Abstract
The genetics of Ewing sarcoma development remain obscure. The incidence of Ewing sarcoma is ten-fold less in Africans as compared to Europeans, irrespective of geographic location, suggesting population-specific genetic influences. Since GGAA-containing microsatellites within key target genes are necessary for Ewing sarcoma-specific EWS/FLI DNA binding and gene activation, and gene expression is positively correlated with the number of repeat motifs in the promoter/enhancer region, we sought to determine if significant polymorphisms exist between African and European populations which might contribute to observed differences in Ewing sarcoma incidence and outcomes. GGAA-microsatellites upstream of two critical EWS/FLI-target genes, NR0B1 and CAV1, were sequenced from subjects of European and African descent. While the characteristics of the CAV1 promoter microsatellites were similar across both populations, the NR0B1 microsatellite in African subjects was significantly larger, harboring more repeat motifs, a greater number of repeat segments, and longer consecutive repeats, than in European subjects. These results are biologically intriguing as NR0B1 was the most highly enriched EWS/FLI bound gene in prior studies, and is absolutely necessary for oncogenic transformation in Ewing sarcoma. These data suggest that GGAA-microsatellite polymorphisms in the NR0B1 gene might influence disease susceptibility and prognosis in Ewing sarcoma in unanticipated ways.
Introduction
Ewing sarcoma is an aggressive bone-associated malignancy with a high propensity to metastasize. Unfortunately, survival rates for patients with recurrent or metastatic disease have remained stagnant at a very dismal 10-20% (1, 2). Despite an evolving understanding of the molecular mechanisms involved in Ewing sarcoma oncogenesis, molecular phenotypes predictive of disease biology and susceptibility are lacking (3). At present, detectable metastatic disease and local recurrence are the most important predictive parameters of a poor clinical outcome (4, 5).
For unknown reasons, considerable ethnic variation exists in the incidence of Ewing sarcoma: the population-specific incidence of Ewing sarcoma in the United States for European, Asian and African populations is 0.155, 0.082 and 0.017, respectively (6), which is consistent with earlier reports (7, 8). The incidence of Ewing sarcoma in African populations is 10-fold less than European populations and this discrepancy is independent of geographic location, suggesting a strong genetic influence for these observations (7). Additionally, in those with the disease, patients of European descent appear to have better clinical outcomes than patients of African, Asian and Hispanic descent (9, 10). To date, no studies have conclusively explained these epidemiological patterns of disease (3).
On a molecular level, Ewing sarcoma is characterized by balanced somatic translocations fusing the EWSR1 gene to a member of the ETS-family of transcription factors, most commonly FLI1 (11, 12). The resultant EWS/FLI fusion product functions as an aberrant transcription factor and a crucial upstream oncoprotein driving tumorigenesis in Ewing sarcoma. Consequently, prior investigations attempting to address the aforementioned ethnic patterns of disease susceptibility have focused on identifying polymorphisms within the EWSR1 locus. Intron 6 of EWSR1 is known to house a high density of Alu repeat insertions, and a truncated allele of Alu repeats has been described in populations of African descent, although at a frequency of only 8% (13). This does not conceptually explain the 10-fold decrease in disease susceptibility in African populations. Furthermore, this Alu polymorphism is located within intron 6, whereas the EWSR1 breakpoint region spans from intron 7 to intron 10 (14, 15). In another study, DuBois et al., genotyped various single nucleotide polymorphisms (SNPs) within the EWSR1 gene in patients with Ewing sarcoma, and compared these to Caucasian and African-American controls; they did not find any statistical association linking SNPs or genetic variation within intron 7 to disease susceptibility or ethnicity (16). In a recent Ewing sarcoma genome-wide association study, Postel-Vinay et al., identified 3 candidate susceptibility loci on chromosomes 1, 10 and 15 using a comprehensive SNP analysis (17). The authors demonstrate a greater frequency of these susceptibility loci in Europeans as compared to Africans. However, none of the identified susceptibility loci were in proximity to the common EWSR1 and FLI breakpoint regions or associated with direct EWS/FLI target genes. Additionally, the oncogenic importance of these recently identified susceptibility loci in the pathogenesis of Ewing sarcoma has yet to be determined. Finally, it does not appear that the differences in SNP frequency will fully account for the population-specific incidence difference in tumor development. Presently, the genetic influences involved in Ewing sarcoma susceptibility and prognosis remain uncertain and alternative explanations warrant further consideration.
Recent microarray datasets have identified various upregulated and downregulated EWS/FL1 targets (18-20). Subsequent chromatin immunoprecipitation and microarray (ChIP-chip) and ChIP followed by next-generation sequencing (ChIP-seq) datasets have demonstrated that EWS/FLI binds with high affinity to a tetra-nucleotide GGAA-microsatellite element embedded within the promoter/enhancer regions of various upregulated targets vital to the process of oncogenic transformation (21-23). These EWS/FLI “microsatellite response elements” are instrumental for EWS/FLI DNA binding and subsequent gene activation (21, 22, 24) and interestingly, an increasing number of GGAA repeats appears to substantially augment EWS/FLI-mediated gene activation (21, 24).
