Abstract
The circadian system synchronizes behavioral and physiologic processes with daily changes in the external light-dark cycle, optimizing energetic cycles with the rising and setting of the sun. Molecular clocks are organized hierarchically, with neural clocks orchestrating the daily switch between periods of feeding and fasting, and peripheral clocks generating 24hr oscillations of energy storage and utilization. Recent studies indicate that clocks respond to nutrient signals, and that high-fat diet influences the period of locomotor activity under free-running conditions, a core property of the clock. A major goal is to identify the molecular basis for the reciprocal relationship between metabolic and circadian pathways. Here, we highlight the role of peptidergic hormones and macromolecules as nutrient signals integrating circadian and metabolic systems.
Circadian Regulation of Metabolic Function
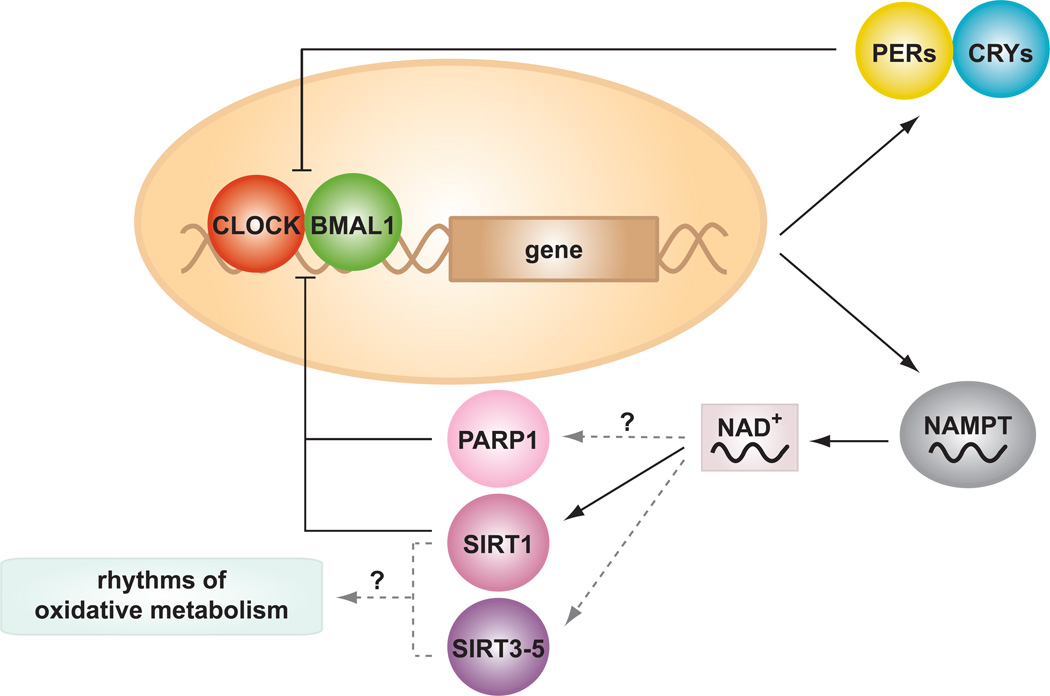
Circadian rhythms (derived from the Latin “circa diem”, which means “about a day”) were first discovered in plants nearly three centuries ago, and have since been observed in nearly all kingdoms of life [1, 2]. The core circadian clock is critical not only in the synchronization of behavioral and physiological processes with the external light-dark cycle, but also in the temporal partitioning of incompatible biochemical and energetic processes, in order to prevent futile energetic cycles such as the separation of oxygenic photosynthesis and nitrogen fixation in plants, or glycolytic and oxidative pathways in vertebrates. Circadian rhythms are generated by a transcription-translation autoregulatory feedback loop which cycles with a periodicity of ~24 hours (reviewed in [3–5]). The positive limb of this loop is driven by the transcriptional activators CLOCK and BMAL1, which activate transcription of the Period (Per) and Cryptochrome (Cry) genes (figure 1), both of which drive the negative limb. Upon reaching a critical threshold concentration, PER and CRY translocate to the nucleus where they inhibit the activity of the CLOCK:BMAL1 heterodimer, thus leading to a decrease in Per and Cry expression. Furthermore, AMPK and Casein Kinase Iεand δ (CKIε/CKIδ) trigger the phosphorylation and ubiquitin-mediated degradation of PER and CRY in the cytoplasm, thus providing a additional layer of regulation of the core clock machinery [6, 7]. The nuclear hormone receptors (NHRs) REV-ERBαand RAR-related orphan receptor alpha (RORα), both transcriptional targets of the CLOCK/BMAL1 complex, are also involved in the regulation of clock genes and comprise a short negative feedback loop that modulates Bmal1 transcription; REV-ERBα represses, while RORα activates, Bmal1 transcription [8, 9].
Figure 1. Circadian control of NAD+ and NAD+-dependent enzymes.
The core circadian clock is composed of a feedback loop involving a series of activators (CLOCK/BMAL1) and repressors (CRYs/PERs) that generate ~24h rhythms of gene transcription. Targets of the circadian clock include genes involved in the production and utilization of nutrient metabolites, including the NAD+ biosynthetic enzyme NAMPT. NAD+ is an important cofactor for the metabolic regulators, SIRT1, SIRT3–5 and PARP1, which may mediate rhythms of oxidative metabolism.
