Abstract
Imaging mass spectrometry (IMS) has evolved to be a promising technology due to its ability to detect a broad mass range of molecular species and create density maps for selected compounds. It is currently one of the most useful techniques to determine the spatial distribution of neuropeptides in cells and tissues. Although IMS is conceptually simple, sample preparation steps, mass analyzers, and software suites are just a few of the factors that contribute to the successful design of a neuropeptide IMS experiment. This review provides a brief overview of IMS sampling protocols, instrumentation, data analysis tools, technological advancements and applications to neuropeptide localization in neurons and endocrine tissues. Future perspectives in this field are also provided, concluding that neuropeptide IMS could revolutionize neuronal network and biomarker discovery studies.
Keywords: Imaging mass spectrometry, MALDI, neurons, neuroendocrine tissues, sampling, neuropeptides
1. Introduction
The actions of neuronal circuits regulate and modify an organism's functions and behaviors. These circuits and their activities are extensively modulated by a diverse array of cell-cell signaling molecules. Neuropeptides represent a major class of chemical messengers in organisms with nervous systems and affect multiple physiological processes and behaviors like hunger, thirst, pain, stress and reproduction [1-8]. Neuropeptides are cleaved from large preprohormones, resulting in biologically active forms after numerous enzymatic processing steps in various orders [9-10]. Resultant neuropeptides can have a diverse range of physical and chemical properties, presenting a significant challenge for their detection, identification and quantitation in neural tissues. This challenge is exacerbated by the low pico- to sub-femtomolar in vivo concentrations of neuropeptides [11-12].
Edman degradation has been conventionally employed to determine neuropeptide sequences, but this technique is relatively slow and requires large sample amounts and extensive purification, increasing the difficulties of sample preparation and characterization. The developments of soft ionization methods, electrospray ionization (ESI) and matrix-assisted laser desorption/ionization (MALDI) [13-14], and sensitive mass analyzers have accelerated neuropeptide discovery. In the past decade, over one thousand neuropeptides have been characterized based on MS analyses from biological tissues [15]. The discovery and identification of such a large number of neuropeptides has demanded the establishment of a database containing final peptide forms from preprohormones. The library's establishment could facilitate the functional characterization of neuropeptides in cells, clusters and organisms. However, tissue homogenization through grinding also results in a dilution of trace level neuropeptides that are present in specific regions or single neurons [16-18], hindering detection. Alternatively, neuropeptide profiling and mapping directly from tissue preserves analyte spatial information and anatomical context. The ability to visualize neuropeptide distributions in cells, clusters and organs also allows the experimenter to determine if neuropeptides from distinct families or multiple isoforms from the same family co-localize in specific regions. Such spatial information about neuropeptides can help elucidate the underlying mechanism of cell-cell signaling interactions via neuropeptide messengers.
Neuropeptide mapping within an organ or cell cluster can be achieved by combining immunohistochemical (IHC) staining with high-resolution microscopy [19-20]. This technique is a powerful tool for neurobiologists, but it provides fairly limited chemical information and often suffers from cross-reactivity. These limitations can be overcome by MALDI- imaging MS (MALDI-IMS), which integrates the high sensitivity and chemical specificity of a mass analyzer with the imaging capability of a MALDI source [21]. MALDI-IMS generates ion density maps of neuropeptides from biological tissues by acquiring mass spectra according to a predefined Cartesian grid. The array of mass spectra is then processed into an image where each pixel corresponds to its mass spectrum (Figure 1). The application of MALDI-IMS to neuropeptide mapping in biological tissues and cells offers several advantages. The technique does not require labeling, which eliminates potential label interference, and can detect unknown molecules from biological surfaces. Furthermore, an image can be generated for each peak in the average mass spectrum, and these images can be combined to produce multiplexed images useful for determining peptide co-localization.
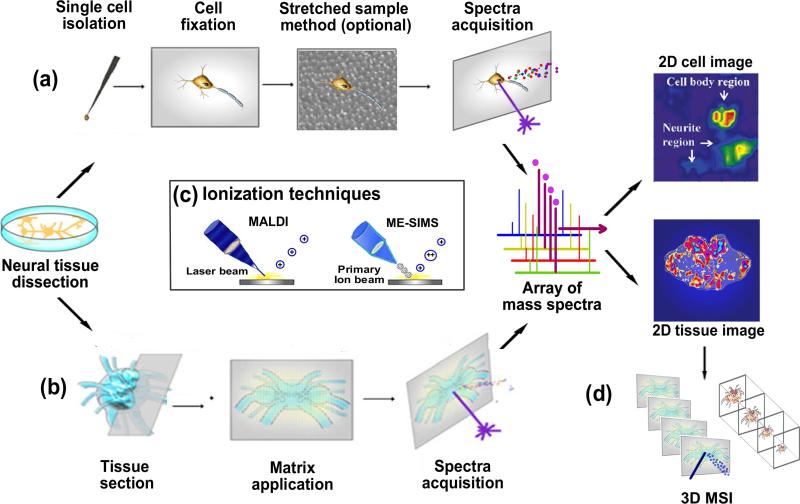
Figure 1.
Overall workflow of IMS methodology including the sampling of neuropeptides from (a) neurons and (b) neural tissues. (a) Neurons of interest are first isolated and transferred onto a target plate, fixed, stretched on Parafilm anchored with beads array (optional), sprayed with matrix and then subjected to IMS acquisition. (b) Sections are acquired from cryosectioning of neural tissue, transferred onto a target plate, washed with organic solvent (optional), sprayed with matrix and then subjected to IMS acquisition. The mass spectrum for each pixel is collected, and this array of spectra is further processed into a representative image for each peak present in the spectra. (c) MALDI is the primary ionization technique employed in neuropeptide imaging, whereas several IMS studies using ME-SIMS exist. (d) Schematic of 3D neuropeptide imaging by stacking the 2D images acquired from serial sections of tissue.
2. Methodology
A typical workflow of MALDI-IMS involves sample preparation, matrix application, mass spectra acquisition, and data analysis (Figure 1). The outcome of an IMS experiment is strongly determined by these elements.
Many publications of MALDI-MS studies targeting neuropeptides in neuroendocrine tissues exist. However, unique sample preparation protocols must be followed to successfully image neuropeptides in single cells, resulting in a limited number of publications [22-23]. Therefore, sample preparation for MALDI-IMS of neuropeptides in cellular domains will be addressed separately from tissue domains.
2.1 Sample preparation
Sample preparation in imaging experiments aims to generate reproducible and reliable MS images directly from tissue sections or cells. In this section, several crucial sample preparation considerations are discussed, including tissue and cell collection, storage, sectioning and pre-treatment prior to matrix application.
2.1.1 Sample preparation for tissues
Proper preparation of a tissue slice must maintain its structural integrity and morphology while avoiding the delocalization and degradation of peptides. The most common procedure to acquire tissue is to snap-freeze it in powdered dry ice, liquid nitrogen or liquid nitrogen-chilled isopentane, etc. and then storing it at -80°C until use [24]. Another method consists of loosely wrapping the tissue in aluminum foil and gently placing it into liquid nitrogen, ice-cold ethanol or isopropanol bath for 30-60 seconds [25]. We recommend the latter method because the longer freezing process avoids possible tissue cracking and fragmentation.
A tissue stabilization step minimizes sample aging effects during storage. The most commonly used preservation technique, formaldehyde-fixed paraffin-embedding (FFPE), causes protein/peptide cross-linking and should be avoided in IMS applications [26]. Irradiation [27] by a microwave oven and heat denaturation by the Stabilizor T1, developed by Denator (Gothenburg, Sweden), can effectively denature active proteolytic enzymes, preventing post mortem degradation [28-29].
Neuropeptide IMS experiments typically require sectioning tissue into 10-20 μm thick slices [30]. Embedding tissues in supporting media allows for easy handling and precise sectioning of tissue samples. However, polymer-containing embedding material, such as optimal cutting temperature (OCT) compound, Tissue-tek and carboxymethylcellulose (CMC), often produce intense interfering background signals and should be avoided [25]. Verhaert et al. published IMS data of a neuropeptide in a cockroach brain embedded in sucrose [31]. Our lab reported gelatin as an alternative embedding compound to provide minimal interfering peaks while preserving tissue integrity [32-33].
Tissue sections are transferred and attached onto a stainless steel conductive plate by thaw-mounting [25]. These metal plates are sometimes replaced with Indium-Tin-Oxide (ITO)-coated conductive glass slides [34-35] to allow microscopic analyses of the sections after IMS analysis. Moreover, nonconductive microscopic glass slides have also been used for successful imaging of tissue samples on an intermediate-pressure MALDI linear ion trap (LIT) mass spectrometer, greatly reducing the cost of supplies [31]. One alternative tissue attachment approach uses adhesive double-sided conductive tape to mount the tissue section to the sample plate [26]. This method is more suitable for low water-content tissues [36].
Washing tissue sections with an organic solvent can lead to increased ion yields and protein/large peptide signals by fixing tissues and removing ion-suppressing salts and lipids [37]. Solvents like ethanol, methanol, acetone, hexane and chloroform have been used for tissue washing to improve protein/large peptide imaging sensitivity [38-39]. However, the washing solvent composition must be tested and optimized for neuropeptides since specific molecule classes often require different treatment. For example, 70% ethanol in water has been reported to remove salts and lipids and enhance neuropeptide signals from medical leech sections [40], rat brain sections [41] and rat and human pituitary tissue sections [42]. However, concerns arise that such washing step might result in diffusion of small, soluble neuropeptides. A new procedure developed by Van hove et al. uses solvent wetted fiber-free paper to enable local washing of tissue sections for IMS applications [43].
