Abstract
Background
Hepatitis B virus (HBV) is one of leading causes of various hepatic diseases including acute and chronic hepatitis, cirrhosis, and hepatocellular carcinoma. Hundreds of million people worldwide are infected by HBV, chronically.
Objectives
This study in conducted to investigate the influence of Hepatitis B virus (HBV) genotypes and type I IFN-αreceptor β subunit (IFNAR2) expression in liver on response to treatment with pegylated IFN-α-2a (Peg-IFN-α-2a) for chronic hepatitis B infection.
Patients and Methods
In this study, 65 eligible patients with chronic hepatitis B disease were enrolled. HBV genotypes of these patients were analyzed by using PCR-RFLP of the surface gene of HBV. The expression of IFNAR2 in the liver was immune histochemically investigated using anti-IFNAR2 antibody. All immune histochemical slides were read semi-quantitatively by image analysis. Chronic hepatitis B patients were treated with Peg-IFN-α2a therapy for a 48-week period and followed up for 24 weeks. Baseline characteristics and sustained viral response (SVR) to Peg-IFN-α-2a therapy were evaluated.
Results
55 % of patients exhibited HBV genotype B and 31.7 % patients exhibited HBV genotypes C infections. After treatment with Peg-IFN-α-2a, SVR was achieved in 66.7 % of patients with HBV genotype B and in 26.3 % of patients with HBV genotype C (P = 0.009). Semiquantitative and the image analysis indicated by gray level values revealed a higher IFNAR2 expression in the group with severe inflammation (P < 0.001). Patients’ high IFNAR2 protein expression had a significant impact on SVR to Peg-IFN-α-2a therapy (P = 0.028).
Conclusions
HBV genotype B and high expression of IFNAR2 in the liver of chronic hepatitis B patients are closely associated with better response to Peg-IFN-α-2a therapy in chronic hepatitis B disease.
Keywords: Hepatitis B, Chronic; IFNAR2 Protein, Human; Peginterferon alfa-2a
1. Background
Hepatitis B virus is one of leading causes of various hepatic diseases including acute and chronic hepatitis, cirrhosis, and hepatocellular carcinoma. Hundreds of million people worldwide are infected by hepatitis B virus (HBV), chronically. HBV infection is one of the most common viral infections in China. The seroepidemiological survey on HBV infection conducted in 2006 showed that hepatitis B surface antigen (HBsAg) carrier rate was 7.18 % in all population with an estimated 93 million HBsAg carriers [1]. Antiviral drugs can be used to inhibit HBV replication and thus reduce the risk of progression to cirrhosis, or liver failure or cancer. Currently, two groups of drugs approved by Federal Drug Administration for treatment of chronic hepatitis B disease consist of the immune modulators such as conventional interferon alpha (IFN-α) and pegylated interferon alpha-2a (Peg-IFN-α-2α), and nucleoside–nucleotide analogs such as lamivudine and adefovir dipivoxil. However, not all patients with chronic hepatitis B infection respond to these treatments. IFN-α elicits antiviral, immune modulatory, anti-proliferative, and gene induction properties through binding to type I interferon receptor (IFNAR) [2]. The effect of treatment of chronic hepatitis B disease with IFN-α is sustained viral suppression with HBeAg seroconversion resulted in improved clinical outcomes. Factors known to influence the response include serum HBV DNA level, alanine aminotransferase (ALT), age , grade of inflammatory activity in liver, and host factorsthat might be genetic (sex, cytokine polymorphism). However, IFN-α treatment has several disadvantages including tediousness and requirement of daily injections with high occurrence of side-effects. In order to reduce side effects and improve the efficacy, PegIFN-α-2α has been developed with one dose per week prescription [3]. Previous studies have shown that Peg-IFN-α2a is superior to IFN-α and lamivudine in the treatment of chronic hepatitis B infection [4]. The use of Peg-IFNα-2a has the advantages of limited treatment course and absence of viral resistance, which allows the use of other antiviral agents in early non-responders. Several studies have also shown that there were a combination of virological factors, host immunological factors, and genetic factors attribute to the response of hepatitis B virus to Peg-IFN-α-2a [5][6][7]. Because no antiviral treatment is perfect, early and accurate prediction of treatment response is helpful to modify therapeutic regimen based on these indications [8]. Pretreatment predictors can help physicians to select the best time of initiation as well as the type of medication to be administered. Correct choice of the first-line potent therapy is essential to achieve long-term SVR. Therefore, identifying host-dependent and virus-related factors would significantly predict sustained response to IFN in patients with chronic hepatitis B disease.
