Abstract
PAX5 rearrangements resulting in the expression of fusion transcripts account for 2–3% of childhood B-cell precursor acute lymphoblastic leukemia. Most PAX5 fusions are rare and many of them have only been described in a couple of, or even only in single, cases. We have identified the third case with a PAX5-AUTS2 fusion, which results from unbalanced t(7;9)(q11.2;p13.2) rearrangements. Our findings substantiate that PAX5-AUTS2 is a recurrent fusion gene in pediatric B-cell precursor acute lymphoblastic leukemia, and we summarize the clinical characteristics of such patients.
Keywords: PAX5-AUTS2, Fusion transcript, Childhood acute lymphoblastic leukemia
1. Introduction
In pediatric B-cell precursor acute lymphoblastic leukemia (BCP-ALL) PAX5 is one of the most frequent targets affected by genetic alterations including deletions, point mutations, and gene rearrangements [1]. PAX5 deletions and PAX5 mutations are found in about 30% and 5–7% of the cases, respectively, and PAX5 rearrangements in 2–3% [1–4]. PAX5 rearrangements either result in bona fide in-frame fusions and the expression of chimeric transcripts [1,3,4] or in PAX5 fusions to genes in opposite orientation, out-of-frame fusions or the expression of truncated isoforms [4].
To date, 16 different in-frame PAX5 fusion partners comprising a heterogeneous group of genes that encode transcription factors, structural proteins, the non-receptor tyrosine kinase JAK2, and several with so far unknown functions have been reported [1,3,4]. A unifying feature of all PAX5 fusion proteins is the retention of the PAX5 paired DNA-binding domain (PD), which is fused to either the entire fusion partner protein or its C-terminal portion [1,3,4].
While many of the PAX5 fusions have so far only been observed in a couple of, or only single, cases, a number of them are more commonly found. The most frequently observed rearrangements are PAX5-ETV6 and PAX5-C20orf112 followed by PAX5-ELN, PAX5-FOXP1, and PAX5-JAK2 each of which has been detected in at least three cases [1,3–5].
We here report the third case with a PAX5-AUTS2 fusion resulting from unbalanced t(7;9)(q11.2;p13.2) rearrangements and summarize the clinical and laboratory characteristics of these patients.
2. Patient and methods
2.1. Case report
A 7-month-old boy presented with 158 × 109/L leucocytes in the peripheral blood (PB) and 91% and 89% blast cell infiltration in the PB and bone marrow (BM), respectively (Table 1). The blast cells showed an L1 morphology and immunophenotyping established the diagnosis of a CD10+ BCP-ALL. Ninety percent of the blast cells were CD19 and CD10 positive and ∼10% weakly positive for cytoplasmic IgM; and they were negative for the myeloid markers CD13, CD15, CD33, and MPO and the T-cell markers cytoplasmic CD3 and surface CD7.
Table 1.
Patient and laboratory data of PAX5-AUTS2 positive cases.
| No. | Age (yrs) | Sex | WBC (×109/L) | Blasts BM | Morphology/phenotype | Karyotype | Reference |
|---|---|---|---|---|---|---|---|
| 1 | 3.1 | M | 171.5 | 93% | L1/B-III | 45,XY,-7,der(9)t(7;9)(q11;p13)[15]/46,XY[1] | Kawamata et al.a[6] |
| 2 | 2.8 | F | 5.6 | 80% | L1/B-III | 45,XX,-7,der(9)t(7;9)(q11;p13),dup(16)(p11p13)[14]/ 46,XX[2] |
Coyaud et al. [7] |
| 3 | 0.6 | M | 158.0 | 89% | L1/B-II/III | 46,XY,t(7;9)(q11;p13)[8] | This work |
BM, bone marrow; WBC, white blood cell count; yrs, years.
Initially in this case no cytogenetic analysis was performed.
Because of a poor prednisone response at day 8 the patient was treated, with informed consent, in the high risk group (HRG) of the ALL-BFM 90 clinical trial. A complete morphologic remission (CR) was achieved after completion of induction therapy; however, 2.1 years after initial diagnosis the patient had a central nervous system (CNS) relapse. The patient achieved a second CR but died of an infectious complication 3.4 years after initial diagnosis. Cytogenetics of the PB showed a 46,XY,t(7;9)(q11;p13) karyotype, and fluorescence in situ hybridization (FISH) indicated a PAX5 rearrangement. The patient's laboratory and clinical parameters are summarized in Tables 1 and 2.
