Abstract
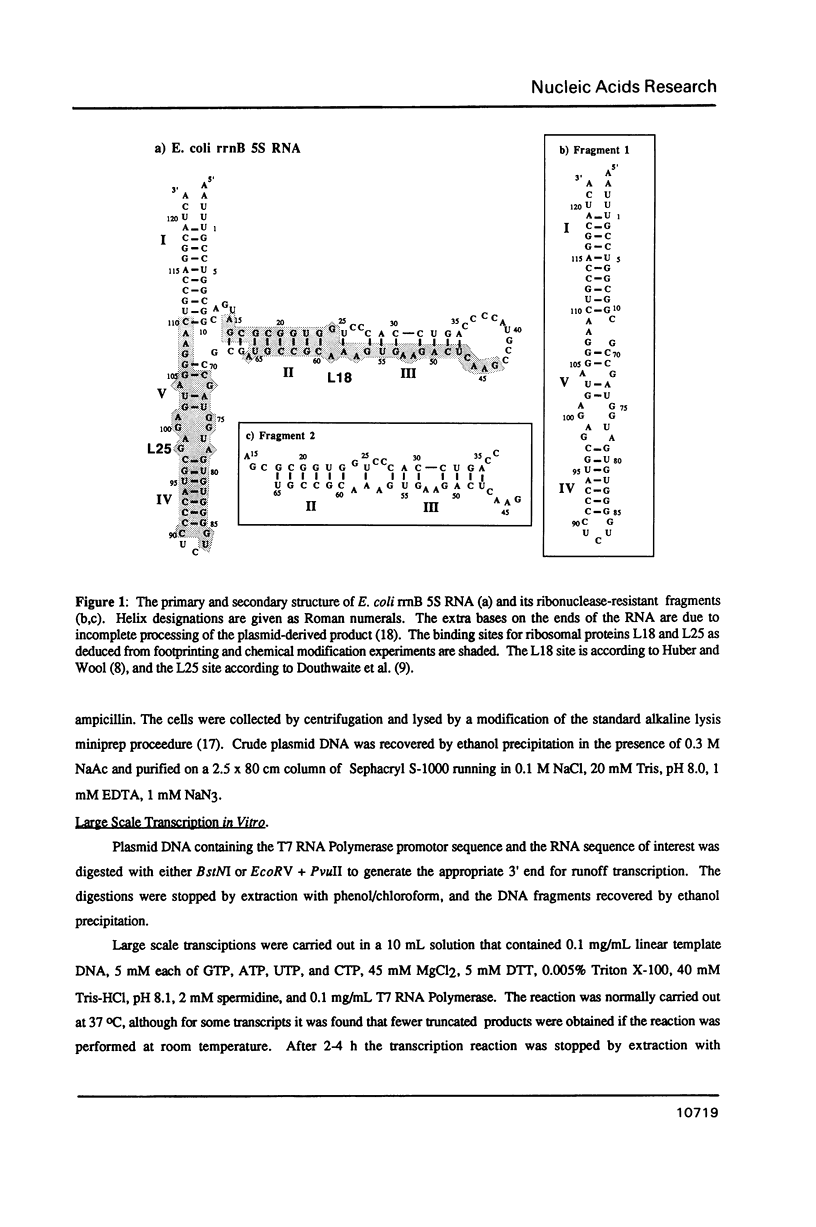
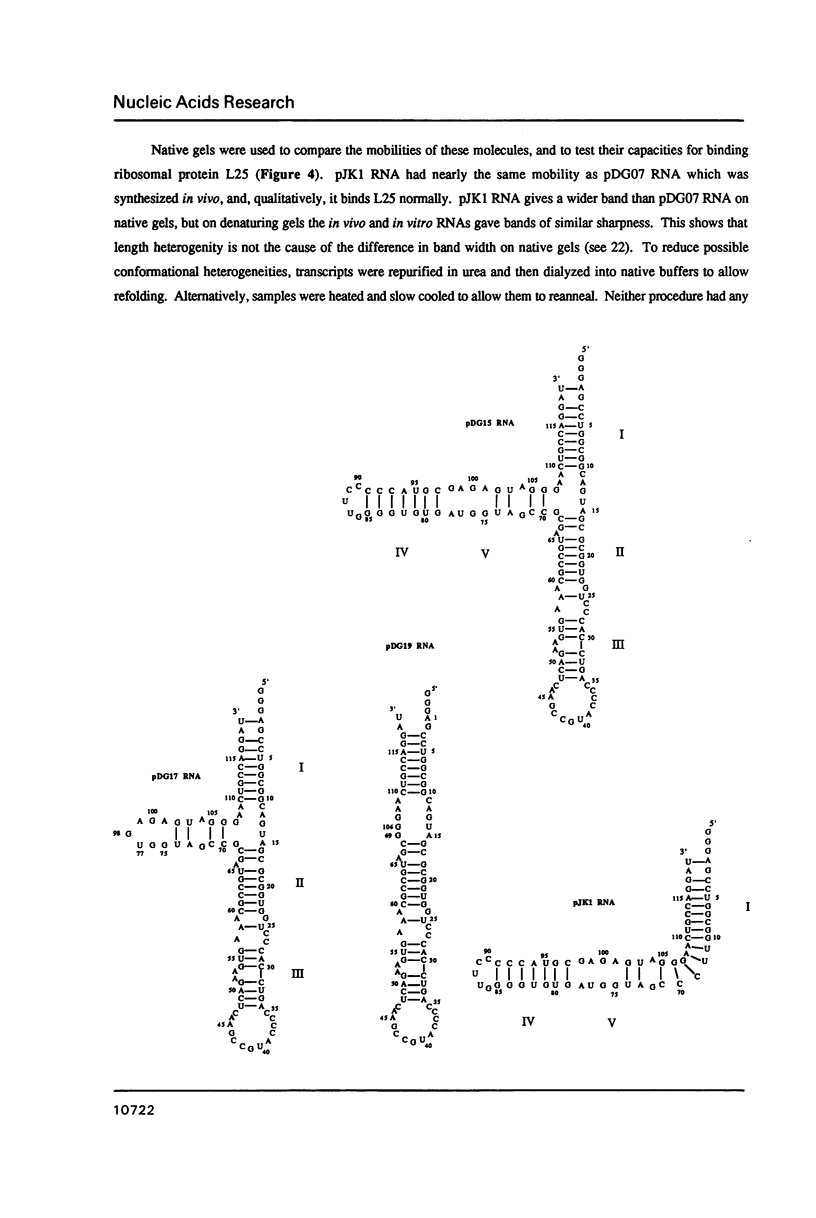
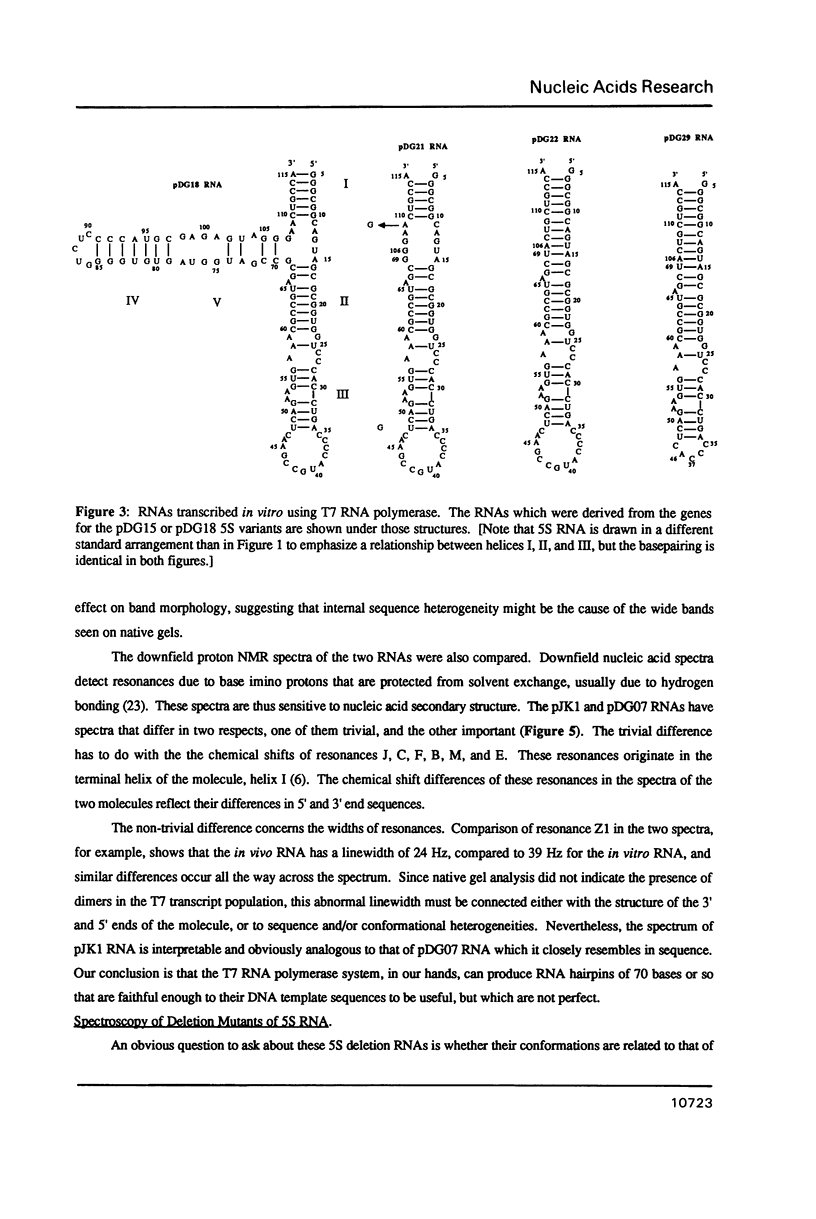
Several deletion variants of E. coli 5S RNA have been constructed and produced either in vivo or in vitro using T7 RNA Polymerase. Their structures and ribosomal protein L18 binding properties have been examined. All of them are similar to wild-type 5S RNA in their helix II-III regions, where L18 binds [Huber, P.W. and Wool, I.G. (1984) Proc. Natl. Acad. Sci. (USA) 81, 322-326; Douthwaite, S., Christensen, A., and Garrett, R.A. (1982) Biochemistry 21, 2313-2320.], by NMR criteria. However, none of the molecules examined that lack the helix IV-helix V stem bind L18 efficiently, even though that portion of 5S RNA is outside the L18 footprint. The L18 binding site is clearly more than a simple hairpin loop.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brosius J. Toxicity of an overproduced foreign gene product in Escherichia coli and its use in plasmid vectors for the selection of transcription terminators. Gene. 1984 Feb;27(2):161–172. doi: 10.1016/0378-1119(84)90137-9. [DOI] [PubMed] [Google Scholar]
- Carey J., Cameron V., de Haseth P. L., Uhlenbeck O. C. Sequence-specific interaction of R17 coat protein with its ribonucleic acid binding site. Biochemistry. 1983 May 24;22(11):2601–2610. doi: 10.1021/bi00280a002. [DOI] [PubMed] [Google Scholar]
- Carey J., Uhlenbeck O. C. Kinetic and thermodynamic characterization of the R17 coat protein-ribonucleic acid interaction. Biochemistry. 1983 May 24;22(11):2610–2615. doi: 10.1021/bi00280a003. [DOI] [PubMed] [Google Scholar]