2.1.2 Sample preparation for cells
Distinct neuropeptide expression patterns have been observed at the distal regions and the soma of a neuron or among adjacent neurons. Because of this chemical heterogeneity, information about neuropeptide identities and locations in individual neurons is valuable to neurobiologists. Determining neuropeptide complements at the cellular level can help researchers understand the role of neuropeptides in intracellular regulation and intercellular communication. MALDI-MS profiling has been applied to cultured/isolated cells to probe neuropeptides from single identified neurons isolated from Lymnaea stagnalis [44-47], Aplysia californica [48-49], Periplaneta americana [50], Ascaris suum [51] and Rattus sp. [52]. However, challenges arise when the goal of an IMS experiment is to examine the distribution of neuropeptides at single-cell resolution. The complex cell culture media must be removed or replaced before MALDI-IMS experiments. No mechanical damage to cells during removal of the extracellular media is acceptable. Moreover, neuropeptides are small, soluble molecules that diffuse quickly out of cells. The original locations of neuropeptides must be preserved in the process of media removal and matrix application. Additionally, the single-cell resolution must not be compromised by the size of the matrix crystals, the diameter of the focused laser beam or the motorized movement of the mass spectrometer's stage. Several technological breakthroughs for IMS have solved the above-mentioned issues and are discussed in 2.3.2.
Mollusks serve as a useful model system when developing and improving sample preparation techniques in single-cell IMS because of their relatively large neurons and simple, well-characterized nervous system. The first single-cell IMS technique was pioneered by the Sweedler group using single cultured cerebral ganglion neurons from Aplysia californica [23]. Their approach to single-cell IMS began with the isolation of single neurons. The ganglion was incubated in protease solution first to remove the sheath. The neurons were then dissected using sharp needles and transferred onto a glass slide. Following dissection, the neurons were placed and cultured in a dish filled with artificial sea water (ASW)-antibiotic solution [23]. For MALDI-MS profiling, neurons can be placed onto a MALDI target plate after briefly rinsing to remove salts. Methods for IMS were investigated to preserve cell morphology, prevent neuropeptide redistribution and replace the cell matrix. A 30% glycerol-ASW mixture was found to substitute ASW-antibiotic media well and stabilize neuron morphology without ion-suppression. After exposure to glycerol for 1-5 minutes, the extracellular media was removed by vacuum suction from the desired cell culture regions [23]. The extracellular media must be optimized for specific types of neurons to preserve cell morphology and prevent neuropeptide redistribution.
MALDI-IMS investigations of neuropeptides in small neurons are challenging. An innovative sample preparation technique, the stretched sample method, was developed by Zimmerman et al. to account for the small size of single neurons [22, 53-55]. Briefly, dissected, individual neurons were placed on a bead monolayer array embedded in a stretchable substrate, Parafilm. After glycerol stabilization, the directional orientation of the cells was marked, and a digital optical image of the cells without stretching was recorded. Then, the substrate was manually stretched and placed on conductive ITO glass slides for further MALDI-IMS analyses [22, 54]. This example highlights new developments in sample preparation enabling single-cell IMS.
2.2 Matrix application
Choosing a matrix and its application method are critical elements in the IMS workflow. An appropriate matrix should provide the highest signal intensity and the largest number of analyte signals with minimal interfering matrix peaks. α-cyano-4-hydroxycinnamic acid (CHCA) and 2,5-dihydroxy benzoic acid (DHB) are commonly used in neuropeptide IMS [25, 32-33, 56]. Mixtures of conventional MALDI matrices and organic bases are known as solid/liquid ionic matrices. These matrices can sometimes improve spectral quality, crystallization and vacuum stability [57-60]. Another technique deposits a thin layer of gold nanoparticles on matrix-sprayed rat brain sections and was reported to increase signal intensity and image quality of the neuropeptide vasopressin [61].
Matrix application is an important step under continuous development. It is desirable to obtain a homogenous layer of analyte-doped matrix crystals smaller than observable tissue features. Small matrix solution droplets favor the acquisition of high spatial resolution images but can reduce extraction efficiency and signal intensity. A recent comparison of matrix coating methods observed that lipid peaks dominated spectra when using a dry matrix spray. Regular, solvent-based matrix coating methods formed larger droplets that efficiently extracted neuropeptides from crustacean brain sections for subsequent detection and imaging demonstrating the tradeoff of sensitivity and spatial resolution [32].
Matrix is generally deposited either as homogeneous layers (spray coating) or discrete spots (microspotting). For spray coating, a pneumatic spray (pneumatic sprayer, airbrush, TM sprayer system from HTX imaging and thin layer chromatography sprayer) or vibrational spray (ImagePrep Device from Bruker Daltonics) can be chosen to apply a uniform layer of small matrix droplets [32-34, 62]. Manual sprayers usually cover tissue with a homogenous matrix layer of medium-sized crystals but suffer from large variability. A robotic spray apparatus, like the TM sprayer, improves the reproducibility of manual spraying while preserving its advantages. Electrospray deposition creates a mist containing small droplets useful for high-resolution imaging studies [61]. However, the coated surface may receive varying amounts of matrix because of differences in electrical insulating properties. Microspotters deposit pl-sized droplets of matrix according to a predefined array and require multiple rounds of spotting for sufficient matrix coverage. Different automatic spotting devices are available and include the desktop inkjet printer, CHIP produced by Shimadzu (equipped with inkjet-style piezonozzles) and the acoustic PORTRAIT reagent multi-spotter [63-65]. Solvent free methods like sublimation or dry coating eliminate the chance of analyte diffusion induced by wet application, but these methods yield relatively low sensitivities for IMS analyses except for lipids because of limited analyte-matrix interactions [66-67]. Nevertheless, Deutskens et al. dry-coated sinapinic acid to rat cerebellum sections and then rehydrated the matrix on the section to maximize the extraction of proteins from samples [68]. This approach is robust and may be useful for high-resolution IMS investigations of neuropeptides.
2.3 IMS acquisition
Technological developments have improved the throughput, robustness, sensitivity, and spatial resolution of IMS. This section discusses aspects related to IMS acquisition in the general IMS workflow.
2.3.1 Instrumentation
Most state-of-the-art mass analyzers can be equipped with a MALDI source with IMS capabilities. Mass analyzer and laser optics advancements have improved IMS measurements in multiple categories. In this section, the new technologies and how they can be applied to IMS of neuropeptides are discussed.
Conventional IMS experiments are based on the microprobe approach. In the “microprobe” mode, the laser beam is focused to a small region from which a single spectrum is acquired. By rastering the surface according to a predefined Cartesian grid, an array of mass spectra is acquired and reconstructed to a cohesive MS image where each pixel contains the data from the corresponding spectrum. The laser beam focus determines the pixel density of a MS image, i.e. the spatial resolution. The majority of commercial instruments have laser spot diameters of ~30-100 μm. Bruker Daltonics has developed a proprietary kHz smartbeam-II MALDI laser integrated with a novel FlashDetector™ on their MALDI-TOF/TOF mass spectrometer, ultrafleXtreme™ [69]. The patented 1 kHz laser allows for focus diameters down to 10 μm and is useful for probing neuropeptides at the cellular level. Special confocal-type objectives designed by Spengler and coworkers offer sub-micron spatial resolution and are promising for future IMS studies of neuropeptides from single cells [70-72].
An alternative approach to increasing spatial resolution is “microscope” MALDI-IMS. This source employs a defocused UV laser beam to envelop a large area, from which a stigmatic projection of ions were desorbed/ionized. The high-quality ion optics transfer and magnify the ion packet produced by the single laser shot, relying on a position-sensitive detector to record the arrival time and position of the ions [73-74]. An increased number of pixels are simultaneously acquired, which dramatically increases the throughput of high resolution imaging. The resulting stigmatic images of highest magnification reported by Luxembourg et al. reached a pixel size of 500 nm and a resolving power of 4 μm [42, 74]. In this configuration the limiting factor for spatial resolution is no longer the size of the desorption/ionization area, but rather, the quality of the ion optics, the ion optical magnification, and the pixel size of the detector. Unfortunately, the method is not widely used because of technical constraints like the need for specialized ion optics, a fast detector and computing software to reconstruct ion images.
The step size of a laser probe affects acquisition time, but it can also be reduced to improve spatial resolution. An oversampling technique reported by the Sweedler group introduced a setup where the incremental movement of the laser beam is smaller than the laser diameter [75]. After sample material is ablated away at the initial spot, ions are desorbed/ionized from a much smaller area with each incremental step. This method does not necessitate any additional hardware and is simple. However, it consumes the total sample, excluding the possibility of sample reanalysis.
Time-of-flight (TOF) analyzers are routine tools in MALDI-IMS applications. TOF analyzers determine mass to charge ratios (m/z) by accelerating desorbed ions to the same kinetic energy and then measuring the time they take to travel through a defined path length [76]. This design provides high sensitivity, wide mass range and fast analysis speed. Moreover, the addition of another TOF tube allows for tandem mass fragmentation and improves the spectral resolution compared to a single TOF instrument [77]. The throughput of IMS benefits from the speed of TOF/TOF. In addition, recent advances in laser technology further reduce the time consumed during IMS experiments. For instance, Applied Biosystems introduced continuous rastering in rows across the sample surface instead of using the conventional and time-consuming “stop-and-go” method for acquiring spectra [78]. Another example is a newly developed MALDI-TOF instrument that significantly shortens acquisition time by utilizing continuous scanning with a 5 kHz laser [79]. Acquisition time was shortened even further by the introduction of a hybrid quadruple time-of-flight mass spectrometer equipped with a short-pulse 20 kHz Nd:YVO4 laser [80]. These developments facilitate high throughput IMS studies since acquisition time is not compromised by the mass analyzer.
