Abstract
Infectious diseases are the leading causes of death worldwide. The development of efficient and low cost prophylactics to prevent pathogenic infection is given high priority in the twenty-first century. Commensal bacteria are largely seen as harmless and can survive symbiotically (in many cases) in niches throughout the human body. Advances in genetic engineering and understanding of pathogenesis have revealed many potential strategies to develop engineered bacteria for prophylaxis purposes: including live vaccines and anti-infective agents. In this review we discuss recent advances and potentialities of prophylaxis with engineered bacteria.
Introduction
Over the past 10 years, there has been an increased emphasis on understanding the important relationship between human-associated microbial communities and the development of disease. The great variability in microbiota populating a single human is derived from a complex product of biological processes, environmental factors and socio-cultural practices. Human-associated microbiomes have been massively characterized through the Human Microbiome Project (HMP) [1–3] by the National Institutes of health and International Human Microbiome Consortium (IHMC). The advancement of metagenomics [4,5] and high-throughput sequencing technologies has expanded the repository of taxonomic and functional human microbiome data. With the increase in available information about which bacterial strains exist cooperatively with humans, there has been interest in harnessing commensal strains to combat potentially pathogenic ones. Commensal bacteria s status as “tolerated” by the host provides an opportunity to prevent infection at the bacterial level [6] without removing helpful strains from the system [7].
Features of commensal bacteria
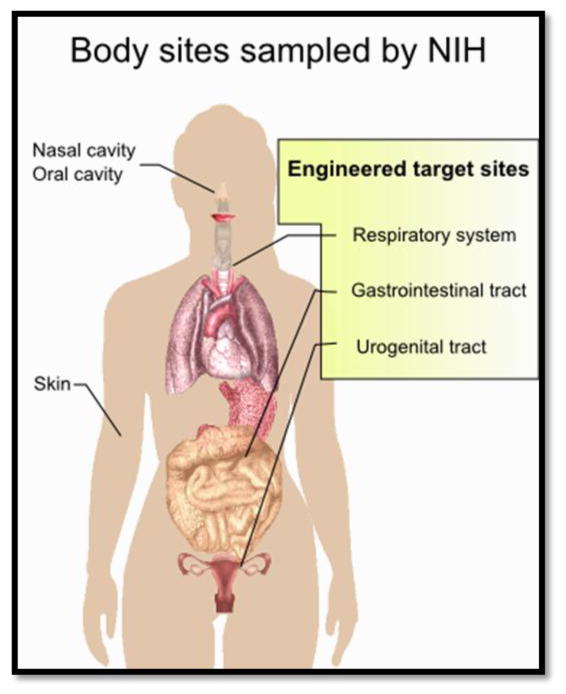
There are trillions of commensal bacterial cells, which outnumber human cells by a factor of 10 [8• •]. Most commensal bacteria live at the mucosal surface of the gastrointestinal, urogenital, oral and respiratory tract (Figure 1). The greatest numbers are found in the digestive tract. They interact with host metabolism, produce metabolites for host physiology and facilitate immune system development. In particular, they act as the first line of defense against most pathogenic infections [9]. In order to populate their niche continually, commensal bacteria must adhere (usually to mucosal surfaces) and outcompete other organisms for available nutrients [10].
Figure 1.
Body sights sampled by the Human Microbiome Project. Engineered commensal bacteria are typically targeted to the respiratory system, gastrointestinal tract and urogenital tract (yellow box).
Advantages of using commensal bacteria as delivery systems
Their ability to colonize the same mucosal surfaces targeted by pathogens has led to research exploring the use of commensal bacteria as delivery systems for various compounds aimed at lowering the risk of infection. Commensals are resistant to gastric and bile acid toxicity, hence they can survive and bypass primary host defenses[11,12]. Introduction of engineered commensal bacteria under circumstances in which they normally colonize would allow them to take up residence within the host and provide longer term protection [9,10]. Some commensal strains (including several lactobacillus subspecies) are generally regarded as safe (GRAS) by the Food and Drug Administration (FDA) and fulfill the criteria of presumed safety developed by European Food Safety Authority [12 ]. With advancements in molecular biology that make transformation of many commensal strains simple and inexpensive combined with genomic profiles that make them well-characterized, the library of potential recombinant approaches using commensal bacteria has grown tremendously over the last 5 years [12 ]. An example can be seen in expression systems that incorporate different antibody fragments into the attB site of lactobacillus strains, making the expression of these fragments in vivo routine [13]. There have been a number of detailed reviews focusing on the field of using recombinant lactic acid bacteria (LAB) as mucosal biotherapeutic agents [11,14–16]. The aim of this review is to update this information and expand the topic area to include other strains of bacteria being used as prophylactics against infection or colonization.
