SUMMARY
The adaptor protein Bcl10 is a critically important mediator of T cell receptor (TCR)-to-NF-κB signaling. Bcl10 degradation is a poorly understood biological phenomenon suggested to reduce TCR activation of NF-κB. Here we have shown that TCR engagement triggers the degradation of Bcl10 in primary effector T cells, but not in naïve T cells. TCR engagement promoted K63-polyubiquitination of Bcl10, causing Bcl10 association with the autophagy adaptor, p62. Paradoxically, p62 binding was required for both Bcl10 signaling to NF-κB and gradual degradation of Bcl10 by autophagy. Bcl10 autophagy was highly selective, as it spared Malt1, a direct Bcl10 binding partner. Blockade of Bcl10 autophagy enhanced TCR activation of NF-κB. Together, these data demonstrate that selective autophagy of Bcl10 is a pathway-intrinsic homeostatic mechanism that modulates TCR signaling to NF-κB in effector T cells. This homeostatic process may protect T cells from adverse consequences of unrestrained NF-κB activation, such as cellular senescence.
INTRODUCTION
Antigen stimulation of the T cell receptor (TCR) initiates a complex signaling cascade, culminating in the initiation of a transcriptional program, which drives T cell proliferation and differentiation. The NF-κB transcription factor is a particularly important target of TCR signaling, playing a central role in driving entry into cell cycle via stimulating transcription of numerous effector molecules, including interleukin-2 (IL-2) (Skaug et al., 2009; Vallabhapurapu and Karin, 2009). The adaptor protein, Bcl10, plays a key role in transmitting signals from the TCR to NF-κB. In the absence of Bcl10, T cells are unable to efficiently proliferate and differentiate in response to TCR engagement (Ruland et al., 2001; Schulze-Luehrmann and Ghosh, 2006; Thome et al., 2010).
Previous studies have suggested that a TCR-dependent mechanism targets Bcl10 for proteolysis, in concert with activation of NF-κB. Although these studies suggested that Bcl10 degradation may be a mechanism to limit TCR signals to NF-κB, the data supporting such a model are limited. Additionally, the molecular mechanism of Bcl10 degradation remains highly controversial, with different groups publishing data supporting diverse degradatory mechanisms (Hu et al., 2006; Lobry et al., 2007; Scharschmidt et al., 2004; Wu and Ashwell, 2008; Zeng et al., 2007). Also, almost all experiments in previous studies were performed with long-term tumor cell lines, such as Jurkat. It is therefore unclear to what extent the phenomenon of Bcl10 degradation is relevant to the biology of primary T cells.
Macroautophagy (henceforth termed autophagy) is a cellular process by which cytosolic constituents are engulfed in double-membrane encapsulated vesicles, followed by delivery to lysosomes for degradation. Autophagy can serve as a survival mechanism under conditions of metabolic starvation and growth factor withdrawal, via non-selective degradation of cytosolic constituents for re-use (Chaturvedi and Pierce, 2009; Lunemann and Munz, 2009). Emerging evidence suggests that autophagy can also target specific proteins for destruction. Specifically, data suggest that this specialized type of autophagy, referred to as selective autophagy, can target specific proteins or organelles for degradation. In general, proteins enter the selective autophagy pathway following post-translational modification with K63-polyubiquitin chains. K63-polyubiquitinated proteins are then bound by p62 (also called SQSTM-1) or by functionally related autophagy adaptor proteins, followed by engulfment in autophagosomes (Kirkin et al., 2009b; Kraft et al., 2010; Levine et al., 2011).
Interestingly, recent data have shown that TCR stimulation induces autophagy (Li et al., 2006), that autophagy contributes to T cell survival and antigen-dependent proliferation (Pua et al., 2007), and that autophagy-deficient T cells produce more interleukin-2 (IL-2) than their wild-type counterparts (Jia et al., 2011). These data suggest that TCR-dependent autophagy limits the production of T cell effector molecules such as IL-2 that are essential mediators of T cell proliferative responses. However, the mechanistic link between TCR-induced autophagy and the modulation of T cell effector responses remains to be determined. In this study, we have established that Bcl10 is degraded via selective autophagy, reducing TCR activation of NF-κB and limiting NF-κB-dependent effector responses. This study therefore implicates selective autophagy as a mechanism that modulates antigen receptor signaling and, more generally, as an intrinsic mechanism that can restrain activation of a ligand-initiated signaling cascade.
RESULTS
Bcl10 associates with autophagosomes following TCR stimulation
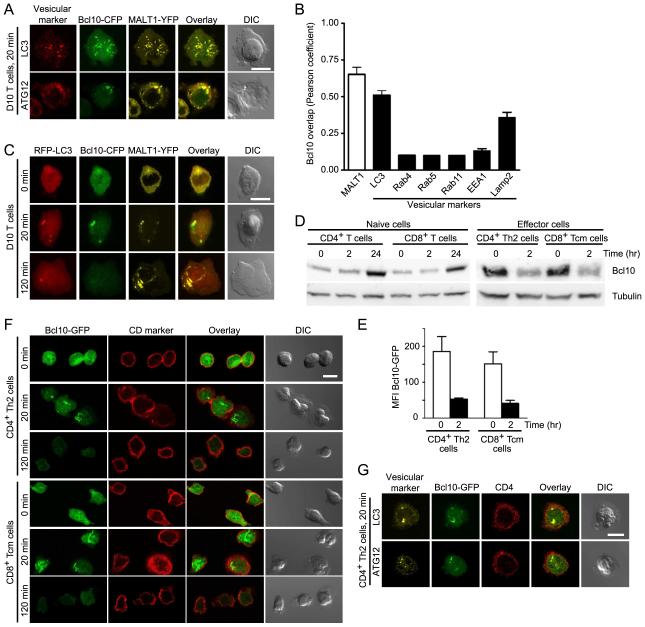
Our previous microscopy studies demonstrated that TCR signaling leads to oligomerization of Bcl10 and Malt1, forming punctate cytosolic structures called POLKADOTS, which may include a vesicular component (Rossman et al., 2006). To identify vesicular structures associated with POLKADOTS, we used anti-CD3 to activate D10 T cells expressing Bcl10-cyan fluorescent protein (CFP) and MALT1-yellow fluorescent protein (YFP). We also stained these T cells with antibodies directed against a variety of intracellular vesicle markers. Confocal microscopy analysis showed that LC3 and ATG12, markers for autophagosomes, closely co-localized with Bcl10 and MALT1 in POLKADOTS (Fig. 1A). Pearson’s co-localization analysis showed higher correlation between Bcl10 and LC3 compared to the other vesicular markers (Fig. 1B). Notably, Lamp2, a lysosomal marker, also showed increased co-localization with POLKADOTS (Fig. 1B).
Figure 1.
T cell activation leads to Bcl10 co-localization with markers of the autophagy-lysosomal degradation system and subsequent Bcl10 degradation. (A) D10 T cells expressing Bcl10-CFP and MALT1-YFP were stimulated with anti-CD3 for 20 min and stained with antibodies to the indicated autophagosome markers, to assess co-localization. (B) Pearson coefficient analysis was calculated by intensity correlation analysis of Bcl10-CFP fluorescence with MALT1-YFP fluorescence (white bar) or with fluorescence from antibodies specific for the indicated vesicular makers (black bars). For Pearson coefficient analysis, at least 30 cells were measured in each group. Values are means, error bars, SEM. (C) D10 T cells expressing RFP-LC3, Bcl10-CFP, and MALT1-YFP were stimulated with anti-CD3 and assessed by confocal microscopy for co-localization. (D) Primary naïve and effector T cells were stimulated with anti-CD3+anti-CD28 for the indicated times. Lysates were analyzed by immunoblotting for Bcl10 degradation. Blots were also probed with anti-α-tubulin as a loading control. Data are representative of two experiments. (E) CD4+ Th2 cells and CD8+ Tcm cells expressing Bcl10-GFP were stimulated for the indicated times with anti-CD3+anti-CD28. Flow cytometry was used to assess Bcl10-GFP degradation. Values are median Bcl10-GFP fluorescence (error bars, SEM), generated from three independent experiments. Representative flow cytometry histograms are shown in Supplemental Fig. 1C. (F,G) Primary effector T cells expressing Bcl10-GFP were stimulated with anti-CD3+anti-CD28 for the indicated times. Cells were stained with anti-CD4 or anti-CD8 to identify cell lineage (F); or with anti-LC3 and anti-ATG12 to assess co-localization with Bcl10-GFP (G). Images are representative of two independent experiments. Scale bar = 10μm. See also Figure S1.