Quality MALDI-IMS experiments result in high spatial resolution images of unambiguously identified compounds. High spatial resolution MS images are quickly generated by TOF instruments, but these analyzers suffer from modest mass resolving powers and mass accuracies. Fourier transform ion cyclotron resonance (FT-ICR) mass analyzers can provide comparable spatial resolutions to a TOF/TOF mass analyzer while offering superior resolution and accuracy for unequivocal discrimination of analytes [81]. Taban et al. compared the performance of MALDI-FT-ICR with MALDI-TOF in neuropeptide imaging from a rat brain section. They observed distinct distribution patterns for three peptides resolved in a 0.5 Da mass window by FT-ICR. In contrast, the TOF mass analyzer merely delivered a combined distribution image due to its limited resolving power [41]. In addition to the ability to image isobaric ions, FT-ICR provides unique ion trapping and storage capabilities. This feature uses an in-cell-accumulation technique reported by Kutz et al. [82] and continuous accumulation of selected ions (CASI) developed by Bruker Daltonics [83] to improve the detection sensitivity of trace amount of analytes. Several multi-stage tandem MS (MSn) techniques, including sustained off-resonance irradiation collision-induced dissociation (SORI-CID), electron capture dissociation (ECD) and infrared multiphoton dissociation (IRMPD), have contributed to the successful identification of neuropeptides and lipids from minute quantities of tissue samples. Despite the advantages, the FT-ICR's primary limitation is the slow scan speed, which is particularly problematic for serial IMS experiments. This can be partially alleviated by using a more powerful magnetic field since Fourier transients become shorter without sacrificing spectral resolution as the ion cyclotron frequency is increased. The current commercially available FT-ICR with the highest magnetic field is 15 Tesla manufactured by Bruker Daltonics [84].
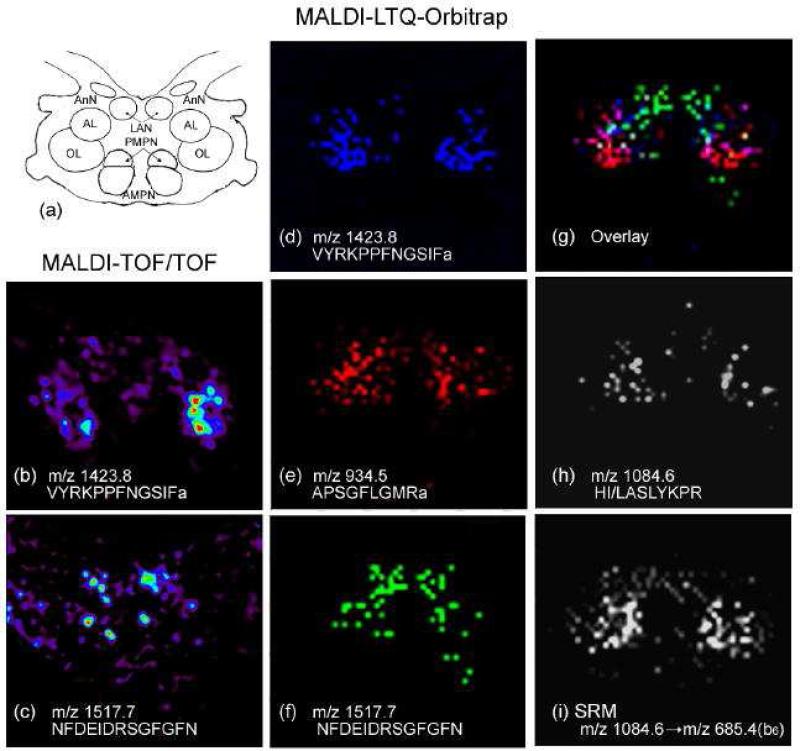
In addition to the FT-ICR, another FT-based mass analyzer, the Orbitrap, manufactured by Thermo Fisher Scientific, provides comparable resolution to an FT-ICR. While an ICR measures the frequencies of ion cyclotron resonance in a magnetic field, the Orbitrap measures the axial oscillation of ions moving back and forth along a spindle-like electrode within an electrostatic field [85]. The oscillation frequency is proportional to the square root of the electrical-field strength, whereas the cyclotron frequency is related to the magnetic field strength [86]. The FT-ICR relies on high-field magnets for high resolving power, consequently increasing cost. Predictably, the Orbitrap's popularity in IMS studies of neuropeptides will be growing due to its excellent performance and relatively low cost compared with FT-ICR. For instance, Chen et al. acquired MS images of neuropeptides VYRKPPFNGSIFamide (m/z 1423.8), APSGFLGMRamide (m/z 934.5) and NFDEIDRSGFGFN (m/z 1517.7) using a MALDI-LTQ-Orbitrap instrument from a section of lobster brain (Figure 2d-f). The MS images obtained with the Orbitrap matched the anatomical structure of the brain (Figure 2a) and images obtained with a TOF/TOF (Figure 2b-c) but offered much higher mass spectral resolution with lower background of noise [87]. Furthermore, a selected reaction monitoring (SRM) MS/MS imaging experiment was conducted on a novel neuropeptide at m/z 1084.6 from lobster brain using the MALDI-LTQ-Orbitrap. Interfering ions were significantly reduced when compared to the image of the precursor ion at m/z 1084.6 (Figure 2h), and the novel peptide was successfully mapped and de novo sequenced as HI/LASLYKPR directly from a complex tissue sample as shown in Figure 2i. A direct comparison among the LTQ-Orbitrap, 7 Tesla FT-ICR and TOF/TOF mass spectrometers also revealed that the high resolution Orbitrap instrument enabled the detection of the greatest number of neuropeptides. Sequence-specific fragmentation conducted by different dissociation techniques such as higher energy collisional induced dissociation (HCD), collisional induced dissociation (CID) and pulse-Q dissociation (PQD) on Orbitrap was valuable in this investigation. Tandem mass spectra acquired on the MALDI-Orbitrap were complementary to the liquid chromatography (LC)-ESI-MS/MS data, providing more information for de novo sequencing of peptides [87]. Another neuropeptide study by Verhaert et al. employed a MALDI-LTQ instrument for IMS of neuropeptides from the corpora cardiaca and allata of Acheta domesticus (the house cricket). The study found complementary sequence information for imaged neuropeptides by using the instrument's tandem MS capabilities [88]. The Orbitrap's high resolving power assisted in differentiating three neuropeptides with accurate masses within 1 Da from one another from American cockroach (Periplaneta americana) neurosecretory organs [31].
Figure 2.
MALDI imaging of neuropeptide localization in the lobster H. americanus brain using both MALDI TOF/TOF and MALDI LTQ Orbitrap mass spectrometers. (a) Schematic drawing of the lobster brain, which contains multiple neuropils, including anterior (AMPN) and posterior medial protocerebral neuropils (PMPN), olfactory lobe (ON), accessory lobe (AL), antenna I neuropil (AnN) and lateral II antenna neuropil (LAN). Ion images of (b) VYRKPPFNGSIFamide (m/z 1423.8) and (c) NFDEIDRSGFGFN (m/z 1517.7) were obtained using a MALDI TOF/TOF. Ion images of multiple known neuropeptides were acquired by MALDI LTQ Orbitrap, including: (d) VYRKPPFNGSIFamide (m/z 1423.8); (e) APSGFLGMRamide (m/z 934.5); (f) NFDEIDRSGFGFN (m/z 1517.7); and (g) overlay of the above three neuropeptides. A novel peptide HI/LASLYKPR (m/z 1084.6) in the lobster brain was also mapped using a MALDI LTQ Orbitrap instrument by (h) precursor ion scanning of m/z 1084.6 and (i) selected reaction monitoring of the transition between m/z 1084.6 and sequence-specific fragment ion m/z 685.4 (b6). Adapted with permission from Ref. [87].
The integration of ion mobility (IM) with MALDI-TOF adds conformational information to MALDI-IMS data. IM-TOF is a two-dimensional gas-phase separation technique that separates analytes according to their m/z and collisional cross-section. Upon desorption/ionization from the tissue surface, ions travel inside an IM drift cell, which has an applied electric field and a carrier buffer gas that opposes ion motion. The migration time of an ion inside the drift cell is a product of the its mass, charge, size and shape, thus allowing separation of isobaric ions based on their collisional cross-section [89-90]. Jackson et al. demonstrated utility of IM in MS imaging studies through an IMS study of a rat brain section [91]. For each MS event per pixel, a MALDI-IM two-dimensional (2D) plot was defined by the ions’ drift time and m/z. Three major compound classes (lipids, peptides, and matrix-related ions) were distributed along individual familial “trend lines” and separated from each other by differences in their drift time. This feature indicates that MALDI-IM-TOF is applicable to the field of neuropeptide imaging, which is often complicated by the interference of high abundance isobaric lipids.
2.3.2 Data analysis
Post-acquisition software, such as BioMap (http://www.maldi-msi.org, available for free to download) and proprietary programs for IMS systems (e.g., FlexImaging from Bruker Daltonics, ImageQuest from Thermo Fisher Scientific and TissueView from Applied Biosystems/MDS), are used to visualize density maps of selected ions. These software packages give the user the ability to adjust color scales, overlay ion density maps, and integrate MS images with histological images. Data acquired with different mass spectrometers can vary, leading to the necessity to combine data for a more complete neuropeptide profile. Unfortunately, processing data between different instruments and data analysis software is difficult. To facilitate the exchange and processing of imaging mass spectrometry data, a standardized format, imzML, has been developed, and a program that converts raw data to the imzML format is available to download at imzml.org [92].
Biomarker discovery studies compare IMS data sets sampled from a control group with those from a treated group. Data-mining software like ClinProTools by Bruker Daltonics enables principal component analysis (PCA) and hierarchical clustering of multiple IMS data sets to extract differentially expressed or distributed molecules and determine candidate biomarkers.