Table 2.
Clinical characteristics and outcome of PAX5-AUTS2 positive cases.
| No. | Clinical trial | CNS disease | MRD day 33 | MRD day 78 | MRD risk | Prednisone response | CR day 33 | Risk group | Relapse | Time to relapse (yrs)a | Outcome | Follow-up (yrs)b |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ALL-BFM 2000 | Yes | 10−4 | Neg | IR | Good | Yes | MRG | 1st BM 2nd BM |
1.4 1.6 |
Dead | 1.7 |
| 2 | ALL-BFM 2000 | No | Neg | Neg | LR | Good | Yes | SRG | / | / | 1st CR | 2.2+ |
| 3 | ALL-BFM 90c | No | ND | ND | ND | Poor | Yes | HRG | CNS | 2.1 | Dead | 3.4 |
BM, bone marrow; CNS, central nervous system; CR, complete morphologic remission; MRD, minimal residual disease; ND, not determined; IR, intermediate risk; LR, low risk; MRG, medium risk group; SRG, standard risk group; HRG, high risk group; yrs, years.
From initial diagnosis.
Follow-up from initial diagnosis to date of death or last follow-up date.
In the ALL-BFM 90 trial MRD was not used as parameter for risk stratification.
2.2. FISH and Reverse Transcription-Polymerase Chain Reaction
FISH using PAX5-specific cosmid probes was conducted as described [3]. RT-PCR for the detection of PAX5-AUTS2 transcripts was performed using primers PAX5ex5-F1 (5′-TACTCCATCAGCGGCATCC-3′) and AUTS2ex8-R1 (5′-TATTGGCTGTGCGACCTGAG-3′) located in exons 5 and 8 of PAX5 and AUTS2, respectively; the amplification product was directly sequenced (Eurofins, MWG, Operon, Germany). The exon nomenclature used corresponds to that of the NCBI reference sequences PAX5 NM_016734.1 and AUTS2 NM_015570.2 (accessed March 2012).
2.3. In silico protein analysis
Protein analysis was performed using BLASTP and COBALT (http://www.ncbi.nlm.nih.gov), PSORTII (http://psort.hgc.jp) and CAST (http://athina.biol.uoa.gr/CAST/). Further information regarding the AUTS2 protein family and posttranslational modifications were obtained from PRINTS (http://www.bioinf.man.ac.uk/) and the PhosphoSitePlus® database (http://www.phosphosite.org).
3. Results and discussion
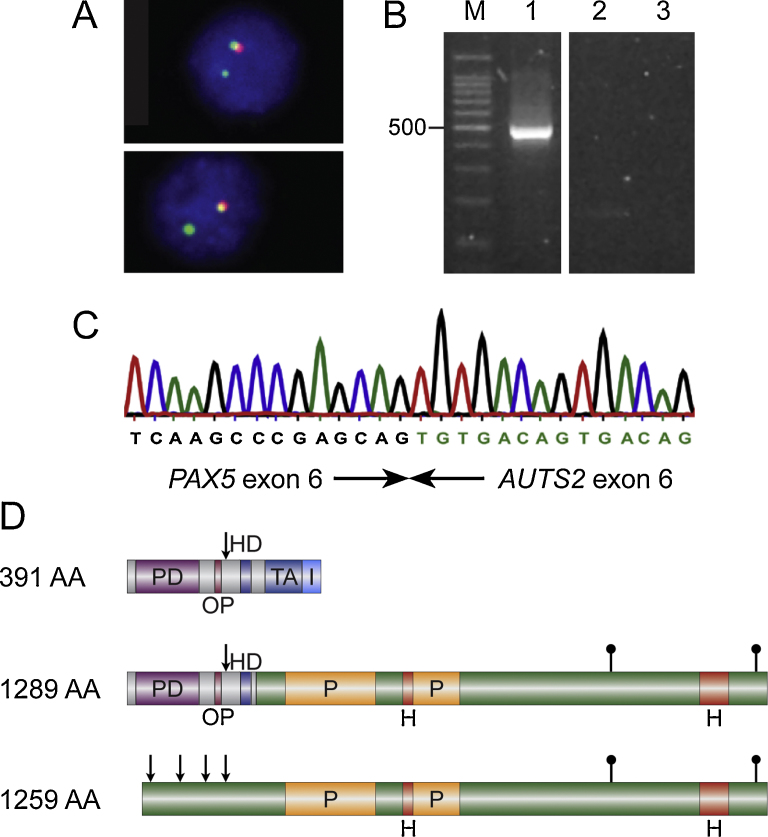
Interphase FISH of a pediatric BCP-ALL with a t(7;9)(q11;p13) using PAX5-specific probes detected a 3′-end deletion of the gene (Fig. 1A) suggesting the presence of a PAX5 fusion with a partner gene located at 7q11. Subsequent RT-PCR revealed the expression of PAX5-AUTS2 transcripts, in which exon 6 of PAX5 was fused to exon 6 of AUTS2 (Fig. 1B and C). Similar in-frame fusions of PAX5 exon 6 to AUTS2 exons 4 or 7 have been previously described [6,7]. Therefore, the breakpoints within AUTS2 appear to be variable but that within PAX5 consistently occurs in intron 6.