To confirm that autophagosomes associate with POLKADOTS, we stably expressed a red fluorescent protein (RFP)-LC3 recombinant protein in D10 cells. At 20 min post-activation, D10 T cells showed induction of punctate cytosolic RFP-LC3 signals, representing TCR-induced autophagosomes (Li et al., 2006). RFP-LC3+ autophagosomes co-localized with cytosolic Bcl10 and MALT1-containing POLKADOTS (Fig. 1C). At 120 min, the Bcl10-CFP signal was lost, reflecting post-activation Bcl-10 degradation (Hu et al., 2006; Lobry et al., 2007; Scharschmidt et al., 2004; Wu and Ashwell, 2008; Zeng et al., 2007), and the remaining MALT1-YFP was coalesced into one or several large aggregated structures. Together, these co-localization data suggest that POLKADOTS interact with vesicles of the autophagy-lysosomal degradation system.
Bcl10 degradation occurs in effector T cells, but not in naïve T cells
To assess whether or not Bcl10 degradation occurs in primary T cells, we purified CD4+ and CD8+ T cells from C57BL/6 mice. Cells were stimulated with anti-CD3+anti-CD28 and analyzed by immunoblotting to measure Bcl10. We observed no change in Bcl10 at 0 and 2 hr post-stimulation and increased Bcl10 by 24 hr stimulation (Fig. 1D). To reconcile this lack of Bcl10 degradation in stimulated naïve primary T cells with the clear Bcl10 degradation observed in D10 T cells by our group (Fig. 1C and data not shown) and in the Jurkat T cell line and additional cell types by others (Hu et al., 2006; Lobry et al., 2007; Scharschmidt et al., 2004; Wu and Ashwell, 2008; Zeng et al., 2007), we hypothesized that Bcl10 degradation might be a function of T cell differentiation, since D10 is a terminally differentiated Th2 cell clone. To test this hypothesis, we in vitro differentiated CD4+ Th2 effector cells and CD8+ Tcm cells (Fig. S1A, B). These differentiated effectors were then re-stimulated with anti-CD3+anti-CD28, and Bcl10 degradation was assessed via immunoblotting. Both CD4+ Th2 cells and CD8+ Tcm cells showed reduced Bcl10 at 2 hr compared to unstimulated T cells (Fig. 1D). As a second assay to confirm Bcl10 degradation in primary effector cells, we expressed Bcl10-GFP in CD4+ Th2 and CD8+ Tcm cells. Flow cytometry analysis showed reduced median fluorescence of Bcl10-GFP at 2 hr post-anti-CD3+anti-CD28 stimulation in both effector cell types (Fig. 1E, Fig. S1C).
We next examined Bcl10 re-distribution in primary effector T cells. We stimulated CD4+ Th2 cells for 0 min and 20 min, followed by antibody staining and confocal microscopy to visualize endogenous Bcl10. This analysis showed that Bcl10 exhibited a diffuse distribution in the absence of stimulation, whereas Bcl10 formed distinct cytosolic clusters following 20 min of anti-CD3+anti-CD28 stimulation (Fig. S1D). Therefore, endogenous Bcl10 and Bcl10-GFP show highly similar redistribution patterns in primary T effector cells.
Using CD4+ Th2 and CD8+ Tcm cells infected with a Bcl10-GFP retrovirus, we then performed an anti-CD3+anti-CD28 stimulation time course. Via confocal microscopy analysis, we observed formation of Bcl10-GFP cytosolic clusters at 20 min, followed by substantial Bcl10-GFP degradation at 2 hr (Fig. 1F). Also, Bcl10-GFP clusters co-localized with the autophagosome markers, LC3 and ATG12 (Fig. 1G), consistent with the D10 T cell phenotype (Fig. 1C). Bcl10-GFP co-localization with ATG12 also occurred in response to D10 T cell stimulation by cognate antigen presented by an antigen presenting cell (APC) (Fig. S1E). Finally, autophagosome association with Bcl10 was not associated with induction of apoptosis or other mechanisms of cell death, as there was minimal activation of caspase 3 (Fig. S1F) in stimulated D10 T cells. Moreover, the fraction of stimulation-induced cell death was <5% during the first 2 hr of anti-CD3 stimulation, increasing to only ~7% by 8 hr post-stimulation (Fig. S1G). Together, the data in Fig. 1 demonstrate that effector, but not naïve, T cells degrade Bcl10 in response to TCR stimulation. Additionally, TCR-dependent Bcl10 degradation is temporally correlated with close spatial association between Bcl10 cytosolic clusters (POLKADOTS) and autophagosomes. These data suggest that POLKADOTS may be a site of Bcl10 degradation via autophagy.
TCR stimulation triggers K63-polyubiquitination of Bcl10 and Bcl10-p62 physical association
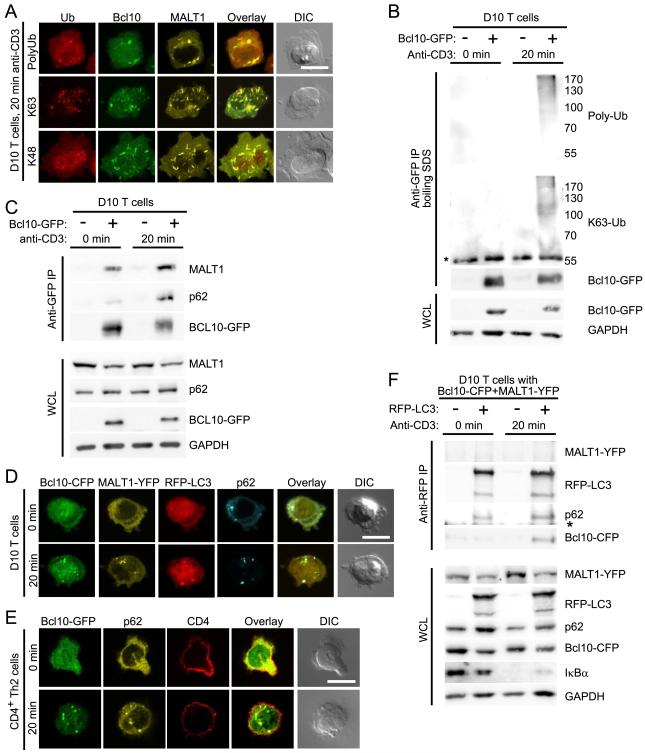
Although signals for selective autophagy of individual proteins are not yet completely understood, both monoubiquitination and K63-polyubiquitination can target proteins for autophagy, whereas K48-polyubiquitinated proteins are typically degraded by the proteasome (Kirkin et al., 2009b; Kraft et al., 2010). A previous study reported that Bcl10 is modified by both K63- and K48-polyubiquitin chains following TCR stimulation (Wu and Ashwell, 2008). To assess whether one or more proteins in POLKADOTS are modified by polyubiquitin, we stained anti-CD3 stimulated D10 T cells with a general antibody against polyubiquitin, as well as with antibodies specific for either K48- or K63-polyubiquitin. Confocal microscopy showed that POLKADOTS contained polyubiquitinated proteins. Furthermore, the K63-polyubiquitin antibody intensely labeled the POLKADOTS, whereas the anti-K48-polyubiquitin staining was primarily confined to the nucleus, demonstrating that proteins within POLKADOTS are primarily modified by K63-polyubiquitin chains (Fig. 2A). To assess whether Bcl10 is directly K63-polyubiquitinated in D10 T cells, we immunoprecipitated Bcl10-GFP (following boiling in SDS), using a GFP antibody. Immunoblotting with both the general polyubiquitin antibody and the K63-polyubiquitin antibody revealed a high molecular weight smear only in anti-CD3 activated D10 cells expressing Bcl10-GFP (Fig. 2B). These data therefore support previous observations that Bcl10 is K63-polyubiquitinated in response to TCR ligation (Wu and Ashwell, 2008).
Figure 2.
K63-polyubiquitinated Bcl10 interacts with p62. (A) D10 T cells expressing Bcl10-CFP and MALT1-YFP were stimulated for 20 min with anti-CD3, followed by staining with anti-polyubiquitin, anti-K48-polyubiquitin, or anti-K63-polyubiquitin. (B) Unstimulated or anti-CD3 stimulated D10 T cells with or without Bcl10-GFP were lysed and heated at 95°C for 5 min with 1% SDS to disrupt protein-protein interactions. Bcl10-GFP was immunoprecipitated using anti-GFP, followed by immunoblot analysis to detect Bcl10-GFP ubiquitination. (C) D10 T cells expressing or not expressing Bcl10-GFP were stimulated with anti-CD3 for the indicated times and lysed. Bcl10-GFP was recovered by immunoprecipitation using anti-GFP, followed by immunoblot analysis with the indicated antibodies to detect Bcl10 binding partners. (D) Confocal microscopy analysis of D10 T cells expressing Bcl10-CFP, MALT1-YFP, RFP-LC3, with 0 or 20 min anti-CD3 stimulation, to assess co-localization between proteins. (E) CD4+ Th2 cells expressing Bcl10-GFP were stimulated for the indicated times with anti-CD3+anti-CD28. Cells were stained with anti-p62 and anti-CD4, and confocal microscopy was used to assess co-localization between Bcl10 and p62. (F) D10 T cells expressing Bcl10-CFP and MALT1-YFP, with or without RFP-LC3, were stimulated for the indicated times with anti-CD3. Cells were lysed and RFP-LC3 was immunoprecipitated, using anti-RFP. Immunoblotting with the indicated antibodies was used to detect LC3 binding partners. (*) indicates position of Ig heavy chain. WCL, whole cell lysates; Ub, ubiquitin. Scale bar = 10μm. Data representative of two (B,C,F) or three (A,D,E) independent experiments. See also Figure S2.