Engineering strategies targeting bacterial infection
Interfering with the regulation of virulence expression
To adapt to different hosts and environments, many pathogenic bacteria sense signals from their own species, other bacteria or surrounding environments and respond by regulating genes to activate virulence traits. Some extra cellular signals that accumulate when colony density is high mediate colony-wide coordinated behavior known as quorum sensing (QS) [17]. QS systems have been implicated in several virulence control pathways, thus making interruption of QS pathways an attractive target for disease prevention. This strategy carries the added benefit of targeting signaling networks of bacteria rather than their viability, which may reduce antibiotic resistance.
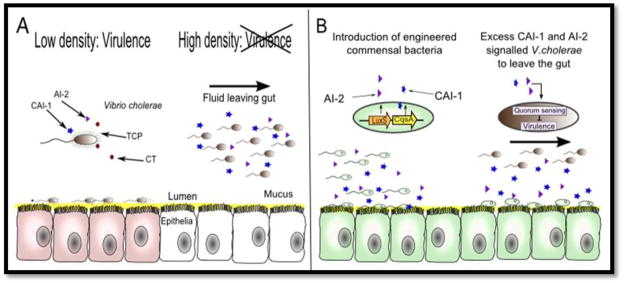
A recent application of this principle was demonstrated by engineering a commensal Escherichia coli strain to inhibit virulence of Vibrio cholerae, the causative pathogen for the diarrheal disease cholera [18• •]. V. cholerae cells can sense their population density by releasing cholera autoinducer (CAI-1) and autoinducer 2 (AI-2) that accumulate in the population as it grows in the upper intestine. At high cell density, V. cholerae detect high concentrations of CAI-1 and AI-2 that lead to repression of virulence factors. To prevent cholerae infection, an E. coli strain naturally producing AI-2 was engineered to express CAI-1 and evaluated in an infant mouse model fed with V. cholerae (Figure 2). Treatment of mice with engineered E. coli significantly decreased V. cholerae colonization in the intestine and effectively prevented disease, without killing the invading organism. In another study, a “nanofactory” composed of multifunctional molecules was engineered to target the QS networks of bacteria [19]. The nanofactory comprises two major modules: an antibody that selectively target bacteria and bind to their cell surfaces, and a fusion protein synthesizing the universal bacterial signal AI-2 when bound to the targeted bacterium. This devise provides a highly precise way to interfere with QS systems in pathogens such as V. cholerae and Bacillus cereus.
Figure 2.
(A) Schematic diagram of how commensal bacteria can be used to speed up infection cycles of V.cholerae. At low cell density, V.cholerae secrete cholera toxin (CT) and toxin coregulated pilus (TCP), which facilitate host ionic imbalance and V. cholerae attachment to host epithelia (red cells), respectively. At high cell density, high concentration of cholerae autoinducer-1 (CAI-1) and autoinducer-2 (AI-2) are present within the colonization site. The virulence genes are turned off by the QS circuit and VC discontinue expressing CT and TCP. V. cholerae detaches and leaves the infected host through the massive efflux of fluid. (B) Engineered commensal bacteria can serve as a prophylactic against cholera by expressing the V. cholerae autoinducers. Excess CAI-1 and AI-2 imitate the high cell density scenario and signal invading V. cholerae to leave the gut before causing damage to the host cells.