NR0B1 and CAV1 are two microsatellite-containing target genes that are highly bound and activated by EWS/FLI. The NR0B1 promoter is the most highly EWS/FLI-bound sequence (21-23) and aberrant expression of this orphan nuclear receptor is absolutely necessary for maintenance of oncogenic transformation in patient-derived Ewing sarcoma cell lines, characterized by anchorage independent growth, and xenograft tumor formation (19, 21, 24). Dysregulated CAV1 expression has been observed in numerous cancer types and is commonly associated with metastatic disease (25). Similar to NR0B1, aberrant expression of CAV1 is necessary for tumorigenesis in Ewing sarcoma (26).
Microsatellite DNA sequences are tandem iterations of simple nucleotide motifs dispersed throughout the genome. The majority of microsatellite DNA is comprised of mono-, di-tri- and tetra-nucleotide repeats and these repetitive elements constitute ~3% of the human genome (27). The repetitive nature of microsatellite DNA renders it more susceptible to mutagenesis and furthermore, the lack of evolutionary pressure on these non-coding regions has licensed an impressive rate of microsatellite polymorphisms in the human population over time (28). Given the mechanistic importance of microsatellite DNA in Ewing sarcoma oncogenesis, we sought to determine if significant polymorphisms of GGAA-microsatellite response elements within key EWS/FLI-target genes may exist in African and European populations that might contribute to observed differences in Ewing sarcoma incidence and outcomes. Given that European populations have a 10-fold greater incidence of Ewing sarcoma relative to African populations, and that EWS/FLI-mediated gene expression is positively correlated with an increasing number of GGAA-repeat motifs, our initial hypothesis was that subjects of European descent would have significantly larger length-polymorphisms of the NR0B1 and CAV1 GGAA-microsatellites as compared to subjects of African descent. Instead, we found a more complicated association between microsatellite characteristics in the populations analyzed.
Methods
DNA Amplification and Cloning
Genomic DNA was isolated from peripheral blood leukocytes or transformed lymphoblastic cell lines of male subjects using established extraction protocols (29, 30). Forward and reverse primers, flanking the NR0B1 and CAV1 GGAA microsatellite loci were designed using promoter sequences obtained from the University of California Santa Cruz Human Genome Browser (http://genome.ucsc.edu/cgi-bin/hgGateway). All PCR amplifications were performed using Pfu polymerase in accordance with established laboratory protocols for microsatellite DNA. Briefly, each 25μl PCR reaction consisted of 40-80ng of genomic DNA, 0.3μM of forward and reverse primers, 1U of Pfu polymerase, 0.8mM of each dNTP and 1x Pfu buffer. Melting was conducted at 94°C for 30 seconds, annealing at 62°C for 30 seconds, and extension at 72°C for 1 minute. Each PCR reaction was cycled 35 times. PCR products were resolved and visualized on a 2% agarose gel, purified, restriction digested and cloned into the pBluescript II KS(+) vector (Stratagene, La Jolla, CA). Competent DH5α E. coli cells were transformed and selected overnight on ampicillin agar. Colonies were counted the following day, with each colony representing an individual PCR-amplification clone. CAV1 is located on chromosome 7 and a high frequency of heterozygosity was anticipated, therefore sequencing using this cloning technique was preferred to ensure accurate detection of both microsatellite alleles. NR0B1 is located on the X chromosome and given all subjects in this study were male, only one allele was anticipated for each subject. However, for measures of consistency, NR0B1 microsatellites were sequenced in an identical fashion.
Microsatellite Sequencing and Analysis
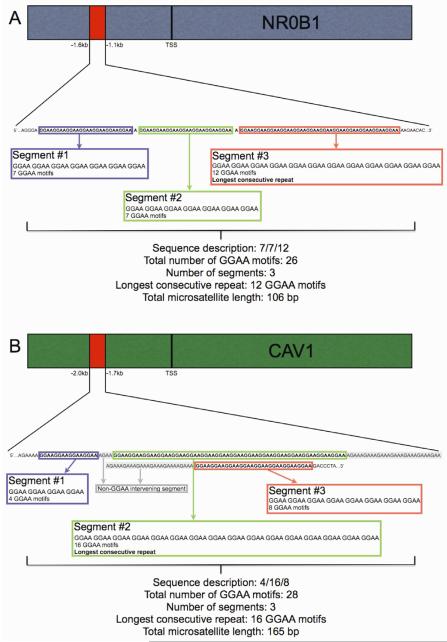
For each African and European subject, a minimum of 10 colonies were recovered, placed in glycerol solution, and commercially sequenced (Beckman Genomics, Danvers, MA). Microsatellite sequences were captured and analyzed from the raw sequence files using a custom PERL program designed to count the overall length of the microsatellite in base pairs (bp), the total number of GGAA motifs, the longest consecutive GGAA repeat, and the number of repeat segments (Figure 1A and 1B). For each subject, data collection was considered complete once a minimum of three clones with a matching sequence were obtained for any given microsatellite allele.
Figure 1.
NR0B1 and CAV1 GGAA-microsatellite sequence characteristics are visualized in panels A and B. The NR0B1 and CAV1 microsatellites are located −1.6 to −1.1kb and −2.0 to −1.7kb upstream from the transcription start site (TSS), respectively in the promoter/enhancer region. Using customized computer software, microsatellite sequences and pre-defined microsatellite characteristics were identified and quantified from the raw sequencing files. The NR0B1 GGAA-microsatellite is characterized by repeat segments separated by a single adenosine nucleotide, whereas the CAV1 GGAA-microsatellite is characterized by numerous repeat segments partitioned by a variable number of AGAA repeat motifs.