Selective autophagy of K63-polyubiquitinated proteins is mediated by one of several known autophagy adaptor proteins. Interestingly, p62 is both an autophagy adaptor and a protein implicated in promoting NF-κB activation downstream of various receptors (Moscat and Diaz-Meco, 2009), including the TCR (Martin et al., 2006). We therefore performed several experiments to assess whether POLKADOTS are sites of interaction between p62 and K63-polyubiquitinated Bcl10. Firstly, we immunoprecipitated Bcl10-GFP from unstimulated and anti-CD3-stimulated D10 T cells. Immunoblotting revealed that while Bcl10-MALT1 association was relatively insensitive to stimulation, the Bcl10-p62 interaction increased following TCR stimulation (Fig. 2C). We next used confocal microscopy to visualize endogenous p62 in D10 T cells expressing Bcl10-CFP, MALT1-YFP, and RFP-LC3. As expected, in unstimulated T cells, p62 showed primarily a punctate distribution (p62 “speckles”), due to the self-aggregation that is induced by the N-terminal PB1 domain (Moscat and Diaz-Meco, 2009). Upon anti-TCR stimulation, we observed p62 speckles co-localizing with autophagosomes (RFP-LC3) and Bcl10+Malt1 POLKADOTS (Fig. 2D). We also confirmed these data via expressing YFP-tagged p62 in D10 T cells and observing co-localization of YFP-p62 and Bcl10-CFP POLKADOTS following anti-CD3 stimulation (Fig. S2A). We obtained similar results with CD4+ Th2 cells, in which we observed cytosolic p62 speckles co-localizing with Bcl10-GFP POLKADOTS, following anti-CD3+anti-CD28 stimulation (Fig. 2E). Endogenous Bcl10 also showed similar co-localization with endogenous p62 in CD4+ Th2 cells (Fig. S2B). Additionally, we observed Bcl10-GFP association with p62 in response to antigen+APC stimulation (Fig. S2C). Finally, antibody staining data showed that endogenous NBR1, another autophagy adaptor which can oligomerize with p62 (Kirkin et al., 2009a), also colocalized with Bcl10-GFP POLKADOTS (Fig. S2D).
Previous studies have suggested that p62 interacts with Malt1 and that Malt1 can be K63-polyubiquitinated in TCR-dependent manner (Duwel et al., 2009; Martin et al., 2006). Thus, published data suggest that polyubiquitination of either Bcl10 or Malt1 (or both proteins) could account for the intense K63-polyubiquitin labeling of POLKADOTS and the association with p62. However, the fact that Bcl10, but not Malt1, was rapidly degraded following TCR activation suggested that only Bcl10 enters the autophagy pathway. Therefore, to ascertain if Bcl10, and not MALT1, interacts with autophagosomes, we immunoprecipitated LC3 from lysates of D10 T cells, using an RFP antibody. As expected, p62 co-immunoprecipitated with RFP-LC3 in lysates from both stimulated and unstimulated T cells, showing that p62 constitutively binds LC3 (Fig. 2F). In contrast, Bcl10 co-immunoprecipitated with RFP-LC3 only after anti-CD3 stimulation. These data suggest that a TCR-dependent process, such as K63-polyubiquitination of Bcl10, is required to promote Bcl10 interaction with p62 and LC3. Importantly, we failed to detect MALT1 in the RFP-LC3 immunoprecipitates, consistent with the observation that there is no appreciable degradation of Malt1 in response to TCR engagement. Because Bcl10 and Malt1 are constitutively associated within T cells (Thome et al., 2010) (see also Fig. 2C), and there was no detectable Malt1 in complexes containing LC3, p62, and Bcl10 (Fig. 2F), our data suggest that a highly selective mechanism targets Bcl10 for association with autophagosomes. Together, the data in Fig. 2 suggest that, in response to TCR signaling, Bcl10 is K63-polyubiquitinated and physically associates with p62 speckles and LC3+ autophagosomes.
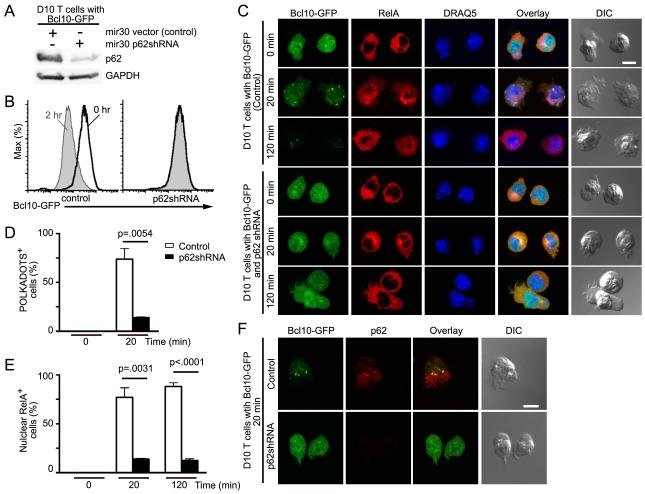
Expression of p62 is required for TCR-dependent Bcl10 degradation
To assess whether p62 plays a role in Bcl10 degradation, D10 cell lines expressing Bcl10-GFP were infected with either the empty shRNA vector or a p62-shRNA (p62-silenced) (Fig. 3A). We stimulated these cell lines with anti-CD3 (0 hr and 2 hr) and quantified Bcl10-GFP via flow-cytometry. While control cells showed a substantial decrease in Bcl10-GFP fluorescence in response to anti-CD3 stimulation, Bcl10-GFP fluorescence remained unchanged in the p62-silenced cells (Fig. 3B). We then performed confocal microscopy experiments to assess NF-κB activation, by identifying cells with RelA nuclear translocation. In the absence of stimulation, control and p62-silenced cells showed diffuse Bcl10 and cytosolic RelA, as expected. Following 20 min of anti-CD3 stimulation, Bcl10 formed POLKADOTS in control cells, while remaining diffusely localized in p62-silenced cells. Also, RelA translocated to the nucleus in control cells, whereas it remained cytosolic in p62-silenced cells. At 120 min, control cells had greatly reduced Bcl10 fluorescence, reflecting Bcl10 degradation, whereas Bcl10 fluorescence remained unchanged in p62-silenced cells, consistent with flow cytometry data. Also, we continued to observe RelA in the nucleus in control cells, whereas RelA remained cytosolic in the p62-silenced cell line (Fig. 3C). Quantification of data in the Fig. 3C experiment revealed a significant decrease in both cells forming Bcl10 POLKADOTS and cells with nuclear RelA in the p62-silenced line. Moreover, those p62-silenced cells that formed Bcl10 POLKADOTS were the same cells that exhibited RelA nuclear translocation (Fig. 3D-E and data not shown). Staining with anti-p62 revealed that the small percentage of p62-silenced cells with Bcl10 POLKADOTS retained detectable amounts of p62 that co-localized with POLKADOTS (Fig. S3A).
Figure 3.
p62 silencing inhibits RelA nuclear translocation and blocks Bcl10 degradation. (A) D10 T cells expressing Bcl10-GFP plus either mir30 shRNA vector or mir30 p62shRNA were analyzed by immunoblotting to detect p62 expression. (B-E) D10 T cells expressing Bcl10-GFP plus mir30 shRNA vector or mir30 p62shRNA were stimulated with anti-CD3 for the indicated times. Cells were analyzed by flow cytometry to detect degradation of Bcl10-GFP (B); or stained with anti-RelA and the DNA dye, DRAQ5, and analyzed by confocal microscopy to assess RelA nuclear translocation (C). Graphs showing percentage of cells in (C) forming Bcl10-GFP clusters (POLKADOTS) (D) or with RelA nuclear translocation (E). (F) D10 T cells expressing Bcl10-GFP plus mir30 shRNA vector or mir30 p62shRNA were stimulated for 20 min with anti-CD3 and stained with anti-p62. p62 silencing and p62-Bcl10 co-localization were assessed by confocal microscopy. Means (+/− SEM) were calculated by counting at least 20 cells from each of three experiments. Scale bar = 10μm. Data are representative of two (A,B) or three (C,F) independent experiments. See also Figure S3.
