Abstract
Proper display of three-dimensional time-of-flight secondary ion mass spectrometry (ToF-SIMS) imaging data of complex, non-flat samples requires a correction of the data in the z-direction. Inaccuracies in displaying three dimensional ToF-SIMS data arise from projecting data from a non-flat surface onto a 2D image plane, as well as possible variations in the sputter rate of the sample being probed. The current study builds on previous studies by creating software written in Matlab to apply the z-correction to entire 3D data sets, the ZCorrectorGUI (available at http://mvsa.nb.uw.edu/). Three dimensional image data sets were acquired from NIH/3T3 fibroblasts by collecting ToF-SIMS images using a dual beam approach (25 keV Bi3+ for analysis cycles and 20 keV C60++ for sputter cycles). The entire data cube was then corrected using the new ZCorrectorGUI software, producing accurate chemical information from single cells in 3D. For the first time, a three dimensional corrected view of a lipid-rich subcellular region, possibly the nuclear membrane, is presented. Additionally, the key assumption of a constant sputter rate throughout the data acquisition was tested using ToF-SIMS and atomic force microscopy (AFM) analysis of the same cells. For the dried NIH/3T3 fibroblasts examined in this study, the sputter rate was found to not change appreciably in x, y or z, and the cellular material was sputtered at approximately 10 nm per 1.25×1013 ions C60++/cm2.
Introduction
Time-of-flight secondary ion mass spectrometry (ToF-SIMS) is a powerful tool that has been previously used to study a wide range of biological materials including (but not limited to): cells and tissues, lipids, DNA, drug eluting stents, explanted biomaterials and decellularized matrix1–15. The unique abilities of ToF-SIMS to acquire a full mass spectrum at high mass resolution with submicron lateral resolution16 makes for a bright future for spectroscopic and imaging analysis of biological materials17. These characteristics are required to visualize sub-cellular features, which may prove powerful for analyzing drug or metabolite behavior within single cells.
In addition to creating two-dimensional chemical maps of the surface, three-dimensional imaging has become feasible with the incorporation of cluster ion sources into ToF-SIMS instruments. This has been facilitated by the increase in yield during analysis provided by primary ions such as Bi3+, and the ability of C60 sputtering to efficiently remove material while minimizing residual damage in the remaining material18–23. A 3D imaging experiment with current ToF-SIMS instrumentation often requires a dual beam approach24–27, where multiple sequential analysis and sputter cycles are carried out until the feature of interest is fully consumed. This mode of analysis is becoming more widely used in the ToF-SIMS community. In 2006, Gillen et al. constructed 3D images of various pharmaceuticals in biocompatible polymers28, and in 2007 Fletcher et al. constructed 3D images of a Xenopus Laevis oocyte3. Soon after, Breitenstein et al. published a view of a single cell in three dimensions24. Recently, Fletcher et al. imaged frozen-hydrated HeLa-M cells in 3D, and were able to localize the rich adenine signal from DNA within the nucleus29. These latter two papers addressed an important challenge in producing realistic 3D images from ToF-SIMS depth profiles, the fact that the data matrix is inverted as the image data is acquired from a 3D object.
The correction of ToF-SIMS data to create a more accurate representation of the original sample is not new to the SIMS community. There are two distinct corrections required. The first corrects a data set acquired from a sample that is three dimensional. The need for this correction is explained in more detail in the following paragraph. The second correction addresses any significant changes to the erosion rate as the sample is sputtered. This may result from nonuniform composition within the sample30, 31, changes to the incident angle of the primary ion beam32, 33, or sputter induced topographic damage34, 35. In 1982 Patkin et al. describe a correction to the intensity of ion images due to differential sputtering in plastic embedded plant tissue during dynamic SIMS experiments31. Relative sputtering rates of 1.5 and 1.2 were calculated for the cells walls and nuclei, compared to the cytoplasm. Wagter et al. used atomic force microscopy (AFM) to correct for initial topography and differences in the sputter rate for different materials in the dynamic SIMS depth profile data of a Zr-2.5%Nb alloy sample36. With this correction the SIMS data was fit to the AFM topographic maps taken before and after the acquisition of the SIMS depth profile. The AFM data was used to calibrate the z-scale of the SIMS data. This approach has been developed further in a series of papers from the Winograd group30, 37, 38. For example, a protocol was developed for correcting the z-axis in a 3D data set obtained from a thin trehalose film with 15-keV Ga+ ions implanted into it38. The region damaged by the implanted Ga+ had a much lower sputter rate that then the surrounding trehalose, which was compensated for with a similar AFM calibration of the SIMS depth profile. In instances where differential sputtering is significant, this type of correction is necessary for the accurate 3D reconstruction and visualization of the data.