The core molecular clock as described above is found in neurons of the hypothalamic suprachiasmic nucleus (SCN) and is essential for entrainment of locomotor activity and the sleep-wake cycle to light. However, a surprising discovery has been that the entire core clock mechanism is also expressed in extra-SCN regions of the brain and in nearly all peripheral tissues [10, 11]. The presence of clocks in metabolic tissues such as liver, adipose, pancreas, and muscle suggests a critical role of circadian oscillations in metabolic physiology. Indeed, mice with both global and tissue-specific loss-of-function mutations in core clock genes display increased susceptibility to metabolic diseases. For example, ClockΔ19/Δ19 mutant mice, which express a dominant negative mutant form of the CLOCK protein, display diet-induced obesity, hyperlipidemia, hepatic steatosis, and diabetes characterized by hypoinsulinemic hyperglycemia [12–14], and global Bmal1−/− mutant mice display impaired glucose tolerance, increased insulin sensitivity, and age-related myopathy and arthropathy [13–17]. The use of tissue-specific circadian mutant mice has further elucidated the physiologic function of molecular clocks. For example, pancreatic beta cell-specific deletion of Bmal1 results in impaired insulin secretion, reduced islet size, and impaired glucose tolerance [13]. Circadian timing also impacts organismal homeostasis through non-cell-autonomous mechanisms, and recent evidence in mice has revealed that simply restricting high-fat feeding to the normal rest period results in exaggerated obesity [18]. Thus, both genetic and environmental factors underlie the linkage between circadian and metabolic homeostasis.
However, just as disruption of the circadian network influences metabolism, it has also been shown that the reciprocal relationship exists, as altered feeding behavior and metabolism affect the clock [19, 20]. For example, mice fed a high-fat diet were found to have alterations in period length and in the expression and cycling of canonical clock genes, of clock-target genes involved in metabolic homeostasis, and of NHRs that regulate the clock itself [19]. Together, these studies reveal a reciprocal relationship between circadian and metabolic regulation that may hold important clues to the development of metabolic disease.
Recent focus has turned towards identifying the key factors or metabolites that might connect circadian transcriptional networks with nutrient sensing pathways. In this regard, the observation that clocks in metabolic peripheral tissues such as liver and adipose can be entrained by food [21, 22] raises several important questions: 1) What are the metabolic ‘sensors’ of feeding and fasting cycles in peripheral cells? 2) How are peripheral clocks entrained by nutrient flux (e.g., lipid vs carbohydrate)? 3) How does the molecular clock influence nutrient sensing at the level of individual tissues and cells?
Interplay between circadian clocks, NAD+ biosynthesis, and sirtuins
Several recent studies have demonstrated that the circadian clock regulates the synthesis of the essential metabolic cofactor nicotinamide adenine dinucleotide (NAD+). Because of its central role in numerous biochemical redox reactions, NAD+ is an attractive candidate as an integrator of circadian rhythms and nutrient sensing pathways. Indeed, CLOCK:BMAL1 directly regulates the transcript encoding the rate-limiting enzyme in the NAD+ salvage pathway, nicotinamide phosphoribosyltransferase (NAMPT), and NAD+ levels likewise display rhythmicity, at least in the liver and in embryonic fibroblasts [23, 24] (Figure 1). Importantly, abrogation of the clock activator arm (CLOCK/BMAL1) leads to reduced NAD+ levels, whereas ablation of clock repressors CRY1/2 leads to elevated NAD+, indicating that the counter-balance between expression of clock activators and repressors (i.e., a functional molecular clock transcriptional network) controls hepatic NAD+ turnover. Moreover, NAD+ oscillations persist in animals maintained in constant darkness, consistent with dependency on the core molecular clock [23, 25].
In addition to its role in biochemical redox reactions, NAD+ is an important cofactor for the Class III histone and protein deacetylase SIRT1, a member of the sirtuin family of NAD+-dependent deacetylases that regulates a range of metabolic processes, including gluconeogenesis, insulin sensitivity, and life span [26–28]. Many of the transcription factors regulated by SIRT1 are involved in the cellular response to stress and nutrient flux, such as Peroxisome Proliferator-activated Receptor Gamma Coactivator 1-alpha (PGC-1α), Forkhead Box Protein O1 (FOXO1), Transducer of Regulated CREB Protein 2 (TORC2),, Sterol Regulatory Element-Binding Protein 1c (SREBP-1c), and Signal Transducer and Activator of Transcription 3 (STAT3) [26–29]. Given that the activity of SIRT1 depends on levels of NAD+, clock-driven oscillations of NAD+ likely contribute to daily rhythms in the multitude of metabolic pathways regulated by SIRT1. Intriguingly, it has been reported that in addition to being regulated by the clock, SIRT1 also regulates clock function itself (28–30) through deacetylation of both BMAL1 and PER2, leading to altered CLOCK/BMAL1 transcriptional activity [30–32] (figure 1). While the direction of effect of SIRT1 on clock activity appears to be complicated, as reports have demonstrated both activation and repression of CLOCK/BMAL1 [30–32] (it is clear that SIRT1 plays in important role in clock function. It is thus possible that circadian-driven oscillation of NAD+ biosynthesis may in turn modulate SIRT1 and CLOCK/BMAL1 activity, thus “priming” the organism to anticipate the daily transitions between nutrient alternans, corresponding with the fasting/feeding and sleep/wake cycle. Consistent with this idea, SIRT1 has been implicated in the metabolic switch between periods of feeding and fasting, as SIRT1 activity increases upon fasting and in response to caloric restriction [33–35]. For example, during fasting, increased SIRT1 activity activates PGC1α, allowing nutrient-deprived cells to transition from glycolytic ATP production to mitochondrial oxidative ATP synthesis [34]. Whether this increase in SIRT1 activity during nutrient deprivation is dependent upon cyclic NAD+ metabolism, and the extent to which the NAD+/SIRT1 pathway acts as a switch coupling metabolic and circadian oscillators, remain questions for further investigation.