To exclude the possibility that the observed phenotypes in the p62-silenced cell line were due to an unrecognized general defect in TCR signal transduction, we assessed phosphorylation of the kinases ERK1 and ERK2, which are activated via a TCR-dependent mechanism that is distinct from the NF-κB cascade (Smith-Garvin et al., 2009). Although TCR-dependent Bcl10 degradation occurred in control cells and not in p62-silenced cells, similar ERK1 and ERK2 phosphorylation was observed in both cell lines (Fig. S3B). Finally, we assessed whether p62 acts upstream or downstream of the IκB kinase complex (IKK), which initiates terminal NF-κB activation via promoting degradation of IκBα. We stimulated D10 T cells or a D10 p62-silenced cell line with anti-CD3 and measured phosphorylation of IKKα+IKKβ and degradation of endogenous Bcl10 by immunoblotting. Whereas unmanipulated D10 T cells exhibited both Bcl10 degradation and robust phosphorylation of the IKK complex in response to anti-CD3, there was little to no IKK phosphorylation or Bcl10 degradation in p62-silenced cells (Fig. S3C). Thus, consistent with previous data (Martin et al., 2006), this experiment shows that p62 acts upstream of IKK in the TCR-to-NF-κB pathway. Taken together, the data in Figs. 3 and S3 show that expression of p62 in T cells is necessary for both TCR-dependent activation of NF-κB and TCR-directed degradation of Bcl10.
Polyubiquitination of Bcl10 is required for Bcl10 degradation
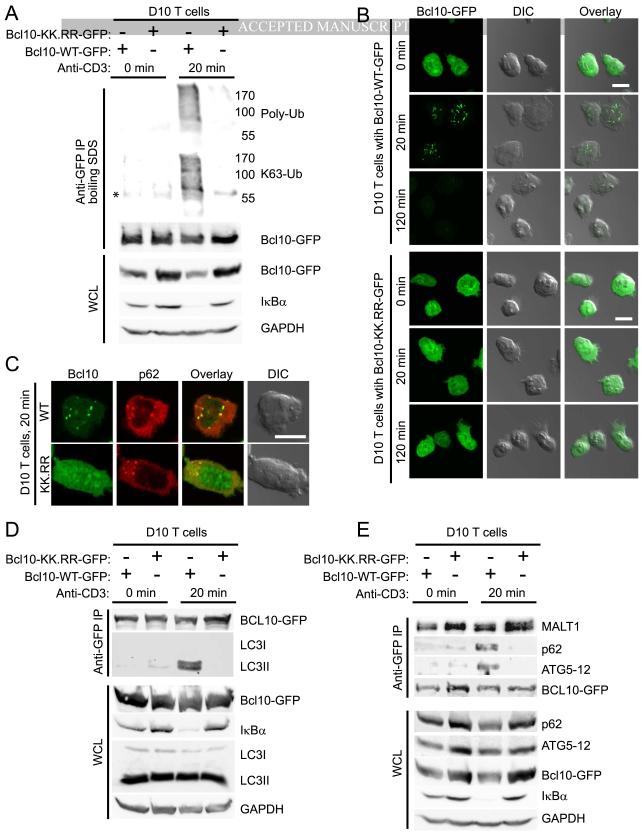
A recent study using Jurkat cells has established that polyubiquitination of human Bcl10 occurs at lysines 31 and 63 (Wu and Ashwell, 2008). Furthermore, blockade of Bcl10 polyubiquitination by mutation of these residues to arginines dramatically inhibits TCR mediated NF-κB activation and Bcl10 degradation (Wu and Ashwell, 2008). To confirm these data, we expressed GFP-tagged wild-type and K31R,K63R (KK.RR) Bcl10 in the D10 Th2 T cell clone. Cells were stimulated for 0 or 20 min with anti-CD3, followed by immunoprecipitation of Bcl10-GFP. Immunoblotting revealed K63-polyubiquitination of wild-type Bcl10 and greatly reduced polyubiquitination of the Bcl10-KK.RR mutant (Fig. 4A). The KK.RR mutant was also deficient in NF-κB activation (as assessed by degradation of IκBα), consistent with previous observations (Wu and Ashwell, 2008).
Figure 4.
Inhibition of Bcl10 K63-polyubiquitination blocks Bcl10 clustering and degradation. (A) Unstimulated or anti-CD3 stimulated D10 T cells expressing either wild-type or the K31R,K63R mutant of Bcl10-GFP (Bcl10-WT-GFP and Bcl10-KK.RR-GFP, respectively) were lysed and heated at 95°C in 1% SDS for 5 min to disrupt protein complexes. Bcl10-GFP was immunoprecipitated using anti-GFP, followed by immunoblotting analysis to detect Bcl10 ubiquitination. (B,C) D10 T cells expressing either Bcl10-WT-GFP or Bcl10-KK.RR-GFP were stimulated for the indicated times with anti-CD3 and analyzed by confocal microscopy. Cells were assessed for redistribution of Bcl10-GFP in POLKADOTS (B) and for Bcl10-p62 co-localization via staining with anti-p62 (C). (D,E) D10 T cells expressing either Bcl10-WT-GFP or Bcl10-KK.RR-GFP were stimulated for the indicated times with anti-CD3, followed by lysis and immunoprecipitation using anti-GFP. Immunobloting was used to detect Bcl10 association with LC3 (D) and with p62 and Atg5-12 (E). (*) indicates position of Ig heavy chain. Scale bar = 10μm. Data are representative of two (A,D,E) or three experiments (B,C). See also Figure S4.
Confocal microscopy analyses revealed that whereas T cells expressing wild-type Bcl10-GFP formed POLKADOTS at 20 min and exhibited substantial loss of Bcl10-GFP fluorescence by 120 min, the KK.RR mutant showed no TCR-dependent change in distribution or fluorescence (Fig. 4B). Examination of the ERK pathway in both cell lines showed equivalent anti-CD3-induced phosphorylation of both ERK1 and ERK2, confirming intact TCR signaling in both cell lines (Fig. S4). Confocal microscopy analysis showed that wild-type Bcl10 co-localized with p62 clusters at 20 min post-stimulation, while the KK.RR mutant failed to translocate to p62 clusters (Fig. 4C). The data in Fig. 4A-C collectively suggest that K63-polyubiquitination of Bcl10 is essential for the Bcl10-p62 interaction and Bcl10 degradation.
To more directly establish a requirement for Bcl10 polyubiquitination for association between p62 and autophagosomes, we immunoprecipitated wild-type and KK.RR Bcl10-GFP, assessing the association of each form of Bcl10 with p62 and the autophagosome membrane proteins, LC3 and Atg5-12. Immunoblotting showed that wild-type Bcl10 associated with p62, LC3, and Atg5-12 in response to anti-CD3 stimulation. However, the polyubiquitination-deficient Bcl10-KK.RR mutant failed to associate with both p62 and autophagosomes in response to stimulation (Fig. 4D, E). Together, the data in Fig. 4 suggest that modification of Bcl10 by K63-polyubiquitination mediates Bcl10 association with p62 and autophagosomes.