To understand the need for correcting a ToF-SIMS image stack created from a 3D object, consider a model hemisphere system. The cyclic sputtering of this sample would be analogous to the peeling of an onion, with a small “layer” of material being removed by each sputter cycle. Due to the fact that the data from this 3D surface is saved in a 2D image slice, a flat image is produced from a non-flat surface. This results in a collection of images where the ensuing “3D” image stack will be upside-down, and assuming a constant sputter rate, have the same shape as the real object. Simply rotating the data stack 180° is not sufficient since information originating from within the cell volume would not be in the correct location. A pixel shift needed to correct this problem was introduced by Breitenstein et al.24. For this correction the upside down contour of the feature being imaged is determined. This contour line is then defined as the new Z=0 line and the data is corrected accordingly. It should be noted that this correction does not take into account any horizontal shifts due to changes in the perceived beam position. However, as shown in the data provided below, these types of shifts, if present, are not significant for the samples used in this work. For an accurate z-correction, the sputter rate through the feature being profiled needs to not change significantly, otherwise an additional correction for differential sputtering must be applied.
The assumption of a constant sputter rate has been used in both of the 3D depth profiling studies of cells that corrected for the non-flat nature of those cells24, 29. Due to the complexity of the cellular structure, it is important to examine the validity of this assumption. Chandra et al. found that there was no noticeable differential sputtering effects in freeze-fractured, freeze-dried NIH/3T3 fibroblasts under dynamic bombardment using Ar+, O2+ and Cs+ primary ions39. It is necessary to determine if this is a valid assumption when C60 is used as the sputter source in ToF-SIMS studies.
The method used to prepare single cells for ToF-SIMS analysis will affect the information obtained from them, and may affect the sputtering issues that are described in this text. There are generally three methods to prepare cells: analyze frozen-hydrated after cryofixation, cryofixation followed by lyophilization, and chemical fixation. Frozen-hydrated analysis best maintains the native structure of the cells29, 40, and also may increase ion yields because of protons donated by the incorporated water matrix41–43. Breitenstein et al. shows that chemical fixation is a viable method to prepare cells in many instances44. The two methods that yield dry cells, cryofixation followed by lyophilization, and chemical fixation with air drying are examined in this study. Dried cells were chosen for comparison of the corrected SIMS data with the AFM topography data since the AFM used in this study does not have a cryogenic stage.
In this study we explore the validity of the assumption of a constant sputter rate in depth profiles of NIH/3T3 cells utilizing C60++ as the sputter source. Furthermore, a new visualization tool, the ZCorrectorGUI, is introduced. This applies a z-axis correction, as described by Breitenstein et al.24, to entire data sets, as well as 3D visualization in the same program. Lastly, we show for the first time, a three dimensional, corrected view of a lipid-rich subcellular feature within a single cell, possibly the nuclear membrane, using ToF-SIMS imaging.
Materials and Methods
Cell Culture
NIH/3T3 fibroblasts we cultured on 1 cm × 1 cm silicon chips45 that were previously cleaned by sequentially sonicating for 5 minutes, 2x each in dichloromethane, acetone and methanol. The cells were grown in Dulbecco’s Modified Eagle Medium (DMEM) (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (Hyclone, Waltham, MA) and 1% antibiotic/antimycotic (Invitrogen, Carlsbad, CA). Cells were seeded at densities between 20,000–40,000 cells and allowed to grow for 24–48 hours, depending on the desired coverage.
Chemical Fixation
Silicon chips with seeded cells were removed from the culture media and gently rinsed for 5 seconds in 150 mM ammonium acetate (Sigma, St. Louis, MO) at pH 7.4.46 The samples were then placed in room temperature 4% ethanol-free formaldehyde (Thermo Scientific, Erie, PA) in 1X phosphate buffered saline (PBS) (EDS Chemicals) for 30 minutes. After fixation the cells were rinsed for 5 seconds, then edges of the silicon chip were touched with a Kimwipe to remove excess media, and the samples were allowed to air dry overnight in a laminar flow hood.
Lyophilization
Silicon chips with seeded cells were removed from the culture media and gently rinsed for 30 seconds in 150 mM ammonium acetate at pH 7.4. Prior to cryofixation the edges of the silicon chip were touched with a Kimwipe to remove excess media. The samples were then plunge frozen in liquid ethane47, and stored in small glass test tubes under liquid nitrogen (LN2). The test tubes were placed into a pre-cooled lyophilization flask, with LN2 still in the tubes, and the flask attached to the vacuum of a manifold type freeze drier (SP Scientific, Warminster, PA). The samples were lyophilized overnight.
