Abstract
The discovery of microRNAs (miRNAs) provides a new and powerful tool for studying the mechanisms, diagnosis and treatments of cancer. In this study, we employed AFFX miRNA expression chips to search for miRNAs that may be aberrantly expressed in gastric cancer tissues and to investigate the potential roles that miRNAs may play in the development and progression of gastric cancer. 14 miRNAs were found to be down-regulated and 2 miRNAs up-regulated in gastric cancer tissues compared to the normal gastric tissues. Among the aberrantly expressed miRNAs, miR-574-3p was selected to further study its expression features and functional roles. Interestingly, the reduced expression of miR-574-3p occurred mainly in the early stages of gastric cancer or in cancers with high level of differentiation, suggesting that it can be used as a marker for a mild case of gastric cancer. Functional study revealed that cell proliferation, migration and invasion were significantly inhibited in miR-574-3p-transfected gastric cancer SGC7901 cells. Computational prediction and experimental validation suggest that Cullin2 may be one of the targets of miR-574-3p. Overall our study suggests that the aberrantly expressed miRNAs may play regulatory and functional roles in the development and progression of gastric cancer.
Keywords: microRNA, gastric cancer, target gene, proliferation, invasion
1. Introduction
Cancer is a polygenic disease processing in many steps, and many genes have been found to be involved in the formation and progression of cancer. Gene alterations play important roles in uncontrollable proliferation, apoptosis evasion, angiogenesis, metastasis and invasion of cancer [1–4]. However, the mechanisms of controlling the expression of the genes are still not fully understood. The discovery of microRNA provides a powerful tool for studying the mechanisms, diagnosis and treatments of cancers [5–6].
MicroRNAs (miRNAs), approximately 19–24 nucleotides in length, are evolutionarily conserved small single-stranded non-coding RNA molecules. It was reported that more than 60% of human protein-coding genes have been under selective pressure to maintain their pairing with miRNAs [7]. They work mainly at post-transcriptional level by binding to the sequences in the 3′ untranslated regions (3′UTR) of their targeted mRNAs resulting in translational repression or gene silencing [7–9]. So far, the regions encoding some miRNAs have been found to be located in cancer-associated genomic regions or at fragile genomic sites [10]. MiRNAs are involved in regulation of wide array of biological processes, such as cell proliferation, differentiation and apoptosis [11–12]. Theses processes are deregulated during the development and progress of a cancer, which may associated with abnormal changes of miRNA. Indeed, aberrant expressions of miRNAs have been studied in several types of human cancers, including lung [13], colon [14], breast [15], prostate [16], hepatocellular [17] and ovarian [18] cancers. They may play roles in the development and progression of cancers similar to those played by oncogenes or tumor suppressor genes [19–20].
Although aberrant expression of miRNAs in gastric cancer has been reported recently, the results are various between groups [21–24]. In addition, the function roles of theses aberrant expressed miRNAs in gastric cancer remain largely unknown and their mRNA targets are barely studied. In order to gain more information on this issue, we employed AFFX miRNA expression chips to study the aberrant expressions of miRNAs and quantitative real-time PCR to identify and confirm miRNAs with aberrant expressions in gastric cancer. Among the aberrantly expressed miRNAs, we chose miR-574-3p to further study their functional roles concerning proliferation, migration and invasion of gastric cancer cells. We also computationally predicted and experimentally validated the target of miR-574-3p.
2. Materials and Methods
2.1. Materials
Trizol was purchased from Invitrogen, and MirVana™ miRNA isolation Kit was purchased from Ambion. AFFX miRNA expression chips and RNA Labeling Kit were purchased from Affymetrix (CA, USA). miRNA cDNA Synthesis Kit, miRNA PrimeScript®RT Enzyme Mix, and SYBR®Premix Ex Taq™ were all purchased from Takara, China. Cell counting kit-8 (CCK-8) was purchased from Dojindo Laboratories (Dojindo, Japan). PVDF membrane was purchased from Bio-Rad (CA, USA). Rabbit anti human CUL2 and mouse anti human β-actin monoclonal antibodies, goat anti-rabbit and goat anti-mouse secondary antibody were purchased from Proteintech Group, Inc (IL, USA).