2. Objectives
The purpose of this study was to determine whether the type I IFN-α receptor β subunit (IFNAR2) expression in liver and HBV genotypes influence the response to PegIFN-α-2a (Pegasus, Roche, Basel, Switzerland) immunotherapy in chronic hepatitis B disease.
3. Patients and Methods
3.1. Subjects
Outpatients from 161 hospital of PLA and Nanfang Hospital of Southern Medical University with chronic hepatitis B disease were screened and included in this study. Our study enrolled 65 eligible patients with chronic hepatitis B infection of which five patients did not finish the treatment as a result of severity of side-effects. Exclusion criteria included patients with infections caused by other viruses, such as hepatitis A, C, D, and E viruses, cytomegalovirus, Epstein-Barr virus, and human immunodeficiency virus, and patients with autoimmune hepatitis, primary biliary cirrhosis, sclerosing cholangitis, metabolic disorders, liver cirrhosis, decompensated liver diseases, a current or past history of alcohol abuse, psychiatric conditions, previous liver transplantation, or evidence of hepatocellular carcinoma, and patients who were taking antiviral drugs or interferon before the biopsy. Patients fulfilling the inclusion and exclusion criteria were offered to participate in the study. Those patients who opted for antiviral therapy started 180 μg Peg-IFN-α-2a subcutaneously per week for 48 weeks and followed up for 24 weeks. At the end of treatment, response to Peg-IFN-α-2a therapy was defined as normalization of serum ALT levels and HBV DNA loss. Continuous response after cessation of treatment in the patients was defined as sustained viral response (SVR). Relapse is defined as undetectable HBV-DNA level in serum and normal ALT at the end of the treatment, but detectable HBV-DNA level in serum or abnormal ALT at the end of follow-up. Non-responders were defined as those with relapse and those who did not attain viral or biologic response at the end of treatment. The study protocol was approved and monitored by the ethics committee of Nanfang and 161 Hospital, and written informed consent was obtained from the patients.
3.2. Serological Tests
Sera HBeAg versus anti-HBeAg and HBsAg versus anti-HBsAg were measured using AxSYM MEI kits (Abbott laboratories). The serum HBV DNA level was determined using Roche real-time light cycle fluorescence PCR with a lower limit of detection of 1 × 103 copies/mL. Serum ALT levels were measured using commercial kits by a Hitachi Automatic Biochemical Analyzer. The remaining of sera were stored at -70 ℃.
3.3. Histological Evaluation
All liver biopsies were performed percutaneously using a 16-gauge needle without or with radiological/ultrasound guidance. Liver biopsy specimens were retrospectively reviewed by an experienced hepatopathologist blinded to patients and clinical status. The liver inflammatory grade and fibrotic stage were evaluated according to modified Knodell histological activity index [9]. Histological extent of liver injury was classified according to the involved area of tissue necrosis and inflammation: G0 no inflammation, G1 mild inflammation, G2 hepatocellular damage or moderate inflammation, G3 severe hepatocellular damage, G4 severe and widespread hepatitis with bridging confluent necrosis.
3.4. Genotyping of HBV
HBV DNA samples extracted from patients sera were subjected to polymerase chain reaction (PCR) amplification. PCR reactions were run by using HBV S gene type-specific primers and then PCR products were subjected to restriction fragment – length polymorphism (RFLP) analysis as previously described [10][11][12].
3.5. IFNAR2 Expression in Liver
The expression of IFNAR2 in the liver was detected by the use of Strept – Avidin–Biotin–Peroxidase Complex (SABC) immune histochemical staining methods. All the slides were allowed to dry at room temperature and rinsed twice with phosphate buffered saline (PBS) solution (pH 7.2), then incubated with blocking solution 3 % of H2O2 for 10 min at room temperature. Non-specific antibody binding was blocked by 1.5 % rabbit serum albumin in PBS solution for 20 min. Slides were incubated overnight at 4℃ with rabbit anti-human IFNAR2 polyclonal antibody( R and D, Minneapolis ,USA), diluted in 1:1000 in PBS. The bridging antibody (biotin-labeled goat anti-rabbit IgG) and SABC complex were diluted to 1:100 and incubated with the specimens for 20 min at room temperature. Finally color development was achieved by the use of diaminobenzidine tetrachloride and the slides were counterstained with hematoxylin. Positive rate (brown color cells) was automatically measured with the biological image analysis system 2000 (Leica, Berlin, Germany).