In addition to SIRT1, recent evidence suggests a role for other NAD+-dependent enzymes in metabolic regulation, each of which may also be influenced by clock-control of NAD+ turnover. For example, the sirtuin orthologs SIRT2 to 7, which are distributed across a number of subcellular compartments (i.e., SIRT2 is cytoplasmic, SIRT3–5 are mitochondrial, SIRT6 is nuclear, and SIRT7 is nucleolar), act on a wide range of proteins including histones, transcription factors, and enzymes involved in intermediary metabolism. Interestingly, activity of the mitochondrial sirtuins SIRT3 to 5 is intimately involved in the adaptive response to fasting [36] For example, SIRT3 deacetylates and activates the fatty acid oxidation enzyme long-chain acyl coenzyme A dehydrogenase (LCAD) in mouse liver in response to a 24 hr fast, thereby facilitating fuel for β-oxidation. Consistently, Sirt3−/− mice display defects in fasting fatty acid oxidation [37]. Similar studies revealed that SIRT3 also deacetylates and activates enzymes involved in other mitochondrial pathways including ketogenesis, urea cycle, electron transport, acetate metabolism, and production of reactive oxygen species [38–42]. While SIRT4 has not been shown to possess deacetylase activity, it has instead been shown to catalyze the NAD+-dependent ADP-ribosylation of glutamate dehydrogenase 1 (GLUD1), an enzyme involved in ammonia detoxification and the citric acid cycle [43]. SIRT5 also plays an important role in urea cycle regulation via deacetylation of carbamoyl phosphate synthetase (CPS1) [44]. Thus, the NAD+-dependent sirtuin superfamily participates in both protein deacetylation and ADP-ribosylation, establishing a rapid means of upregulating mitochondrial energy production during nutrient deprivation. The potential control of SIRT3–5 activity through clock-mediated NAD+ biosynthesis suggests a possible mechanism by which circadian oscillations of feeding and fasting metabolic pathways are controlled.
Finally, recent studies have also implicated the NAD+-dependent ADP-ribosyltransferase PARP-1 as a mediator of the circadian clock. When activated by NAD+, liver PARP-1 binds to and ADP-ribosylates CLOCK, thus altering the interaction of CLOCK/BMAL1 with DNA and the clock repressor proteins PER and CRY [45] (figure 1). While still at an early stage of investigation, the possibility that clock control of NAD+ biosynthesis may mediate the function of sirtuins and of PARP not only within nuclei, but also within other subcellular compartments, remains an intriguing question. Together, these studies raise the possibility that circadian control of NAD+ metabolism couples internal energetic cycles with oscillation in the external nutrient environment.
A role for cellular redox and ROS in regulating circadian clocks
Whereas only recently has the circadian clock been tied to cycles of NAD+ metabolism, a role for cellular redox flux in regulating circadian rhythms was initially proposed by McKnight and coworkers in 2001, who demonstrated that the oxidation state of NAD(P) cofactors influences the binding of BMAL1 complexes to DNA [46]. Increased levels of oxidized NAD+ and NADP+ lead to decreased binding of CLOCK/BMAL1 and the Neuronal PAS Domain-containing Protein 2 (NPAS2) /BMAL1 to DNA, suggesting that the redox state per se may entrain molecular clock activity, and in turn couple circadian and metabolic cycles.
Two recent studies have further demonstrated that the redox state of peroxiredoxin proteins exhibit properties of a self-sustained circadian oscillator, controlling circadian rhythms of metabolic proteins even in the absence of ongoing gene transcription [47, 48]. Circadian rhythms of metabolic enzyme activities in anucleate mature human red blood cells have been observed for decades [49–51]. However, only recently has a potential mechanism for these rhythms been uncovered. O’Neill et al. demonstrated that the redox state of peroxiredoxin antioxidant proteins oscillates with a periodicity of ~24 hours. Oscillating peroxiredoxin redox state affects its antioxidant activity, generating self-sustained rhythms of cellular redox status (i.e., NADH/NADPH levels), which may in turn generate rhythms of metabolic enzyme activity [47]. Similar to clock-dependent circadian rhythms, these oscillations are temperature-entrainable and temperature-compensated. Independent studies in the single-celled alga Ostreococcus tauri identified ~24h rhythms of peroxiredoxin oxidation, which were sustained in the absence of functional molecular clock components [48]. These studies indicate that certain metabolic oscillations exhibit circadian properties independent of active gene transcription rhythms, and raise the possibility that other nutrients may act independently of transcriptional control to generate rhythms of metabolic pathways.
Crosstalk between circadian clocks and nuclear hormone receptors
REV-ERBs and RORs
Emerging evidence has also recently indicated that NHRs and their cognate ligands may also act as ‘nutrient sensors’, coupling circadian and metabolic pathways. NHRs represent a large family of homologous transcription factors that contain both DNA and ligand binding domains that bind a wide range of metabolic intermediates, including the classical endocrine hormones (e.g. GR, TR, ER), lipid- and steroid-derived molecules (e.g. PPARs, LXR, RORs), xenobiotics (e.g. PXR), and other metabolites (REV-ERBs, RARs) [52]. Also, many NHRs are classified as orphan receptors since their endogenous ligands have not been identified (e.g. ERRα). Of note, in addition to direct NHR transcriptional regulation of Bmal1 via REV-ERB and ROR [8, 9, 52], (more than half of the ~50 known NHRs display circadian oscillations in metabolic tissues such as liver, adipose, and skeletal muscle [8, 53–56], demonstrating a complex feedback loop, whereby the clock both regulates and is regulated by NHRs.
Indeed, mice lacking REV-ERBα display a shortened circadian period in constant darkness [9], as well as several metabolic abnormalities including altered lipid and bile metabolism [57, 58]. These observations are consistent with the recent discovery of coordinated binding of REV-ERBα to lipogenic targets in the hepatic cistrome [59]. Likewise, RORα-deficient (staggerer) mice also display altered circadian rhythms and are predisposed to several aging-related phenotypes, including athlerosclerosis [52, 60–62]. Therefore, NHRs directly integrate nutrient signals with transcriptional regulation of the clock [63].