Bacteriocin producing system
In other work that took advantage of a pathogens QS system, commensal E. coli was engineered to detect signals from a pathogen and to produce an antimicrobial peptide, a bacteriocin, upon detection [20• •]. Specifically, Pseudomonas aeruginosa has a LasI/LasR system: LasI produces 3-oxo-C12 homoserine lactone (HSL) to activate LasR that leads to the expression of virulence factors. Based on this QS mechanism, Saeidi et al. engineered an E. coli strain with three vital components targeting P. aeruginosa: a sensing device expressing LasR to detect 3-oxo-HSL, a synthesis device encoding the bacteriocin pyocin S5 under control of the luxR promoter, and a discharge device encoding E7 lysis protein to release pyocin S5. When P. aeruginosa was co-cultured with the engineered E. coli at a ratio of 1:4, their growth and biofilm formation was inhibited by 99% and 90%, respectively, demonstrating the utility of harnessing bacterial sensing and producing capabilities against pathogens.
Targeting toxins or adhesions
Bacterial adhesion, toxin formation or secretion systems can be potential targets for therapeutics as they are all equally important for establishing infection [21]. Paton et al. have engineered E. coli strains producing chimeric surface lipopolysaccharides that are capable of neutralizing, on contact, shiga-toxin secreted by pathogenic E. coli or cholera toxin secreted by V. cholerae in vivo [22,23]. Probiotic lactic acid bacteria (LAB) have been engineered to secrete either pathogenic adhesion proteins or flagellin in order to competitively inhibit adhesion of pathogens such as Listeria monocytogenes or Salmonella enterica [24,25]. These studies aimed to engineer commensal bacteria to outcompete for valuable surface space in the host s intestine, forcing pathogens to pass through the gastrointestinal tract without detrimental effects. Although in vitro tests showed promising results, equipping probiotic strains with the ability to strongly adhere to epithelium may impose the risk of causing an enteric inflammatory responses [26].
Antigen development
Attenuated pathogens or LAB have been engineered to function as vaccine delivery vehicles against pathogens including Yersinia pseudotuberculosis [27], Salmonella Typhimurium [28,29], and Pneumococcal nasopharyngeal [30,31]. One particular study by Mohamadzadeh et al. showed that a recombinant LAB vaccine could be as effective as a traditional injected vaccine in an animal model [32•]. In this study, oral administration of mice with Lactobacillus acidophilus secreting dendritic cell-targeted antigens afforded equal protection against lethal Bacillus anthracis as traditional antigens injected with alum. By enhancing the expression of this antigen in L. gasseri, 100% protection of mice against anthrax was achieved [33]. These studies demonstrated great potential to apply LAB vaccines in a more clinically relevant setting.
Engineering strategies targeting virus infection
Target on virus-host fusion site
Prophylactic engineered bacteria have been used to colonize the urogenital tract and secret therapeutic agents that target specific viruses [34]. Most recently, Lactobacillus jensenii was engineered by Lageneur et al [35• •] to secrete cyanovirin-N (CV-N) and tested in a repetitive vaginal simian HIV-challenged Chinese rhesus macaque model [36]. This strain colonized the lower vaginal tract for up to 6 weeks and produced CV-N in situ without observable inflammatory responses compared to the controls. The infection rate was reduced by 62.9% in the monkeys colonized by L. jensenii when compared to the control population [35]. An important aspect of this approach was the use of L. jensenii, which naturally colonizes the vaginal wall of human women and may therefore be more effective in humans than it was in monkeys. It should be noted that there are differences between simian and human HIV; however, this approach still holds promise in that changes to L. jensenii (that could include modified CV-N) would be relatively simple to engineer provided they were needed or available.
It is sometimes necessary or advantageous to design vaccines against heterologous viral challenge. RANTES ( Regulated upon Activation, Normal T-cell Expressed, and Secreted) and C1C5 RANTES belong to members of chemokines that recognize HIV-1 receptor protein, such as CCR5 [37•]. When co-cultured with CD4+ T cells and macrophages, engineered L. jensenii synthesizing both RANTES and C1C5 RANTES minimized infection by different clades of HIV strains. While RANTES was effective in co-culture models, it is expected that its degradation in vivo would limit its efficacy. Recombinant Caulobacter crescentus was also shown to neutralize HIV infection, in this case by surface displaying antibody proteins that block the fusion between HIV and its host [38,39].