Fig. 1.
Molecular genetic analysis of PAX5-AUTS2 positive case. (A) FISH using PAX5-specific cosmids showing a 3′-end deletion: 5′-end-specific clone (green signals); 3′-end-specific clone (red signals). (B) RT-PCR using primers located in PAX5 exon 5 and AUTS2 exon 8 resulting in amplification of PAX5-AUTS2 fusion transcripts. M, molecular weight marker 100 bp ladder (Promega); lane 1, patient No. 3; lane 2, normal control; lane 3, no template control. (C) Sequence chromatogram of the PAX5-AUTS2 fusion junction showing fusion between exon 6 of PAX5 and exon 6 of AUTS2. (D) Schematic representation of the structure of PAX5 (top) and AUTS2 (bottom) wild-type proteins as well as the putative consensus chimeric protein (middle). PD, paired domain; OP, octapeptide; HD, homeodomain; TA, transactivation domain; I, inhibitory domain; H, histidine-rich regions; P, proline-rich region; arrows indicate nuclear localization signal (NLS); filled lollipops represent serine phosphorylation sites.
Since PAX5 and AUTS2 are transcribed in the same – centromere to telomere – orientation, a reciprocal translocation may generate the fusion gene. However, in two cases due to loss of the der(7) chromosome the translocation was unbalanced (Table 1). This is also reflected by the detection of one of the cases by SNP arrays (No. 1; Table 1), which can only indicate fusion genes in case of unbalanced rearrangements [6]. Karyotyping of this previously published case now confirmed loss of the der(7). The cytogenetically unbalanced translocations in two of the PAX5-AUTS2 cases and loss of the PAX5 3′-end in the third one underline the observation that in the majority of PAX5-rearranged cases the reciprocal fusion transcripts are not expressed [3,4] and that, thus, the PAX5-partner fusions drive leukemogenesis [1].
The putative consensus chimeric protein deduced from the fusion transcripts not only contains the PAX5 PD, which is typically retained in all PAX5 fusion proteins [1,3,4], but also the octapeptide motif, the nuclear localization signal (NLS), and the homeodomain, which are fused to the C-terminal regions of AUTS2 (Fig. 1D). With the exception of PAX5-ETV6, in which only the PAX5 PD is maintained, in several PAX5 fusions the breakpoint occurs in intron 5, therefore, also the NLS is present [1,4]. In the remaining PAX5 fusions the breakpoints are located within further downstream introns resulting in the retention of additional conserved protein domains [1,4]. Thus, PAX5-AUTS2 may belong to a subtype of PAX5 fusion proteins whose function might be distinct from those that retain only the PD.
AUTS2, which is expressed in the developing brain, has been described as autism susceptibility candidate gene 2 and its mutation has also been linked to autosomal dominant mental retardation [8]. The nuclear protein AUTS2 belongs to the fibrosin-1-like protein family, which consists of AUTS2, FBRS (fibrosin), and FBRSL1 (fibrosin-like 1), which share a so-called FIBROSIN1LPF 5-element fingerprint signature. Moreover, AUTS2 is highly conserved among species and harbors several putative NLSs, two proline- alternating with two histidine-rich regions, and two potential serine phosphorylation sites (Fig. 1D); the function of the latter sites has, however, not been analyzed in detail yet. In the putative PAX5-AUTS2 chimeric protein almost the entire AUTS2 protein, except for the region containing the NLSs, is fused to PAX5 which retains its NLS (Fig. 1D).