3. Biological applications of IMS of neuropeptides
3.1 IMS of neuropeptides at the organ level
IMS was used before being applied to protein profiling and imaging from biological surfaces. Secondary ion mass spectrometry (SIMS)-IMS mapped inorganic small molecules in material science applications for decades until Caprioli et al. developed MALDI-IMS to map larger biomolecules from tissue sections [21]. The successful application of MALDI-IMS to larger peptides and proteins contributed to its extension into signaling peptide localization in neuroendocrine organs. Verhaert et al. used a MALDI LTQ-Orbitrap to image secretory neuropeptides from the American cockroach (Periplaneta americana) [31]. They also identified and differentiated neuropeptides in Acheta domesticus (the house cricket) from lipids by examining the diagnostic tandem MS fragments of the two classes [88]. Similarly, Dekeyser et al. investigated the differential distribution of neuropeptides in the brain and pericardial organ from the crab Cancer borealis [33]. Over 30 neuropeptides from 10 neuropeptide families were detected using a MALDI-TOF/TOF instrument and characterized by in situ fragmentation to verify peptide identities. Spatial relationships between multiple neuropeptide isoforms of the same family and the relative distributions of neuropeptide families were elucidated in this study. Intriguingly, both studies found that neuropeptides appeared to be more differentially localized than phospholipids.
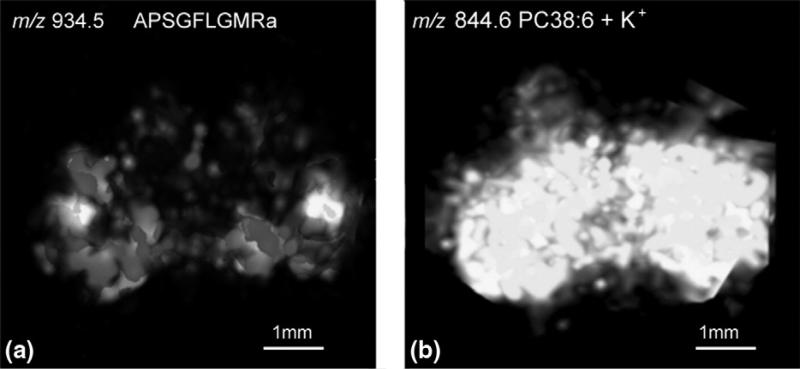
The chemical heterogeneity of neuropeptides in complex neural tissues like the brain necessitates the interrogation of neuropeptide expression patterns in a tissue volume. Three-dimensional (3D) images of neuropeptides reconstructed from 2D images of serial sections deliver contextual information to planar mapping and lower the possibility of neglecting small anatomical structures. Chen et al. established a protocol to examine the volumetric distribution of various neuropeptides from a crustacean brain [32]. By using seven sequential sections with an equal interval of 132 μm on the z-axis of the brain, over twenty neuropeptides belonging to eight families were mapped throughout 3D structures. A reconstructed 3D image of a crustacean neuropeptide Cancer borealis tachykinin-related peptide (CabTRP) 1a is shown in Figure 3a, displaying high abundance on the surface of olfactory lobes in the brain. In contrast, a 3D representation of phosphatidylcholine 38:6 (Figure 3b) shows that it is distributed relatively evenly throughout the whole brain structure. Another example shows the 3D distribution of an RFamide-like peptide SMPSLRLRFa. These results demonstrate that 3D imaging reveals more detailed molecular distribution data than 2D imaging because of the extra dimension. A comprehensive review about 3D IMS applications is a good resource for further reading [93].
Figure 3.
(a) 3D reconstructed ion image of CabTRP 1a by MALDI IMS prepared using regular matrix coating. (b) 3D reconstructed ion image of lipid PC 38:6 by MALDI IMS prepared using dry matrix spraying technique for lipid detection. Adapted with permission from Ref. [32].
Researchers are exploring new ionization techniques for neuropeptide IMS studies. Although SIMS does not yield intact peptide or protein signals above 1 kDa from biological samples, it can produce submicron MS images [94]. Recently, an integrative study was reported, mapping neuropeptides, metabolites and lipids from rat spinal cord, with colocalization patterns being revealed [95]. This multi-faceted MS platform provides an overall picture of molecules utilized by the nervous system. To enhance the desorption/ionization yield of SIMS for neuropeptide imaging, a hybrid technique named matrix-enhanced (ME)-SIMS was developed [58, 61, 96]. The matrix significantly enhanced intensity values, leading to the imaging of three peptides from the cerebral ganglia of the freshwater snail Lymnaea stagnalis at a spatial resolution of 3μm [94].
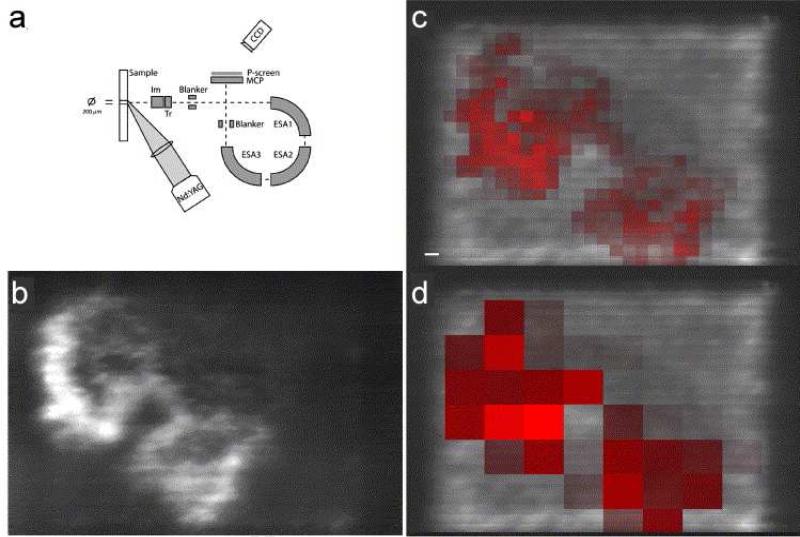
Advancements in sample-handling and mass spectrometers have allowed imaging at cellular-length scales in several studies. As previously described, Altelaar and Luxembourg et al. developed the microscope approach and modified a triply focusing TOF for this purpose (Figure 4a) [42, 74]. In this instrument, desorbed ions within a large defocused laser spot were stigmatically transmitted through the mass analyzer and focused onto a position-sensitive detector. The resulting MS images had a pixel size of 500 nm and an effective resolving power of 4 μm. Numerous pituitary neuropeptides were examined at the same magnification, producing MS images with superior spatial resolution [42]. Figure 4b shows a high resolution MS image obtained with the microscope approach of the diacetylated neuropeptide α- melanocyte stimulating hormone (MSH) from a mouse pituitary section. In comparison, the digitally resampled microprobe images have a pixel size of 80 μm × 80 μm (red) (Figure 4c) and 250 μm × 250 μm (red) (Figure 4d) and lack the resolution required to resolve anatomical features on a cellular scale. High spatial resolution images of other neuropeptides like oxytocin and vasopressin from pituitary gland sections were also shown, highlighting the ability of label-free IMS to deliver images at a biologically relevant length scale. A review by Altelaar et al. describing the detailed configuration and protocol of this microscope approach is worth reading [56]. Interestingly, neuropeptides of mouse pituitary gland section were again imaged in another proof-of-principle experiment aiming to improve spatial resolution. In that study, a linear ion trap Orbitrap was coupled with an in-house developed atmospheric pressure MALDI imaging ion source equipped with a highly focused laser. MS images of neuropeptides, such as oxytocin, vasopressin, copeptin etc. were obtained with a cellular resolution of 5 μm [72]. Nevertheless, these hardware modifications have not been widely practiced because of their complexity.
Figure 4.
Instrumentation of the stigmatic mass microscope and a comparison with microprobe IMS. (a) Schematic representation of the Physical Electronics Trift II mass spectrometer. (b) The stigmatic ion image of the neuropeptide α-MSH (diacetylated) is compared to the distribution of the diacetylated α-MSH in microprobe imaging, depicted with a resampled pixel size of (c) 80 μm × 80 μm (red) and (d) 250 μm × 250 μm (red) and overlaid on the stigmatic TIC image. Scale bar is 100 μm. Adapted with permission from Ref. [42].
Jurchen et al. introduced an oversampling technique by making the raster increment of the sample stage movement smaller than the diameter of the laser beam to achieve a single-cell spatial resolution [75]. This method overcomes the limit of resolution imposed by laser spot size without changing the laser optics of the instrument. Another approach to improve spatial resolution is the stretched sample method [22, 52-54]. The method was employed on the abdominal ganglia of Aplysia californica by adhering the section to a glass bead array anchored to a stretchable membrane and then stretching the membrane to extend the size of the ganglia in two dimensions. This preparation fragmented the tissue to thousands of spatially isolated bead islands, leading to the restriction of analyte redistribution and sufficient extraction of neuropeptides during matrix application.
3.2 IMS of neuropeptides at the cellular level
Invertebrates are useful model systems when developing sample preparation techniques for neuropeptide IMS studies of single cells. Many invertebrate organisms have been characterized, including the freshwater snail (Lymnaea stagnalis) [44-47], sea mollusk (Aplysia californica and Aplysia vaccaria) [48-49, 97], crayfish (Orconectes limosus and Procambarus clarkii) [98-99], cockroach (Periplaneta americana) [31, 50, 100], ticks (Ixodes ricinus and Boophilus microplus) [101], and parasites nematode (Ascaris suum) [51]. Using vesicles sampled from the exocrine atrial gland of Aplysia californica as a model, Rubakhin et al. detected a wide range of bioactive peptides within individual vesicles (1–2 μm) [102]. Later, they optimized a MS-friendly glycerol fixation protocol and reported different neuropeptide complements probed from the dendrites and soma of an isolated intact neuron from Aplysia [23].