Antigen development
Lei and coworkers developed a recombinant Lactococcuslactis-hemagglutinin (HA) vaccine against influenza H5N1 in the form of edible enteric-coated mini capsules. 5/5 mice that were fed with capsule-secreted-HA were found to survive after 14 days [40]. Furthermore, the same group engineered L. lactis by combining recombinant avian influenza virus HA1 and adjuvant cholera toxin subunit-B. Elevated levels of IgG, IFN gamma and fecal IgA were observed after 10 days and 100% protection was provided to mice treated with HA1 and CTB after a lethal H5N1 virus challenge [41].
Challenges and future perspectives
That recombinant commensal bacterial prevention of disease is in its infancy can be seen by the relatively low number of preclinical studies in animal models reported in the literature [42]. It is clear that there are several challenges to overcome before this approach is more commonplace. First, an engineered bacterial strain needs to be introduced into its host without compromising the activity and integrity of either the bacteria or the host. Optimally, the prophylactic system will be engineered to only target infectious agents, and it can be re-introduced or eliminated as needed. However, the complex nature of commensal microbial communities in the human body makes it very difficult to predict long term behavior of engineered cells [43]. Their ability to occupy a niche at an effective density is as important as their engineered functionality. In addition, it is possible that the prophylactic system might produce other signals that may impair microbial homeostasis within host, possibly resulting in unintended shifts in enteric ecology. Furthermore, there is a risk of horizontal gene transfer between both bacteria in the intestine and between bacteria and intestinal epithelial cells. Biocontainment strategies should be developed to reduce dissemination of recombinant strains into the environment [44]. Lastly, it is unclear that whether these therapeutic systems will engender any evolutionary pressure on the target pathogens in the long run. In order to address such challenges, greater understanding of the complex relationships between host, symbiotic microbes, and invading pathogens is needed. Continued insight into bacterial pathogenesis in humans will provide potential targets for new antimicrobial agents [45,46]. Rational design of therapeutics might possibly benefit from progress in synthetic biology, which aims to facilitate construction of more complicated, clinically applicable circuits in commensal bacteria [47,48].
Conclusion
Currently there is a compelling need to develop new therapeutics against infectious disease due to pathogenic resistance to antibiotics and a lack of efficacious vaccines. The advantage of using commensal bacteria to address these problems lies mainly in their protected niche within the human body. An important step towards utilizing the advantages afforded by commensal bacteria is the demonstration of their protective efficacy in animal models (Table 1). Engineered therapeutic and prophylactic commensal bacteria have great potential to be a clinical reality in the near future.
Table 1.
Recent examples of engineering commensal bacteria for prevention of infection challenge
| Target | Vehicle | Mechanism | Outcome | Reference |
|---|---|---|---|---|
| Vibrio cholerae | E. coli | Interfere with qurom sensing system to repress toxin expression | Increased survival rate in infant mice model challenged with V. cholerae by 92% | [18• •] |
| Pseudomonas aeruginosa | E. coli | Synthesize and release pyocin | Reduced viable P. aeruginosa cells and their biofilm formation by 99% and 90% respectively | [20• •] |
| Listeria monocytogenes | L. paracasei | Express Listeria adhesion protein | Reduced L. monocytogenes translocation and cytotoxicity in Caco-2 cells by 46% and 79% respectively | [24] |
| Bacillus anthracis | L. acidophilus | Express B. anthracis protective antigen targeted to dentric cells | Demonstrated equal protection as purified antigen offered. Increased survival of mice challenged with B. anthracis | [32•, 33] |
| Yersinia pseudotuberculosis | L. lactis | Secret low-calcium response V antigen against Yersinia infections | Increased survival of mice against both oral and systematic Y pseudotuberculosis. infections | [27] |
| Salmonella Typhimurium | Bifidobacterium longum | Secret Salmonella-flagellin antigen | Increased survival of mice challenged with Salmonella Typhimurium | [25] |
| Pneumococcal nasopharyngeal | L. casei | Secret pneumococcal surface protein C | Reduced pneumococcal colonization in mice | [30] |
| Pneumococcal nasopharyngeal | L. lactis | Secret pneumococcal protective protein A | Both live and inactivated LAB vaccine induced protective immunity in mice against pneumococcal challenges | [31] |
| HIV-1 | L. jensenii | Secrete RANTES and C1C5 RANTES | Inhibited HIV-1 infection in CD4+ T cells and macrophages | [37 ] |
| Chimeric simian/HIV | L. jensenii | Express HIV-1 entry inhibitor cyanovirin-N | Reduced infection rate by 62.9% in Chinese rhesus macaque model | [35 ] |
| Influenza H5N1 | L. lactis | Display H5N1 HA antigen with or without cholera toxin subunit B | 100% survival rate for mice fed with pgsA-HA1 and CTB | [40, 41] |
| HIV-1 | Caulobacter crescentus | Display antigens such as protein G together with CD4/MIP1alpha on S- layer | Enhanced in HIV-1 neutralization | [38, 39] |
Highlights.