Regarding the clinical course, all three PAX5-AUTS2 positive patients were assigned to different risk arms of the respective clinical trials. They all achieved a CR after completion of induction therapy; however, two of the patients experienced an early relapse and both patients died, one from an infectious complication in second CR and one from progressive leukemia after a further relapse. The third patient remains in first CR for more than two years after diagnosis (Table 2).
The general rarity of recurrent PAX5 fusion gene positive BCP-ALL precludes any conclusions regarding their prognostic value. Yet, although the number of patients is very small, it is interesting to note that two of the three PAX5-AUTS2 patients had a CNS involvement either at the time of diagnosis or relapse and two patients experienced early relapses tentatively indicating that PAX5-AUTS2 positive cases may have a rather poor outcome.
In summary, the description of now three cases with a PAX5-AUTS2 fusion confirms its recurrence in pediatric BCP-ALL and emphasizes the importance of functional studies of PAX5 fusion genes to understand the pathogenesis of this rare disease.
Conflict of interest
All authors declare no conflicts of interest.
Acknowledgments
This work was supported by a grant from the Austrian Science Fund (FWF P21554-B19 to S.S.) and the St. Anna Kinderkrebsforschung.
Authors’ contributions: DD and KN conducted experiments and analyzed data; JB and GP performed cytogenetic analysis; AM and AA are responsible for the clinical data; SS supervised the study and drafted the manuscript. All authors approved the final manuscript.
References
- 1.Medvedovic J., Ebert A., Tagoh H., Busslinger M. Pax5: a master regulator of B cell development and leukemogenesis. Adv Immunol. 2011;111:179–206. doi: 10.1016/B978-0-12-385991-4.00005-2. [DOI] [PubMed] [Google Scholar]
- 2.Familiades J., Bousquet M., Lafage-Pochitaloff M., Bene M.C., Beldjord K., De Vos J. PAX5 mutations occur frequently in adult B-cell progenitor acute lymphoblastic leukemia and PAX5 haploinsufficiency is associated with BCR-ABL1 and TCF3-PBX1 fusion genes: a GRAALL study. Leukemia. 2009;23(11):1989–1998. doi: 10.1038/leu.2009.135. [DOI] [PubMed] [Google Scholar]
- 3.Nebral K., Denk D., Attarbaschi A., Konig M., Mann G., Haas O.A. Incidence and diversity of PAX5 fusion genes in childhood acute lymphoblastic leukemia. Leukemia. 2009;23(1):134–143. doi: 10.1038/leu.2008.306. [DOI] [PubMed] [Google Scholar]
- 4.Coyaud E., Struski S., Prade N., Familiades J., Eichner R., Quelen C. Wide diversity of PAX5 alterations in B-ALL: a Groupe Francophone de Cytogenetique Hematologique study. Blood. 2010;115(15):3089–3097. doi: 10.1182/blood-2009-07-234229. [DOI] [PubMed] [Google Scholar]
- 5.Pichler H., Moricke A., Mann G., Teigler-Schlegel A., Niggli F., Nebral K. Prognostic relevance of dic(9;20)(p11;q13) in childhood B-cell precursor acute lymphoblastic leukaemia treated with Berlin–Frankfurt–Munster (BFM) protocols containing an intensive induction and post-induction consolidation therapy. Br J Haematol. 2010;149(1):93–100. doi: 10.1111/j.1365-2141.2009.08059.x. [DOI] [PubMed] [Google Scholar]
- 6.Kawamata N., Ogawa S., Zimmermann M., Niebuhr B., Stocking C., Sanada M. Cloning of genes involved in chromosomal translocations by high-resolution single nucleotide polymorphism genomic microarray. Proc Natl Acad Sci USA. 2008;105(33):11921–11926. doi: 10.1073/pnas.0711039105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coyaud E., Struski S., Dastugue N., Brousset P., Broccardo C., Bradtke J. PAX5-AUTS2 fusion resulting from t(7;9)(q11.2;p13.2) can now be classified as recurrent in B cell acute lymphoblastic leukemia. Leukemia Res. 2010;34(12):e323–e325. doi: 10.1016/j.leukres.2010.07.035. [DOI] [PubMed] [Google Scholar]
- 8.Bedogni F., Hodge R.D., Nelson B.R., Frederick E.A., Shiba N., Daza R.A. Autism susceptibility candidate 2 (Auts2) encodes a nuclear protein expressed in developing brain regions implicated in autism neuropathology. Gene Expr Patterns. 2010;10(1):9–15. doi: 10.1016/j.gep.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]