TCR-dependent autophagy is a mechanism of Bcl10 degradation
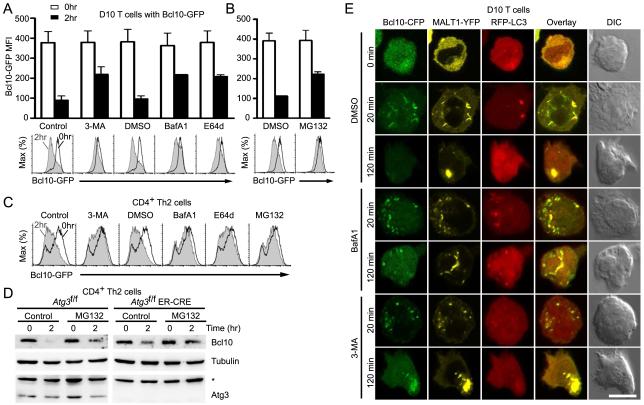
To specifically assess the effect of the autophagy-lysosomal degradation pathway on Bcl10 degradation, we employed three pharmacological agents that block this pathway at differing points and by distinct mechanisms: 3-methyladenine (3-MA), a type III PI3-kinase inhibitor that suppresses autophagosome formation (Seglen and Gordon, 1982); Bafilomycin A1 (BafA1), which blocks the H+ATPase pump, thereby preventing acidification of lysosomes and/or lysosome-autophagosome fusion (Klionsky et al., 2008); and E64d, an inhibitor of lysosomal proteases (Wilcox and Mason, 1992). Following pretreatment of cells with inhibitors, we used flow cytometry to quantify Bcl10-GFP at 0 hr and 2 hr post anti-CD3 stimulation. There was no effect of inhibitors on Bcl10-GFP in the absence of stimulation. Following anti-CD3 stimulation, control cells pre-treated with vehicle exhibited a clear decrease in Bcl10-GFP median fluorescence intensity (MFI), reflecting degradation of Bcl10-GFP, while all three drug treatments inhibited Bcl10 degradation (Fig. 5A). The partial effect of all three drugs on Bcl10 proteolysis suggested that the proteasome may also contribute to Bcl10 degradation. Indeed, p62 can direct ubiquitinated proteins to both the autophagy-lysosome pathway and the proteasome (Moscat et al., 2009; Seibenhener et al., 2004). To assess the potential role of the proteasomal pathway on Bcl10 degradation, we pre-treated D10 T cells with the proteasomal inhibitor MG132, followed by 0 hr or 2 hr anti-CD3 stimulation. Flow-cytometry analysis revealed a partial block of Bcl10 degradation, similar to the effects of autophagy inhibitors (Fig. 5B). Analogous effects on Bcl10 degradation were observed when the same inhibitors were used with C57BL/6 Th2 cells expressing Bcl10-GFP (Fig. 5C).
Figure 5.
Blockade of autophagy prevents TCR-dependent Bcl10 degradation. D10 T cells (A,B) expressing Bcl10-GFP were pre-treated with distilled water (control), DMSO (vehicle control), the autophagy inhibitor 3-methyladenine (3-MA), or the lysosomal degradation inhibitors Bafilomycin A1 (BafA1) and E64d (A) or the proteasomal inhibitor MG132 (B), and stimulated for the indicated times with anti-CD3. Bcl10-GFP degradation was assessed by flow cytometry. Histograms are representative data from one experiment and bar graphs are mean (+/−SEM.) of Bcl10-GFP median fluorescence intensity (MFI) from three independent experiments. (C) The experiment of (A) and (B) was repeated using primary Th2 cells. (D) Primary Th2 cells expressing (Atg3f/f) or not expressing ATG3 (Atg3f/f ER-Cre) were pretreated with vehicle (control) or MG132, followed anti-CD3+anti-CD28 stimulation for the indicated times. Endogenous Bcl10, tubulin, and ATG3 were detected by immunoblotting. Asterisk indicates non-specific band. (E) D10 T cells expressing Bcl10-CFP, MALT1-YFP and RFP-LC3 were pre-treated with DMSO (control), BafA1, or 3-MA, followed by anti-CD3 stimulation. Cells were imaged by confocal microscopy to assess co-localization between proteins. Data are representative of two independent experiments. Scale bar = 10μm.
To confirm the specificity of the chemical inhibitors of autophagy, we employed a genetic approach. Specifically, we in vitro differentiated Th2 cells from mice in which the essential autophagy gene, Atg3, can be conditionally inactivated (Jia and He, 2011). Atg3 deletion was induced in Th2 cells via 4-hydroxytamoxifen (4-OHT) treatment. Following 2 hr stimulation with anti-CD3+anti-CD28, we compared the degree of Bcl10 degradation between WT control (Atg3f/f) and Atg3-deleted (Atg3f/f ER-Cre) cells, in the presence and absence of MG132 (Fig. 5D). Whereas control cells (Atg3f/f) exhibited robust degradation of endogenous Bcl10, much less Bcl10 degradation occurred in Th2 cells lacking ATG3. Moreover, while MG132 treatment partially blocked Bcl10 degradation in the Atg3f/f control cells, the residual Bcl10 degradation in cells lacking ATG3 was completely or nearly completely blocked by MG132 treatment. These data provide strong genetic evidence that TCR-mediated degradation of endogenous Bcl10 is controlled by autophagy, with a contribution from the proteasomal system.
TCR-dependent autophagy is responsible for separating Bcl10 from Malt1
To assess the effect of autophagy inhibitors on the subcellular distribution of Bcl10, we pretreated D10 T cells expressing Bcl10-CFP, MALT1-YFP and RFP-LC3 with 3-MA or BafA1. We then stimulated cells with anti-CD3 for 20 min or 2 hr, followed by confocal microscopy analysis. Consistent with data in Figs. 1-2, untreated controls exhibited co-localization between Bcl10- and Malt1-containing POLKADOTS and LC3 puncta at 20 min post-stimulation (Fig. 5E). By 120 min post-stimulation, control cells showed depletion of Bcl10-GFP, and concentration of the remaining Malt1 in large aggregates. Cells treated with BafA1 exhibited a phenotype indistinguishable from controls at 20 min. However, at 120 min, Bcl10 remained closely associated with LC3+ puncta in BafA1-treated cells. Importantly, most of the cellular Malt1 no longer co-localized with the Bcl10 puncta, forming aggregates within the cell that were clearly distinct from the Bcl10+LC3+ aggregates. In the case of 3-MA treatment, we observed normal formation of Bcl10- and Malt1-containing POLKADOTS at 20 min, but very weak formation of LC3+ autophagosomes, consistent with expectations. At 120 min, there was little evidence of Bcl10 degradation, and Bcl10 remained tightly associated with the large Malt1 aggregate.
Combined with the data of Figs. 1-4, the data in Fig. 5E suggest a sequential process by which Bcl10 becomes associated with p62, captured by LC3+ autophagosomes, and degraded via the autophagy-lysosome pathway: TCR stimulation leads to K63-polyubiquitination of Bcl10. Via the ubiquitin-binding (UBA) domain of p62, Bcl10 becomes concentrated at p62 speckles, and the resulting p62-Bcl10 clusters efficiently promote signal transduction to IKK. Simultaneously, via interaction with the LC3 interaction region (LIR) of p62, autophagosomes assemble at p62 speckles, engulfing K63-polyubiquitinated Bcl10, modulating signals from Bcl10 to IKK. Bcl10 is then degraded within autophagosomes (with contributions by the proteasome), leaving aggregates of Malt1 that coalesce over time.
The autophagy of Bcl10 was mechanistically associated with the physical separation of Bcl10 and Malt1, as demonstrated by the existence of spatially distinct Bcl10+LC3+ puncta and larger aggregates containing Malt1 (and not Bcl10), following to BafA1 treatment. In other words, in both untreated and BafA1-treated cells, autophagosomes engulf Bcl10, without capturing Malt1. BafA1 treatment inhibits a later degradative step, blocking autophagosome-lysosome fusion and/or lysosomal acidification, thereby leaving Bcl10 enclosed within LC3+ vesicles at 120 min. By contrast, when autophagosome formation is blocked by 3-MA, Bcl10 is not captured from the p62 speckles by autophagosomes. Thus, in the absence of autophagosome formation, Bcl10 and Malt1 are never physically separated, remaining co-localized during the coalescence of Malt1 aggregates. Overall, the data in Fig. 5E suggest that autophagy is an essential component of the mechanism that separates Bcl10 from Malt1, enabling the selective TCR-dependent degradation of Bcl10.
Inhibition of autophagy enhances TCR mediated NF-κB activation
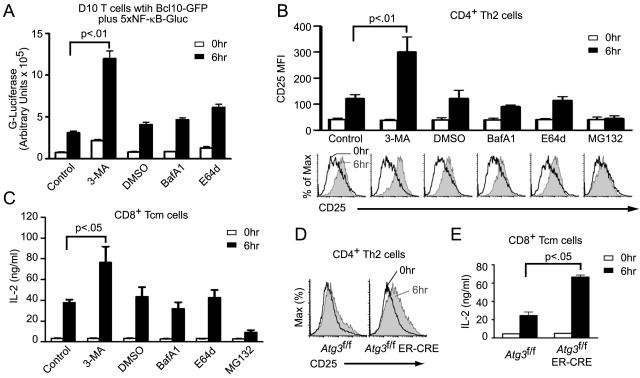
A prediction of the above model is that 3-MA treatment or Atg3 deletion should leave a signaling-competent form of Bcl10 associated with Malt1 and p62 at late time points following TCR engagment. In contrast, BafA1 and E64d treatment should not block the autophagy of Bcl10, but rather its later degradation by the lysosome. Consequently, the Bcl10 that becomes engulfed within autophagosomes in BafA1 and E64d-treated cells should be unable to signal to NF-κB because it is not accessible to downstream cytoplasmic signaling partners. Thus, 3-MA treatment and Atg3 deletion should enhance signaling to NF-κB, whereas BafA1 and E64d treatment should have no impact on signaling to NF-κB. We tested these predictions in several independent assays. First, we generated a D10 cell line expressing both Bcl10-GFP and a Gaussia luciferase reporter gene under control of an NF-κB-dependent promoter. After drug pre-treatment, we stimulated this cell line using anti-CD3. Prior to anti-CD3 stimulation, there was generally no effect of the inhibitors, although we noted a modest increase in basal NF-κB activity following treatment with the autophagy inhibitor, 3-MA. Following 6 hr of anti-CD3 stimulation, 3-MA pre-treated cells exhibited significantly increased NF-κB activity (approximately 3-fold greater than control). In contrast, lysosomal inhibitors did not significantly impact NF-κB activity in anti-CD3-treated cells (Fig. 6A).