Advantages of using commensal bacteria as delivery systems.
Different strategies were used to engineer commensal bacteria to target bacterial or viral infection through inhibiting virulence expression, producing bacteriocins, neutralizing toxins, preventing adhesion, blocking fusion sites, and enhancing the immune response of host cells.
Current challenges and its future perspectives were also discussed.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Yih-Lin Goh, Email: yg259@cornell.edu.
HongFei He, Email: hh379@cornell.edu.
John C. March, Email: jcm224@cornell.edu.
References
- 1.Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, Bonazzi V, McEwen JE, Wetterstrand KA, Deal C, Baker CC, et al. The NIH Human Microbiome Project. Genome Res. 2009;19(12):2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blaser MJ. Harnessing the power of the human microbiome. Proc Natl Acad Sci U S A. 2010;107(14):6125–6126. doi: 10.1073/pnas.1002112107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Proctor LM. The Human Microbiome Project in 2011 and beyond. Cell Host Microbe. 2011;10(4):287–291. doi: 10.1016/j.chom.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464 (7285):59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuczynski J, Lauber CL, Walters WA, Parfrey LW, Clemente JC, Gevers D, Knight R. Experimental and analytical tools for studying the human microbiome. Nat Rev Genet. 2012;13(1):47–58. doi: 10.1038/nrg3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bagnoli F, Baudner B, Mishra RPN, Bartolini E, Fiaschi L, Mariotti P, Nardi-Dei V, Boucher P, Rappuoli R. Designing the Next Generation of Vaccines for Global Public Health. OMICS: A Journal of Integrative Biology. 2011;15 (9):545–566. doi: 10.1089/omi.2010.0127. [DOI] [PubMed] [Google Scholar]
- 7.Hill DA, Hoffmann C, Abt MC, Du Y, Kobuley D, Kirn TJ, Bushman FD, Artis D. Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol. 2010;3(2):148–158. doi: 10.1038/mi.2009.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8••.Tlaskalova-Hogenova H, Stepankova R, Kozakova H, Hudcovic T, Vannucci L, Tuckova L, Rossmann P, Hrncir T, Kverka M, Zakostelska Z, Klimesova K, et al. The role of gut microbiota (commensal bacteria) and the mucosal barrier in the pathogenesis of inflammatory and autoimmune diseases and cancer: contribution of germ-free and gnotobiotic animal models of human diseases. Cellular & Molecular Immunology. 2011;8(2):110–120. doi: 10.1038/cmi.2010.67. This is a comprehensive overview of the relationship between gut microbiota and their roles in the pathogenesis of different inflammatory, autoimmune disease and cancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wardwell LH, Huttenhower C, Garrett WS. Current concepts of the intestinal microbiota and the pathogenesis of infection. Curr Infect Dis Rep. 2011;13(1):28–34. doi: 10.1007/s11908-010-0147-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Villena J, Oliveira MLS, Ferreira PCD, Salva S, Alvarez S. Lactic acid bacteria in the prevention of pneumococcal respiratory infection: Future opportunities and challenges. International Immunopharmacology. 2011;11 (11):1633–1645. doi: 10.1016/j.intimp.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 11.Wells JM, Mercenier A. Mucosal delivery of therapeutic and prophylactic molecules using lactic acid bacteria. Nature Reviews Microbiology. 2008;6(5):349–362. doi: 10.1038/nrmicro1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12••.Wells J. Mucosal Vaccination and Therapy with Genetically Modified Lactic Acid Bacteria. In: Doyle MP, Klaenhammer TR, editors. Annual Review of Food Science and Technology. Vol. 2.2. 2011. pp. 423–445. This review illustrated the use of recombinant LAB as mucosal delivery systems. They discussed the potential of vaccines for addressing different diseases, and also discussed some of their concerns and future perspectives. [DOI] [PubMed] [Google Scholar]