3.6. Statistical Analysis
The clinical quantitative data were reported as mean ± SD, and categorical variables were expressed together with frequency and percentages. The intergroup differences of quantitative variables were verified by Student’s t-test, while the differences of categorical values were analyzed by χ2 test. A multivariate analysis with a forward stepwise logistic regression model including all factors that had a P value of 0.05 in univariate analysis was then performed to identify factors predict an SVR, independently. Odds ratio (OR) with 95 % confidence interval (95 % CI) was calculated. All analyses were conducted using SPSS version 15.0 (SPSS, Chicago, IL, USA). P-values were set at 0.05 (P < 0.05) for statistically significant difference.
4. Results
4.1. Characteristics of the Patients
65 patients were enrolled and received peg-IFN-α-2a. Of these patients, five did not completed treatment because of severe adverse effects, such as depression (n = 2), irritability (n = 1), thyroiditis (n = 2); thus 60 patients who completed the study were included in the final analysis. Distribution of HBV genotypes in patients with chronic hepatitis B disease and base-line characteristics of HBV DNA genotypes were detected with PCR-RFLP and shown in Figure 1. Distribution of different HBV genotypes in patients with chronic hepatitis B disease was shown in Table 1. Genotype B was found in 33 patients (55.0 %), genotype C in 19 (31.7 %), mixed genotype (B + C) in three (5.0 %), genotypes D in three (5.0 %), and two subtypes were not recognized. Genotypes B and C were distinguished as major genotypes in this area. The baseline clinical data of patients with HBV genotypes B and C were compared in Table 2. No significant differences in male: female ratio, mean age, baseline ALT, and HBV DNA levels was observed between the two groups when classified by HBV genotype.
Figure 1.
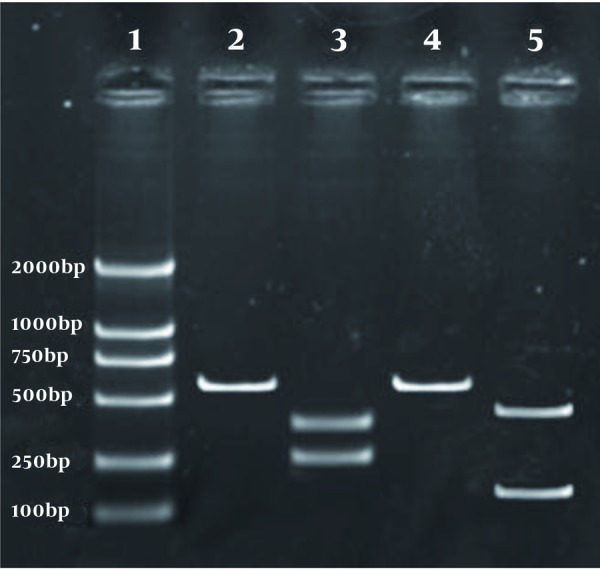
PCR-Based RFLP Analysis of HBV Genotypes (Lane 1: DNA size markers (100, 250, 500, 700, 1000, 2000 bp), Lane 2: undigested B genotype PCR product, Lane 3: StyI digestion patterns in B genotype (253bp and 332bp), Lane 4: undigested C genotype PCR product, Lane 5: BsrI digestion patterns in C genotype (126bp and 459bp))
Table 1. Distribution of Different Hepatitis B Virus Genotypes in Patients With Chronic Hepatitis B.
| Patients, No. (%) | |
| B | 33 (55.0) |
| C | 19 (31.7) |
| B + C | 3 (5.0) |
| D | 3 (5.0) |
Table 2. Baseline Characteristics of 52 Patients With Chronic Hepatitis B Virus Infection, Classified by Hepatitis B Virus Genotypes.