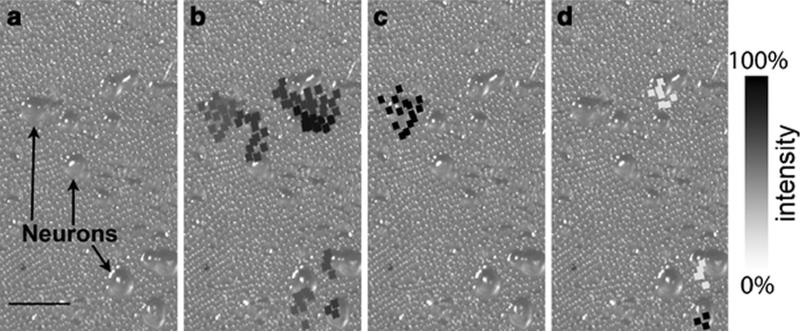
Although the majority of developments pursuing high spatial resolution involved neuropeptides at the organ domains, these methods are readily applicable to neuropeptide imaging at the cellular domain. One example of the techniques initially developed for tissue imaging and later extended to single-cell imaging is the stretched sample method [22, 55]. Zimmerman et al. described the protocol for stretching cultured cells before IMS and tandem MS [22]. Figure 5a shows an optical image of the cell culture before stretching, in which several cell bodies are visible. Upon acquisition of IMS data, ion images were reconstructed based on bead position and overlaid onto the initial sample image shown in Figures 5b-d. The ion image at m/z 635 (Figure 5b) was co-registered with the majority of cell somas in the optical image, demonstrating minimal analyte redistribution and the precise location of the ion in cell bodies. Figures 5c and d display MS images of the two most intense ion signals. The identification of the putative peptides was accomplished by accurate mass matching and in situ MS/MS fragmentation of the putative neuropeptides. The development of improved micro-sampling protocols in combination with continuous advancements of high spatial resolution techniques should enable the sub-cellular mapping of neuropeptides from single cells with MALDI-IMS in the near future.
Figure 5.
Overlaid optical and reconstructed ion images created by the stretched sample method from a neuronal culture from different ganglia of Aplysia central nervous system. About 1200 total beads were probed. (a) An optical image of the sample showing the cultured neurons on a substrate of borosilicate glass beads embedded in a Parafilm M layer. In (b)–(d), the reconstructed ion images overlay the original optical image to show the selected analyte distributions, with (b) showing m/z 635, (c) m/z 1238, and (d) m/z 1540, which have been assigned as a membrane lipid, FRFa peptide, and pedal peptide, respectively. The squares represent the bead positions that have the appropriate m/z signal with their grey color matching the intensity scale bar on the right. In each image, the intensity has been scaled to the maximum level. The analyte distributions are confined to individual cell somas in many cases. Scale bar is 500 μm. Adapted with permission from Ref. [22].
4. Future Perspective
IMS is poised to become a powerful tool to map thousands of molecules from biological surfaces simultaneously, ranging from proteins and peptides to metabolites and pharmaceutical compounds. Various developments and improvements in imaging acquisition speed, spatial resolution and spectral resolution are occurring, removing technical limitations for the emerging applications of IMS to biological sciences. The utilization of IMS to neuropeptide mapping has produced impressive results. A wealth of data regarding neuropeptide distribution and differential processing has been obtained via IMS experiments studying single cells and whole organisms. Neuropeptide distribution and assignments from IMS data could be cross-referenced with other imaging techniques like immunohistochemical (IHC) staining or in situ hybridization and standard bioanalytical approaches applied to tissue or cell homogenates. The TArgeted multiplex MS IMaging (TAMSIM) technique demonstrated the usefulness of IMS coupled with IHC using human tissue sections [103]. TAMSIM, developed by Thiery et al., employs standard IHC procedures to target specific molecules using multiple antibodies with attached photocleavable tags pertaining to discrete masses [103-104]. The mass tags are released without the need of matrix upon laser irradiation, producing MS images for the mass of each tag corresponding to the target molecules [104]. This multiplex imaging method can map multiple endogenous proteins below a mass spectrometer's detection limit with relatively high resolution.
Since no prior knowledge of molecular identities is required for IMS applications, it may become a standard biomarker discovery tool. Minerva et al. evaluated the capability of MALDI-MS to study peptide expression patterns in the mouse pancreas under normal and pathological conditions. The study identified a peptide marker, the C-peptide of insulin, that was dramatically up-regulated in obese mice [105]. Sköld et al observed a significant decrease of a large neuron-specific peptide, PEP-19, in a mouse model of Parkinson's disease using MALDI-IMS [106]. Hanrieder et al. quantified relative abundances of striatal neuropeptides in a rat model of L-DOPA-induced dyskinesia (LID) and revealed several upregulated dynorphins and enkephalins in the dorsolateral striatum of high-dyskinetic rats [107]. Two prodynorphin derived peptides, dynorphin B and α-neoendorphin, were positively associated with LID severity. These findings underscore the critical role MALDI-IMS plays in the study of neuropeptide dynamics during neurological diseases. With IMS, a better understanding of the molecular pathology of various diseases such as cancer, neurodegenerative diseases and viral infections can be obtained.
Several challenges impede the progress of IMS. Advanced computational tools and bioinformatics approaches must be developed to keep up with the ever-increasing raw data file size from high resolution IMS experiments as well as to extract biologically relevant information from these large collections of MS spectra. Additionally, techniques must be refined and commercialized to enable IMS experiments to be performed at the cellular level. Furthermore, MALDI IMS was viewed as an invasive sampling technique until ambient desorption and ionization methods like desorption electrospray ionization (DESI) or direct analysis in real time (DART) were developed [108, 109]. These ambient ion sources circumvent limitations imposed by vacuum operational conditions, and it is predicted that these sampling techniques will enable the transition from ex vivo to in vivo imaging analyses of molecules in living systems. However, these ambient ionization techniques currently suffer from poor sensitivity for relatively large biomolecules like peptides and proteins. Recent surface-ionization methods like laserspray ionization (LSI) and matrix-assisted inlet ionization (MAII) produce multiply charged ions under atmospheric pressure (AP) ionization conditions [110-112]. One proof-of-principle experiment generated a crude image of peptides from mouse brain sections using LSI in transmission geometry, assigning peptide identities by in situ fragmentation [110]. This AP ionization technique provides the first example of multiply charged peptides produced from tissues and demonstrated its capability in IMS applications. Future developments of IMS technologies will increase the versatility of this technique and improve its utility to the field of neuroscience.
Highlights.
General strategies and workflow for imaging mass spectrometry (IMS) are presented.
The current status and progress in IMS of neuropeptide (NP) analysis are reviewed.
Sampling protocols, instrumentation, and software for IMS of NPs are discussed.
Examples of neuropeptide imaging at the organ and cellular levels are highlighted.
Future directions highlight key developments in IMS and the impact on NP studies.