2.2. Patients and specimens
Gastric cancer tissues and their adjacent normal tissues were collected from 20 patients with primary gastric cancer during their initial cancer-removal surgery. All samples were snap-frozen within 10 minutes after excision and stored at −196°C in liquid nitrogen. The written informed consent was obtained from each patient at the First Hospital and the Second Hospital of Jilin University, Changchun, China where the specimens were obtained. The research was approved by the Chinese IRB overseeing human subjects at the First Hospital and the Second Hospital of Jilin University and by the Institutional Review Board (IRB) at the University of Georgia, USA.
The histological subtype and pathologic stage of each tumor were determined according to the World Health Organization (WHO) and the tumor, lymph node and metastasis (TNM) classification system. The clinicpathological characteristics of the tumor tissues from these patients are summarized in Table 1.
Table 1.
Clinicopathological characteristics of the patients
| Clinical parameters | Number of cases |
|---|---|
| Gender | |
| Male | 12 |
| Female | 8 |
| Age | |
| < 40 | 13 |
| ≥40 | 7 |
| Level of differentiation | |
| Poorly differentiated | 7 |
| Moderately differentiated | 8 |
| Highly differentiated | 5 |
| Clinical stage | |
| I–II | 11 |
| III–IV | 9 |
| Lymph node metastasis | |
| Positive | 8 |
| Negative | 12 |
2.3. MicroRNA preparation
Total RNA from tissue samples was extracted using the Trizol RNA, and its content was analyzed using the UV2800 ultraviolet spectrophotometer (UNIC, NY, USA) having the A260/A280 ratio between 1.8 to 2.0, representing that the RNA samples are highly purified and not degraded. The miRNA was isolated from the total RNA and purified using the mirVana ™ miRNA isolation Kit.
2.4. MicroRNA microarrays
AFFX miRNA expression chips were used to perform the miRNA expression assay. Total RNA from 16 cancer tissues and their adjacent normal tissues was isolated and miRNAs were labeled using the RNA Labeling Kit. The labeled miRNA hybridization solution was added to the chip for hybridization for 17 hours at 45°C, 60 rpm. After being washed and stained using the GeneChip® Fluidics Station 450 (Affymetrix), the chip was inserted into the Affymetrix autoloader carousel using an appropriate fluidics script and scanned with the GeneChip® Scanner 3000 with GeneChip® Operating Software.
2.5. Quantitative real-time PCR
SYBR Green quantitative real-time PCR was used to measure the content of miRNA and mRNA in tissue and cell. We first obtained cDNA using One Step PrimeScript® miRNA cDNA Synthesis Kit. 1μg total RNA, 2μl miRNA PrimeScript®RT Enzyme Mix, 10 μl miRNA Reaction Buffer Mix and 2μl 0.1% BSA were mixed, incubated in 37°C, water-bath for 60 min. The reaction was inactivated by dipping the reaction tube into 85°C water-bath for 5 seconds. The real-time PCR was prepared with the SYBR®Premix Ex Taq™ quantitative augmentation reaction system. The total reaction volume was 20μl including 10μl SYBR® Premix Ex Taq™II (2×, 2 μl reverse transcription reaction products, 0.4μl ROX Reference Dye (50×), 0.8μl Uni-miR qPCR Primer (10μM), 0.8μl purposely primer (10μM). The PCR was performed in the ABI PRISM 7300 Fast Real-Time PCR System (Ambion) at 95°C for 30sec, then at 95°C for 5 sec and 60°C for 31 sec for 40 cycles. The primers used for the real-time PCR are shown in Table 2. All the primers were synthesized by Shanghai ShengGong Biotechnology Company. The results of real-time PCR is expressed as 2−ΔΔCt, ΔCt (for miRNA: measured Ct subtracting the internal control U6 Ct; for mRNA: measured Ct subtracting the GAPDH Ct), or ΔΔCt (cancer ΔCt subtracting the normal ΔCt), with the lower the values of 2−ΔΔCt, the lower the expression level of the mRNA or miRNA.
Table 2.