| HBV a Genotype B (n = 33) | HBV Genotype C (n=19) | P value | |
| Gender, No. | NS a | ||
| Male | 23 | 10 | |
| Female | 10 | 9 | |
| Age, y, mean ± SD | 31.5 ± 7.5 | 31.6 ± 9 | NS |
| ALT a level, U/L, mean ± SD | 163.5 ± 97.5 | 134.0 ± 59.6 | NS |
| HBeAg a positivity, No. (%) | 20 (60.6) | 14 (73.7) | NS |
| HBV DNA levels, log10copies/ml, mean ± SD | 9.3 ± 3.1 | 9.1 ± 1.9 | NS |
a Abbreviations: ALT: Alanine Aminotransferase, HBeAg: Hepatitis B e Antigen, NS: Not Significant
4.2. Response of HBV Genotypes B and C to Peg-IFN-α-2a Therapy
Patients with HBV genotype C exhibited significantly lower SVR to Peg-IFN-α-2a, only 26.3 %, whereas 66.7 % of patients with genotype B manifested SVR to Peg-IFN-α-2a (P = 0.009) (Table 3).
Table 3. Response to Interferon-α Therapy in Patients With Different Genotypes.
a Abbreviations: NSVR, not sustained viral response; SVR, sustained viral response
4.3. IFNAR2 Protein Expression Correlates With Liver Inflammatory Grade
IFNAR2 detected mainly in cytoplasm or cytomembrane of hepatocytes was shown in Figure 2. The IFNAR2 gray scale in G1, G2, and G3 were (5.34 ± 1.66), (6.52 ± 1.56), and (26.68 ± 4.94), respectively. There was a significant difference between G1 and G3, or G2 and G3 (P < 0.001).
Figure 2.
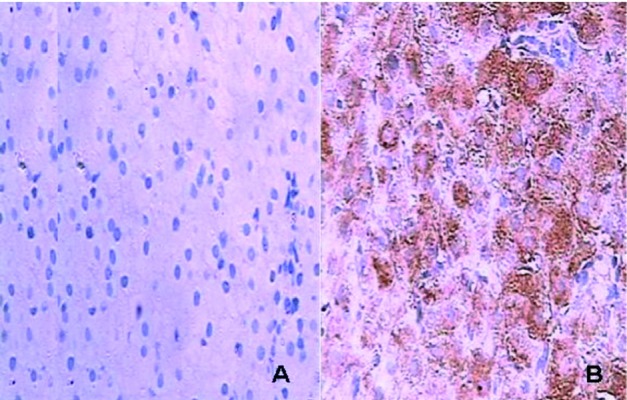
Immunostaining of IFNAR2 in Liverz (A: No background staining was observed in the negative control experiments B: IFNAR2 expressed in cytoplasm and cytomembrane of hepatocytes)
4.4. SVR to Peg-IFN-α-2a Therapy and Factors Affecting SVR
Overall, 30 patients (46.2 % by intention-to-treat analysis and 50.0 % by per protocol analysis) achieved SVR. Only those who completed the treatment were included for further analyses to identify factors affecting SVR. Patients with high IFNAR2 protein expression showed a significant impact on patient’s SVR to Peg-IFN-α-2a therapy (P = 0.028). The baseline clinical data of 60 patients with chronic hepatitis B infection are compared in Table 3. No significant differences in male: female ratio, mean age, baseline ALT, and HBV DNA levels was observed between the two groups when classified by HBeAg status. Patients with HBeAg positive sera exhibited significantly lower SVR to Peg-IFN-α-2a compared to serum negative HBeAg patients (33.3 % vs. 61.1 %, P = 0.035) (Table 4). Baseline factors associated with SVR suc h as IFNAR2 expression in the liver, ALT level, gender, age, HBeAg status, HBV genotype, and HBV DNA level were included in multiple logistic regression analysis model. This model identified three variables independently associated with increased SVR. They include IFNAR2 expression in the liver (OR = 3.80; 95 % CI: 2.54-5.70; P < 0.05), HBV-DNA level < 105copies/mL (OR = 1.70; 95 % CI: 1.14-2.56; P < 0.05), and ALT level (OR = 1.05; 95 % CI: 1.03-1.08; P = 0.036) (Table 5).