Acknowledgement
Preparation of this manuscript was supported in part by National Science Foundation (CHE-0957784), National Institutes of Health through grants 1R01DK071801 and R56DK071801. L.L. acknowledges an H. I. Romnes Faculty Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gundlach AL, Burazin TCD, Larm JA. Distribution, regulation and role of hypothalamic galanin systems: renewed interest in a pleiotropic peptide family. Clin Exp Pharmacol Physiol. 2001;28:100–5. doi: 10.1046/j.1440-1681.2001.03411.x. [DOI] [PubMed] [Google Scholar]
- 2.Jensen J. Regulatory peptides and control of food intake in non-mammalian vertebrates. Comp Biochem Phys A. 2001;128:471–9. doi: 10.1016/s1095-6433(00)00329-9. [DOI] [PubMed] [Google Scholar]
- 3.Levin ER, Hu RM, Rossi M, Pickart M. Arginine vasopressin stimulates atrial-natriuretic-peptide gene-expression and secretion from rat diencephalic neurons. Endocrinology. 1992;131:1417–23. doi: 10.1210/endo.131.3.1380442. [DOI] [PubMed] [Google Scholar]
- 4.Okada Y, Tsuda Y, Bryant SD, Lazarus LH. Endomorphins and related opioid peptides. Vitam Horm. 2002;65:257–79. doi: 10.1016/s0083-6729(02)65067-8. [DOI] [PubMed] [Google Scholar]
- 5.Kieffer BL, Gaveriaux-Ruff C. Exploring the opioid system by gene knockout. Prog Neurobiol. 2002;66:285–306. doi: 10.1016/s0301-0082(02)00008-4. [DOI] [PubMed] [Google Scholar]
- 6.Stefano GB, Fricchione GL, Goumon Y, Esch T. Pain, immunity, opiate and opioid compounds and health. Med Sci Monit. 2005;11:Ms47–Ms53. [PubMed] [Google Scholar]
- 7.Audsley N, Weaver RJ. Neuropeptides associated with the regulation of feeding in insects. Gen Comp Endocrinol. 2009;162:93–104. doi: 10.1016/j.ygcen.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 8.Mercier AJ, Friedrich R, Boldt M. Physiological functions of FMRFamide-like peptides (FLPs) in crustaceans. Microsc Res Tech. 2003;60:313–24. doi: 10.1002/jemt.10270. [DOI] [PubMed] [Google Scholar]
- 9.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2:953–71. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 10.Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–16. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 11.Li LJ, Sweedler JV. Peptides in the Brain: Mass spectrometry-based measurement approaches and challenges. Annu Rev Anal Chem. 2008;1:451–83. doi: 10.1146/annurev.anchem.1.031207.113053. [DOI] [PubMed] [Google Scholar]
- 12.Kendrick KM. Microdialysis measurement of in vivo neuropeptide release. J Neurosci Methods. 1990;34:35–46. doi: 10.1016/0165-0270(90)90040-m. [DOI] [PubMed] [Google Scholar]
- 13.Karas M, Bachmann D, Bahr U, Hillenkamp F. Matrix-assisted ultraviolet-laser desorption of nonvolatile Compounds. Int J Mass Spectrom. 1987;78:53–68. [Google Scholar]
- 14.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Electrospray ionization for mass-spectrometry of large biomolecules. Science. 1989;246:64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 15.Hummon AB, Amare A, Sweedler JV. Discovering new invertebrate neuropeptides using mass spectrometry. Mass Spectrom Rev. 2006;25:77–98. doi: 10.1002/mas.20055. [DOI] [PubMed] [Google Scholar]
- 16.Defelipe J. Neocortical neuronal diversity: chemical heterogeneity revealed by colocalization studies of classic neurotransmitters, neuropeptides, calcium-binding proteins, and cell-surface molecules. Cereb Cortex. 1993;3:273–89. doi: 10.1093/cercor/3.4.273. [DOI] [PubMed] [Google Scholar]
- 17.Mckelvy JF, Blumberg S. Inactivation and metabolism of neuropeptides. Annu Rev Neurosci. 1986;9:415–32. doi: 10.1146/annurev.ne.09.030186.002215. [DOI] [PubMed] [Google Scholar]
- 18.Takeda Y, Takeda J, Smart BM, Krause JE. Regional distribution of neuropeptide-gamma and other Tachykinin peptides derived from the Substance-P gene in the rat. Regul Peptides. 1990;28:323–33. doi: 10.1016/0167-0115(90)90030-z. [DOI] [PubMed] [Google Scholar]
- 19.Broberger C, Johansen J, Johansson C, Schalling M, Hokfelt T. The neuropeptide Y agouti gene-related protein (AGRP) brain circuitry in normal, anorectic, and monosodium glutamate-treated mice. P Natl Acad Sci USA. 1998;95:15043–8. doi: 10.1073/pnas.95.25.15043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chronwall BM, Dimaggio DA, Massari VJ, Pickel VM, Ruggiero DA, Odonohue TL. The anatomy of neuropeptide Y-containing neurons in rat brain. Neuroscience. 1985;15:1159–81. doi: 10.1016/0306-4522(85)90260-x. [DOI] [PubMed] [Google Scholar]
- 21.Caprioli RM, Farmer TB, Gile J. Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal Chem. 1997;69:4751–60. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- 22.Zimmerman TA, Rubakhin SS, Sweedler JV. MALDI mass spectrometry imaging of neuronal cell cultures. J Am Soc Mass Spectrom. 2011;22:828–36. doi: 10.1007/s13361-011-0111-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rubakhin SS, Greenough WT, Sweedler JV. Spatial profiling with MALDI MS: distribution of neuropeptides within single neurons. Anal Chem. 2003;75:5374–80. doi: 10.1021/ac034498+. [DOI] [PubMed] [Google Scholar]
- 24.Jehl B, Bauer R, Dorge A, Rick R. The use of propane-isopentane mixtures for rapid freezing of biological specimens. J Microsc. 1981;123:307–9. doi: 10.1111/j.1365-2818.1981.tb02475.x. [DOI] [PubMed] [Google Scholar]
- 25.Schwartz SA, Reyzer ML, Caprioli RM. Direct tissue analysis using matrix-assisted laser desorption/ionization mass spectrometry: practical aspects of sample preparation. J Mass Spectrom. 2003;38:699–708. doi: 10.1002/jms.505. [DOI] [PubMed] [Google Scholar]
- 26.Lemaire R, Desmons A, Tabet JC, Day R, Salzet M, Fournier I. Direct analysis and MALDI imaging of formalin-fixed, paraffin-embedded tissue sections. J Proteome Res. 2007;6:1295–305. doi: 10.1021/pr060549i. [DOI] [PubMed] [Google Scholar]
- 27.Che FY, Lim J, Pan H, Biswas R, Fricker LD. Quantitative neuropeptidomics of microwave-irradiated mouse brain and pituitary. Mol Cell Proteomics. 2005;4:1391–405. doi: 10.1074/mcp.T500010-MCP200. [DOI] [PubMed] [Google Scholar]
- 28.Sköld K, Svensson M, Norrman M, Sjogren B, Svenningsson P, Andrén PE. The significance of biochemical and molecular sample integrity in brain proteomics and peptidomics: Stathmin 2-20 and peptides as sample quality indicators. Proteomics. 2007;7:4445–56. doi: 10.1002/pmic.200700142. [DOI] [PubMed] [Google Scholar]
- 29.Svensson M, Boren M, Sköld K, Falth M, Sjogren B, Andersson M, et al. Heat stabilization of the tissue proteome: a new technology for improved proteomics. J Proteome Res. 2009;8:974–81. doi: 10.1021/pr8006446. [DOI] [PubMed] [Google Scholar]
- 30.Crossman L, McHugh NA, Hsieh YS, Korfmacher WA, Chen JW. Investigation of the profiling depth in matrix-assisted laser desorption/ionization imaging mass spectrometry. Rapid Commun Mass Spectrom. 2006;20:284–90. doi: 10.1002/rcm.2259. [DOI] [PubMed] [Google Scholar]
- 31.Verhaert PPM, Strupat K, Conaway M. Imaging of similar mass neuropeptides in neuronal tissue by enhanced resolution MALDI MS with an Ion Trap-Orbitrap™ hybrid instrument. Methods Mol Biol. 2010;656:433–49. doi: 10.1007/978-1-60761-746-4_25. [DOI] [PubMed] [Google Scholar]
- 32.Chen RB, Hui LM, Sturm RM, Li LJ. Three dimensional mapping of neuropeptides and lipids in crustacean brain by mass spectral imaging. J Am Soc Mass Spectrom. 2009;20:1068–77. doi: 10.1016/j.jasms.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeKeyser SS, Kutz-Naber KK, Schmidt JJ, Barrett-Wilt GA, Li LJ. Imaging mass spectrometry of neuropeptides in decapod crustacean neuronal tissues. J Proteome Res. 2007;6:1782–91. doi: 10.1021/pr060603v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chaurand P, Norris JL, Cornett DS, Mobley JA, Caprioli RM. New developments in profiling and imaging of proteins from tissue sections by MALDI mass spectrometry. J Proteome Res. 2006;5:2889–900. doi: 10.1021/pr060346u. [DOI] [PubMed] [Google Scholar]
- 35.Chaurand P, Schwartz SA, Billheimer D, Xu BGJ, Crecelius A, Caprioli RM. Integrating histology and imaging mass spectrometry. Anal Chem. 2004;76:1145–55. doi: 10.1021/ac0351264. [DOI] [PubMed] [Google Scholar]
- 36.Kawamoto T. Use of a new adhesive film for the preparation of multi-purpose fresh-frozen sections from hard tissues, whole-animals, insects and plants. Arch Histol Cytol. 2003;66:123–43. doi: 10.1679/aohc.66.123. [DOI] [PubMed] [Google Scholar]
- 37.Kaletas BK, van der Wiel IM, Stauber J, Dekker LJ, Guzel C, Kros JM, et al. Sample preparation issues for tissue imaging by imaging MS. Proteomics. 2009;9:2622–33. doi: 10.1002/pmic.200800364. [DOI] [PubMed] [Google Scholar]
- 38.Lemaire R, Wisztorski M, Desmons A, Tabet JC, Day R, Salzet M, et al. MALDI-MS direct tissue analysis of proteins: improving signal sensitivity using organic treatments. Anal Chem. 2006;78:7145–53. doi: 10.1021/ac060565z. [DOI] [PubMed] [Google Scholar]