Time-of-flight Secondary Ion Mass Spectrometry
ToF-SIMS experiments were performed using an ION-TOF TOF.SIMS 5–100 (ION-TOF GmbH, Münster, Germany) equipped with two ion sources. A liquid metal ion gun (LMIG) was used to generate a pulsed 25 keV Bi3+ beam, whereas an electron impact gun was used to generate a 20 keV C60++ beam. Both beams hit the target at an angle of 45°. For the 3D data sets, the Bi3+ beam was rastered over a constant area (varies between data sets) and centered inside a 500 μm × 500 μm C60++ crater. For the 3D data sets, a high spatial resolution (HSR) mode was used to acquire the images, while a high mass resolution bunched mode was used to acquire spectra intermittently for peak assignments. The mass resolution (m/ Δm) for C2H3+ was roughly 300 and 5000 for the imaging and spectra modes, respectively. Target currents were measured before each data set using a Faraday cup. The Bi3+ current was typically 0.04–0.05 pA with an 80.1 ns pulse width for the imaging mode, while the C60++ current varied between data sets. The C60 sputtering time was varied, depending on the current, to achieve a constant sputter dose of 1.25 × 1013 ions/cm2 for each sputter cycle. Analysis cycles using a Bi3+ sputter dose of 5.8 × 1011 ions/cm2 was used to acquire images (256×256 pixels) at each slice.
Atomic Force Microscopy
To measure the height of the dried cells before ToF-SIMS analysis, a Dimension ICON (Bruker, Santa Barbara, CA) atomic force microscope (AFM) was used in ScanAsyst® mode in air with a silicon nitride, ScanAsyst-Air tip. Raw AFM data was imported into the NanoScope Analysis software and a second order plane fit was applied to all images. No further modifications were applied to the images.
Data Handling/ ZcorrectorGUI
Peak intensity images from peaks known to correspond to components of cells were exported from the IONTOF Version 4 program (ION-TOF GmbH, Münster, Germany) as .bif files and saved in a folder. Those files were then imported into the ZCorrectorGUI (Dan Graham Ph.D, NESAC/BIO, University of Washington) toolbox written for MATLAB (MathWorks, Natick, MA).
The ZCorrectorGUI is a graphical user interface (GUI) for MATLAB that incorporates a similar “z-correction” pixel shift as described in24, as well as 2D and 3D image display capabilities that allow visualization of a corrected data cube. The ZCorrectorGUI can import image files exported from the IONTOF software as .bif (Version 4) or .bif6 (Version 6) files. For non-IONTOF users, the data can be loaded from the Matlab workspace once it is imported into Matlab as properly organized data matrices (see the tutorials available at http://mvsa.nb.uw.edu/node/340). Any combination of masses can be viewed in the corrected state and examined in 3D. Three color overlays can be created in the corrected and 3D views. In addition transparency values can be varied to examine selected components within the 3D volume.
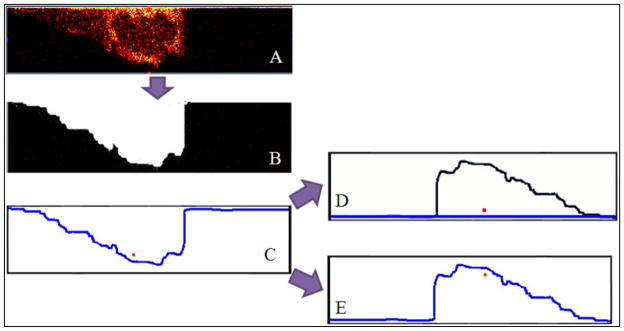
The ZcorrectorGUI shifts the pixels vertically to their correct position by first defining a new z=0 boundary, then moving every pixel to the correct location based on this new boundary. A schematic of this process is shown in Figure 1. Figure 1A shows a XZ slice through a cell depicting the m/z 58+ peak intensities with no correction, showing the first analysis cycle at the top and the last analysis cycle at the bottom. The slice is upside-down but some sub-cellular features can be seen. To correct the z-axis of this image, the program thresholds the total counts image for a given slice (B) and finds the line at the interface defining the new z=0 (blue line) (C). The distance between the new z=0 line and a given pixel is measured, and after flattening of the new z=0 line, the pixel is correctly placed at the measured distance from the flattened z=0 line (D). As a comparison, the result from simply rotating the image slice 180° can be seen in E, where the highlighted pixel (in red) is in the incorrect location. This pixel shift method is applied to all slices in the data cube, so for a data set with 10, 256×256 pixel images, the above process will occur 10 × 256 × 256 (655,360) times.
Figure 1.
A flow diagram of the z-correction process. (A) XZ slice from a cell, showing the m/z 58+peak intensities. (B) Thresholded image of the total counts image from the same data set as A. (C) An outline of the thresholded image is created. This is the new z=0 line. (D) Results when the z-correction is properly implemented. The z=0 line is flat, and the red pixel is in its proper position. (E) Results from flipping the image in C 180° without the z-correction. The location of the red pixel is wrong.