Interestingly, the metabolic transcriptional regulator PGC1α acts as a coactivator for RORα and β at the Bmal1 promoter [64]. Consistently, mice lacking PGC1α display disrupted clock gene oscillations, abnormal locomotor activity, and slight changes in free-running period length [64, 65], in addition to defects in mitochondrial oxidative metabolic pathways such as respiration and fatty acid oxidation [66–68]. PGC1α also acts as a coactivator for the metabolic orphan nuclear hormone receptor Estrogen Related Receptor α (ERRα), which has recently been shown to display circadian rhythms of transcriptional activity and is directly regulated by the molecular clock [69–72]. Finally, activity of PGC1α is dependent upon NAD+-dependent deacetylation by SIRT1, so it is tempting to speculate that clock regulation of NAD+ may in turn drive an additional metabolic feedback loop involving PGC1α [29, 64]. Several studies implicate RORα as a lipid sensor, and x-ray crystallography evidence indicates that cholesterol and oxysterol function as RORα ligands [73, 74]. RORα activates lipogenic pathways, including expression of the transcription factor SREBP1c [75]. Interestingly, REV-ERBα controls the circadian expression of Insulin-induced Gene 2 (Insig2), a negative regulator of SREBP1c activity [57]. Thus, sterols may also play an important role in circadian clock oscillations, and in turn, in the temporal control of lipogenesis.
Finally, heme is an additional metabolite that may act as a metabolic signal coupling metabolic and circadian oscillations. Heme, a cofactor for oxidative reactions including mitochondrial oxidative phosphorylation, is present in a complex with REV ERBα and promotes its activity as a transcriptional repressor [55, 76, 77]. Thus, heme may represent an additional node linking environmental nutrient flux with circadian oscillation [55, 78, 79].
Glucocorticoid-regulated NHRs
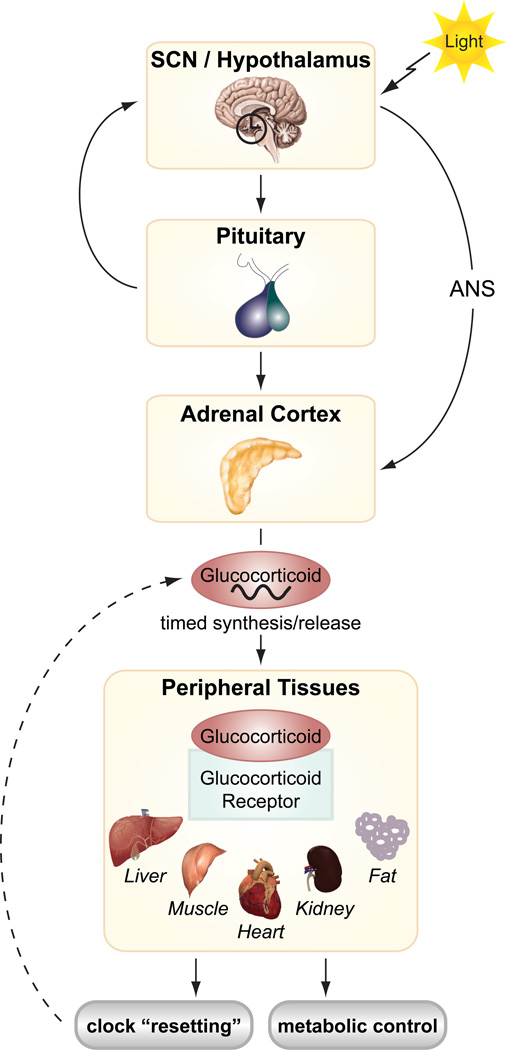
Glucocorticoids (GC) and the glucocorticoid receptor (GR) in peripheral tissues have also emerged as an important link between circadian clocks and feeding/fasting cycles [80]. GCs are steroid hormones produced in the adrenal cortex that participate in adaptive pathways during fasting in liver, adipose, and skeletal muscle. GCs exhibit well-characterized circadian oscillations, arising through relays from the SCN to the hypothalamic-pituitary-adrenal (HPA) axis (figure 2). Light entrains the GC rhythm, generating a zenith in the early morning in humans [81, 82]. While the detailed mechanisms have not yet been fully defined, several studies have suggested that GCs in turn affect clock activity in peripheral tissues and may act as important mediators between central and peripheral clocks [56]. Pharmacological activation of the GR by dexamethasone resets the clock in several peripheral tissues, including liver, kidney, and heart, and GR has been shown to regulate Rev-erbα and Per1 expression [83–85] (figure 2). GR may also regulate the clock through direct binding to PER2 [86]. Furthermore, in liver, GR has been proposed to be a key physiological entrainment signal of clock oscillations, leading to coupling of the food- and SCN-derived resetting cues [83, 87]. In fact, it has been proposed that SCN-controlled GC release can oppose food entrainment of peripheral clocks, and liver-specific GR knockout mice display accelerated phase shifting in response to restricted daytime feeding [87]. Since GC signaling is not present within the SCN [83], this pathway may be essential for altering circadian clock oscillations in peripheral metabolic tissues in response to stress and changes in nutrient availability.
Figure 2. Coordination of central and peripheral clocks by glucocorticoids.
Light-derived cues stimulate hormonal signaling from hypothalamus/SCN to the adrenal cortex (via the autonomic nervous system (ANS) or via the pituitary gland), producing daily rhythms of glucocorticoid release. Glucocorticoids activate the glucocorticoid receptor in peripheral tissues, which ‘resets’ the molecular clocks and metabolic pathways.