Sequences of primers and miRNA mimics
| Name | Primer or miRNA mimics sequences |
|---|---|
| U6-F | CGCTTCGGCAGCACATATACTA |
| U6-R | CGCTTCACGAATTTGCGTGTCA |
| miR-31 | AGGCAAGATGCTGGCATAGCT |
| miR-574-3p | CACGCTCATGCACACACCCCACA |
| GAPDH-F | AGAAGGCTGGGGCTCATTTG |
| GAPDH-R | AGGGGCCATCCACAGTCTTC |
| CUL2-F | GAGTGCGTTGGATAAGGCCCT |
| CUL2-R | CTCTGTCATCCCTTTCGCTGACT |
| RXRA-F | CGCCATCTTTGACAGGGTGCT |
| RXRA-R | GGTTCGAGAGCCCCTTGGAGT |
| miR-574-3p mimics sense | CACGUCAUGCACACACCCACA |
| miR-574-3p mimics antisense | UGUGGGUGUGUGCUGGCGUG |
| Negtive control sense | UUCUCCGAACGUGUCACGUTT |
| Negtive control antisense | ACGUGACACGUUCGGAGAATT |
2.6. Cell culture and transient miRNA transfection
Human gastric cancer cell line SGC7901 was purchased from American Type Culture Collection (ATCC). The cells were cultured in RPMI1640 medium (Gibco, NY, USA) containing 10% Fetal Bovine Serum, 1.5 mM L-Glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin, and incubated at 37°C with 5% CO2.
To investigate the effect of miR-574-3p on cell proliferation, cell migration and invasion, and CUL2 protein expression, the SGC7901 cells were transiently infected with miR-574-3p mimics. We used another RNA duplex, a non-homologous to any human genome sequences, as a negative control (NC). The miR-574-3p mimics and the NC were synthesized by GenePharma (Shanghai, China). All pyrimidine nucleotides in the miR-574-3p mimics and NC were substituted by their 2′-O-methyl analogs to improve stability of the RNA duplex. The sequences of the miR-574-3p mimics and the NC are shown in Table 2. Transient transfection of the miRNA mimics was carried out with the Lipofectamine 2000 kit (Invitrogen, CA, USA) in accordance with the manufacturer’s procedure. The final concentration of the miRNA mimics for each transfection is 100 nM. The miR-574-3p level in the trransfected SGC7901 cells was measured by real-time PCR.
2.7. Cell proliferation assay
The cell proliferation was measured with the CCK-8. The count of the living cells was performed following the manufacturer’s protocol. Briefly, after the miR-574-3p mimics or the NC transiently transfected SGC7901 cells were cultured in 96-well plates at a density of 2×104 cells/well under the culture conditions described above for 48 or 72 hours, each well was added with 10μl CCK-8 solution incubating for another 1 hour. The optical density (OD) value of the cells in each well was measured at 450 nm with the enzyme-linked immunosorbent assay plate reader (Bio-Rad, CA, USA).
2.8. Cell invasion and migration assays
The invasive and the migration potential of the transfected SGC7901 cells were evaluated by transwell cell matrigel invasion and migration assay. The invasion assay was performed with transwell chambers (8 μm pore; Corning, NY, USA) coated with matrigel (BD Biosciences, Bedford, MA). The cells were starved overnight in the RPMI1640 medium containing 0.1% FBS and added to the upper chambers in a 24 wells plate at a concentration of 2×104/chamber. The lower chambers were filled with 500μl RPMI1640 medium supplemented with 10% FBS. After 48 hours of incubation, the filters on the transwell chambers were washed with PBS, fixed with methanol for 1 min, and stained with the 0.5% crystal violet. The cells on the underside of the filters were counted under a light microscope (Olympus, Japan) at 100× magnification. The results for each membrane are an average counting of five visual fields. Cell migration assay procedure was similar to the method described above except the transwell chambers were not coated with the matrigel and the starved cells were incubated for 24 hours. After cotton swabs removal of the non-migratory cells on the upper face of the filters, the migrated cells on the underside of the filters of the chambers were fixed, stained and counted.