Results and Discussion
ZcorrectorGUI
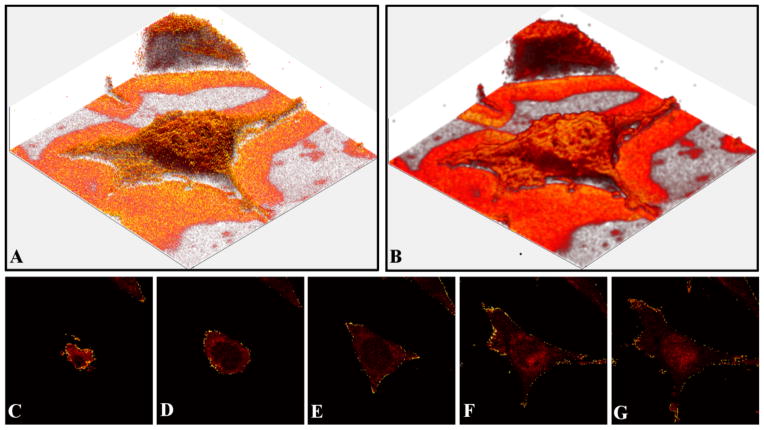
Figure 2 shows data from a single 3T3 fibroblast that was chemically fixed with 4% paraformaldehyde and air dried. There are 56 slices in the data set, each image is 86×86 μm2 and consists of 256×256 pixels. In 2A, the cell is visualized by plotting the intensity of the lipid peak at m/z 58+. In Figure 2B the same data is plotted after smoothing. The data was smoothed using the Matlab ‘smooth3’ function using the default (3,3,3) box filter. This function uses a standard box filter to smooth the data by replacing the data in each voxel with the mean of the data within the box size chosen. These images only depict the outer lipid membrane of the cell, but there are methods that can be used to “see inside” past the plasma membrane. The first method is depicted in Figure 2C–G, which shows selected corrected XY slices of the m/z 58+ peak intensities for the same cell. These corrected slices are analogous to an x-ray computed tomography (CT) scan as they represent the information in the horizontal plane at that position. This allows the users to determine accurately where in the z-direction a target component, such as a metabolite or drug, is located. The ZcorrectorGUI is also capable of displaying the corrected data in the YZ or XZ planes. In addition, movies can be created that visualize slicing through the corrected data in any of these planes.
Figure 2.
Visualization options provided by the ZcorrectorGUI. The data set consists of 56 images. Each image contained 256×256 pixels and was 86×86 μm2 in size. (A) An image of the m/z 58+ signals from a corrected data set of a single NIH/3T3 fibroblast. (B) The same data from A, after smoothing. (C–G) A series of corrected XY images of the m/z 58+peak intensities. With the corrected data, one can move through the cell vertically in a similar fashion as a CT scan moves through tissue. C is at the top of the cell, while G is near the bottom of the cell.
Another way to visualize components within the cell is to use a 3-D image similar to those shown in Figures 2A and B, but to make the entire data set semi-transparent. A similar method was used to map the adenine signal in HeLa-M cells recently29. The results using the ZcorrectorGUI can be seen in Figure 3, where the m/z 58+ signal is displayed in all images. In A the data was first smoothed and then made translucent by changing the alpha value (a measure of the total opacity of the image: 0 = fully transparent, 1 = fully opaque) to 0.02. Here a lipid rich region can be seen within the nucleus, which is possibly a nucleolus. In B only the voxels with an intensity of 1–25 are displayed, and the alpha value was set to 0.04. For C, only the voxels with an intensity of 3 are displayed, and the alpha value is 0.25. A spherical feature consisting of lipids, likely the nuclear membrane, is clearly visible. This ability to display voxels with their intensity specified by the user is useful because it allows internal features to be visualized. To our knowledge this is the first reported 3D images of a nuclear membrane-like component using a MS imaging technique.
Figure 3.
Visualization of the m/z 58+ signal from a single NIH/3T3 fibroblast. The data was corrected in the ZcorrectorGUI and the transparency was adjusted within the GUI to allow the visualization of the sub-cellular lipid-rich feature, most likely the nuclear membrane. There are 56 images in the data set, with each image 86×86 μm2 in size and containing 256×256 pixels. (A) m/z 58+ image, data smoothed, 0.02 alpha value (a measure of transparency). (B) m/z 58+ image, only the voxels with intensity values between 1–25 are plotted, alpha value 0.04. (C) m/z 58+ image, only voxels with an intensity value of 3 are plotted, alpha value 0.25.