- Christiansen J., Douthwaite S. R., Christensen A., Garrett R. A. Does unpaired adenosine-66 from helix II of Escherichia coli 5S RNA bind to protein L18? EMBO J. 1985 Apr;4(4):1019–1024. doi: 10.1002/j.1460-2075.1985.tb03733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davanloo P., Rosenberg A. H., Dunn J. J., Studier F. W. Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2035–2039. doi: 10.1073/pnas.81.7.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douthwaite S., Christensen A., Garrett R. A. Binding site of ribosomal proteins on prokaryotic 5S ribonucleic acids: a study with ribonucleases. Biochemistry. 1982 May 11;21(10):2313–2320. doi: 10.1021/bi00539a007. [DOI] [PubMed] [Google Scholar]
- Gewirth D. T., Abo S. R., Leontis N. B., Moore P. B. Secondary structure of 5S RNA: NMR experiments on RNA molecules partially labeled with nitrogen-15. Biochemistry. 1987 Aug 11;26(16):5213–5220. doi: 10.1021/bi00390a047. [DOI] [PubMed] [Google Scholar]
- Gewirth D. T., Moore P. B. Effects of mutation on the downfield proton nuclear magnetic resonance spectrum of the 5S RNA of Escherichia coli. Biochemistry. 1987 Sep 8;26(18):5657–5665. doi: 10.1021/bi00392a012. [DOI] [PubMed] [Google Scholar]
- Göringer H. U., Wagner R. Construction and functional analysis of ribosomal 5S RNA from Escherichia coli with single base changes in the ribosomal protein binding sites. Biol Chem Hoppe Seyler. 1986 Aug;367(8):769–780. doi: 10.1515/bchm3.1986.367.2.769. [DOI] [PubMed] [Google Scholar]