Figure 6.
Blockade of autophagy enhances TCR-mediated NF-κB activation. (A) D10 T cells expressing Bcl10-GFP plus an integrated 5×NF-κB Gaussia luciferase reporter were pre-treated with indicated inhibitors. Cells were stimulated with anti-CD3 for the indicated times, and supernatants were harvested. Graph shows mean secreted luciferase activity (+/− SEM). (B) CD4+ Th2 cells were pre-treated with the indicated compounds and stimulated with anti-CD3+anti-CD28 for the indicated times. Cells were analyzed by flow cytometry to measure induction of CD25 expression after gating on the CD4 positive population (>90%). Histograms are representative data from one experiment and bar graphs are means (+/− SEM) of anti-CD25 fluorescence generated from three independent experiments. (C) The experiment of (B) was repeated with CD8+ Tcm cells, and secreted IL-2 was measured by XTT assay. Bar graphs are means, +/− SEM. (D) Primary Th2 cells expressing (Atg3f/f) or not expressing ATG3 (Atg3f/f ER-Cre) were stimulated for 6 hr with anti-CD3+anti-CD28 and analyzed by flow cytometry for CD25 expression. (E) The experiment of (D) was repeated with CD8+ Tcm cells, and secreted IL-2 was measured by XTT assay. Bar graphs are means, +/− SEM. Data are representative of two independent experiments (C-E).
As a second approach, we pre-treated in vitro differentiated Th2 cells with 3-MA, BafA1, E64d, and MG132. Cells were treated for 0 hr or 6 hr with anti-CD3+anti-CD28 and assessed by flow cytometry for expression of CD25, a gene highly dependent on NF-κB activation and Bcl10 expression (Ballard et al., 1988; Kingeter and Schaefer, 2008; Ruland et al., 2001). While 3-MA pre-treated cells showed a significantly higher increase in anti-CD3-induced CD25 expression compared to control cells, the increase in CD25 expression following BafA1 and E64d pre-treatment was indistinguishable from controls (Fig. 6B). MG132, in addition to inhibiting Bcl10 degradation, also prevents proteasomal degradation of IκBα, thereby inhibiting the terminal step leading to NF-κB activation (Palombella et al., 1994). As expected, there was no upregulation of CD25 in MG132 pre-treated cells (Fig. 6B). As a third approach, we measured IL-2 secretion from CD8+ Tcm cells. Following pre-treatment with the same inhibitors employed in Fig. 6B, CD8+ Tcm cells were stimulated with anti-CD3+anti-CD28 for 0 hr or 6 hr, and the amount of IL-2 (also highly NF-κB and Bcl10 dependent (Hoyos et al., 1989; Kingeter and Schaefer, 2008; Ruland et al., 2001)) was measured in cell supernatants. Similar to the results of Fig. 6B, treatment with the lysosomal degradation inhibitors, BafA1 and E64d, had little impact on IL-2 secretion, whereas inhibition of NF-κB activation by MG132 treatment prevented IL-2 secretion. Only inhibition of autophagy by 3-MA led to increased IL-2 production in response to anti-CD3 stimulation (Fig. 6C).
To confirm that the 3-MA data truly reflect inhibition of autophagy and not an off-target effect of this drug, we also tested the effect of Atg3 deletion on anti-CD3+anti-CD28 induction of CD25 expression by Th2 cells as well as the production of IL-2 by Tcm cells (Fig 6D-E). These data confirmed that cells lacking ATG3 (Atg3f/f ER-Cre) showed higher expression of CD25 and greater production of IL-2 than WT control cells (Atg3f/f). Taken together, the data in Fig. 6 show that TCR signaling to NF-κB is augmented by inhibition of the autophagy-lysosomal degradation pathway prior to autophagosome formation. Inhibition of later steps (autophagosome-lysosome fusion and lysosomal proteolytic activity) has no impact on NF-κB signaling. These data are therefore consistent with the predictions described above.
DISCUSSION
In this study, we have shown that selective autophagy is a major mechanism of TCR-dependent degradation of Bcl10. Our data are consistent with our previous fluorescence resonance energy transfer (FRET) study, showing a time-dependent dissociation of Bcl10 and Malt1 specifically within the POLKADOTS structures (Rossman et al., 2006). Indeed, Malt1, a constitutive binding partner of Bcl10, is not destroyed in this autophagy process, but remains in aggregates which coalesce over time into a much larger structure (Rossman et al., 2006). Previous data suggest that Bcl10 phosphorylation plays a role in dissociating Bcl10-Malt1 complexes (Wegener et al., 2006). However, even if Bcl10 phosphorylation has a profound effect on the avidity of the Bcl10-Malt1 interaction, it is unclear how Malt1 avoids engulfment by autophagosomes while remaining localized at a site that cannot be distinguished (by conventional confocal microscopy) from the site of autophagosome engulfment of Bcl10. How, then, is Bcl10 selectively targeted for autophagy, when both proteins are clearly in close proximity to p62 during the capture of Bcl10 by autophagosomes? This complex mechanistic question will require much future work.
Importantly, we found that p62 is crucial both for Bcl10-dependent NF-κB activation and for termination of Bcl10 signaling, via promoting Bcl10 autophagy. As our previous FRET data indicated that Bcl10 signaling partners are enriched at POLKADOTS (Rossman et al., 2006), K63-polyubiquitination of Bcl10 may promote association with and activation of downstream signaling partners by creating Bcl10 clusters that serve as cytosolic signaling platforms. Via the p62-LC3 association, autophagy depleted K63-polyubiquitinated Bcl10 from these signaling platforms, reducing activating signals to NF-κB. It will be important to determine how the formation of Bcl10 signaling complexes and the termination of signaling by Bcl10 autophagy are orchestrated to yield the observed kinetics of NF-κB activation and deactivation downstream of TCR engagement (Kingeter et al., 2010).
Additionally, we found that Bcl10 degradation is confined to effector T cells. This observation may reflect the fact that naïve T cells, in contrast to effector T cells, require an extended period of TCR signaling to successfully proliferate (Iezzi et al., 1998). Thus, the observed increase in Bcl10 expression in naïve T cells may be necessary for these cells to achieve the precise amount of NF-κB activation required for successful completion of G1. Conversely, in effector T cells, the degradation of Bcl10 may be necessary to temporally limit stimulation to the time period during which NF-κB signals promote T cell activation and cell cycle entry. This homeostatic mechanism may furthermore protect effector T cells from undesired consequences of inappropriate activation of NF-κB in later phases of the cell cycle, such as induction of senescence (Zhi et al., 2011) or apoptosis (Iezzi et al., 1998).
A growing body of data implicates selective autophagy in diverse cellular processes such as degradation of toxic protein aggregates and processing antigens for presentation to T cells (Kirkin et al., 2009b; Kraft et al., 2010; Levine et al., 2011). However, we are aware of only one previous study of higher eukaryotes that has identified a role of selective autophagy in directly regulating a signal transduction pathway, as part of normal cellular physiology. A recent publication suggests that selective autophagy of disheveled (Dvl) negatively regulates the Wnt pathway (Gao et al., 2010). Multiple aspects of this regulatory mechanism are strikingly similar to the NF-κB regulatory pathway we have described. For example, the autophagy of Dvl is facilitated by p62, and binding to p62 requires Dvl ubiquitination. Additionally, degradation of Dvl is partially sensitive to MG132, indicating that the proteasome can participate in degradation of ubiquitinated Dvl.
There is also a notable difference between the autophagy-dependent destruction of Dvl and Bcl10. Specifically, data suggest that the destruction of Dvl is initiated by cell stressors, such as starvation, and is therefore not triggered as an integral process within the Wnt pathway. By contrast, in the case of TCR-to-NF-κB signaling, our data indicate that antigen signaling through the TCR triggers both activation of NF-κB and the selective autophagy of Bcl10, inhibiting NF-κB activation. In other words, these findings strongly suggest that Bcl10 autophagy is an integrated homeostatic mechanism allowing the TCR to very precisely control the magnitude and/or kinetics of NF-κB activation in effector T cells. The existence of such a regulatory mechanism is consistent with our recent observation that the TCR-to-NF-κB cascade is digital, with the amount of NF-κB activation in individual T cells being remarkably invariant, regardless of the intensity of TCR signals (Kingeter et al., 2010). In general terms, our data demonstrate that selective autophagy can serve as a built-in homeostatic mechanism by which a receptor precisely controls the extent of activation of a target transcription factor.