Other circadian metabolite-regulated NHRs
While many additional NHRs are rhythmically expressed, less is known about the metabolites that regulate these factors and whether they might affect the circadian clock. For example, retinoic acid was shown to shift Per2 rhythmicity in muscle cells through the retinoic acid-responsive NHRs RAR and RXR [54, 88]. Members of the PPAR family of NHRs also directly regulate clock gene expression. PPARα is a positive regulator of both Bmal1 and Rev-erbα in liver [54, 63, 86]. Likewise, synthesis of several PPAR proteins, including PPARαγδ are regulated by the circadian clock [63]. Ligands for PPARs include fatty acids and eicosanoids, including the diurnally-regulated gut metabolite oleylethanolamide (OEA). OEA is synthesized in the small intestine during the rest period and suppresses food intake in a PPARα-dependent manner [89, 90]. Thus, OEA acts as another nutrient signal that may feedback to the clock within either local tissues or as a circulating signal to regulate circadian behaviors. In addition, PPARγ regulates Bmal1 expression in blood vessels and helps maintain daily rhythms of blood pressure and heart rate [91]. PPARγ is regulated by the prostaglandins, and it is thus possible that oscillations of these molecules are essential for the maintenance of circadian rhythms in the vasculature. Metabolite profiling studies will be necessary to determine the importance of other lipid-derived PPAR ligands as effectors of the molecular clock.
Circadian regulation of metabolic peptide hormones
In addition to NHRs, several metabolic peptide hormones, such as leptin and ghrelin, display circadian oscillations that may act to coordinate nutrient signals from peripheral tissues with central behavioral outputs (e.g. feeding, physical activity) to maintain whole body energy homeostasis. Several studies have implicated the rhythmic release of adipocyte-derived hormone leptin and stomach-derived hormone ghrelin as important for relaying food cues to the brain to establish food anticipatory behaviors [92–97]. For example, humans display increased levels of leptin at night, contributing to nocturnal appetite suppression by hypothalamic neurons [98]. In contrast, ghrelin levels increase before meal times and have been suggested to play a role in food anticipatory behavior [99]. Although the mechanisms controlling the rhythmic release of leptin and ghrelin from peripheral tissues remain unclear, it is known that their effects can modulate the SCN-derived clock [92, 96]. As a potential inverse to the GC/GR pathway, these hormones may play a key role in providing information from peripheral food-responsive clocks to the central pacemaker (SCN).
Concluding remarks
Recent publications have shed light on the molecular pathways connecting circadian rhythms and metabolism. One of the most intriguing aspects of this connection is the reciprocal nature of the crosstalk between nutrient sensing pathways and the clock. Coupling of circadian and metabolic oscillators also raises intriguing evolutionary questions. That certain biochemical oscillations exist in cells in the absence of transcription (e.g. peroxiredoxin oxidation in human red blood cells and algae), and that circadian phosphorylation cycles of cyanobacteria can be reproduced in vitro [47, 48, 100, 101], support the hypothesis that metabolic oscillators represent the most ancient biological clock [102]. From this vantage point, the emergence of clocks encoded by an autoregulatory transcription feedback loop in plants, fungi and metazoa, may represent a more recent adaptation in the evolution of circadian systems. However, it is important to note that even in cyanobacteria, most biochemical oscillators are coupled to transcriptional oscillators —so the question is a bit of a “chicken or the egg” puzzle [1]. Indeed, the diverse pathologies associated with circadian disruption in mammalian clock mutants highlights the interdependence of metabolic and circadian systems and provides new avenues to consider the impact of sleep and circadian timing on disorders including obesity, cardiovascular disease and diabetes mellitus [12].
In summary, two central questions remain in understanding the mechanism through which molecular clocks contribute to metabolic homeostasis (1) the identity of metabolic ‘sensors’ that coordinate circadian cycles within and between peripheral tissues across the feeding/fasting cycle, and (2) the mechanism through which cell autonomous and non-autonomous signals are integrated within peripheral tissues to produce coherent oscillation in physiologic systems. One approach has been to examine classical neuroendocrine factors on a candidate basis, while unbiased metabolomics surveys have also been deployed to identify metabolic-circadian coupling mechanisms [103]. One short-term benefit to emerge from metabolomics profiling may be the development of surrogate biomarkers for circadian time and in so doing, advance our ability to tell time in experimental and clinical settings. As we further explore the role of peripheral clocks in metabolic homeostasis, it will be important to dissect the patterns of oscillations of not only gene transcripts, but also metabolites and hormones. Circadian clocks represent an ensemble of oscillators both within the brain and in peripheral tissues, and it is the phasic synchrony of these multiple tissue clocks that gives rise to overt rhythms of behavior and physiology necessary for metabolic health.