Sputter rate of single cells
The most critical assumption made when applying this z-correction to the data is that the sputter rate of the sample is homogenous throughout the sample in the x, y and z directions. A cell is a complicated biological system with many components, including organelles, the cytoskeleton network, and nucleoli. The sputter rate of these materials may be different from one another. Other assumptions include an easy to find contrast between the feature of interest and the background, as well as a flat substrate that exhibits minimal sputtering. To determine the validity of the assumption of a constant sputter rate, AFM height profiles and corrected ToF-SIMS cross-sections of the same cell were compared. If these data do not closely agree, then differential sputtering is present, and an additional correction must be implemented to produce an accurate 3D reconstruction of the cell..
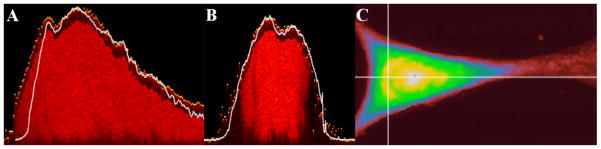
An overlay of an AFM height profile and a corrected XZ ToF-SIMS cross section of a single cryofixed, lyophilized fibroblast is shown in Figure 4A (This is a separate cell/dataset from the one shown in Figures 2 and 3). The AFM height profile and overlay with the corrected YZ ToF-SIMS cross section that run perpendicular to the slice in Figure 4A is shown in Figure 4B. The horizontal and vertical lines in Figure 4C define the slices used for the AFM height profiles and the ToF-SIMS corrected cross-sections seen in 4A and B. The AFM height profiles were scaled to match the corrected ToF-SIMS cross-sections. If the sputter rate were completely homogeneous throughout the data acquisition, then the AFM and ToF-SIMS data along any line should match exactly. In Figures 4A and B, there is good agreement between the AFM and ToF-SIMS data, indicating that the change in sputter rate throughout the acquisition of the data set is small under the experimental conditions used in this study. In Figure 4A, the ZCorrectorGUI was able to appropriately correct for most of the topography, except towards the left side of the cell where the ToF-SIMS data extends out further on the left side than the AFM data, possibly due to shadowing during C60 sputtering. However, the excellent agreement between the ToF-SIMS and AFM profiles over the majority of the cell indicates that the assumption of a constant sputter rate is valid.
Figure 4.
A comparison of corrected ToF-SIMS and AFM data from the same cell. (A) An overlay of a corrected slice from ToF-SIMS showing the distribution of all the detected ions and the height profile acquired with AFM along the same line (horizontal line in C). The AFM height profile is white and the ToF-SIMS data is red. (B) An overlay of a corrected slice from ToF-SIMS showing the distribution of all the detected ions and the height profile acquired with AFM along the same line (vertical line in C). The AFM height profile is white and the ToF-SIMS data is red. (C) The AFM height image. Image is 50×90 μm2. Intensity scale changes from red to white as the height increases from 0 to 1.8 μm.
A different cell that was prepared by chemical fixation with 4% formaldehyde followed by air drying is shown in the Supplemental Information, Figure S.1. The AFM and ToF-SIMS results for this cell are in good agreement, similar to the results for the lyophilized cell in Figure 4. Thus, for both lyophilized and chemically fixed NIH 3T3 fibroblasts, while there probably are likely some changes to the sputter rate, the assumption of a constant sputter rate for the z-correction appears to be valid.
This agrees with data from Chandra et al. who used direct current Ar+, Cs+, and O2+ beams to etch freeze-dried NIH/3T3 fibroblasts and found no evidence of differential sputtering between the nucleus and cytoplasm39. This data is also consistent with the independence of sputter rate from density, which was shown in a recent study by Muramoto et al. for C60 sputtering during the depth profiling of thin organic films48. If density had a significant effect on the sputter rate, the higher density nucleoli would be expected to have a different sputter rate compared to the less dense cytoplasm.
With some cells, as shown in Figure S.2, it was observed that there is a small salt residue present after all of the organic material had been removed by sputtering. This is the same data set displayed in Figure 4. Figure S.2 A–C shows the summation of the Na+ and K+ signal at slices 90, 132, and 141 respectively. These are raw, uncorrected images 86×86 μm2 in size and containing 256×256 pixels. Figures S.2 D–F show the summation of the major lipid components at the same slices, and G–I are the total ion signal at these slices. The salt signal is still present in two small horizontal bands in slice 141, well after all of the organic material has been removed. A possible explanation for this is that this substance is underneath the cell and only becomes detectable after the cellular material above it has been etched away. The ammonium acetate rinse used on this sample is used to remove excess surface salts, but likely would not remove any salt crystals underneath the cells. Another explanation is that salt species may build up with increasing C60 dose during the profile due to the lower sputter yield of these ions compared to the organic cellular material. Based on the similarities in the overlays in Figures 4 and S.1 as well as the TOC graphic, this does not appear to be the case. The intracellular Na+ and K+ signals from the cell in Figure 4 were rather homogenous in x and y across all slices, and did not appear to increase with depth (data not shown). If salt accumulation was decreasing the average sputter rate with increasing C60 dose, and leading to the formation of the salt residue, then the residue should be present in a much larger area of the cell, not only the two small regions shown in Figure S.2 C. It would especially be expected to form under the area of the tallest portion of the cell due to the most salt accumulation, which was not observed. Lastly, the cell shown in Figure 4 had roughly double salt content of the cell in Figure S.1 (data not shown), and the average sputter rate for both cells was determined to be very similar. All of the points suggest that salt accumulation is not significantly affecting the average sputter rate of dried single cells using 20 keV C60++ as the sputter source, and that any leftover salt residue originates from underneath the cell.