- 39.Seeley EH, Oppenheimer SR, Mi D, Chaurand P, Caprioli RM. Enhancement of protein sensitivity for MALDI imaging mass spectrometry after chemical treatment of tissue sections. J Am Soc Mass Spectrom. 2008;19:1069–77. doi: 10.1016/j.jasms.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meriaux C, Arafah K, Tasiemski A, Wisztorski M, Bruand J, Boidin-Wichlacz C, et al. Multiple changes in peptide and lipid expression associated with regeneration in the nervous system of the medicinal leech. Plos One. 2011;6 doi: 10.1371/journal.pone.0018359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taban IM, Altelaar AFM, Van der Burgt YEM, McDonnell LA, Heeren RMA, Fuchser J, et al. Imaging of peptides in the rat brain using MALDI-FTICR mass spectrometry. J Am Soc Mass Spectrom. 2007;18:145–51. doi: 10.1016/j.jasms.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 42.Altelaar AFM, Taban IM, McDonnell LA, Verhaert PDEM, de Lange RPJ, Adan RAH, et al. High-resolution MALDI imaging mass spectrometry allows localization of peptide distributions at cellular length scales in pituitary tissue sections. Int J Mass Spectrom. 2007;260:203–11. [Google Scholar]
- 43.Van Hove ERA, Smith DF, Fornai L, Glunde K, Heeren RMA. An alternative paper based tissue washing method for mass spectrometry imaging: localized washing and fragile tissue analysis. J Am Soc Mass Spectrom. 2011;22:1885–90. doi: 10.1007/s13361-011-0203-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jimenez CR, Li KW, Dreisewerd K, Spijker S, Kingston R, Bateman RH, et al. Direct mass spectrometric peptide profiling and sequencing of single neurons reveals differential peptide patterns in a small neuronal network. Biochemistry. 1998;37:2070–6. doi: 10.1021/bi971848b. [DOI] [PubMed] [Google Scholar]
- 45.Jimenez CR, Vanveelen PA, Li KW, Wildering WC, Geraerts WPM, Tjaden UR, et al. Neuropeptide expression and processing as revealed by direct matrix-assisted laser desorption/ionization mass spectrometry of single neurons. J Neurochem. 1994;62:404–7. doi: 10.1046/j.1471-4159.1994.62010404.x. [DOI] [PubMed] [Google Scholar]
- 46.Li KW, Hoek RM, Smith F, Jimenez CR, Vanderschors RC, Vanveelen PA, et al. Direct peptide rrofiling by mass spectrometry of single identified neurons reveals complex neuropeptide-processing pattern. J Biol Chem. 1994;269:30288–92. [PubMed] [Google Scholar]
- 47.Li KW, Jimenez CR, Vanveelen PA, Geraerts WPM. Processing and Targeting of a molluscan egg-laying peptide prohormone as revealed by mass-spectrometric peptide fingerprinting and peptide sequencing. Endocrinology. 1994;134:1812–9. doi: 10.1210/endo.134.4.8137747. [DOI] [PubMed] [Google Scholar]
- 48.Garden RW, Moroz LL, Moroz TP, Shippy SA, Sweedler JV. Excess salt removal with matrix rinsing: direct peptide profiling of neurons from marine invertebrates using matrix-assisted laser desorption ionization time-of-flight mass spectrometry. J Mass Spectrom. 1996;31:1126–30. doi: 10.1002/(SICI)1096-9888(199610)31:10<1126::AID-JMS403>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 49.Li LJ, Garden RW, Romanova EV, Sweedler JV. In situ sequencing of peptides from biological tissues and single cells using MALDI-PSD/CID analysis. Anal Chem. 1999;71:5451–8. doi: 10.1021/ac9907181. [DOI] [PubMed] [Google Scholar]
- 50.Neupert S, Schattschneider S, Predel R. Allatotropin-related peptide in cockroaches: identification via mass spectrometric analysis of single identified neurons. Peptides. 2009;30:489–94. doi: 10.1016/j.peptides.2008.10.023. [DOI] [PubMed] [Google Scholar]
- 51.Jarecki JL, Andersen K, Konop CJ, Knickelbine JJ, Vestling MM, Stretton AO. Mapping neuropeptide expression by mass spectrometry in single dissected identified neurons from the dorsal ganglion of the nematode Ascaris suum. ACS Chem Neurosci. 2010;1:505–19. doi: 10.1021/cn1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rubakhin SS, Sweedler JV. Characterizing peptides in individual mammalian cells using mass spectrometry. Nat Protoc. 2007;2:1987–97. doi: 10.1038/nprot.2007.277. [DOI] [PubMed] [Google Scholar]
- 53.Tucker KR, Serebryannyy LA, Zimmerman TA, Rubakhin SS, Sweedler JV. The modified-bead stretched sample method: development and application to MALDI-MS imaging of protein localization in the spinal cord. Chem Sci. 2011;2:785–95. doi: 10.1039/C0SC00563K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zimmerman TA, Rubakhin SS, Romanova EV, Tucker KR, Sweedler JV. MALDI mass spectrometric imaging using the stretched sample method to reveal neuropeptide distributions in aplysia nervous tissue. Anal Chem. 2009;81:9402–9. doi: 10.1021/ac901820v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zimmerman TA, Monroe EB, Sweedler JV. Adapting the stretched sample method from tissue profiling to imaging. Proteomics. 2008;8:3809–15. doi: 10.1002/pmic.200800331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Altelaar AFM, Luxembourg SL, McDonnell LA, Piersma SR, Heeren RMA. Imaging mass spectrometry at cellular length scales. Nat Protoc. 2007;2:1185–96. doi: 10.1038/nprot.2007.117. [DOI] [PubMed] [Google Scholar]
- 57.Armstrong DW, Zhang LK, He LF, Gross ML. Ionic liquids as matrixes for matrix-assisted laser desorption/ionization mass spectrometry. Anal Chem. 2001;73:3679–86. doi: 10.1021/ac010259f. [DOI] [PubMed] [Google Scholar]
- 58.Fitzgerald JJD, Kunnath P, Walker AV. Matrix-enhanced secondary ion mass spectrometry (ME-SIMS) using room temperature ionic liquid matrices. Anal Chem. 2010;82:4413–9. doi: 10.1021/ac100133c. [DOI] [PubMed] [Google Scholar]
- 59.Lemaire R, Tabet JC, Ducoroy P, Hendra JB, Salzet M, Fournier I. Solid ionic matrixes for direct tissue analysis and MALDI imaging. Anal Chem. 2006;78:809–19. doi: 10.1021/ac0514669. [DOI] [PubMed] [Google Scholar]
- 60.Tholey A, Zabet-Moghaddam M, Heinzle E. Quantification of peptides for the monitoring of protease-catalyzed reactions by matrix-assisted laser desorption/ionization mass spectrometry using ionic liquid matrixes. Anal Chem. 2006;78:291–7. doi: 10.1021/ac0514319. [DOI] [PubMed] [Google Scholar]
- 61.Altelaar AFM, Klinkert I, Jalink K, de Lange RPJ, Adan RAH, Heeren RMA, et al. Gold-enhanced biomolecular surface imaging of cells and tissue by SIMS and MALDI mass spectrometry. Anal Chem. 2006;78:734–42. doi: 10.1021/ac0513111. [DOI] [PubMed] [Google Scholar]
- 62.Schuerenberg M, Luebbert C, Deininger SO, Ketterlinus R, Suckau D. MALDI tissue imaging: mass spectrometric localization of biomarkers in tissue slices. Nat Methods. 2007;4:iii–iv. [Google Scholar]
- 63.Aerni HR, Cornett DS, Caprioli RM. Automated acoustic matrix deposition for MALDI sample preparation. Anal Chem. 2006;78:827–34. doi: 10.1021/ac051534r. [DOI] [PubMed] [Google Scholar]
- 64.Baluya DL, Garrett TJ, Yost RA. Automated MALDI matrix deposition method with inkjet printing for imaging mass spectrometry. Anal Chem. 2007;79:6862–7. doi: 10.1021/ac070958d. [DOI] [PubMed] [Google Scholar]
- 65.Franck J, Arafah K, Barnes A, Wisztorski M, Salzet M, Fournier I. Improving tissue preparation for matrix-assisted laser desorption ionization mass spectrometry imaging. Part 1: Using microspotting. Anal Chem. 2009;81:8193–202. doi: 10.1021/ac901328p. [DOI] [PubMed] [Google Scholar]
- 66.Hankin JA, Barkley RM, Murphy RC. Sublimation as a method of matrix application for mass spectrometric imaging. J Am Soc Mass Spectrom. 2007;18:1646–52. doi: 10.1016/j.jasms.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Puolitaival SM, Burnum KE, Cornett DS, Caprioli RM. Solvent-free matrix dry-coating for MALDI imaging of phospholipids. J Am Soc Mass Spectrom. 2008;19:882–6. doi: 10.1016/j.jasms.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Deutskens F, Yang JH, Caprioli RM. High spatial resolution imaging mass spectrometry and classical histology on a single tissue section. J Mass Spectrom. 2011;46:568–71. doi: 10.1002/jms.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schafer R. UltrafleXtreme: redefining MALDI-TOF-TOF mass spectrometry performance. LC GC Eur. 2009:26–7. [Google Scholar]
- 70.Koestler M, Kirsch D, Hester A, Leisner A, Guenther S, Spengler B. A high-resolution scanning microprobe matrix-assisted laser desorption/ionization ion source for imaging analysis on an ion trap/Fourier transform ion cyclotron resonance mass spectrometer. Rapid Commun Mass Spectrom. 2008;22:3275–85. doi: 10.1002/rcm.3733. [DOI] [PubMed] [Google Scholar]
- 71.Römpp A, Guenther S, Schober Y, Schulz O, Takats Z, Kummer W, et al. Histology by mass spectrometry: label-free tissue characterization obtained from high-accuracy bioanalytical imaging. Angew Chem Int Edit. 2010;49:3834–8. doi: 10.1002/anie.200905559. [DOI] [PubMed] [Google Scholar]
- 72.Guenther S, Römpp A, Kummer W, Spengler B. AP-MALDI imaging of neuropeptides in mouse pituitary gland with 5 mm spatial resolution and high mass accuracy. Int J Mass Spectrom. 2011;305:228–37. [Google Scholar]
- 73.Klerk LA, Altelaar AFM, Froesch M, McDonnell LA, Heeren RMA. Fast and automated large-area imaging MALDI mass spectrometry in microprobe and microscope mode. Int J Mass Spectrom. 2009;285:19–25. [Google Scholar]