Statistical methods
A power calculation was prospectively determined using preliminary data assessing length-polymorphisms of the NR0B1 GGAA microsatellite in African and Europeans. The mean microsatellite length in these populations ranged from 322 – 342 bp with a pooled standard deviation of 45 base pairs. Using a two-sided t-test it was determined that a 100 subjects from each population would provide sufficient statistical power to detect an average difference of 20 base pairs in the overall microsatellite lengths between groups (α = 0.05, β = 0.2).
Given the anticipated heterozygosity of the CAV1 microsatellite, European and African datasets for both genes were analyzed using a linear mixed effects model (“R” statistical computing software, version 2.8.0), fit with a random effect for the alleles of each subject and a fixed effect for the population grouping variable (European vs. African). An F-test was used to compare the population variance of the data points between European and African groups and a Chi-square test was used in the analysis of proportions of the NR0B1 data. For all statistical measures, significance is defined as p < 0.05.
Results
Population demographics and cloning results
Genomic DNA from 104 European and 106 African subjects was available for assessment, respectively. African DNA samples were collected from various continental Africa ethnic populations. The Kenyan and Mbuti Pygmy samples were obtained as purified DNA and transformed cell lines, respectively, from the Coriell Institute (Camden, New Jersey, USA). European DNA samples were obtained from unrelated individuals of European descent locally in the state of Utah. The present-day Utah population remains genetically diverse and comparable to the genetic constitution of its ancestral European populations, despite a unique Mormon heritage (31). Demographics of the study participants are summarized in Table 1. Sequence data of the NROB1 and CAV1 GGAA microsatellites were obtained for 98 and 94 European and 105 and 104 African subjects, respectively. The missing data points were felt to be a result of a combination of polymorphisms at the primer binding sites and suboptimal DNA quality in a small number of samples.
Table 1.
African and European population demographics
| n | Collection location |
Ethnicity | Linguistic group | |
|---|---|---|---|---|
| Africans | ||||
| Alur | 9 | Dem. Rep. Congo |
Alur | Nilotic |
| Hema | 16 | Dem. Rep. Congo |
Hema | Nilotic |
| Mbuti Pygmy | 4 | Dem. Rep. Congo |
Pygmy | Nilo-saharan |
| Nande | 18 | Dem. Rep. Congo |
Nande | Bantu |
| South African | 29 | South Africa | Nguni, Pedi/Sotho, Tsonga, Tswana, Xhoso |
Bantu |
| Kenyan | 30 | Webuye, Kenya | Luhya | Bantu |
| Total | 106 | |||
|
| ||||
| Europeans | ||||
| Utah | 104 | Utah, USA | Northern and Western European |
Indo-European |
| Total | 104 | |||
Abbreviations: Dem. Rep. Congo – Democratic Republic of the Congo; USA – United States of America.
The primary microsatellite characteristics measured for each subject were the total number of GGAA motifs, the largest consecutive GGAA repeat segment, the number of repeat segments, and the overall microsatellite length (in base pairs; bp), including intervening base pairs and/or non-GGAA repetitive elements. Figures 1A and 1B illustrate the GGAA-microsatellite sequence characteristics of NR0B1 and CAV1, respectively. The NR0B1 microsatellite is characterized by a series of consecutive GGAA motifs separated by single adenosine base substitutions, whereas consecutive GGAA motifs in the CAV1 microsatellite are partitioned by a variable number of intervening tandem ‘AGAA’ motifs.
Consequently, the total microsatellite length of the NR0B1 gene is directly proportional to the number of GGAA motifs, whereas the CAV1 microsatellite length is dependent on the number of GGAA and AGAA motifs. The number of intervening AGAA motifs was variable for each subject. NROB1 is located on the X chromosome, and as expected, virtually all European and African subjects had a single allele. Two NR0B1 alleles were observed in 6 of 98 sequenced European subjects and 1 of 105 Africans. Another highly polymorphic microsatellite near the androgen receptor (Xq12) showed only one allele in the European individuals (data not shown) suggesting that the additional NR0B1 alleles observed in 6 of 98 Europeans may represent a duplication/amplification of the NR0B1 microsatellite on the X chromosome, or alternatively, may represent mosaicism from a somatic mutation, although this was not further analyzed. CAV1 is located on chromosome 7 and 56.4% of European samples and 65.4% of African samples were heterozygous for this locus. A high degree of heterozygosity within microsatellite loci is a common observation in African populations, which is supported by our findings (32).