It was previously shown for organic delta-layers that the sputtering yield and depth resolution decreased with increasing C60 ion dose. The degradation in depth resolution was believed to be primarily caused by the development of topography on the sample surface49. A similar diminishment of the sputter yield may also be occurring here. However, the data shown in Figure 4 shows that there appears to be little change detected in the sputter rate as a function of depth. This cell type already has inherent roughness prior to sputtering, and subsequent C60 etch cycles do not significantly increase this native topography (See Figure S.3). Additionally, to minimize the damage caused the Bi analysis beam, low Bi ion doses compared to the C60 sputter dose were used26.
Varying the angle of incidence of the primary ion with the surface has been shown to affect the sputter yield both experimentally32 and computationally33. It follows that the sputter yield from topographically complex surfaces such as cells may also be affected. While this could play a small role in the data presented here, based on Figures 4 and S.1 as well as the TOC graphic, it does not appear to result in significant changes to the sputter rate. This could be due to the fact that the topography of the cell is sufficiently rough so any sputter rate differences are averaged out across the cell surface.
The average sputter rate of single NIH/3T3 fibroblasts can be calculated by dividing the number of sputter cycles needed to remove all of the cellular material by the maximum height of the cell, as measured by AFM. The cell featured in Figure 4 was considered removed after 131 slices when the lipid signal was no longer detected, while the maximum AFM measured height of the cell was 1412 nm. This yields an average sputter rate of 10.8 nm per 1.25 ×1013 ions/cm2 C60++. For the cell featured in Figure S.1, the ToF-SIMS data set consisted of 88 slices and the maximum AFM measured height was 905 nm, making the average sputter rate 10.3 nm per 1.25 ×1013 ions/cm2 C60++. Although only two cells are used here, the calculated sputter rates are in good agreement. There seems to be little difference between the average sputter rates in dried cells that were prepared in two different ways.
Although the sputter rate appears to not change significantly for any x-y-z positions in these cells, this may not hold for every biological sample and every preparation method. For example, frozen-hydrated NIH/3T3 fibroblasts will probably exhibit preferential sputtering due to the large sputter yield of the ice compared to the organic material50. It also remains to be seen if differential sputter rates are observed in tissue sections sputtered by C60 bombardment. Tissue sections may have a more heterogeneous microstructure than single cells which could lead to different components sputtering at distinct rates, which has been observed in plant cells31.
Conclusion
A new GUI for MATLAB is introduced that performs a “z-correction” pixel shift on a data matrix, and allows for new visualization methods in two and three dimensions. Using this new GUI, for the first time, a three dimensional corrected view of a lipid-rich subcellular region, possibly the nuclear membrane, is presented. An important assumption for this z-correction is that sputter rate is constant within the sample. We show, on different dried cells prepared by either cryofixation with freeze drying or chemical fixation with air drying, that this is a valid assumption. While there may be small changes in the sputter rate at any given position due to differential sputtering, they do not appear to be significant enough to warrant a further correction to the data. The average sputter rate of dried NIH/3T3 fibroblasts cells was approximately 10 nm per 1.25×1013 ions C60++/cm2. The tools developed in this study provide improved visualization of target chemistries within single cells and will be useful for other biomedically relevant three dimensional systems. The ZCorrectorGUI can be downloaded for free at: http://mvsa.nb.uw.edu/
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the funding and facilities provided by the National ESCA and Surface Analysis Center for Biomedical Problems (NESAC/BIO) through grant EB-002027 from the National Institutes of Health. The authors thank Prof. Buddy Ratner for the use of his cell culture facilities, as well as Prof. Cecilia Giachelli for use of the lyophilizer. The authors also thank Dr. Jim Hull for assistance with acquiring the atomic force microscopy data.