2.9. Expression of CUL2 mRNA and protein
We employed the real-time PCR and Western blotting to observe the effect of miR-574-3p on the expression of CUL2. We have described the method of the real-time PCR above. For the Western blotting, the transfected SGC7901 cells were lysed in a protein extraction buffer [0.5 mol/L Tris.Cl (pH 7.4), 150mmol/L NaCl, 0.1 mmol/l ethylene diamine tetra aceticacid (EDTA) (pH 7.0), 1 mmol/l phenylmethylsulfonyl fluoride (PMSF), 2.5 mg/ml Aprotinin, 1 mmol/l dithiothreitol (DTT), 1% Triton X-100, 1% Sodium deoxycholate, 1% SDS]. The supernatants were collected after centrifugation (10000 rpm, 10 min, 4°C), and loaded (30 μg protein) on a 10% of SDS-PAGE. After separated, the proteins in the gel were transferred onto a PVDF membrane. The membrane was incubated in 5% milk blocking solution for 2h at room temperature, and then incubated with CUL2 or β-actin monoclonal antibody (1:1000) at room temperature for 1h. After reacting with goat anti-rabbit or anti-mouse secondary antibody (1:1000), the membrane was covered completely with an equal amount of enhancer and peroxide solution from an ECL Plus kit (Beyotime, China) for 2 min and exposed to the film. The density of the bands was quantified with the Image Tool (version 3.0).
2.10. Data processing and statistical analysis
The target genes of specific miRNAs were predicted using two prediction algorithms, TargetScan (http://www.targetscan.org/) and miRDB (http://mirdb.org/cgi-bin/search.cgi). miRNA expression levels were summarized based on the raw probe intensities using the Robust Multichip Average procedure in the APT package (Affymetrix). Quantile normalization was done on both the cancer and the normal datasets. miRNAs with the maximum normalized signal intensity in both the cancer and reference samples lower than 4 were removed. Only miRNAs with at least 1.5 fold change between the cancer and the matching reference tissues, along with their P < 0.05 for the detected changes after adjustment for multiple tests using R, were considered for further analyses. A t-test was used for assessing the statistical significance of each differential expression analysis result. P-value < 0.05 was considered as being significant.
3. Results
3.1. Aberrant expression of miRNAs in gastric cancer
miRNA microarray was used to obtain the miRNA expression profiles of gastric cancer. 16 miRNAs were found to be aberrantly expressed, of which 14 were down-regulated and 2 up-regulated in gastric cancer tissues comparing with their adjacent normal tissues. Among these miRNAs, up-regulation of miR-455-3p and down-regulation of miR-574-3p, miR-1207-5p, miR-486-5p and miR-768-5p in gastric cancer have not been reported to the best of our knowledge (Table 3 and Figure 1).
Table 3.
Expression changes of miRNAs in gastric cancer
| Down-regulated | Up-regulated |
|---|---|
| hsa-let-7g | has-miR-455-3p |
| hsa-miR-200b | hsa-miR-34a |
| hsa-miR-768-3p | |
| hsa-let-7d | |
| hsa-miR-104-3p | |
| hsa-miR-27b | |
| hsa-miR-1207-5p | |
| hsa-miR-663 | |
| hsa-miR-486-5p | |
| hsa-miR-222 | |
| hsa-miR-574-3p | |
| hsa-miR-768-5p | |
| hsa-miR-378 | |
| hsa-miR-31 |
Figure 1. miRNA expression signature for gastric cancer.
The tree on the left was constructed using a hierarchical clustering [25] based on the probe signal intensity after normalization and log2 transformation. The miRNAs clustered together have similar expression patterns. The heatmap on the right shows the expression patterns of the 16 differentially expressed miRNAs in 16 gastric cancer tissues and their reference tissues, which are color-coded, with red denoting high and green the low expression level.
3.2. Validation of miRNAs expression in gastric cancer tissues
To confirm the accuracy of our chip results, we chose miR-31, miR-574-3p, miR-455-3p and miR-768-5p for further experimental validation using quantitative real-time PCR because of their obvious fold changes on the miRNA microarray analysis. It was showed that miR-31, miR-574-3p and miR-768-5p expressed at lower levels and miR-455-3p expressed at a higher level in gastric cancer tissues compared to their matching references (P<0.01) (Figure 2), which were consistent with the microarray data (Figure 1).
Figure 2. Validdation of miRNA expressions in gastric cancer.
(A) The expression levels of miR-31, miR-574-3p, miR-455-3p and miR-768-5p from 20 gastric cancer and their adjacent normal tissues were measured using SYBR Green quantitative real-time PCR. (B) The fold changes of miRNA expression in gastric cancer tissues compared to normal tissues. RNA U6 was used as an internal control. The experiments were repeated 3 times and similar results were obtained. All results, normalized to the U6 mRNA expression, were expressed as 2−ΔΔCt (ΔCt=CtmiRNAs−CtU6, ΔΔCt = ΔCt tumor − ΔCt normal). The results showed by the mean value ± SEM, ** p<0.01 vs. normal tissues.