Overall, our data establish a molecular mechanism whereby signaling to NF-κB is negatively modulated by TCR-induced autophagy of Bcl10. Our data suggest that Bcl10 degradation is mediated by p62-dependent selective targeting of K63-polyubiquitinated Bcl10 for entry into the autophagy-lysosomal proteolysis system. Combined with the recent report of regulation of the Wnt pathway by a similar selective autophagy process, our data suggest that selective autophagy may be a common mechanism for modulation of diverse signaling pathways. Indeed, while this manuscript was in revision, a report was published describing an autophagy mechanism that limits inflammasome activation in macrophages by a very similar p62- and polybiquitin-mediated process (Shi et al., 2012).
EXPERIMENTAL PROCEDURES
Cell lines and reagents
D10 T cells were maintained as previously described (Rossman et al., 2006). Pre-treatments with inhibitors were as follows: 3-methyladenine (MP Biomedicals) in distilled water (1mM),16 hr; Bafilomycin A1 (Enzo Life Sciences) in DMSO (100nM), 1 hr; E64d (Cayman chemicals) in DMSO (10μg/ml), 16 hr; MG132 in DMSO (20μM), 1 hr. Antibodies are detailed in Supplemental Methods.
CD8+ Tcm and CD4+ Th2 cell differentiation
Lymph nodes and spleens were collected from C57BL/6, Atg3f/f, or Atg3f/f ER-Cre mice (Jia and He, 2011). Negative bead sorting of CD4+ and CD8+ T cells, CD8+ Tcm cell differentiation, and CD4+ Th2 cell differentiation were performed as described (Fitch et al., 2006; Kingeter and Schaefer, 2008; Manjunath et al., 2001). Deletion of Atg3 via 4-OHT treatment was as described (Jia and He, 2011). All mouse experiments were approved by the Uniformed Services University Institutional Animal Care and Use Committee.
Flow-cytometry for detection of Bcl10-GFP degradation
D10 T cells or CD4+ Th2 cells or CD8+ Tcm cells (5 × 105) expressing Bcl10-GFP were stimulated with plate bound anti-CD3 (D10) or anti-CD3+anti-CD28 (primary T cells), pelleted by centrifugation, and fixed using 3% formaldehyde. Flow cytometry was performed using BD LSRII flow-cytometer, as described (Kingeter and Schaefer, 2008). Data were analyzed using Flowjo software (TreeStar).
Confocal Microscopy and Pearson’s co-localization analysis
Fixation and staining of T cells was as previously described (Rossman et al., 2006). Confocal images were obtained using a 40× 1.4 NA oil objective, a Zeiss 710 NLO microscope, and Zen software. LSM files were exported to Adobe Photoshop for cropping and adjusting contrast levels. POLKADOTS were scored through visual observation of Bcl10 clusters. Nuclear RelA was scored with visual observation of RelA signal overlapping with nuclear DRAQ5 signal. Quantification of co-localization was performed using the Intensity Correlation Analysis plugin in MBF ImageJ (www.macbiophotonics.ca/imagej). 30 cells in each group were analyzed to generate the Pearson’s correlation coefficient with a range of +1 (perfect correlation) to −1 (perfect exclusion).
Immunoprecipitation and Immunoblotting
D10 T cells were stimulated as described (Rossman et al., 2006). Cells were incubated on ice for 30 min in a lysis buffer (Wu et al., 2006) with 1× Halt protease inhibitor cocktail (Pierce Biotechnology) and 5 mM N-ethylmaleimide. Pre-cleared lysates were incubated with anti-GFP (MP Biomedicals) or anti-RFP (Rockland) overnight at 4°C. Antibody-antigen complexes were then captured using protein G sepharose beads (GE). Bcl10 ubiquitination was detected as described (Wu and Ashwell, 2008). Briefly, after cell lysis, 1%SDS was added to the lysates and heated for 5 min at 95° C. Lysates were diluted to a concentration of 0.1% SDS, followed by overnight immunoprecipitation with anti-GFP. Immunoblotting was performed as described (Rossman et al., 2006).
Gaussia Luciferase and XTT assays
For NF-κB luciferase assays, supernatants from 5×105 cells/condition were collected at 6 hr post-stimulation and Luciferase activity was measured using a Gaussia Luciferase kit (Targeting Systems). For IL-2 assays, supernatants from 5×105 cells/condition were collected at 6 hr post-stimulation and added to 96-well plates loaded with 5000 HT-2 cells/well. HT-2 cells were grown for 48 hr, followed by addition of XTT to measure viability (Biotium). Absorbance (OD) was measured using a spectrometer at 490 nm. Measured OD was converted to ng/ml IL-2 by linear regression, employing a standard curve and Graph Pad Prism 4.0.
Statistical analysis
For Fig. 3, p-values were calculated using an unpaired two-tailed t-test. For Fig. 6A-C, and E, p-values for the difference in mean between controls and treated cells were calculated by one-way ANOVA with Dunnett’s Multiple comparison post-test.
Supplementary Material
HIGHLIGHTS.
TCR-dependent Bcl10 degradation occurs in effector T cells, but not naïve T cells
Bcl10 is degraded via TCR-dependent selective autophagy
p62 is a key mediator of NF-κB activation and Bcl10 autophagy
Selective autophagy of Bcl10 limits TCR activation of NF-κB
ACKNOWLEDGMENTS
The authors thank J. Latoche for care of mice, C. Olsen (Uniformed Services University) for consultation regarding statistics, D. McDaniel for microscopy support, R. Tsien for fluorescent protein cDNAs, M. Schell for imaging advice, and C.-Z. Giam, E. Mitre, and A. Snow for critical reading of the manuscript. Supported by grants from the US National Institutes of Health (AI057481 to BCS and AI074944 and AI073947 to YWH), the Center for Neuroscience and Regenerative Medicine (CNRM) (to BCS), and a pre-doctoral fellowship (to SP) from the American Heart Association.