CAV1 GGAA-microsatellite polymorphisms are similar in Europeans and Africans
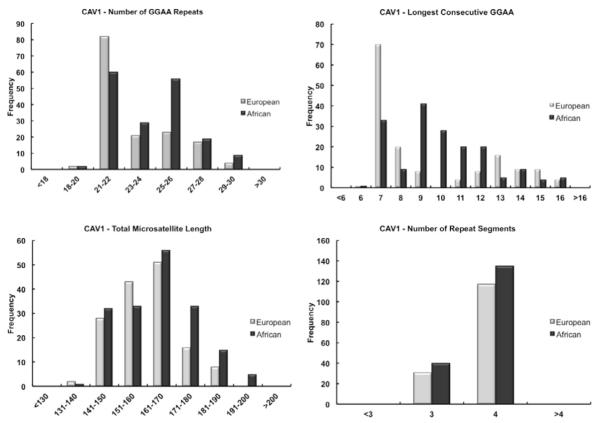
Considerable length-polymorphisms of the CAV1 GGAA-microsatellite were observed in both Europeans and Africans, confirming this microsatellite is highly polymorphic within both populations. The distributions of the primary microsatellite characteristics for CAV1 in both populations are illustrated in Figure 2. The statistical means for each microsatellite characteristic are summarized in Table 2. All measured CAV1 microsatellite characteristics were very similar when comparing the European and African groups; the average microsatellite length of both populations differed by only 4 base pairs and similarly, the total number of repeats differed by one GGAA motif. While these differences were statistically significant, this is more reflective of the number of data points used in the analysis, as these small differences are unlikely to be biologically-relevant.
Figure 2.
Histogram plots demonstrating the distribution of the CAV1 GGAA microsatellite characteristics in European and African populations. The data plots for the total number of repeats (Panel A), the total microsatellite length (Panel C) and the number of repeat segments (Panel D) show very similar distributions comparing European and African populations.
Table 2.
NR0B1 and CAV1 microsatellite characteristics
| European | African | p-value | |
|---|---|---|---|
| NR0B1 | |||
|
| |||
| Microsatellite length (bp) | 97.9 (±46) Range (65-246) |
129.8 (±62) Range (57-298) |
p < 0.0001 |
| Number of GGAA motifs | 24.0 (±11) Range (16-60) |
31.8 (±15) Range (14-72) |
p < 0.0001 |
| Longest consecutive repeat | 10.9 (±1) Range (8-16) |
12.0 (±2) Range (8-21) |
p < 0.0001 |
| Number of repeat segments | 3.0 (±1) Range (2-7) |
3.7 (±2) Range (2-9) |
p = 0.0006 |
|
| |||
| CAV1 | |||
|
| |||
| Microsatellite length (bp) | 160.5 (±11) Range (138-190) |
164.5 (±13) Range (134-194) |
p = 0.0035 |
| Number of GGAA motifs | 23.2 (±2) Range (20-29) |
24.1 (±2) Range (20-30) |
p = 0.016 |
| Longest consecutive repeat | 9.4 (±3) Range (6-16) |
10.0 (±2) Range (6-16) |
p = 0.05 |
| Number of repeat segments | 3.8 (±0.4) Range (3-4) |
3.8 (±0.4) Range (3-4) |
p = 0.68 |
Data presented mean (± standard deviation)
NR0B1 GGAA-microsatellites exhibit polymorphic differences between European and African populations
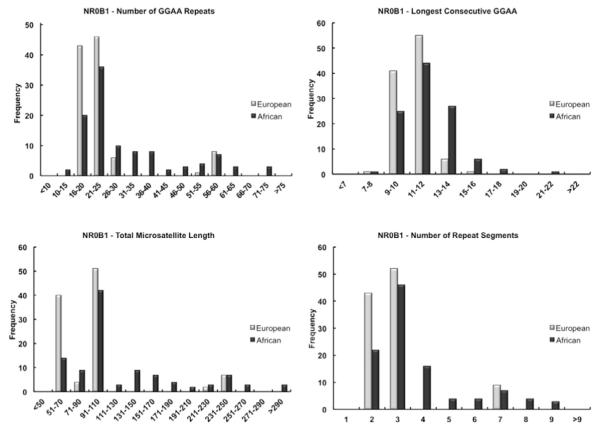
Similar to CAV1, the NR0B1 microsatellite was also highly polymorphic within both populations (Table 2). Within the African population, there was considerably more variance in the distribution of all NR0B1 microsatellite characteristics as compared to Europeans (p<0.01, F-test). In contrast to the CAV1 data, considerable population differences were observed for the mean values of the NROB1 microsatellite: compared to the Europeans, African subjects had a greater number of GGAA repeats, a longer total microsatellite length, more repeat segments, and longer consecutive GGAA repeats (Table 2). Figure 3 illustrates the distribution of the NR0B1 microsatellite characteristics in Europeans and Africans, highlighting the distribution of the African data towards larger repeat numbers and microsatellite lengths. European NR0B1 microsatellites on the other hand, are more tightly clustered around smaller sequences with less GGAA motifs and smaller consecutive repeat segments. It is important to note that the distribution of both NR0B1 and CAV1 microsatellite data observed in Figures 2 and 3 does not fit a normal distribution and in light of this finding, all comparative statistics were assessed in duplicate using a Mann-Whitney U test (“R” statistical computing software, version 2.8.0), yielding virtually identical p-values and significance as depicted in Table 2.
Figure 3.
Histogram plots demonstrating the distribution of the NR0B1 GGAA microsatellite characteristics in European and African populations. Panel A illustrates the total number of repeats, showing a predominant clustering of the European data around shorter sequences consisting of 16-25 repeats, whereas the African data is much more disperse with a greater distribution towards larger repeat numbers. Similar trends are depicted for the total microsatellite length (Panel C) and the number of repeat segments (Panel D).