- 74.Luxembourg SL, Mize TH, McDonnell LA, Heeren RMA. High-spatial resolution mass spectrometric imaging of peptide and protein distributions on a surface. Anal Chem. 2004;76:5339–44. doi: 10.1021/ac049692q. [DOI] [PubMed] [Google Scholar]
- 75.Jurchen JC, Rubakhin SS, Sweedler JV. MALDI-MS imaging of features smaller than the size of the laser beam. J Am Soc Mass Spectrom. 2005;16:1654–9. doi: 10.1016/j.jasms.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 76.Campana JE. Time-of-flight mass spectrometry: a historical overview. Anal Instrum. 1987;16:1–14. [Google Scholar]
- 77.Vestal ML, Campbell JM. Tandem time-of-flight mass spectrometry. Method Enzymol. 2005;402:79–108. doi: 10.1016/S0076-6879(05)02003-3. [DOI] [PubMed] [Google Scholar]
- 78.Simmons DA. Improved MALDI-MS imaging performance using continuous laser rastering. MDS Analytical Technologies. 2008 [Google Scholar]
- 79.Spraggins JM, Caprioli R. High-speed MALDI-TOF imaging mass spectrometry: rapid ion image acquisition and considerations for next generation instrumentation. J Am Soc Mass Spectrom. 2011;22:1022–31. doi: 10.1007/s13361-011-0121-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Trim PJ, Djidja MC, Atkinson SJ, Oakes K, Cole LM, Anderson DMG, et al. Introduction of a 20 kHz Nd:YVO4 laser into a hybrid quadruple time-of-flight mass spectrometer for MALDI-MS imaging. Anal Bioanal Chem. 2010;397:3409–19. doi: 10.1007/s00216-010-3874-6. [DOI] [PubMed] [Google Scholar]
- 81.Marshall AG, Hendrickson CL, Jackson GS. Fourier transform ion cyclotron resonance mass spectrometry: A primer. Mass Spectrom Rev. 1998;17:1–35. doi: 10.1002/(SICI)1098-2787(1998)17:1<1::AID-MAS1>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 82.Kutz KK, Schmidt JJ, Li LJ. In situ tissue analysis of neuropeptides by MALDI FTMS in-cell accumulation. Anal Chem. 2004;76:5630–40. doi: 10.1021/ac049255b. [DOI] [PubMed] [Google Scholar]
- 83.Fuchser J, Cornett S, Becker M. High Resolution Molecular imaging of pharmaceuticals at therapeutic levels. Bruker Daltonik GmbH. 2008 [Google Scholar]
- 84.Arnaud CH. High-res mass spec. Chem Eng News. 2010;88:10–15. [Google Scholar]
- 85.Makarov A. Electrostatic axially harmonic orbital trapping: A high-performance technique of mass analysis. Anal Chem. 2000;72:1156–62. doi: 10.1021/ac991131p. [DOI] [PubMed] [Google Scholar]
- 86.Hu QZ, Noll RJ, Li HY, Makarov A, Hardman M, Cooks RG. The Orbitrap: a new mass spectrometer. J Mass Spectrom. 2005;40:430–43. doi: 10.1002/jms.856. [DOI] [PubMed] [Google Scholar]
- 87.Chen RB, Jiang XY, Conaway MCP, Mohtashemi I, Hui LM, Viner R, et al. Mass spectral analysis of neuropeptide expression and distribution in the nervous system of the lobster Homarus americanus. J Proteome Res. 2010;9:818–32. doi: 10.1021/pr900736t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Verhaert PD, Conaway MCP, Pekar TM, Miller K. Neuropeptide imaging on an LTQ with vMALDI source: The complete ‘all-in-one’ peptidome analysis. Int J Mass Spectrom. 2007;260:177–84. [Google Scholar]
- 89.Hoaglund CS, Valentine SJ, Sporleder CR, Reilly JP, Clemmer DE. Three-dimensional ion mobility TOFMS analysis of electrosprayed biomolecules. Anal Chem. 1998;70:2236–42. doi: 10.1021/ac980059c. [DOI] [PubMed] [Google Scholar]
- 90.Wyttenbach T, Bowers MT. Gas-phase conformations: the ion mobility/ion chromatography method. Top Curr Chem. 2003;225:207–32. [Google Scholar]
- 91.Jackson SN, Ugarov M, Egan T, Post JD, Langlais D, Schultz JA, et al. MALDI-ion mobility-TOFMS imaging of lipids in rat brain tissue. J Mass Spectrom. 2007;42:1093–8. doi: 10.1002/jms.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Römpp AST, Hester A, Klinkert I, Both JP, Heeren RM, Stöckli M, Spengler B. imzML: imaging mass spectrometry markup language: a common data format for mass spectrometry imaging. Methods Mol Biol. 2011;696:205–24. doi: 10.1007/978-1-60761-987-1_12. [DOI] [PubMed] [Google Scholar]
- 93.Ye H, Greer T, Li LJ. From pixel to voxel: a deeper view of biological tissue by 3D mass spectral imaging. Bioanalysis. 2011;3:313–32. doi: 10.4155/bio.10.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Altelaar AFM, van Minnen J, Jimenez CR, Heeren RMA, Piersma SR. Direct molecular imaging of Lymnaea stagnalis nervous tissue at subcellular spatial resolution by mass spectrometry. Anal Chem. 2005;77:735–41. doi: 10.1021/ac048329g. [DOI] [PubMed] [Google Scholar]
- 95.Monroe EB, Annangudi SR, Hatcher NG, Gutstein HB, Rubakhin SS, Sweedler JV. SIMS and MALDI MS imaging of the spinal cord. Proteomics. 2008;8:3746–54. doi: 10.1002/pmic.200800127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.McDonnell LA, Piersma SR, Altelaar AFM, Mize TH, Luxembourg SL, Verhaert PDEM, et al. Subcellular imaging mass spectrometry of brain tissue. J Mass Spectrom. 2005;40:160–8. doi: 10.1002/jms.735. [DOI] [PubMed] [Google Scholar]
- 97.Li LJ, Garden RW, Floyd PD, Moroz TP, Gleeson JM, Sweedler JV, et al. Egg-laying hormone peptides in the Aplysiidae family. J Exp Biol. 1999;202:2961–73. doi: 10.1242/jeb.202.21.2961. [DOI] [PubMed] [Google Scholar]
- 98.Redeker V, Toullec JY, Vinh J, Rossier J, Soyez D. Combination of peptide profiling by matrix-assisted laser desorption/ionization time of flight mass spectrometry and immunodetection on single glands or cells. Anal Chem. 1998;70:1805–11. doi: 10.1021/ac971309c. [DOI] [PubMed] [Google Scholar]
- 99.Yasuda A, Yasuda-Kamatani Y, Nozaki M, Nakajima T. Identification of GYRKPPFNGSIFamide (crustacean-SIFamide) in the crayfish Procambarus clarkii by topological mass spectrometry analysis. Gen Comp Endocrinol. 2004;135:391–400. doi: 10.1016/j.ygcen.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 100.Neupert S, Predel R. Mass spectrometric analysis of single identified neurons of an insect. Biochem Biophys Res Commun. 2005;327:640–5. doi: 10.1016/j.bbrc.2004.12.086. [DOI] [PubMed] [Google Scholar]
- 101.Neupert S, Predel R, Russell WK, Davies R, Pietrantonio PV, Nachman RJ. Identification of tick periviscerokinin, the first neurohormone of Ixodidae: single cell analysis by means of MALDI-TOF/TOF mass spectrometry. Biochem Bioph Res Commun. 2005;338:1860–4. doi: 10.1016/j.bbrc.2005.10.165. [DOI] [PubMed] [Google Scholar]
- 102.Rubakhin SS, Garden RW, Fuller RR, Sweedler JV. Measuring the peptides in individual organelles with mass spectrometry. Nat Biotechnol. 2000;18:172–5. doi: 10.1038/72622. [DOI] [PubMed] [Google Scholar]
- 103.Thiery G, Shchepinov MS, Southern EM, Audebourg A, Audard V, Terris B, et al. Multiplex target protein imaging in tissue sections by mass spectrometry-TAMSIM. Rapid Commun Mass Spectrom. 2007;21:823–9. doi: 10.1002/rcm.2895. [DOI] [PubMed] [Google Scholar]
- 104.Thiery G, Anselmi E, Audebourg A, Darii E, Abarbri M, Terris B, et al. Improvements of TArgeted multiplex mass spectrometry IMaging. Proteomics. 2008;8:3725–34. doi: 10.1002/pmic.200701150. [DOI] [PubMed] [Google Scholar]
- 105.Minerva L, Clerens S, Baggerman G, Arckens L. Direct profiling and identification of peptide expression differences in the pancreas of control and ob/ob mice by imaging mass spectrometry. Proteomics. 2008;8:3763–74. doi: 10.1002/pmic.200800237. [DOI] [PubMed] [Google Scholar]
- 106.Sköld K, Svensson M, Nilsson A, Zhang XQ, Nydahl K, Caprioli RM, et al. Decreased striatal levels of PEP-19 following MPTP lesion in the mouse. J Proteome Res. 2006;5:262–9. doi: 10.1021/pr050281f. [DOI] [PubMed] [Google Scholar]
- 107.Hanrieder J, Ljungdahl A, Falth M, Mammo SE, Bergquist J, Andersson M. L-DOPA-induced dyskinesia is associated with regional increase of striatal dynorphin peptides as elucidated by imaging mass spectrometry. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M111.009308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wiseman JM, Ifa DR, Zhu YX, Kissinger CB, Manicke NE, Kissinger PT, et al. Desorption electrospray ionization mass spectrometry: Imaging drugs and metabolites in tissues. P Natl Acad Sci USA. 2008;105:18120–5. doi: 10.1073/pnas.0801066105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Osuga J, Konuma K. Applications of direct analysis in real time (DART) mass spectrometry. J Syn Org Chem Jpn. 2011;69:171–5. [Google Scholar]
- 110.Inutan ED, Richards AL, Wager-Miller J, Mackie K, McEwen CN, Trimpin S. Laserspray ionization, a new method for protein analysis directly from tissue at atmospheric pressure with ultrahigh mass resolution and electron transfer dissociation. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Trimpin S, Inutan ED, Herath TN, McEwen CN. Laserspray ionization, a new atmospheric pressure MALDI method for producing highly charged gas-phase ions of peptides and proteins directly from solid solutions. Mol Cell Proteomics. 2010;9:362–7. doi: 10.1074/mcp.M900527-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Richards AL, Lietz CB, Wager-Miller JB, Mackie K, Trimpin S. Imaging mass spectrometry in transmission geometry. Rapid Commun Mass Spectrom. 2011;25:815–20. doi: 10.1002/rcm.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]