Footnotes
The views expressed are those of the authors and do not necessarily reflect those of the Uniformed Services University of the Health Sciences or the Department of Defense. The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ballard DW, Bohnlein E, Lowenthal JW, Wano Y, Franza BR, Greene WC. HTLV-I tax induces cellular proteins that activate the kappa B element in the IL-2 receptor alpha gene. Science. 1988;241:1652–1655. doi: 10.1126/science.241.4873.1652. [DOI] [PubMed] [Google Scholar]
- Chaturvedi A, Pierce SK. Autophagy in immune cell regulation and dysregulation. Curr Allergy Asthma Rep. 2009;9:341–346. doi: 10.1007/s11882-009-0050-1. [DOI] [PubMed] [Google Scholar]
- Duwel M, Welteke V, Oeckinghaus A, Baens M, Kloo B, Ferch U, Darnay BG, Ruland J, Marynen P, Krappmann D. A20 negatively regulates T cell receptor signaling to NF-kappaB by cleaving Malt1 ubiquitin chains. J Immunol. 2009;182:7718–7728. doi: 10.4049/jimmunol.0803313. [DOI] [PubMed] [Google Scholar]
- Fitch FW, Gajewski TF, Hu-Li J. Production of TH1 and TH2 cell lines and clones. Curr Protoc Immunol. 2006 doi: 10.1002/0471142735.im0313s72. Chapter 3, Unit 3 13. [DOI] [PubMed] [Google Scholar]
- Gao C, Cao W, Bao L, Zuo W, Xie G, Cai T, Fu W, Zhang J, Wu W, Zhang X, Chen YG. Autophagy negatively regulates Wnt signalling by promoting Dishevelled degradation. Nat Cell Biol. 2010;12:781–790. doi: 10.1038/ncb2082. [DOI] [PubMed] [Google Scholar]
- Hoyos B, Ballard DW, Bohnlein E, Siekevitz M, Greene WC. Kappa B-specific DNA binding proteins: role in the regulation of human interleukin-2 gene expression. Science. 1989;244:457–460. doi: 10.1126/science.2497518. [DOI] [PubMed] [Google Scholar]
- Hu S, Du MQ, Park SM, Alcivar A, Qu L, Gupta S, Tang J, Baens M, Ye H, Lee TH, et al. cIAP2 is a ubiquitin protein ligase for BCL10 and is dysregulated in mucosa-associated lymphoid tissue lymphomas. J Clin Invest. 2006;116:174–181. doi: 10.1172/JCI25641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iezzi G, Karjalainen K, Lanzavecchia A. The duration of antigenic stimulation determines the fate of naive and effector T cells. Immunity. 1998;8:89–95. doi: 10.1016/s1074-7613(00)80461-6. [DOI] [PubMed] [Google Scholar]
- Jia W, He YW. Temporal regulation of intracellular organelle homeostasis in T lymphocytes by autophagy. J Immunol. 2011;186:5313–5322. doi: 10.4049/jimmunol.1002404. [DOI] [PubMed] [Google Scholar]
- Jia W, Pua HH, Li QJ, He YW. Autophagy regulates endoplasmic reticulum homeostasis and calcium mobilization in T lymphocytes. J Immunol. 2011;186:1564–1574. doi: 10.4049/jimmunol.1001822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingeter LM, Paul S, Maynard SK, Cartwright NG, Schaefer BC. Cutting edge: TCR ligation triggers digital activation of NF-kappaB. J Immunol. 2010;185:4520–4524. doi: 10.4049/jimmunol.1001051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingeter LM, Schaefer BC. Loss of protein kinase C theta, Bcl10, or Malt1 selectively impairs proliferation and NF-kappa B activation in the CD4+ T cell subset. J Immunol. 2008;181:6244–6254. doi: 10.4049/jimmunol.181.9.6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V, Lamark T, Sou YS, Bjorkoy G, Nunn JL, Bruun JA, Shvets E, McEwan DG, Clausen TH, Wild P, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009a;33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009b;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Elazar Z, Seglen PO, Rubinsztein DC. Does bafilomycin A1 block the fusion of autophagosomes with lysosomes? Autophagy. 2008;4:849–950. doi: 10.4161/auto.6845. [DOI] [PubMed] [Google Scholar]
- Kraft C, Peter M, Hofmann K. Selective autophagy: ubiquitin-mediated recognition and beyond. Nat Cell Biol. 2010;12:836–841. doi: 10.1038/ncb0910-836. [DOI] [PubMed] [Google Scholar]
- Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Capan E, Zhao Y, Zhao J, Stolz D, Watkins SC, Jin S, Lu B. Autophagy is induced in CD4+ T cells and important for the growth factor-withdrawal cell death. J Immunol. 2006;177:5163–5168. doi: 10.4049/jimmunol.177.8.5163. [DOI] [PubMed] [Google Scholar]
- Lobry C, Lopez T, Israel A, Weil R. Negative feedback loop in T cell activation through IkappaB kinase-induced phosphorylation and degradation of Bcl10. PNAS. 2007;104:908–913. doi: 10.1073/pnas.0606982104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunemann JD, Munz C. Autophagy in CD4+ T-cell immunity and tolerance. Cell Death Differ. 2009;16:79–86. doi: 10.1038/cdd.2008.113. [DOI] [PubMed] [Google Scholar]
- Manjunath N, Shankar P, Wan J, Weninger W, Crowley MA, Hieshima K, Springer TA, Fan X, Shen H, Lieberman J, von Andrian UH. Effector differentiation is not prerequisite for generation of memory cytotoxic T lymphocytes. J Clin Invest. 2001;108:871–878. doi: 10.1172/JCI13296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin P, Diaz-Meco MT, Moscat J. The signaling adapter p62 is an important mediator of T helper 2 cell function and allergic airway inflammation. Embo J. 2006;25:3524–3533. doi: 10.1038/sj.emboj.7601250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell. 2009;137:1001–1004. doi: 10.1016/j.cell.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscat J, Diaz-Meco MT, Wooten MW. Of the atypical PKCs, Par-4 and p62: recent understandings of the biology and pathology of a PB1-dominated complex. Cell Death Differ. 2009;16:1426–1437. doi: 10.1038/cdd.2009.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palombella VJ, Rando OJ, Goldberg AL, Maniatis T. The ubiquitin-proteasome pathway is required for processing the NF-kappa B1 precursor protein and the activation of NF-kappa B. Cell. 1994;78:773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- Pua HH, Dzhagalov I, Chuck M, Mizushima N, He YW. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J Exp Med. 2007;204:25–31. doi: 10.1084/jem.20061303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman JS, Stoicheva NG, Langel FD, Patterson GH, Lippincott-Schwartz J, Schaefer BC. POLKADOTS are foci of functional interactions in T-Cell receptor-mediated signaling to NF-kappaB. Mol Biol Cell. 2006;17:2166–2176. doi: 10.1091/mbc.E05-10-0985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruland J, Duncan GS, Elia A, del Barco Barrantes I, Nguyen L, Plyte S, Millar DG, Bouchard D, Wakeham A, Ohashi PS, Mak TW. Bcl10 is a positive regulator of antigen receptor-induced activation of NF-kappaB and neural tube closure. Cell. 2001;104:33–42. doi: 10.1016/s0092-8674(01)00189-1. [DOI] [PubMed] [Google Scholar]
- Scharschmidt E, Wegener E, Heissmeyer V, Rao A, Krappmann D. Degradation of Bcl10 induced by Tcell activation negatively regulates NF-kappaB signaling. Molecular and Cellular Biology. 2004;24:3860–3873. doi: 10.1128/MCB.24.9.3860-3873.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze-Luehrmann J, Ghosh S. Antigen-receptor signaling to nuclear factor kappa B. Immunity. 2006;25:701–715. doi: 10.1016/j.immuni.2006.10.010. [DOI] [PubMed] [Google Scholar]
- Seglen PO, Gordon PB. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proceedings of the National Academy of Sciences of the United States of America. 1982;79:1889–1892. doi: 10.1073/pnas.79.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibenhener ML, Babu JR, Geetha T, Wong HC, Krishna NR, Wooten MW. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol Cell Biol. 2004;24:8055–8068. doi: 10.1128/MCB.24.18.8055-8068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C-S, Shenderov K, Huang N-N, Kabat J, Abu-Asab M, Fitzgerald KA, Sher A, Kehrl JH. Activation of autophagy by inflammatory signals limits IL-1[beta] production by targeting ubiquitinated inflammasomes for destruction. Nat Immunol. 2012;13:255–263. doi: 10.1038/ni.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaug B, Jiang X, Chen ZJ. The role of ubiquitin in NF-kappaB regulatory pathways. Annu Rev Biochem. 2009;78:769–796. doi: 10.1146/annurev.biochem.78.070907.102750. [DOI] [PubMed] [Google Scholar]
- Smith-Garvin JE, Koretzky GA, Jordan MS. T cell activation. Annual review of immunology. 2009;27:591–619. doi: 10.1146/annurev.immunol.021908.132706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thome M, Charton JE, Pelzer C, Hailfinger S. Antigen receptor signaling to NF-kappaB via CARMA1, BCL10, and MALT1. Cold Spring Harb Perspect Biol. 2010;2:a003004. doi: 10.1101/cshperspect.a003004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- Wegener E, Oeckinghaus A, Papadopoulou N, Lavitas L, Schmidt-Supprian M, Ferch U, Mak TW, Ruland J, Heissmeyer V, Krappmann D. Essential role for IkappaB kinase beta in remodeling Carma1-Bcl10-Malt1 complexes upon T cell activation. Mol Cell. 2006;23:13–23. doi: 10.1016/j.molcel.2006.05.027. [DOI] [PubMed] [Google Scholar]
- Wilcox D, Mason RW. Inhibition of cysteine proteinases in lysosomes and whole cells. Biochem J. 1992;285(Pt 2):495–502. doi: 10.1042/bj2850495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CJ, Ashwell JD. NEMO recognition of ubiquitinated Bcl10 is required for T cell receptor-mediated NF-kappaB activation. Proc Natl Acad Sci U S A. 2008;105:3023–3028. doi: 10.1073/pnas.0712313105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CJ, Conze DB, Li T, Srinivasula SM, Ashwell JD. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-kappaB activation [corrected] Nat Cell Biol. 2006;8:398–406. doi: 10.1038/ncb1384. [DOI] [PubMed] [Google Scholar]
- Zeng H, Di L, Fu G, Chen Y, Gao X, Xu L, Lin X, Wen R. Phosphorylation of Bcl10 negatively regulates T-cell receptor-mediated NF-kappaB activation. Mol Cell Biol. 2007;27:5235–5245. doi: 10.1128/MCB.01645-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi H, Yang L, Kuo YL, Ho YK, Shih HM, Giam CZ. NF-kappaB hyper-activation by HTLV-1 tax induces cellular senescence, but can be alleviated by the viral anti-sense protein HBZ. PLoS Pathog. 2011;7:e1002025. doi: 10.1371/journal.ppat.1002025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.