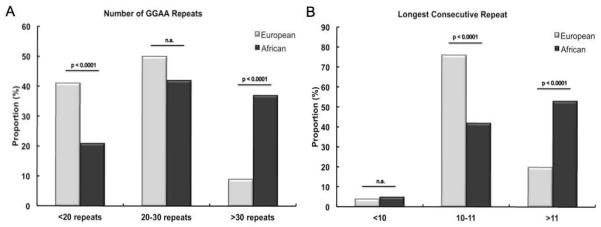
The most frequent observation in the NR0B1 GGAA-microsatellite for both European and African groups was a sequence characterized by 3 repeat segments, 24 GGAA motifs and a total microsatellite length of 98 base pairs. The anatomy of this modal sequence closely resembles the NR0B1 microsatellite observed in various Ewing sarcoma cell lines (21, 24). Based on these observations, the NR0B1 microsatellite data was further stratified into three categories: alleles of 20-30 GGAA motifs (3 repeat segments), alleles with less than 20 GGAA motifs (2 repeat segments) and alleles with greater than 30 GGAA motifs (>3 repeat segments) (Figure 4A). For NR0B1, both European and African groups had a similar proportion of 3-segment microsatellites (p=0.49, Chi-square). Interestingly, Africans had a significantly greater proportion of NR0B1 microsatellites containing > 3 repeat segments as compared to Europeans (36% versus 9%), and significantly less 2-segment microsatellites (19% versus 41%) (p< 0.001, Chi-square). The most frequent observation in both ethnic groups for the longest number of consecutive GGAA repeats was 11; therefore the NR0B1 data were stratified into repeats characterized by <10 motifs, >11 motifs and 10-11 motifs (Figure 4B). Greater than 50% of the African NR0B1 microsatellites housed a consecutive repeat of > 11 GGAA motifs, while ~20% of the European population had microsatellites that large (p<0.001, Chi-square).
Figure 4.
Stratification of the NR0B1 microsatellite data illustrating the proportion of European and African microsatellite sequences above and below the modal distribution of the total number of GGAA repeats (panel A) and the longest consecutive repeat sequence (panel B).
Discussion
Microsatellite DNA is scattered throughout genome and these repetitive sequences are highly polymorphic across ethnically distinct populations. GGAA-containing microsatellites have recently been identified as EWS/FLI-binding sites and response elements by numerous investigators (21-24, 33). With data also suggesting EWS/FLI-mediated gene expression is positively influenced by an increasing number of repeats, this novel mechanistic role of microsatellite DNA in Ewing sarcoma oncogenesis inspires alternative theories to explore the genetic influences governing disease epidemiology and biology. This is important, as presently the genetic factors governing Ewing sarcoma susceptibility and prognosis remain obscure. The 10-fold difference in the incidence of Ewing sarcoma in African populations, irrespective of geographic location suggests a strong genetic influence, and we therefore hypothesized that polymorphic differences in these microsatellite elements may provide an alternative molecular explanation for the observed ethnic patterns of susceptibility.
The data presented here clearly show that EWS/FLI-microsatellite response elements within the NR0B1 and CAV1 promoters are highly polymorphic in both European and African populations. The NR0B1 microsatellite was especially polymorphic where the number of repeats ranged from 16-60 and 14-72 in Europeans and Africans, respectively. We have also shown that the NR0B1 microsatellite in a population of African subjects is significantly larger, houses more repeat motifs, contains a greater number of repeat segments and longer consecutive repeats. Additionally, nearly 40% of African NR0B1 microsatellites have > 30 GGAA repeats as compared to 9% in Europeans. Measurements of the NR0B1 microsatellite also demonstrated significantly more genetic variance within the African population, whereas European microsatellite data was more tightly clustered around smaller, 2 or 3 segment repeats ranging from 16-25 repeat motifs.
Compared to European and Asian populations, increased genetic diversity in African populations has been observed for many microsatellite loci (32), Alu insertion polymorphisms (34), and mitochondrial DNA (35). Our results are therefore somewhat expected, although in the context of EWS/FLI-mediated oncogenesis, these observed differences are biologically intriguing. Prior in vitro studies suggest an increasing number of GGAA motifs enhances EWS/FLI-mediated gene activation in key microsatellite containing target genes (21, 24) and since Europeans have a 10-fold greater susceptibility to Ewing sarcoma, our preliminary hypothesis favored larger microsatellites in Europeans. The data presented here challenge our initial hypothesis and forge several possible interpretations: Firstly, it is still possible that larger repeats in the NR0B1 promoter confer greater EWS/FLI-mediated oncogenic potential and consequently, individuals with larger repeats would have more aggressive disease. While this does not explain the relative infrequency of Ewing sarcoma in African populations, a recent epidemiological study of 1700 patients diagnosed with Ewing sarcoma demonstrated significantly lower rates of overall survival in populations of African descent compared to Europeans (10). Although not controlling for possible confounding variables such as access to health care resources or socioeconomic issues, these observations would support the hypothesis that the larger NR0B1 microsatellite may confer a worse clinical outcome in those who develop the disease.