- 13.Cruz Martin M, Pant N, Ladero V, Gunaydin G, Andersen KK, Alvarez B, Martinez N, Alvarez MA, Hammarstrom L, Marcotte H. Integrative Expression System for Delivery of Antibody Fragments by Lactobacilli. Applied and Environmental Microbiology. 2011;77(6):2174–2179. doi: 10.1128/AEM.02690-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daniel C, Roussel Y, Kleerebezem M, Pot B. Recombinant lactic acid bacteria as mucosal biotherapeutic agents. Trends in Biotechnology. 2011;29(10):499–508. doi: 10.1016/j.tibtech.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 15.Bermudez-Humaran LG. Lactococcus lactis as a live vector for mucosal delivery of therapeutic proteins. Hum Vaccin. 2009;5(4):264–267. doi: 10.4161/hv.5.4.7553. [DOI] [PubMed] [Google Scholar]
- 16.Bermudez-Humaran LG, Kharrat P, Chatel JM, Langella P. Lactococci and lactobacilli as mucosal delivery vectors for therapeutic proteins and DNA vaccines. Microb Cell Fact. 2011;10(Suppl 1 S4) doi: 10.1186/1475-2859-10-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Waters CM, Bassler BL. Quorum sensing: Cell-to-cell communication in bacteria. Annual Review of Cell and Developmental Biology. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 18••.Duan FP, March JC. Engineered bacterial communication prevents Vibrio cholerae virulence in an infant mouse model. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(25):11260–11264. doi: 10.1073/pnas.1001294107. This study demonstrated that engineered E. coli can be utilized to interfere with quorum sensing system of V. cholerae to prevent infection. It was one of the first papers to use bacterial QS against a pathogen in an animal model. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernandes R, Roy V, Wu HC, Bentley WE. Engineered biological nanofactories trigger quorum sensing response in targeted bacteria. Nature Nanotechnology. 2010;5(3):213–217. doi: 10.1038/nnano.2009.457. [DOI] [PubMed] [Google Scholar]
- 20• •.Saeidi N, Wong CK, Lo TM, Nguyen HX, Ling H, Leong SSJ, Poh CL, Chang MW. Engineering microbes to sense and eradicate Pseudomonas aeruginosa, a human pathogen. Mol Syst Biol. 2011;7:521–532. doi: 10.1038/msb.2011.55. This paper described a synthetic network that enabled E. coli to detect and kill P. aeruginosa by producing pyocin. It shows the potential of using bacterial sensing and responding for preventing infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rasko DA, Sperandio V. Anti-virulence strategies to combat bacteria-mediated disease. Nature Reviews Drug Discovery. 2010;9(2):117–128. doi: 10.1038/nrd3013. [DOI] [PubMed] [Google Scholar]
- 22.Paton AW, Jennings MP, Morona R, Wang H, Focareta A, Roddam LF, Paton JC. Recombinant probiotics for treatment and prevention of enterotoxigenic Escherichia coli diarrhea. Gastroenterology. 2005;128(5):1219–1228. doi: 10.1053/j.gastro.2005.01.050. [DOI] [PubMed] [Google Scholar]
- 23.Focareta A, Paton JC, Morona R, Cook J, Paton AW. A recombinant probiotic for treatment and prevention of cholera. Gastroenterology. 2006;130(6):1688–1695. doi: 10.1053/j.gastro.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 24.Koo OK, Amalaradjou MAR, Bhunia AK. Recombinant Probiotic Expressing Listeria Adhesion Protein Attenuates Listeria monocytogenes Virulence In Vitro. Plos One. 2012;7(1):e29277. doi: 10.1371/journal.pone.0029277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanchez B, Lopez P, Gonzalez-Rodriguez I, Suarez A, Margolles A, Urdaci MC. A flagellin-producing Lactococcus strain: interactions with mucin and enteropathogens. Fems Microbiology Letters. 2011;318(2):101–107. doi: 10.1111/j.1574-6968.2011.02244.x. [DOI] [PubMed] [Google Scholar]