References
- 1.Jones EA, Lockyer NP, Vickerman JC. Int J Mass spectrom. 2007;260:146–57. [Google Scholar]
- 2.Klerk LA, Dankers PYW, Popa ER, Bosman AW, Sanders ME, Reedquist KA, Heeren RMA. Anal Chem. 2010;82:4337–43. doi: 10.1021/ac100837n. [DOI] [PubMed] [Google Scholar]
- 3.Fletcher JS, Lockyer NP, Vaidyanathan S, Vickerman JC. Anal Chem. 2007;79:2199–206. doi: 10.1021/ac061370u. [DOI] [PubMed] [Google Scholar]
- 4.Magnusson YK, Friberg P, Sjovall P, Malm J, Chen Y. Obesity. 2008;16:2745–53. doi: 10.1038/oby.2008.424. [DOI] [PubMed] [Google Scholar]
- 5.Sjovall P, Johansson B, Belazi D, Stenvinkel P, Lindholm B, Lausmaa J, Schalling M. Appl Surf Sci. 2008;255:1177–80. [Google Scholar]
- 6.Baker MJ, Zheng L, Winograd N, Lockyer NP, Vickerman JC. Langmuir. 2008;24:11803–10. doi: 10.1021/la802582f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baio JE, Cheng F, Ratner DM, Stayton PS, Castner DG. J Biomed Mater Res. 2011:97A. doi: 10.1002/jbm.a.33025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wagner MS, Castner DG. Langmuir. 2001;17:4649–60. [Google Scholar]
- 9.Wagner MS, Horbett TA, Castner DG. Langmuir. 2003;19:1708–15. [Google Scholar]
- 10.May CJ, Canavan HE, Castner DG. Anal Chem. 2004;76:1114–22. doi: 10.1021/ac034874q. [DOI] [PubMed] [Google Scholar]
- 11.Lee CY, Gamble LJ, Grainger DW, Castner DG. Biointerphases. 2006;1:82–92. doi: 10.1116/1.2219110. [DOI] [PubMed] [Google Scholar]
- 12.Lee CY, Harbers GM, Grainger DW, Gamble LJ, Castner DG. J Am Chem Soc. 2007;129:9429–38. doi: 10.1021/ja071879m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belu A, Mahoney C, Wormuth K. J Controlled Release. 2008;126:111–21. doi: 10.1016/j.jconrel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 14.Fisher GL, Belu AM, Mahoney CM, Wormuth K, Sanada N. Anal Chem. 2009;81:9930–40. doi: 10.1021/ac901587k. [DOI] [PubMed] [Google Scholar]
- 15.Barnes CA, Brison J, Michel R, Brown BN, Castner DG, Badylak SF, Ratner BD. Biomaterials. 2011;32:137–43. doi: 10.1016/j.biomaterials.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gunnarsson A, Kollmer F, Sohn S, Hook F, Sjovall P. Anal Chem. 2010;82:2426–33. doi: 10.1021/ac902744u. [DOI] [PubMed] [Google Scholar]
- 17.Fletcher JS. Analyst. 2009;134:2204–15. doi: 10.1039/b913575h. [DOI] [PubMed] [Google Scholar]
- 18.Wong SCC, Hill R, Blenkinsopp P, Lockyer NP, Weibel DE, Vickerman JC. Appl Surf Sci. 2003;203–204:219–22. [Google Scholar]
- 19.Weibel D, Wong S, Lockyer N, Blenkinsopp P, Hill R, Vickerman JC. Anal Chem. 2003;75:1754–64. doi: 10.1021/ac026338o. [DOI] [PubMed] [Google Scholar]
- 20.Benguerba M, Brunelle A, Della-Negra S, Depauw J, Joret H, Le Beyec Y, Blain MG, Schweikert EA, Assayag GB, Sudraud P. Nuc Inst Met Phys Res Sec B. 1991;62:8–22. [Google Scholar]
- 21.Gillen G, Roberson S. Rapid Commun Mass Spectrom. 1998;12:1303–12. doi: 10.1002/(SICI)1097-0231(19981015)12:19<1303::AID-RCM330>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 22.Boussofiane-Baudin K, Bolbach G, Brunelle A, Della-Negra S, Hakansson P, Le Beyec Y. HAL - CCSd - CNRS. 1994. [Google Scholar]
- 23.Cheng J, Wucher A, Winograd N. J Phys Chem B. 2006;110:8329–36. doi: 10.1021/jp0573341. [DOI] [PubMed] [Google Scholar]
- 24.Breitenstein D, Rommel CE, Mollers R, Wegener J, Hagenhoff B. Angew Chem Int Ed. 2007;46:5332–35. doi: 10.1002/anie.200604468. [DOI] [PubMed] [Google Scholar]