One of the challenges in understanding the origins of Ewing sarcoma is that a poorly defined, permissive cellular/genetic environment is required for EWS/FLI-mediated transformation (36, 37). With this in mind, an alternative hypothesis is that in the setting of endogenous EWS/FLI, the larger NR0B1 GGAA-microsatellite observed in Africans may result in NR0B1 protein levels incompatible with oncogenic transformation, for example, due to toxicity of highly-overexpressed protein. This could prevent subsequent clonal proliferation of cells harboring chromosomal translocations characteristic of Ewing sarcoma, therefore rendering African populations less susceptible to tumorigenesis.
On the other hand, it is possible that beyond a critical limit, an increasing number of GGAA repeats actually impairs EWS/FLI-mediated gene activation. The NR0B1 GGAA microsatellite has been sequenced in a variety of Ewing sarcoma cell lines and the number of GGAA motifs ranged from 17 – 26 (24). In this regard, the NR0B1 microsatellite data in the European population more closely parallels the various patient-derived cell lines investigated. However, this may simply be a result of a sampling bias, as Ewing sarcoma cell lines are statistically less likely to have originated from patients of African descent given the low incidence in this population. A recent ChIP-seq dataset, reported by Patel et al., specifically looked at the relationship of EWS/FLI-occupancy of GGAA-microsatellite containing target genes and observed maximal enrichment at microsatellites of 14 tandem GGAA motifs (23). In their series, enrichment precipitously decreased as the number of consecutive repeats approached 20. Perhaps, the relationship of microsatellite size and transcriptional activation is not linear, but rather greatest over a narrow range of repeat motifs or combinations of particular microsatellite characteristics that optimize stoichiometric requirements for EWS/FLI and co-factor occupancy. For example, the smaller 2 and 3 segment repeats more commonly observed in the European population may afford a more ideal configuration for EWS/FLI occupancy and gene activation compared to massive 60-70 repeat segments more frequently observed in Africans.
Amalgamating the interpretations of these data, it is possible that within the NR0B1 GGAA-microsatellite, a “sweet spot” exists, where a narrow, defined range of GGAA motifs is required for optimal transcriptional regulation of NR0B1 or facilitating NR0B1 protein levels permissive of EWS/FLI-mediated cell proliferation and tumorigenesis.
In the present study, we quantified 4 microsatellite characteristics including the total number of repeats, the longest consecutive repeat, the number of repeat segments and overall microsatellite length. If these observed polymorphisms are biologically relevant, it remains unclear whether EWS/FLI-mediated gene activation is dependent on the total number of repeats or the number of consecutive repeats. Presently, there is in vitro evidence to suggest both microsatellite characteristics influence EWS/FL1 DNA binding and gene activation. In a report by Gangwal et al., EWS/FLI mediated gene expression increased exponentially in synthetic expression constructs ranging from 4-7 consecutive repeats (21). Quantifying NR0B1 mRNA levels in various Ewing sarcoma cell lines demonstrated a linear increase in gene expression in cell lines with an increasing number of GGAA motifs in the NR0B1 promoter (24). Although, the precise constitution of these microsatellites as it pertains to gene expression was not further scrutinized in the latter study, this data would suggest NR0B1 gene expression is dependent on an increasing number of total GGAA repeats.
A weakness of the present study is that the microsatellite analysis performed here was limited to only two candidate genes known to be upregulated by EWS/FLI. In our recent ChIP-chip dataset, a GGAA-microsatellite response element was observed in 12 of the top 134 (9%) direct EWS/FLI targets (21). Interestingly, a GGAA-microsatellite response element was only observed in up-regulated targets. NR0B1 and CAV1 were selected based on the high relative EWS/FLI-enrichment of their respective promoter elements and the necessity of these genes for EWS/FLI-mediated oncogenesis (19, 21, 24, 26). The NR0B1 promoter was the most highly enriched EWS/FLI target (21) and dysregulation of CAV1 has been implicated in numerous other models of oncogenesis (25). We observed significant inter-ethnic polymorphisms of the NR0B1 microsatellite, but not the CAV1 microsatellite and it is possible that these observations are limited to the NR0B1 microsatellite. African populations have roughly 20% greater microsatellite diversity than Europeans (32), which in conjunction with the results presented here, warrant the characterization of other microsatellite containing genes involved in Ewing sarcomagenesis. Additionally, it is important to note that 34/106 DNA samples from the African population were obtained from transformed lymphoblastic cell lines. A potential concern is that microsatellite elements may be unstable in these transformed cell lines. To address this potential concern, European and African datasets were compared with and without the inclusion of the transformed cell line data and the population differences remained highly significant favoring larger NR0B1 microsatellites in the African population (data not shown). Finally, the DNA samples used in this study were obtained from healthy individuals without a diagnosis of cancer and therefore our results cannot be directly associated with epidemiological parameters or clinical outcomes. Regardless, the exploratory nature of this study was successful in identifying significant inter-ethnic polymorphisms of the NR0B1 GGAA-microsatellite in two populations with well-established differences in Ewing sarcoma susceptibility and clinical outcome. Consequently, these results certainly justify future functional investigations designed to further delineate how the anatomic constitution of these microsatellite sequences influence EWS/FLI-mediated tumorigenesis.