Table 4. Baseline Characteristic and Sustained Viral Response to Peg-IFN-α-2a of 60 Patients With Chronic HBV Infection, Classified by HBeAg Status.
| HBeAg a Negative (n = 36) | HBeAg Positive (n = 24) | P value | |
| Gender, No. | NS a | ||
| Male | 23 | 13 | |
| Female | 13 | 11 | |
| Age, y, mean ± SD | 32.5 ± 7.0 | 31.8 ± 9.5 | NS |
| ALT a level, U/L, mean ± SD | 164.5 ± 90.3 | 136.0 ± 79.5 | NS |
| HBV DNA levels, log10copies/ml, mean ± SD | 9.4 ± 3.0 | 9.2 ± 2.9 | NS |
| SVR a | 22.36 | 8.24 | 0.035 |
a Abbreviations: ALT, alanine aminotransferase; HBeAg, Hepatitis B e Antigen; NS, Not Significant; SVR, Sustained Viral Response
Table 5. Independent Predictors of Sustained Viral Response Identified by Multivariate Analysis.
| OR (95 % CI) | P value | |
| IFNAR2 | 3.80 (2.54-5.7) | < 0.05 |
| HBV DNA level < 105 copies/ml | 1.75 (1.14-2.56) | < 0.05 |
| ALT level | 1.05 (1.03-1.08) | < 0.05 |
5. Discussion
Chronic hepatitis B is a potentially severe form of liver disease that often progresses to cirrhosis, hepatic decompensating, and hepatocellular carcinoma [13]. Active-viral replication is always associated with rapid disease progression. As a consequence, optimal therapy with antiviral drugs may achieve maximal virologic suppression to improve the clinical outcome. The present study aimed to investigate the role of HBV genotypes and IFNAR2 in response to Peg-IFN-α-2a in Chinese patients with hepatitis B. Until now, based on an intergroup divergence of 8 % or more in the complete nucleotide sequence, HBV can be classified into nine genotypes (A-I) [14][15][16][17]. It has been shown that HBV genotypes have a characteristic geographic distribution [18]. In addition, several lines of evidence indicate that there are different sustained responses to conventional interferon in patients with different genotypes. Studies suggested that genotype A is associated with a higher rate of IFN-α-induced HBeAg seroconversion than genotype D [18], and genotype B is associated with a higher rate of IFN-α-induced HBeAg clearance than genotype C [19]. Similarly, our study revealed that HBV genotype C was associated with a lower rate of antiviral response to Peg-IFN-α-2a treatment in Chinese patients with chronic hepatitis B disease compared to genotype B (26.3% vs. 66.7 %, P = 0.009). Further studies with enlarged sample sizes are needed to validate our results and the prevalence of mixed HBV genotype B and C infections and their implications for prognosis and treatment. In addition to HBV genotype factor, our study investigated whether expression levels of IFNAR2 in the liver of chronic hepatitis B patients are correlated with the response to Peg-IFN-α-2a therapy. IFNs are known to bind to receptors on the cell surface that facilitate activation of Janus kinase/signal transduction pathway that result in activation of other transcription pathways [20][21]. IFNAR mRNA expression in liver of patients with chronic hepatitis B infection has been shown to be related to the response to IFN-α therapy. Absent or low intrahepatic IFNAR mRNA levels are related to a poor response to IFN-α [22][23]. Our study demonstrated that expression of IFNR2 protein had a close correlation with hepatic inflammation. Moreover, the majority of responders with chronic hepatitis B exhibited a high level of IFNR2 protein expression prior to treatment, which conforms to other previous studies [24][25]. To the best of our knowledge, this is the first study performed in China to determine both viral genotype and expression level of host IFNR2 that are closely correlated with the response to Peg-IFN-α-2a therapy in patients with chronic hepatitis B disease. In addition to IFNAR2, there are numerous genes involved in interferon-alpha pathway. Future studies are needed to identify those genes involved in Peg-IFN-α-2a therapy. HBeAg-negative in chronic hepatitis B disease usually represents at later phases of disease progression. HBeAg clearance may follow an exacerbation of hepatitis manifested by ALT level elevation [26]. Progression to cirrhosis or liver cancer was frequently observed in these patients [27]. Antiviral treatment, either by long-term nucleoside– nucleotide analogs or administration of IFN, is therefore necessary to inhibit course of disease. However, viral responses to standard IFN or nucleoside–nucleotide therapy were occurred in low rates among these patients. In our study, SVR to Peg-IFN-α-2a was achieved in approximately 50% of HBeAg-negative patients, with a significant rates compared to HBeAg-positive patients (33.3 % vs. 61.1 %). Therefore, Peg-IFN is a better choice for HBeAg-negative patients.