- 25.Brison J, Benoit DSW, Muramoto S, Robinson M, Stayton PS, Castner DG. Surf Interface Anal. 2011;43:354–57. doi: 10.1002/sia.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brison J, Muramoto S, Castner DG. J Phys Chem C. 2010;114:5565–73. doi: 10.1021/jp9066179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muramoto S, Brison J, Castner DG. Surf Interface Anal. 2011;43:58–61. doi: 10.1002/sia.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gillen G, Fahey A, Wagner M, Mahoney C. Appl Surf Sci. 2006;252:6537–41. [Google Scholar]
- 29.Fletcher JS, Rabbani S, Henderson A, Lockyer NP, Vickerman JC. Rapid Commun Mass Spectrom. 2011;25:925–32. doi: 10.1002/rcm.4944. [DOI] [PubMed] [Google Scholar]
- 30.Wucher A, Cheng J, Zheng L, Willingham D, Winograd N. Appl Surf Sci. 2008;255:984–86. [Google Scholar]
- 31.Patkin AJ, Chandra S, Morrison GH. Anal Chem. 1982;54:2507–10. [Google Scholar]
- 32.Ryan KE, Smiley EJ, Winograd N, Garrison BJ. Appl Surf Sci. 2008;255:844–46. [Google Scholar]
- 33.Kozole J, Winograd N. Appl Surf Sci. 2008;255:886–89. doi: 10.1016/j.apsusc.2008.05.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fisher GL, Dickinson M, Bryan SR, Moulder J. Appl Surf Sci. 2008;255:819–23. [Google Scholar]
- 35.Shard AG, Green FM, Brewer PJ, Seah MP, Gilmore IS. J Phys Chem B. 2008;112:2596–605. doi: 10.1021/jp077325n. [DOI] [PubMed] [Google Scholar]
- 36.Wagter ML, Clarke AH, Taylor KF, vanderHeide PAW, McIntyre NS. Surf Interface Anal. 1997;25:788–89. [Google Scholar]
- 37.Wucher A, Cheng J, Zheng L, Winograd N. Anal Bioanal Chem. 2009;393:1835–42. doi: 10.1007/s00216-008-2596-5. [DOI] [PubMed] [Google Scholar]
- 38.Wucher A, Cheng J, Winograd N. Anal Chem. 2007;79:5529–39. doi: 10.1021/ac070692a. [DOI] [PubMed] [Google Scholar]
- 39.Chandra S, Ausserer WA, Morrison GH. J Microsc Oxf. 1987;148:223–39. doi: 10.1111/j.1365-2818.1987.tb02869.x. [DOI] [PubMed] [Google Scholar]
- 40.Fletcher JS, Vickerman JC. Anal Bioanal Chem. 396:85–104. doi: 10.1007/s00216-009-2986-3. [DOI] [PubMed] [Google Scholar]
- 41.Roddy TP, Cannon DM, Ostrowski SG, Ewing AG, Winograd N. Anal Chem. 2003;75:4087–94. doi: 10.1021/ac026353t. [DOI] [PubMed] [Google Scholar]
- 42.Piehowski PD, Kurczy ME, Willingham D, Parry S, Heien ML, Winograd N, Ewing AG. Langmuir. 2008;24:7906–11. doi: 10.1021/la800292e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lanekoff I, Kurczy ME, Hill R, Fletcher JS, Vickerman JC, Winograd N, Sjovall P, Ewing AG. Anal Chem. 82:6652–59. doi: 10.1021/ac101243b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Breitenstein D, Rommel CE, Stolwijk J, Wegener J, Hagenhoff B. Appl Surf Sci. 2008;255:1249–56. [Google Scholar]
- 45.Wittig A, Wiemann M, Fartmann M, Kriegeskotte C, Arlinghaus HF, Zierold K, Sauerwein W. Microsc Res Tech. 2005;66:248–58. doi: 10.1002/jemt.20159. [DOI] [PubMed] [Google Scholar]
- 46.Berman ESF, Fortson SL, Checchi KD, Wu L, Felton JS, Wu KJJ, Kulp KS. J Am Soc Mass Spectrom. 2008;19:1230–36. doi: 10.1016/j.jasms.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 47.Steinbrecht RZK. Cryotechniques in Biological Electron Microscopy. 1987. [Google Scholar]
- 48.Muramoto S, Brison J, Castner DG. Anal Chem. 2012;84:365–72. doi: 10.1021/ac202713k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shard AG, Green FM, Brewer PJ, Seah MP, Gilmore IS. J Phys Chem B. 2008;112:2596–605. doi: 10.1021/jp077325n. [DOI] [PubMed] [Google Scholar]
- 50.Russo MF, Szakal C, Kozole J, Winograd N, Garrison BJ. Anal Chem. 2007;79:4493–98. doi: 10.1021/ac070105l. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.