3.3. Differential expressions of miR-574-3p in different grades and stages of gastric cancer
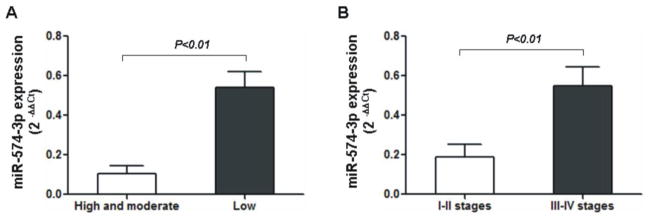
We noted that the expression level of miR-574-3p was significantly reduced in cancer tissues with higher levels of differentiation (P < 0.01); and a similar observation was made on early stages of the cancer (P < 0.01) (Figure 3). The expression level of miR-574-3p in gastric cancer was not affected by the gender and the age of the patients (data not shown).
Figure 3. Expression levels of miR-574-3p in different cancer types and at different stages of gastric cancer.
The expression levels of miR-574-3p in gastric cancer tissues were measured using real-time PCR. (A). Expression levels of miR-574-3p in cancer tissues with high and moderate level of differentiation (n=7) versus those with low level of differentiation (n=13). (B). Expression levels of miR-574-3p in I–II stages (n= 9) versus III–IV stages (n= 11) of the cancer patients. The data were expressed as 2−ΔΔCt (ΔCt=CtmiRNAs−CtU6, ΔΔCt = ΔCt tumor − ΔCt normal), and showed by the mean value ± SEM. The statistical significance values were calculated using t-test.
3.4. Prediction of the potential targets of miR-574-3p
MiRNAs modulate gene expression by interacting with their target mRNAs resulting in mRNA degradation and/or translational repression. To find out the potential targets of miR-574-3p, we performed bioinformatics prediction analysis, based on our previous published data [25]. The prediction of the target genes for miR-574-3p was performed using TargetScan and miRBD. Among the 31 predicted targets of miR-574-3p, RXRA and CUL2 showed high potentials since both RXRA and CUL2 were predicted by both programs, while others were only predicted by one of the programs (Figure 4A). In addition, we also noted from our previous study on 160 sets of gastric cancer tissue mRNA microarrays [25] that only RXRA and CUL2 out of the 807 up-regulated mRNAs were predicted to be the targets of miR-574-3p (Figure 4A). We validated the results of the microarray assays by performing real-time PCR. The results showed that expression levels of RXRA and CUL2 were indeed increased in gastric cancer tissues (Figure 4B).
Figure 4. Prediction of targets of miR-574-3p.
(A). Target prediction for miR-574-3p. Venn diagram shows overlaps between predicted target mRNAs by two prediction algorithms and results of mRNA microarrays. (B). Expression levels of RXRA and CUL2 in gastric cancer tissues. The data were expressed as 2−ΔΔCt (ΔCt=CtRARA/CUL2−CtGAPDH, ΔΔCt = ΔCt tumor − ΔCt normal), and showed by the mean value ± SEM. The statistical significance values were computed using a t-test.
3.5. Negatively regulation of CUL2 by miR-574-3p
CUL2 mRNA expression was significantly reduced in the SGC7901 cells transfected with miR-574-3p mimics (P<0.01), which was 40.1% of that in the Mock group (Figure 5A). However, the miR-574-3p mimics transfection had no influence on RXRA mRNA expression in the SGC7901 cells (Figure 5A). In addition, as shown in the Figure 5B, miR-574-3p mimics significantly decreased CUL2 protein levels to 44.4% of Mock group (P<0.01) in the SGC7901 cells. All these data indicated that CUL2 could be negatively regulated by miR-574-3p.
Fig. 5. mRNA and protein expression of RXRA and CUL2 in SGC7901 cells trasfected with miR-574-3p mimics.
(A). RXRA and CUL2 mRNA expression. (B). CUL2 protein expression. Gastric cancer cell line SGC7901 cells were transiently transfected with 100 nM miR-574-3p mimics for 48 hours and subjected to mRNA expression assay by real-time PCR or protein expression assay by Western blotting, respectively. GAPDH and β-actin were used as an internal loading control. A reproducible result was obtained in three independent experiments. Mock: tranfection reagent control. NC: RNA duplex negative control. The results are mean values ± SD, and the statistical significance values were computed using a t-test. * p<0.05, ** p<0.01 vs. Mock.