Acknowledgements
We thank Ravi Allada, Shin-ichiro Imai, Joseph S. Takahashi, Fred W. Turek, and members of the Bass lab for helpful input and discussions. C.B. Peek is supported by the NIH fellowship grant F32 DK092034-01. K.M. Ramsey received support from NIDDK grant T32 DK007169. J. Bass is supported by NIH grants P01AG011412, R01HL097817-01, and 1R01DK090625-01A1, the American Diabetes Association, Chicago Biomedical Consortium Searle Funds, the Juvenile Diabetes Research Foundation, and the University of Chicago Diabetes Research and Training Center (grant P60 DK020595). J. Bass is a member of the scientific advisory board of ReSet Therapeutics Inc. and has received support from Amylin Pharmaceuticals.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Paranjpe DA, Sharma VK. Evolution of temporal order in living organisms. J Circadian Rhythms. 2005;3:7. doi: 10.1186/1740-3391-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pittendrigh CS. Temporal organization: reflections of a Darwinian clock-watcher. Annual review of physiology. 1993;55:16–54. doi: 10.1146/annurev.ph.55.030193.000313. [DOI] [PubMed] [Google Scholar]
- 3.Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010;330:1349–1354. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takahashi JS, et al. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 6.Lamia KA, et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326:437–440. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Etchegaray JP, et al. Casein kinase 1 delta regulates the pace of the mammalian circadian clock. Mol Cell Biol. 2009;29:3853–3866. doi: 10.1128/MCB.00338-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Asher G, Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 2011;13:125–137. doi: 10.1016/j.cmet.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 9.Preitner N, et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 10.Schibler U, et al. Peripheral circadian oscillators in mammals: time and food. J Biol Rhythms. 2003;18:250–260. doi: 10.1177/0748730403018003007. [DOI] [PubMed] [Google Scholar]
- 11.Dibner C, et al. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annual review of physiology. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- 12.Marcheva B, et al. Clock genes and metabolic disease. J Appl Physiol. 2009;107:1638–1646. doi: 10.1152/japplphysiol.00698.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marcheva B, et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466:571–572. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turek FW, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andrews JL, et al. CLOCK and BMAL1 regulate MyoD and are necessary for maintenance of skeletal muscle phenotype and function. Proc Natl Acad Sci U S A. 107:19090–19095. doi: 10.1073/pnas.1014523107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kondratov RV, et al. Early aging and age-related pathologies in mice deficient in BMAL1, the core component of the circadian clock. Genes Dev. 2006;20:1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bunger MK, et al. Progressive arthropathy in mice with a targeted disruption of the Mop3/Bmal-1 locus. Genesis. 2005;41:122–132. doi: 10.1002/gene.20102. [DOI] [PubMed] [Google Scholar]
- 18.Arble DM, et al. Circadian timing of food intake contributes to weight gain. Obesity (Silver Spring) 2009;17:2100–2102. doi: 10.1038/oby.2009.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kohsaka A, et al. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007;6:414–421. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 20.Barnea M, et al. High-fat diet delays and fasting advances the circadian expression of adiponectin signaling components in mouse liver. Endocrinology. 2009;150:161–168. doi: 10.1210/en.2008-0944. [DOI] [PubMed] [Google Scholar]
- 21.Damiola F, et al. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirota T, Fukada Y. Resetting mechanism of central and peripheral circadian clocks in mammals. Zoolog Sci. 2004;21:359–368. doi: 10.2108/zsj.21.359. [DOI] [PubMed] [Google Scholar]
- 23.Ramsey KM, et al. Circadian Clock Feedback Cycle Through NAMPT-Mediated NAD+ Biosynthesis. Science. 2009;324:651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakahata Y, et al. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sahar S, et al. Altered behavioral and metabolic circadian rhythms in mice with disrupted NAD+ oscillation. Aging (Albany NY) 3:794–802. doi: 10.18632/aging.100368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saunders LR, Verdin E. Sirtuins: critical regulators at the crossroads between cancer and aging. Oncogene. 2007;26:5489–5504. doi: 10.1038/sj.onc.1210616. [DOI] [PubMed] [Google Scholar]
- 27.Donmez G, Guarente L. Aging and disease: connections to sirtuins. Aging Cell. 9:285–290. doi: 10.1111/j.1474-9726.2010.00548.x. [DOI] [PubMed] [Google Scholar]
- 28.Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodgers JT, et al. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 30.Nakahata Y, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grimaldi B, et al. Chromatin remodeling, metabolism and circadian clocks: the interplay of CLOCK and SIRT1. Int J Biochem Cell Biol. 2009;41:81–86. doi: 10.1016/j.biocel.2008.08.035. [DOI] [PubMed] [Google Scholar]
- 32.Asher G, et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 33.Rodgers JT, Puigserver P. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc Natl Acad Sci U S A. 2007;104:12861–12866. doi: 10.1073/pnas.0702509104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodgers JT, et al. Metabolic adaptations through the PGC-1 alpha and SIRT1 pathways. FEBS Lett. 2008;582:46–53. doi: 10.1016/j.febslet.2007.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y, et al. A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature. 2008;456:269–273. doi: 10.1038/nature07349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang JY, et al. Mitochondrial sirtuins. Biochim Biophys Acta. 2010;1804:1645–1651. doi: 10.1016/j.bbapap.2009.12.021. [DOI] [PubMed] [Google Scholar]
