scholar]
- Chen RQ, Yang QK, Lu BW, Yi W, Cantin G, Chen YL, Fearns C, Yates JR, 3rd, Lee JD. CDC25B mediates rapamycin-induced oncogenic responses in cancer cells. Cancer Res. 2009;69:2663–2668. doi: 10.1158/0008-5472.CAN-08-3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba S, Tokuhara M, Morita EH, Abe S. TTP at Ser245 phosphorylation by AKT is required for binding to 14-3-3. J Biochem. 2009;145:403–409. doi: 10.1093/jb/mvn178. [DOI] [PubMed] [Google Scholar]
- Dubouloz F, Deloche O, Wanke V, Cameroni E, De Virgilio C. The TOR and EGO protein complexes orchestrate microautophagy in yeast. Mol Cell. 2005;19:15–26. doi: 10.1016/j.molcel.2005.05.020. [DOI] [PubMed] [Google Scholar]
- Dunlevy JR, Berryhill BL, Vergnes JP, SundarRaj N, Hassell JR. Cloning, chromosomal localization, and characterization of cDNA from a novel gene, SH3BP4, expressed by human corneal fibroblasts. Genomics. 1999;62:519–524. doi: 10.1006/geno.1999.5994. [DOI] [PubMed] [Google Scholar]
- Duran A, Amanchy R, Linares JF, Joshi J, Abu-Baker S, Porollo A, Hansen M, Moscat J, Diaz-Meco MT. p62 is a key regulator of nutrient sensing in the mTORC1 pathway. Mol Cell. 2011;44:134–146. doi: 10.1016/j.molcel.2011.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erpel T, Superti-Furga G, Courtneidge SA. Mutational analysis of the Src SH3 domain: the same residues of the ligand binding surface are important for intra- and intermolecular interactions. EMBO J. 1995;14:963–975. doi: 10.1002/j.1460-2075.1995.tb07077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findlay GM, Yan L, Procter J, Mieulet V, Lamb RF. A MAP4 kinase related to Ste20 is a nutrient-sensitive regulator of mTOR signalling. Biochem J. 2007;403:13–20. doi: 10.1042/BJ20061881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fingar DC, Blenis J. Target of rapamycin (TOR): an integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene. 2004;23:3151–3171. doi: 10.1038/sj.onc.1207542. [DOI] [PubMed] [Google Scholar]
- Gao M, Kaiser CA. A conserved GTPase-containing complex is required for intracellular sorting of the general amino-acid permease in yeast. Nat Cell Biol. 2006;8:657–667. doi: 10.1038/ncb1419. [DOI] [PubMed] [Google Scholar]
- Goberdhan DC, Boyd CA. mTOR: dissecting regulation and mechanism of action to understand human disease. Biochem Soc Trans. 2009;37:213–216. doi: 10.1042/BST0370213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JM, Jeong SJ, Park MC, Kim G, Kwon NH, Kim HK, Ha SH, Ryu SH, Kim S. Leucyl-tRNA Synthetase Is an Intracellular Leucine Sensor for the mTORC1-Signaling Pathway. Cell. 2012;149 doi: 10.1016/j.cell.2012.02.044. in press. [DOI] [PubMed] [Google Scholar]
- Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, Tokunaga C, Avruch J, Yonezawa K. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110:177–189. doi: 10.1016/s0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose E, Nakashima N, Sekiguchi T, Nishimoto T. RagA is a functional homologue of S. cerevisiae Gtr1p involved in the Ran/Gsp1-GTPase pathway. J Cell Sci. 1998;111(Pt 1):11–21. doi: 10.1242/jcs.111.1.11. [DOI] [PubMed] [Google Scholar]
- Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS Lett. 2010;584:1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, Oppliger W, Jenoe P, Hall MN. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell. 2002;10:457–468. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3:542–545. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- Musacchio A, Saraste M, Wilmanns M. High-resolution crystal structures of tyrosine kinase SH3 domains complexed with proline-rich peptides. Nat Struct Biol. 1994;1:546–551. doi: 10.1038/nsb0894-546. [DOI] [PubMed] [Google Scholar]
- Narita M, Young AR, Arakawa S, Samarajiwa SA, Nakashima T, Yoshida S, Hong S, Berry LS, Reichelt S, Ferreira M, et al. Spatial coupling of mTOR and autophagy augments secretory phenotypes. Science. 2011;332:966–970. doi: 10.1126/science.1205407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan C, Gnad F, Olsen JV, Mann M. Quantitative phosphoproteome analysis of a mouse liver cell line reveals specificity of phosphatase inhibitors. Proteomics. 2008;8:4534–4546. doi: 10.1002/pmic.200800105. [DOI] [PubMed] [Google Scholar]
- Panchamoorthy G, Fukazawa T, Stolz L, Payne G, Reedquist K, Shoelson S, Songyang Z, Cantley L, Walsh C, Band H. Physical and functional interactions between SH2 and SH3 domains of the Src family protein tyrosine kinase p59fyn. Mol Cell Biol. 1994;14:6372–6385. doi: 10.1128/mcb.14.9.6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi T, Hirose E, Nakashima N, Ii M, Nishimoto T. Novel G proteins, Rag C and Rag D, interact with GTP-binding proteins, Rag A and Rag B. J Biol Chem. 2001;276:7246–7257. doi: 10.1074/jbc.M004389200. [DOI] [PubMed] [Google Scholar]
- Selman C, Tullet JM, Wieser D, Irvine E, Lingard SJ, Choudhury AI, Claret M, Al-Qassab H, Carmignac D, Ramadani F, et al. Ribosomal protein S6 kinase 1 signaling regulates mammalian life span. Science. 2009;326:140–144. doi: 10.1126/science.1177221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamji AF, Nghiem P, Schreiber SL. Integration of growth factor and nutrient signaling: implications for cancer biology. Mol Cell. 2003;12:271–280. doi: 10.1016/j.molcel.2003.08.016. [DOI] [PubMed] [Google Scholar]
- Tosoni D, Puri C, Confalonieri S, Salcini AE, De Camilli P, Tacchetti C, Di Fiore PP. TTP specifically regulates the internalization of the transferrin receptor. Cell. 2005;123:875–888. doi: 10.1016/j.cell.2005.10.021. [DOI] [PubMed] [Google Scholar]
- Um SH, Frigerio F, Watanabe M, Picard F, Joaquin M, Sticker M, Fumagalli S, Allegrini PR, Kozma SC, Auwerx J, Thomas G. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature. 2004;431:200–205. doi: 10.1038/nature02866. [DOI] [PubMed] [Google Scholar]
- Vander Haar E, Lee SI, Bandhakavi S, Griffin TJ, Kim DH. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat Cell Biol. 2007;9:316–323. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Yan L, Mieulet V, Burgess D, Findlay GM, Sully K, Procter J, Goris J, Janssens V, Morrice NA, Lamb RF. PP2A T61 epsilon is an inhibitor of MAP4K3 in nutrient signaling to mTOR. Mol Cell. 2010;37:633–642. doi: 10.1016/j.molcel.2010.01.031. [DOI] [PubMed] [Google Scholar]
- Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H-ATPase. Science. 2011;334:678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.