3.6. Proliferation, migration and invasion of SGC7901 cells transfected with miR-574-3p
We evaluated the effect of miR-574-3p on proliferation, migration and invasion ability of SGC7901 gastric cell line. The CCK-8 results showed that the number of SGC7901 cells was reduced to 80.4% (p<0.01) or 74.5% (p<0.01) of those in the Mock group at 48 or 72 hours after transfecting with miR-574-3p mimics (Figure 6A). Cell migration and invasion assay performed in transwell chambers showed that the migration and the invasion of the miR-574-3p-transfected SGC7901 cells were declined to 62.1% and 36.3% of those in the Mock group, respectively (P<0.01, and Fig 6B and C).
Fig. 6. Inhibition of proliferation, migration and invasion of SGC7901 cells by miR-574-3p mimics transfection.
(A). Cell proliferation in miR-574-3p transfectants. SGC7901 gastric cancer cells transiently transfected with 100 nM miR-574-3p mimics. At 48 and 72h after the transfection, the cells viabilities were detected by CCK8 assay. (B–C). Cell migration and invasion in miR-574-3p transfectants. SGC7901 gastric cancer cells transfected with miR-574-3p mimics for 24h, and then subjected to transwell migration (24h) and matrigel invasion (48h) assays. Blank, Mock and NC represented SGC7901 cells transfected with nothing, transfection reagent and RNA duplex negative control. Representative quantification (left) and photographs (right) are shown. Magnification: ×100. The results are mean values ± SD, and the statistical significance values were computed using a t-test. **, p<0.01 vs. Mock.
4. Discussion
In the study, we demonstrated with miRNA microarray that some miRNAs were aberrantly expressed in gastric cancer tissues, specifically, 14 miRNAs were down-regulated and 2 miRNAs were up-regulated. Among them, the miRNAs that had more than 4-fold changes in expression were chosen or further experimental validation using real-time PCR including miR-574-3p, miR-31, miR-455-3p and miR-768-3p. The data obtained from the real-time PCR validated that from the miRNA microarray assay. Distinct miRNA expression can be used as signature for diagnosis, prognosis and treatment response of cancer. Moreover, studying profile of miRNA expression in cancer may provide better understanding of molecular pathways contributing to cancer development and progression since miRNAs can affect a wide variety of molecular pathways.
We noted that the reduced expression levels of miR-574-3p were mainly found in early stages of as well as in highly differentiated gastric cancer. This has never been reported before, to the best of our knowledge. These observations suggest that miR-574-3p could be used as a marker for detection of early stages of gastric cancer or diagnosis of less aggressive gastric cancer. The results also led to the speculation that the dynamic changes of the targets proteins for miR-574-3p may play different functional roles at different developmental stages of gastric cancer, as well as in gastric cancers with different levels of differentiation. Aberrant expressions of miR-574-3p were also reported in other cancer types. Recent studies showed that miR-574-3p is down-regulated in bladder cancer [26], and up-regulated, however, in the urine of prostate cancer patients [27] and in the serum of hepatocellular carcinoma patients [28] were reported. The different expression patterns of miR-574-3p may reflect different regulation mechanisms of tissue or circulating miRNAs. Also, the differences of tumor malignancy in patients and tissue specificity could influence the expression of the relevant miRNA.
We predicted by TargetScan and miRDB programs and found that RXRA and CUL2 could be potential targets of miR-574-3p. Experimental study validated that the mRNA and protein expression of CUL2, not RXRA, was down-regulated in the miR-574-3p mimics-transfected gastric cancer SGC7901 cells which indirectly demonstrated the target of miR-574-3p was CUL2. The fundamental function of miRNAs in regulation of their target genes is cleavage of mRNA and/or inhibition of protein synthesis depending on the degree of complementarity with the sequence in the mRNA 3′UTR [29]. Our study suggests that miR-574-3p may play its functional roles through the inhibition of the expression of CLU2 gene. CUL2 is a cytoplastic protein and a component of the ElonginB/C-CUL2-RBX-1-Von Hippel-Lindau (VHL) complex [30], which is formed when cells become hypoxic, and is required for normal vasculogenesis and involved in cell cycle regulation [31]. Our study suggests that the dynamic changes in the expression levels of CUL2 may reflect different requirement for CUL2 across gastric cancers at different development stages and with different levels of differentiation to better serve gastric cancer cells to survive.