- 37.Hirschey MD, et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 464:121–125. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hallows WC, et al. Sirt3 promotes the urea cycle and fatty acid oxidation during dietary restriction. Molecular cell. 2011;41:139–149. doi: 10.1016/j.molcel.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shimazu T, et al. SIRT3 deacetylates mitochondrial 3-hydroxy-3-methylglutaryl CoA synthase 2 and regulates ketone body production. Cell Metab. 2010;12:654–661. doi: 10.1016/j.cmet.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Someya S, et al. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell. 2010;143:802–812. doi: 10.1016/j.cell.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hallows WC, et al. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci U S A. 2006;103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwer B, et al. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci U S A. 2006;103:10224–10229. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haigis MC, et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 44.Nakagawa T, Guarente L. Urea cycle regulation by mitochondrial sirtuin, SIRT5. Aging (Albany NY) 2009;1:578–581. doi: 10.18632/aging.100062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Asher G, et al. Poly(ADP-ribose) polymerase 1 participates in the phase entrainment of circadian clocks to feeding. Cell. 2010;142:943–953. doi: 10.1016/j.cell.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 46.Rutter J, et al. Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science. 2001;293:510–514. doi: 10.1126/science.1060698. [DOI] [PubMed] [Google Scholar]
- 47.O'Neill JS, Reddy AB. Circadian clocks in human red blood cells. Nature. 2011;469:498–503. doi: 10.1038/nature09702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O'Neill JS, et al. Circadian rhythms persist without transcription in a eukaryote. Nature. 2011;469:554–558. doi: 10.1038/nature09654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cornelius G, Rensing L. Daily rhythmic changes in Mg2+-dependent ATPase activity in human red blood cell membranes in vitro. Biochemical and biophysical research communications. 1976;71:1269–1272. doi: 10.1016/0006-291x(76)90791-9. [DOI] [PubMed] [Google Scholar]
- 50.Hartman H, Ashkenazi I. Circadian changes in membrane properties of human red blood cells in vitro, as measured by a membrane probe. FEBS Lett. 1976;67:161–163. doi: 10.1016/0014-5793(76)80356-0. [DOI] [PubMed] [Google Scholar]
- 51.Brok-Simoni F, et al. The diurnal rhythm of enzymes in human red cells. Br J Haematol. 1976;32:601–607. doi: 10.1111/j.1365-2141.1976.tb00964.x. [DOI] [PubMed] [Google Scholar]
- 52.Sonoda J, et al. Nuclear receptors: decoding metabolic disease. FEBS Lett. 2008;582:2–9. doi: 10.1016/j.febslet.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang X, et al. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 54.Yang X, et al. Nuclear receptors, metabolism, and the circadian clock. Cold Spring Harb Symp Quant Biol. 2007;72:387–394. doi: 10.1101/sqb.2007.72.058. [DOI] [PubMed] [Google Scholar]
- 55.Duez H, Staels B. Rev-erb-alpha: an integrator of circadian rhythms and metabolism. J Appl Physiol. 2009;107:1972–1980. doi: 10.1152/japplphysiol.00570.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Teboul M, et al. How nuclear receptors tell time. J Appl Physiol. 2009;107:1965–1971. doi: 10.1152/japplphysiol.00515.2009. [DOI] [PubMed] [Google Scholar]
- 57.Le Martelot G, et al. REV-ERBalpha participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biol. 2009;7:e1000181. doi: 10.1371/journal.pbio.1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Coste H, Rodriguez JC. Orphan nuclear hormone receptor Rev-erbalpha regulates the human apolipoprotein CIII promoter. J Biol Chem. 2002;277:27120–27129. doi: 10.1074/jbc.M203421200. [DOI] [PubMed] [Google Scholar]
- 59.Feng D, et al. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 331:1315–1319. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mamontova A, et al. Severe atherosclerosis and hypoalphalipoproteinemia in the staggerer mouse, a mutant of the nuclear receptor RORalpha. Circulation. 1998;98:2738–2743. doi: 10.1161/01.cir.98.24.2738. [DOI] [PubMed] [Google Scholar]
- 61.Akashi M, Takumi T. The orphan nuclear receptor RORalpha regulates circadian transcription of the mammalian core-clock Bmal1. Nat Struct Mol Biol. 2005;12:441–448. doi: 10.1038/nsmb925. [DOI] [PubMed] [Google Scholar]
- 62.Sato TK, et al. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43:527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 63.Li S, Lin JD. Molecular control of circadian metabolic rhythms. J Appl Physiol. 2009;107:1959–1964. doi: 10.1152/japplphysiol.00467.2009. [DOI] [PubMed] [Google Scholar]
- 64.Liu C, et al. Transcriptional coactivator PGC-1alpha integrates the mammalian clock and energy metabolism. Nature. 2007;447:477–481. doi: 10.1038/nature05767. [DOI] [PubMed] [Google Scholar]
- 65.Grimaldi B, Sassone-Corsi P. Circadian rhythms: metabolic clockwork. Nature. 2007;447:386–387. doi: 10.1038/447386a. [DOI] [PubMed] [Google Scholar]
- 66.Wu Z, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 67.Lehman JJ, et al. The transcriptional coactivator PGC-1alpha is essential for maximal and efficient cardiac mitochondrial fatty acid oxidation and lipid homeostasis. Am J Physiol Heart Circ Physiol. 2008;295:H185–H196. doi: 10.1152/ajpheart.00081.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Spiegelman BM. Transcriptional control of mitochondrial energy metabolism through the PGC1 coactivators. Novartis Found Symp. 2007;287:60–63. discussion 63–69. [PubMed] [Google Scholar]
- 69.Laganiere J, et al. A polymorphic autoregulatory hormone response element in the human estrogen-related receptor alpha (ERRalpha) promoter dictates peroxisome proliferator-activated receptor gamma coactivator-1alpha control of ERRalpha expression. J Biol Chem. 2004;279:18504–18510. doi: 10.1074/jbc.M313543200. [DOI] [PubMed] [Google Scholar]
- 70.Huss JM, et al. Peroxisome proliferator-activated receptor coactivator-1alpha (PGC-1alpha) coactivates the cardiac-enriched nuclear receptors estrogen-related receptor-alpha and -gamma. Identification of novel leucine-rich interaction motif within PGC-1alpha. J Biol Chem. 2002;277:40265–40274. doi: 10.1074/jbc.M206324200. [DOI] [PubMed] [Google Scholar]