- Göringer H. U., Wagner R. Does 5S RNA from E. coli have a pseudoknotted structure? Nucleic Acids Res. 1986 Sep 25;14(18):7473–7485. doi: 10.1093/nar/14.18.7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber P. W., Wool I. G. Nuclease protection analysis of ribonucleoprotein complexes: use of the cytotoxic ribonuclease alpha-sarcin to determine the binding sites for Escherichia coli ribosomal proteins L5, L18, and L25 on 5S rRNA. Proc Natl Acad Sci U S A. 1984 Jan;81(2):322–326. doi: 10.1073/pnas.81.2.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kime M. J., Gewirth D. T., Moore P. B. Assignment of resonances in the downfield proton spectrum of Escherichia coli 5S RNA and its nucleoprotein complexes using components of a ribonuclease-resistant fragment. Biochemistry. 1984 Jul 17;23(15):3559–3568. doi: 10.1021/bi00310a027. [DOI] [PubMed] [Google Scholar]
- Kime M. J., Moore P. B. Nuclear Overhauser experiments at 500 MHz on the downfield proton spectra of 5S ribonucleic acid and its complex with ribosomal protein L25. Biochemistry. 1983 May 24;22(11):2622–2629. doi: 10.1021/bi00280a005. [DOI] [PubMed] [Google Scholar]
- Kime M. J., Moore P. B. Nuclear Overhauser experiments at 500 MHz on the downfield proton spectrum of a ribonuclease-resistant fragment of 5S ribonucleic acid. Biochemistry. 1983 May 24;22(11):2615–2622. doi: 10.1021/bi00280a004. [DOI] [PubMed] [Google Scholar]
- Kime M. J., Moore P. B. Physical evidence for a domain structure in Escherichia coli 5 S RNA. FEBS Lett. 1983 Mar 7;153(1):199–203. doi: 10.1016/0014-5793(83)80147-1. [DOI] [PubMed] [Google Scholar]
- Krug M., de Haseth P. L., Uhlenbeck O. C. Enzymatic synthesis of a 21-nucleotide coat protein binding fragment of R17 ribonucleic acid. Biochemistry. 1982 Sep 14;21(19):4713–4720. doi: 10.1021/bi00262a030. [DOI] [PubMed] [Google Scholar]