Our functional study demonstrated that miR-574-3p over-expression inhibited the proliferation, migration and invasion ability of gastric cancer cells. Its down- regulation in gastric cancer may be associated with development and progression of the cancer. Indeed, a recent study suggests that miR-574-3p plays a tumor suppressor role in bladder cancer (BC) cells [26]. MiRNAs regulate biology function by degradation of target mRNAs, single miRNA can repress the production of hundreds of proteins [29]. MiR-574-3p has been known to target mesoderm development candidate 1 (MESDC1) mRNA directly, subsequently inhibited the proliferation, migration and invasion ability and induced cell apoptosis in BC cells [26]. However, it is not clear whether the suppressor role of miR-574-3p in gastric cancer cells is played by targeting CUL2 directly, further studies are need for validation.
Highlights.
14 miRNAs down-regulated and 2 miRNAs up-regulated in gastric cancer tissues.
Reduced expression of miR-574-3p occurred in the early stage of gastric cancer.
Cullin2 could be negatively regulated by miR-574-3p.
miR-574-3p inhibited the proliferation, migration and invasion of SGC7901 cells
Acknowledgments
We thank the physicians in the First Hospital of Jilin University for providing the specimens used in this study. We also thank Dr. Ming Yang, Department of Immunological Laboratory, Jilin University, for his technical help. This work was supported in part by the National Institutes of Health (1R01GM075331), a “Distinguished Scholar” grant from the Georgia Cancer Coalition, and seed funding from the University of Georgia and in part by Research Fund for the Innovation Program of Higher Education of China and National Basic Research Program of China (973 program, 2011CB512003) and National Natural Science Foundation of China (30872415 and 81071424).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tamura G. Alterations of tumor suppressor and tumor-related genes in the development and progression of gastric cancer. World J Gastroenterol. 2006;12:192–8. doi: 10.3748/wjg.v12.i2.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kwon MJ, Kim SH, Jeong HM, Jung HS, Kim SS, Lee JE, et al. Claudin-4 overexpression is associated with epigenetic derepression in gastric carcinoma. Lab Invest. 2011;91:1652–67. doi: 10.1038/labinvest.2011.117. [DOI] [PubMed] [Google Scholar]
- 3.Ma D, Liu M, Wang AP, Yang H. Cycloxygenase-2 is essential for the survival and proliferation of gastric cancer cells. Cell Biochem Biophys. 2011;61:637–41. doi: 10.1007/s12013-011-9249-6. [DOI] [PubMed] [Google Scholar]
- 4.Xu L, Wang F, Liu H, Xu XF, Mo WH, Xia YJ, et al. Increased expression of cellular repressor of E1A-stimulated gene (CREG) in gastric cancer patients: a mechanism of proliferation and metastasis in cancer. Dig Dis Sci. 2011;56:1645–55. doi: 10.1007/s10620-010-1510-0. [DOI] [PubMed] [Google Scholar]
- 5.Tricoli JV, Jacobson JW. MicroRNA: Potential for Cancer Detection, Diagnosis, and Prognosis. Cancer Res. 2007;67:4553–5. doi: 10.1158/0008-5472.CAN-07-0563. [DOI] [PubMed] [Google Scholar]
- 6.Liu X, Liu L, Xu Q, Wu P, Zuo X, Ji A. MicroRNA as a novel drug target for cancer therapy. Expert Opin Biol Ther. 2012;12:573–80. doi: 10.1517/14712598.2012.671293. [DOI] [PubMed] [Google Scholar]
- 7.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–14. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 10.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bueno MJ, Pérez de Castro I, Malumbres M. Control of cell proliferation pathways by microRNAs. Cell Cycle. 2008;7:3143–8. doi: 10.4161/cc.7.20.6833. [DOI] [PubMed] [Google Scholar]