In conclusion, we have presented data demonstrating considerable length-polymorphisms of the NR0B1 and CAV1 GGAA microsatellite in both European and African populations. Additionally, we have shown that the average NR0B1 GGAA microsatellite is substantially larger in all parameters measured in African populations. Given the oncogenic importance of these GGAA microsatellite response elements, our data certainly validates the need to further assess additional microsatellite containing genes in Ewing sarcoma and more precisely understand how microsatellite polymorphisms influence disease susceptibility, EWS/FL1-mediated tumorigenesis and clinical outcomes.
Acknowledgements
The authors thank Kevin Jones for critical reading of the manuscript. MJM acknowledges support from the Orthopaedic Research and Education Fund (OREF) and SLL acknowledges support from the NIH/NCI via R01 CA140394. The authors also acknowledge support from the Huntsman Cancer Institute via P30CA042014.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Grier HE, Krailo MD, Tarbell NJ, et al. Addition of ifosfamide and etoposide to standard chemotherapy for Ewing’s sarcoma and primitive neuroectodermal tumor of bone. N Engl J Med. 2003;348:694–701. doi: 10.1056/NEJMoa020890. [DOI] [PubMed] [Google Scholar]
- 2.Balamuth NJ, Womer RB. Ewing’s sarcoma. Lancet Oncol. 2010;11:184–92. doi: 10.1016/S1470-2045(09)70286-4. [DOI] [PubMed] [Google Scholar]
- 3.Randall RL, Lessnick SL, Jones KB, et al. Is There a Predisposition Gene for Ewing’s Sarcoma? J Oncol. 2010;2010:397632. doi: 10.1155/2010/397632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cotterill SJ, Ahrens S, Paulussen M, et al. Prognostic factors in Ewing’s tumor of bone: analysis of 975 patients from the European Intergroup Cooperative Ewing’s Sarcoma Study Group. J Clin Oncol. 2000;18:3108–14. doi: 10.1200/JCO.2000.18.17.3108. [DOI] [PubMed] [Google Scholar]
- 5.Maheshwari AV, Cheng EY. Ewing sarcoma family of tumors. J Am Acad Orthop Surg. 2010;18:94–107. doi: 10.5435/00124635-201002000-00004. [DOI] [PubMed] [Google Scholar]
- 6.Jawad MU, Cheung MC, Min ES, et al. Ewing sarcoma demonstrates racial disparities in incidence-related and sex-related differences in outcome: an analysis of 1631 cases from the SEER database, 1973-2005. Cancer. 2009;115:3526–36. doi: 10.1002/cncr.24388. [DOI] [PubMed] [Google Scholar]
- 7.Polednak AP. Primary bone cancer incidence in black and white residents of New York State. Cancer. 1985;55:2883–8. doi: 10.1002/1097-0142(19850615)55:12<2883::aid-cncr2820551231>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 8.Parkin DM, Stiller CA, Draper GJ, et al. The international incidence of childhood cancer. Int J Cancer. 1988;42:511–20. doi: 10.1002/ijc.2910420408. [DOI] [PubMed] [Google Scholar]
- 9.Lee J, Hoang BH, Ziogas A, et al. Analysis of prognostic factors in Ewing sarcoma using a population-based cancer registry. Cancer. 2010;116:1964–73. doi: 10.1002/cncr.24937. [DOI] [PubMed] [Google Scholar]
- 10.Worch J, Matthay KK, Neuhaus J, et al. Ethnic and racial differences in patients with Ewing sarcoma. Cancer. 2010;116:983–8. doi: 10.1002/cncr.24865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delattre O, Zucman J, Melot T, et al. The Ewing family of tumors--a subgroup of small-round-cell tumors defined by specific chimeric transcripts. N Engl J Med. 1994;331:294–9. doi: 10.1056/NEJM199408043310503. [DOI] [PubMed] [Google Scholar]
- 12.Janknecht R. EWS-ETS oncoproteins: the linchpins of Ewing tumors. Gene. 2005;363:1–14. doi: 10.1016/j.gene.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 13.Zucman-Rossi J, Batzer MA, Stoneking M, et al. Interethnic polymorphism of EWS intron 6: genome plasticity mediated by Alu retroposition and recombination. Hum Genet. 1997;99:357–63. doi: 10.1007/s004390050372. [DOI] [PubMed] [Google Scholar]
- 14.Zucman J, Delattre O, Desmaze C, et al. Cloning and characterization of the Ewing’s sarcoma and peripheral neuroepithelioma t(11;22) translocation breakpoints. Genes Chromosomes Cancer. 1992;5:271–7. doi: 10.1002/gcc.2870050402. [DOI] [PubMed] [Google Scholar]
- 15.Plougastel B, Zucman J, Peter M, et al. Genomic structure of the EWS gene and its relationship to EWSR1, a site of tumor-associated chromosome translocation. Genomics. 1993;18:609–15. doi: 10.1016/s0888-7543(05)80363-5. [DOI] [PubMed] [Google Scholar]
- 16.Dubois SG, Goldsby R, Segal M, et al. Evaluation of polymorphisms in EWSR1 and risk of Ewing sarcoma: A report from the childhood cancer survivor study. Pediatr Blood Cancer. 2011 doi: 10.1002/pbc.23263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Postel-Vinay S, Veron AS, Tirode F, et al. Common variants near TARDBP and EGR2 are associated with susceptibility to Ewing sarcoma. Nat Genet. 2012 doi: 10.1038/ng.1085. [DOI] [PubMed] [Google Scholar]