In conclusion, HBV genotype B and high expression of IFNAR2 in liver are closely associated with better response to Peg-IFN-α-2a therapy among Chinese patients with chronic hepatitis B disease. Therefore, recognition of these patients before suitable antiviral therapy is very important to judge on the treatment outcome and prognosis.
Acknowledgments
None Declared.
Footnotes
Implication for health policy/practice/research/medical education: Hepatitis B Virus genotype B and high expression of interferon alpha receptor β subunit may be considered as a predictor of response to interferon antiviral therapy in HBV patients and study of this article is recommended to the hepatologists, virologists, gastroentestinalist and other researchers.
Please cite this paper as: Fan HB, Guo YB, Zhu YF, Chen AS, Zhou MX, Li Z, et al. Hepatitis B Virus Genotype B and High Expression of Interferon Alpha Receptor β Subunit are Associated With Better Response to Pegylated Interferon Alpha 2a in Chinese Patients With Chronic Hepatitis B Infection. Hepat Mon. 2012;12(5): 333-8. DOI: 10.5812/hepatmon.6173
Authors’ Contribution: All authors were involved in all steps of manuscript preparation.
Financial Disclosure: None Declared.
Funding Support: None Declared.
References
- 1.Lu FM, Zhuang H. Prevention of hepatitis B in China: achievements and challenges. Chin Med J (Engl). 2009;122(24):2925–7. [PubMed] [Google Scholar]
- 2.Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5(5):375–86. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 3.Hui AY, Chan HL, Cheung AY, Cooksley G, Sung JJ. Systematic review: treatment of chronic hepatitis B virus infection by pegylated interferon. Aliment Pharmacol Ther. 2005;22(6):519–28. doi: 10.1111/j.1365-2036.2005.02616.x. [DOI] [PubMed] [Google Scholar]
- 4.Ratnam D, Dev A, Nguyen T, Sundararajan V, Harley H, Cheng W, et al. The Efficacy and Tolerability of Pegylated Interferon-alpha-2a in Chronic Hepatitis B: A Multicenter Clinical Experience. J Gastroenterol Hepatol. 2011:[Epub ahead of print]. doi: 10.1111/j.1440-1746.2011.07051.x. [DOI] [PubMed] [Google Scholar]
- 5.Takkenberg B, Terpstra V, Zaaijer H, Weegink C, Dijkgraaf M, Jansen P, et al. Intrahepatic response markers in chronic hepatitis B patients treated with peginterferon alpha-2a and adefovir. J Gastroenterol Hepatol. 2011;26(10):1527–35. doi: 10.1111/j.1440-1746.2011.06766.x. [DOI] [PubMed] [Google Scholar]
- 6.Tian Y, Chen WL, Ou JH. Effects of interferon-alpha/beta on HBV replication determined by viral load. PLoS Pathog. 2011;7(7):e1002159. doi: 10.1371/journal.ppat.1002159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tseng TC, Yu ML, Liu CJ, Lin CL, Huang YW, Hsu CS, et al. Effect of host and viral factors on hepatitis B e antigen-positive chronic hepatitis B patients receiving pegylated interferon-alpha-2a therapy. Antivir Ther. 2011;16(5):629–37. doi: 10.3851/IMP1841. [DOI] [PubMed] [Google Scholar]
- 8.Keeffe EB, Zeuzem S, Koff RS, Dieterich DT, Esteban-Mur R, Gane EJ, et al. Report of an international workshop: Roadmap for management of patients receiving oral therapy for chronic hepatitis B. Clin Gastroenterol Hepatol. 2007;5(8):890–7. doi: 10.1016/j.cgh.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 9.Knodell RG, Ishak KG, Black WC, Chen TS, Craig R, Kaplowitz N, et al. Formulation and application of a numerical scoring system for assessing histological activity in asymptomatic chronic active hepatitis. Hepatology. 1981;1(5):431–5. doi: 10.1002/hep.1840010511. [DOI] [PubMed] [Google Scholar]