- 12.Cho WC. OncomiRs: the discovery and progress of microRNAs in cancers. Mol Cancer. 2007;6:60. doi: 10.1186/1476-4598-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nasser MW, Datta J, Nuovo G, Kutay H, Motiwala T, Majumder S, et al. Down-regulation of micro- RNA-1 (miR-1) in lung cancer. Suppression of tumorigenic property of lung cancer cells and their sensitization to doxorubicin-induced apoptosis by miR-1. J Biol Chem. 2008;283:33394–405. doi: 10.1074/jbc.M804788200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Tazawa H, Tsuchiya N, Izumiya M, Nakagama H. Tumorsuppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci USA. 2007;104:15472–7. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–8. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 16.Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L, et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities, Nat. Med. 2008;14:1271–7. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 17.Yao J, Liang L, Huang S, Ding J, Tan N, Zhao Y, et al. MicroRNA-30d promotes tumor invasion and metastasis by targeting Galphai2 in hepatocellular carcinoma. Hepatology. 2010;51:846–56. doi: 10.1002/hep.23443. [DOI] [PubMed] [Google Scholar]
- 18.Yang H, Kong W, He L, Zhao JJ, O’Donnell JD, Wang J, et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68:425–33. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
- 19.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–70. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 20.Lee YS, Dutta A. MicroRNAs: small but potent oncogenes or tumor suppressors. Curr Opin Investig Drugs. 2006;7:560–4. [PubMed] [Google Scholar]
- 21.Katada T, Ishiguro H, Kuwabara Y, Kimura M, Mitui A, Mori Y, et al. microRNA expression profile in undifferentiated gastric cancer. Int J Oncol. 2009;34:537–42. [PubMed] [Google Scholar]
- 22.Guo J, Miao Y, Xiao B, Huan R, Jiang Z, Meng D, et al. Differential expression of microRNA species in human gastric cancer versus non-tumorous tissues. J Gastroenterol Hepatol. 2009;24:652–7. doi: 10.1111/j.1440-1746.2008.05666.x. [DOI] [PubMed] [Google Scholar]
- 23.Yao Y, Suo AL, Li ZF, Liu LY, Tian T, Ni L, et al. MicroRNA profiling of human gastric cancer. Mol Med Report. 2009;2:963–70. doi: 10.3892/mmr_00000199. [DOI] [PubMed] [Google Scholar]
- 24.Brenner B, Hoshen MB, Purim O, David MB, Ashkenazi K, Marshak G, et al. MicroRNAs as a potential prognostic factor in gastric cancer. World J Gastroenterol. 2011;17:3976–85. doi: 10.3748/wjg.v17.i35.3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui J, Chen Y, Chou WC, Sun L, Chen L, Suo J, et al. An integrated transcriptomic and computational analysis for biomarker identification in gastric cancer. Nucleic Acids Res. 2010;39:1197–207. doi: 10.1093/nar/gkq960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tatarano S, Chiyomaru T, Kawakami K, Enokida H, Yoshino H, Hidaka H, et al. Novel oncogenic function of mesoderm development candidate 1and its regulation by MiR-574-3p in bladder cancer cell lines. Int J Oncol. 2012;40:951–9. doi: 10.3892/ijo.2011.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bryant RJ, Pawlowski T, Catto JW, Marsden G, Vessella RL, Rhees B, et al. Changes in circulating microRNA levels associated with prostate cancer. Br J Cancer. 2012;106:768–74. doi: 10.1038/bjc.2011.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gui J, Tian Y, Wen X, Zhang W, Zhang P, Gao J, et al. Serum microRNA characterization identifies miR-885-5p as a potential marker for detecting liver pathologies. Clin Sci (Lond) 2011;120:183–93. doi: 10.1042/CS20100297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr Opin Cell Biol. 2009;21:452–60. doi: 10.1016/j.ceb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 30.Iwai K, Yamanaka K, Kamura T, Minato N, Conaway RC, Conaway JW, et al. Identification of the von Hippel-lindau tumor-suppressor protein as part of an active E3 ubiquitin ligase complex. Proc Natl Acad Sci U S A. 1999;96:12436–41. doi: 10.1073/pnas.96.22.12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maeda Y, Suzuki T, Pan X, Chen G, Pan S, Bartman T, et al. CUL2 is required for the activity of hypoxia-inducible factor and vasculogenesis. J Biol Chem. 2008;283:16084–92. doi: 10.1074/jbc.M710223200. [DOI] [PMC free article] [PubMed] [Google Scholar]