- Leontis N. B., Moore P. B. NMR evidence for dynamic secondary structure in helices II and III of the RNA of Escherichia coli. Biochemistry. 1986 Jul 1;25(13):3916–3925. doi: 10.1021/bi00361a027. [DOI] [PubMed] [Google Scholar]
- Lin S. Y., Riggs A. D. Lac repressor binding to non-operator DNA: detailed studies and a comparison of eequilibrium and rate competition methods. J Mol Biol. 1972 Dec 30;72(3):671–690. doi: 10.1016/0022-2836(72)90184-2. [DOI] [PubMed] [Google Scholar]
- Pace B., Matthews E. A., Johnson K. D., Cantor C. R., Pace N. R. Conserved 5S rRNA complement to tRNA is not required for protein synthesis. Proc Natl Acad Sci U S A. 1982 Jan;79(1):36–40. doi: 10.1073/pnas.79.1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peattie D. A., Douthwaite S., Garrett R. A., Noller H. F. A "bulged" double helix in a RNA-protein contact site. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7331–7335. doi: 10.1073/pnas.78.12.7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieler T., Erdmann V. A. Three-dimensional structural model of eubacterial 5S RNA that has functional implications. Proc Natl Acad Sci U S A. 1982 Aug;79(15):4599–4603. doi: 10.1073/pnas.79.15.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romaniuk P. J., Lowary P., Wu H. N., Stormo G., Uhlenbeck O. C. RNA binding site of R17 coat protein. Biochemistry. 1987 Mar 24;26(6):1563–1568. doi: 10.1021/bi00380a011. [DOI] [PubMed] [Google Scholar]
- Spierer P., Bogdanov A. A., Zimmermann R. A. Parameters for the interaction of ribosomal proteins L5, L18, and L25 with 5S RNA from Escherichia coli. Biochemistry. 1978 Dec 12;17(25):5394–5398. doi: 10.1021/bi00618a012. [DOI] [PubMed] [Google Scholar]