- 18.Smith R, Owen LA, Trem DJ, et al. Expression profiling of EWS/FLI identifies NKX2.2 as a critical target gene in Ewing’s sarcoma. Cancer Cell. 2006;9:405–16. doi: 10.1016/j.ccr.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 19.Kinsey M, Smith R, Lessnick SL. NR0B1 is required for the oncogenic phenotype mediated by EWS/FLI in Ewing’s sarcoma. Mol Cancer Res. 2006;4:851–9. doi: 10.1158/1541-7786.MCR-06-0090. [DOI] [PubMed] [Google Scholar]
- 20.Tirode F, Laud-Duval K, Prieur A, et al. Mesenchymal stem cell features of Ewing tumors. Cancer Cell. 2007;11:421–9. doi: 10.1016/j.ccr.2007.02.027. [DOI] [PubMed] [Google Scholar]
- 21.Gangwal K, Sankar S, Hollenhorst PC, et al. Microsatellites as EWS/FLI response elements in Ewing’s sarcoma. Proc Natl Acad Sci U S A. 2008;105:10149–54. doi: 10.1073/pnas.0801073105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guillon N, Tirode F, Boeva V, et al. The oncogenic EWS-FLI1 protein binds in vivo GGAA microsatellite sequences with potential transcriptional activation function. PLoS One. 2009;4:e4932. doi: 10.1371/journal.pone.0004932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patel M, Simon JM, Iglesia MD, et al. Tumor-specific retargeting of an oncogenic transcription factor chimera results in dysregulation of chromatin and transcription. Genome Res. 2011 doi: 10.1101/gr.125666.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia-Aragoncillo E, Carrillo J, Lalli E, et al. DAX1, a direct target of EWS/FLI1 oncoprotein, is a principal regulator of cell-cycle progression in Ewing’s tumor cells. Oncogene. 2008;27:6034–43. doi: 10.1038/onc.2008.203. [DOI] [PubMed] [Google Scholar]
- 25.Williams TM, Lisanti MP. Caveolin-1 in oncogenic transformation, cancer, and metastasis. Am J Physiol Cell Physiol. 2005;288:C494–506. doi: 10.1152/ajpcell.00458.2004. [DOI] [PubMed] [Google Scholar]
- 26.Tirado OM, Mateo-Lozano S, Villar J, et al. Caveolin-1 (CAV1) is a target of EWS/FLI-1 and a key determinant of the oncogenic phenotype and tumorigenicity of Ewing’s sarcoma cells. Cancer Res. 2006;66:9937–47. doi: 10.1158/0008-5472.CAN-06-0927. [DOI] [PubMed] [Google Scholar]
- 27.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 28.Eckert KA, Hile SE. Every microsatellite is different: Intrinsic DNA features dictate mutagenesis of common microsatellites present in the human genome. Mol Carcinog. 2009;48:379–88. doi: 10.1002/mc.20499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jorde LB, Bamshad MJ, Watkins WS, et al. Origins and affinities of modern humans: a comparison of mitochondrial and nuclear genetic data. Am J Hum Genet. 1995;57:523–38. doi: 10.1002/ajmg.1320570340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watkins WS, Ricker CE, Bamshad MJ, et al. Patterns of ancestral human diversity: an analysis of Alu-insertion and restriction-site polymorphisms. Am J Hum Genet. 2001;68:738–52. doi: 10.1086/318793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McLellan T, Jorde LB, Skolnick MH. Genetic distances between the Utah Mormons and related populations. Am J Hum Genet. 1984;36:836–57. [PMC free article] [PubMed] [Google Scholar]
- 32.Jorde LB, Rogers AR, Bamshad M, et al. Microsatellite diversity and the demographic history of modern humans. Proc Natl Acad Sci U S A. 1997;94:3100–3. doi: 10.1073/pnas.94.7.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo W, Gangwal K, Sankar S, et al. GSTM4 is a microsatellite-containing EWS/FLI target involved in Ewing’s sarcoma oncogenesis and therapeutic resistance. Oncogene. 2009;28:4126–32. doi: 10.1038/onc.2009.262. [DOI] [PubMed] [Google Scholar]
- 34.Tishkoff SA, Dietzsch E, Speed W, et al. Global patterns of linkage disequilibrium at the CD4 locus and modern human origins. Science. 1996;271:1380–7. doi: 10.1126/science.271.5254.1380. [DOI] [PubMed] [Google Scholar]
- 35.Vigilant L, Stoneking M, Harpending H, et al. African populations and the evolution of human mitochondrial DNA. Science. 1991;253:1503–7. doi: 10.1126/science.1840702. [DOI] [PubMed] [Google Scholar]
- 36.Toomey EC, Schiffman JD, Lessnick SL. Recent advances in the molecular pathogenesis of Ewing’s sarcoma. Oncogene. 2010;29:4504–16. doi: 10.1038/onc.2010.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kovar H. Context matters: the hen or egg problem in Ewing’s sarcoma. Semin Cancer Biol. 2005;15:189–96. doi: 10.1016/j.semcancer.2005.01.004. [DOI] [PubMed] [Google Scholar]