- 71.Schreiber SN, et al. The estrogen-related receptor alpha (ERRalpha) functions in PPARgamma coactivator 1alpha (PGC-1alpha)-induced mitochondrial biogenesis. Proc Natl Acad Sci U S A. 2004;101:6472–6477. doi: 10.1073/pnas.0308686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dufour CR, et al. Genomic convergence among ERRalpha, PROX1, and BMAL1 in the control of metabolic clock outputs. PLoS Genet. 2011;7:e1002143. doi: 10.1371/journal.pgen.1002143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jetten AM. Retinoid-related orphan receptors (RORs): critical roles in development, immunity, circadian rhythm, and cellular metabolism. Nucl Recept Signal. 2009;7:e003. doi: 10.1621/nrs.07003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kallen J, et al. Crystal structure of the human RORalpha Ligand binding domain in complex with cholesterol sulfate at 2.2 A. J Biol Chem. 2004;279:14033–14038. doi: 10.1074/jbc.M400302200. [DOI] [PubMed] [Google Scholar]
- 75.Lau P, et al. The orphan nuclear receptor, RORalpha, regulates gene expression that controls lipid metabolism: staggerer (SG/SG) mice are resistant to diet-induced obesity. J Biol Chem. 2008;283:18411–18421. doi: 10.1074/jbc.M710526200. [DOI] [PubMed] [Google Scholar]
- 76.Yin L, et al. Rev-erb alpha, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318:1786–1789. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- 77.Yin L, et al. Nuclear receptor Rev-erbalpha: a heme receptor that coordinates circadian rhythm and metabolism. Nucl Recept Signal. 2010;8:e001. doi: 10.1621/nrs.08001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Duez H, Staels B. Rev-erb alpha gives a time cue to metabolism. FEBS Lett. 2008;582:19–25. doi: 10.1016/j.febslet.2007.08.032. [DOI] [PubMed] [Google Scholar]
- 79.Kojetin DJ, Burris TP. A role for rev-erbalpha ligands in regulation of adipogenesis. Curr Pharm Des. 17:320–324. doi: 10.2174/138161211795164211. [DOI] [PubMed] [Google Scholar]
- 80.Dickmeis T, Foulkes NS. Glucocorticoids and circadian clock control of cell proliferation: at the interface between three dynamic systems. Mol Cell Endocrinol. 331:11–22. doi: 10.1016/j.mce.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 81.Oster H, et al. The circadian rhythm of glucocorticoids is regulated by a gating mechanism residing in the adrenal cortical clock. Cell Metab. 2006;4:163–173. doi: 10.1016/j.cmet.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 82.Chung S, et al. Adrenal peripheral oscillator in generating the circadian glucocorticoid rhythm. Ann N Y Acad Sci. 2011;1220:71–81. doi: 10.1111/j.1749-6632.2010.05923.x. [DOI] [PubMed] [Google Scholar]
- 83.Balsalobre A, et al. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289:2344–2347. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- 84.Torra IP, et al. Circadian and glucocorticoid regulation of Rev-erbalpha expression in liver. Endocrinology. 2000;141:3799–3806. doi: 10.1210/endo.141.10.7708. [DOI] [PubMed] [Google Scholar]
- 85.Yamamoto T, et al. Acute physical stress elevates mouse period1 mRNA expression in mouse peripheral tissues via a glucocorticoid-responsive element. J Biol Chem. 2005;280:42036–42043. doi: 10.1074/jbc.M509600200. [DOI] [PubMed] [Google Scholar]
- 86.Schmutz I, et al. The mammalian clock component PERIOD2 coordinates circadian output by interaction with nuclear receptors. Genes Dev. 2010;24:345–357. doi: 10.1101/gad.564110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Le Minh N, et al. Glucocorticoid hormones inhibit food-induced phase-shifting of peripheral circadian oscillators. EMBO J. 2001;20:7128–7136. doi: 10.1093/emboj/20.24.7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.McNamara P, et al. Regulation of CLOCK and MOP4 by nuclear hormone receptors in the vasculature: a humoral mechanism to reset a peripheral clock. Cell. 2001;105:877–889. doi: 10.1016/s0092-8674(01)00401-9. [DOI] [PubMed] [Google Scholar]
- 89.Fu J, et al. Oleylethanolamide regulates feeding and body weight through activation of the nuclear receptor PPAR-alpha. Nature. 2003;425:90–93. doi: 10.1038/nature01921. [DOI] [PubMed] [Google Scholar]
- 90.Rodriguez de Fonseca F, et al. An anorexic lipid mediator regulated by feeding. Nature. 2001;414:209–212. doi: 10.1038/35102582. [DOI] [PubMed] [Google Scholar]
- 91.Wang N, et al. Vascular PPARgamma controls circadian variation in blood pressure and heart rate through Bmal1. Cell Metab. 2008;8:482–491. doi: 10.1016/j.cmet.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Huang W, et al. Circadian rhythms, sleep, and metabolism. J Clin Invest. 2011;121:2133–2141. doi: 10.1172/JCI46043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ahima RS, et al. Postnatal leptin surge and regulation of circadian rhythm of leptin by feeding. Implications for energy homeostasis and neuroendocrine function. J Clin Invest. 1998;101:1020–1027. doi: 10.1172/JCI1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kalsbeek A, et al. The suprachiasmatic nucleus generates the diurnal changes in plasma leptin levels. Endocrinology. 2001;142:2677–2685. doi: 10.1210/endo.142.6.8197. [DOI] [PubMed] [Google Scholar]
- 95.Kalra SP, et al. Rhythmic, reciprocal ghrelin and leptin signaling: new insight in the development of obesity. Regul Pept. 2003;111:1–11. doi: 10.1016/s0167-0115(02)00305-1. [DOI] [PubMed] [Google Scholar]
- 96.Mistlberger RE. Neurobiology of food anticipatory circadian rhythms. Physiol Behav. 2011;104:535–545. doi: 10.1016/j.physbeh.2011.04.015. [DOI] [PubMed] [Google Scholar]
- 97.Yildiz BO, et al. Alterations in the dynamics of circulating ghrelin, adiponectin, and leptin in human obesity. Proc Natl Acad Sci U S A. 2004;101:10434–10439. doi: 10.1073/pnas.0403465101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sinha MK, et al. Nocturnal rise of leptin in lean, obese, and non-insulin-dependent diabetes mellitus subjects. J Clin Invest. 1996;97:1344–1347. doi: 10.1172/JCI118551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cummings DE, et al. A preprandial rise in plasma ghrelin levels suggests a role in meal initiation in humans. Diabetes. 2001;50:1714–1719. doi: 10.2337/diabetes.50.8.1714. [DOI] [PubMed] [Google Scholar]
- 100.Nakajima M, et al. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308:414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- 101.Tu BP, McKnight SL. The yeast metabolic cycle: insights into the life of a eukaryotic cell. Cold Spring Harb Symp Quant Biol. 2007;72:339–343. doi: 10.1101/sqb.2007.72.019. [DOI] [PubMed] [Google Scholar]
- 102.Tu BP, McKnight SL. Metabolic cycles as an underlying basis of biological oscillations. Nat Rev Mol Cell Biol. 2006;7:696–701. doi: 10.1038/nrm1980. [DOI] [PubMed] [Google Scholar]
- 103.Minami Y, et al. Measurement of internal body time by blood metabolomics. Proc Natl Acad Sci U S A. 2009;106:9890–9895. doi: 10.1073/pnas.0900617106. [DOI] [PMC free article] [PubMed] [Google Scholar]