- 26.Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134(2):577–594. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- 27.Daniel C, Sebbane F, Poiret S, Goudercourt D, Dewulf J, Mullet C, Simonet M, Pot B. Protection against Yersinia pseudotuberculosis infection conferred by a Lactococcus lactis mucosal delivery vector secreting LcrV. Vaccine. 2009;27(8):1141–1144. doi: 10.1016/j.vaccine.2008.12.022. [DOI] [PubMed] [Google Scholar]
- 28.Wolfenden RE, Layton SL, Wolfenden AD, Khatiwara A, Gaona-Ramirez G, Pumford NR, Cole K, Kwon YM, Tellez G, Bergman LR, Hargis BM. Development and evaluation of candidate recombinant Salmonella-vectored Salmonella vaccines. Poult Sci. 2010;89(11):2370–2379. doi: 10.3382/ps.2010-00702. [DOI] [PubMed] [Google Scholar]
- 29.Yamamoto S, Wada J, Katayama T, Jikimoto T, Nakamura M, Kinoshita S, Lee KM, Kawabata M, Shirakawa T. Genetically modified Bifidobacterium displaying Salmonella-antigen protects mice from lethal challenge of Salmonella Typhimurium in a murine typhoid fever model. Vaccine. 2010;28 (41):6684–6691. doi: 10.1016/j.vaccine.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 30.Hernani Mde L, Ferreira PC, Ferreira DM, Miyaji EN, Ho PL, Oliveira ML. Nasal immunization of mice with Lactobacillus casei expressing the pneumococcal surface protein C primes the immune system and decreases pneumococcal nasopharyngeal colonization in mice. FEMS Immunol Med Microbiol. 2011;62(3):263–272. doi: 10.1111/j.1574-695X.2011.00809.x. [DOI] [PubMed] [Google Scholar]
- 31.Vintini E, Villena J, Alvarez S, Medina M. Administration of a probiotic associated with nasal vaccination with inactivated Lactococcus lactis-PppA induces effective protection against pneumoccocal infection in young mice. Clin Exp Immunol. 2010;159(3):351–362. doi: 10.1111/j.1365-2249.2009.04056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32•.Mohamadzadeh M, Duong T, Sandwick SJ, Hoover T, Klaenhammer TR. Dendritic cell targeting of Bacillus anthracis protective antigen expressed by Lactobacillus acidophilus protects mice from lethal challenge. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(11):4331–4336. doi: 10.1073/pnas.0900029106. This study demonstrated that vaccination with the LAB vaccine provided same protection as purified antigen against B. anthracis infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mohamadzadeh M, Durmaz E, Zadeh M, Pakanati KC, Gramarossa M, Cohran V, Klaenhammer TR. Targeted expression of anthrax protective antigen by Lactobacillus gasseri as an anthrax vaccine. Future Microbiology. 2010;5 (8):1289–1296. doi: 10.2217/fmb.10.78. [DOI] [PubMed] [Google Scholar]
- 34.Fahey JV, Bodwell JE, Hickey DK, Ghosh M, Muia MN, Wira CR. New approaches to making the microenvironment of the female reproductive tract hostile to HIV. Am J Reprod Immunol. 2011;65(3):334–343. doi: 10.1111/j.1600-0897.2010.00949.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35••.Lagenaur LA, Sanders-Beer BE, Brichacek B, Pal R, Liu X, Liu Y, Yu R, Venzon D, Lee PP, Hamer DH. Prevention of vaginal SHIV transmission in macaques by a live recombinant Lactobacillus. Mucosal Immunology. 2011;4(6):648–657. doi: 10.1038/mi.2011.30. The authors demonstrated that recombinant L. jensenii expressing an HIV-1 entry inhibitor, cyanovirin-N was able to colonize the vaginal tract for up to 6 weeks. Macaque models treated with repeated vaginal challenges were used and a 63% reduction in infection was observed in macaques treated with recombinant L. jensenii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu RR, Cheng AT, Lagenaur LA, Huang W, Weiss DE, Treece J, Sanders-Beer BE, Hamer DH, Lee PP, Xu Q, Liu Y. A Chinese rhesus macaque (Macaca mulatta) model for vaginalLactobacilluscolonization and live microbicide development. Journal of Medical Primatology. 2009;38(2):125–136. doi: 10.1111/j.1600-0684.2008.00316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37•.Vangelista L, Secchi M, Liu X, Bachi A, Jia L, Xu Q, Lusso P. Engineering of Lactobacillus jensenii To Secrete RANTES and a CCR5 Antagonist Analogue as Live HIV-1 Blockers. Antimicrobial Agents and Chemotherapy. 2010;54(7):2994–3001. doi: 10.1128/AAC.01492-09. This paper illustrated a proof of concept for the active expression of anti-HIV-1 CCR5 antagonist, RANTES and C1C5 RANTES from recombinant L. jensenii. Both antagonists were shown to inhibit HIV-1 infection on CD4+ T cells and macrophages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duval M, Lewis CJ, Nomellini JF, Horwitz MS, Smit J, Cavacini LA. Enhanced neutralization of HIV by antibodies displayed on the S-layer of Caulobacter crescentus. Antimicrob Agents Chemother. 2011;55(12):5547–5552. doi: 10.1128/AAC.00509-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nomellini JF, Li C, Lavallee D, Shanina I, Cavacini LA, Horwitz MS, Smit J. Development of an HIV-1 specific microbicide using Caulobacter crescentus S-layer mediated display of CD4 and MIP1alpha. PLoS One. 2010;5 (4):e10366. doi: 10.1371/journal.pone.0010366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lei H, Xu Y, Chen J, Wei X, Lam DM. Immunoprotection against influenza H5N1 virus by oral administration of enteric–coated recombinant Lactococcus lactis mini-capsules. Virology. 2010;407(2):319–324. doi: 10.1016/j.virol.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 41.Lei H, Sheng Z, Ding Q, Chen J, Wei X, Lam DMK, Xu Y. Evaluation of Oral Immunization with Recombinant Avian Influenza Virus HA1 Displayed on the Lactococcus lactis Surface and Combined with the Mucosal Adjuvant Cholera Toxin Subunit B. Clinical and Vaccine Immunology. 2011;18(7):1046–1051. doi: 10.1128/CVI.00050-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burrill DR, Boyle PM, Silver PA. A New Approach to an Old Problem: Synthetic Biology Tools for Human Disease and Metabolism. Cold Spring Harbor Symposia on Quantitative Biology. 2011 doi: 10.1101/sqb.2011.76.010686. [DOI] [PubMed] [Google Scholar]
- 43.Baker M. Better living through microbes. Nat Biotechnol. 2005;23(6):645–647. doi: 10.1038/nbt0605-645. [DOI] [PubMed] [Google Scholar]
- 44.Lee P. Biocontainment strategies for live lactic acid bacteria vaccine vectors. Bioeng Bugs. 2010;1(1):75–77. doi: 10.4161/bbug.1.1.10594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Curtis MM, Sperandio V. A complex relationship: the interaction among symbiotic microbes, invading pathogens, and their mammalian host. Mucosal Immunol. 2011;4(2):133–138. doi: 10.1038/mi.2010.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hughes DT, Terekhova DA, Liou L, Hovde CJ, Sahl JW, Patankar AV, Gonzalez JE, Edrington TS, Rasko DA, Sperandio V. Chemical sensing in mammalian host-bacterial commensal associations. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(28):12734–12734. doi: 10.1073/pnas.1002551107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weber W, Fussenegger M. Emerging biomedical applications of synthetic biology. Nature Reviews Genetics. 2012;13(1):21–35. doi: 10.1038/nrg3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruder WC, Lu T, Collins JJ. Synthetic Biology Moving into the Clinic. Science. 2011;333(6047):1248–1252. doi: 10.1126/science.1206843. [DOI] [PubMed] [Google Scholar]