- 10.Sugauchi F, Kumada H, Sakugawa H, Komatsu M, Niitsuma H, Watanabe H, et al. Two subtypes of genotype B (Ba and Bj) of hepatitis B virus in Japan. Clin Infect Dis. 2004;38(9):1222–8. doi: 10.1086/382885. [DOI] [PubMed] [Google Scholar]
- 11.Tanaka Y, Orito E, Yuen MF, Mukaide M, Sugauchi F, Ito K, et al. Two subtypes (subgenotypes) of hepatitis B virus genotype C: A novel subtyping assay based on restriction fragment length polymorphism. Hepatol Res. 2005;33(3):216–24. doi: 10.1016/j.hepres.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 12.Zeng GB, Wen SJ, Wang ZH, Yan L, Sun J, Hou JL. A novel hepatitis B virus genotyping system by using restriction fragment length polymorphism patterns of S gene amplicons. World J Gastroenterol. 2004;10(21):3132–6. doi: 10.3748/wjg.v10.i21.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ganem D, Prince AM. Hepatitis B virus infection--natural history and clinical consequences. N Engl J Med. 2004;350(11):1118–29. doi: 10.1056/NEJMra031087. [DOI] [PubMed] [Google Scholar]
- 14.Arauz-Ruiz P, Norder H, Robertson BH, Magnius LO. Genotype H: a new Amerindian genotype of hepatitis B virus revealed in Central America. J Gen Virol. 2002;83(Pt 8):2059–73. doi: 10.1099/0022-1317-83-8-2059. [DOI] [PubMed] [Google Scholar]
- 15.Olinger CM, Jutavijittum P, Hubschen JM, Yousukh A, Samountry B, Thammavong T, et al. Possible new hepatitis B virus genotype, southeast Asia. Emerg Infect Dis. 2008;14(11):1777–80. doi: 10.3201/eid1411.080437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stuyver L, De Gendt S, Van Geyt C, Zoulim F, Fried M, Schinazi RF, et al. A new genotype of hepatitis B virus: complete genome and phylogenetic relatedness. J Gen Virol. 2000;81(Pt 1):67–74. doi: 10.1099/0022-1317-81-1-67. [DOI] [PubMed] [Google Scholar]
- 17.Tran TT, Trinh TN, Abe K. New complex recombinant genotype of hepatitis B virus identified in Vietnam. J Virol. 2008;82(11):5657–63. doi: 10.1128/JVI.02556-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin CL, Kao JH. The clinical implications of hepatitis B virus genotype: Recent advances. J Gastroenterol Hepatol. 2011;26 (Suppl 1):123–30. doi: 10.1111/j.1440-1746.2010.06541.x. [DOI] [PubMed] [Google Scholar]
- 19.Wai CT, Chu CJ, Hussain M, Lok AS. HBV genotype B is associated with better response to interferon therapy in HBeAg(+) chronic hepatitis than genotype C. Hepatology. 2002;36(6):1425–30. doi: 10.1053/jhep.2002.37139. [DOI] [PubMed] [Google Scholar]
- 20.Aaronson DS, Horvath CM. A road map for those who don't know JAK-STAT. Science. 2002;296(5573):1653–5. doi: 10.1126/science.1071545. [DOI] [PubMed] [Google Scholar]
- 21.Muller U, Steinhoff U, Reis LF, Hemmi S, Pavlovic J, Zinkernagel RM, et al. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264(5167):1918–21. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 22.Gong QM, Kong XF, Yang ZT, Xu J, Wang L, Li XH, et al. Association study of IFNAR2 and IL10RB genes with the susceptibility and interferon response in HBV infection. J Viral Hepat. 2009;16(9):674–80. doi: 10.1111/j.1365-2893.2009.01130.x. [DOI] [PubMed] [Google Scholar]
- 23.Ishimura N, Fukuda R, Fukumoto S. Relationship between the intrahepatic expression of interferon-alpha receptor mRNA and the histological progress of hepatitis C virus-associated chronic liver diseases. J Gastroenterol Hepatol. 1996;11(8):712–7. doi: 10.1111/j.1440-1746.1996.tb00319.x. [DOI] [PubMed] [Google Scholar]
- 24.Ferreira PR, Tenore Sde B. Response predictors to treatment with pegylated interferon in chronic hepatitis B. Braz J Infect Dis. 2010;14(5):519–25. doi: 10.1590/S1413-86702010000500017. [DOI] [PubMed] [Google Scholar]
- 25.Fujiwara D, Hino K, Yamaguchi Y, Ren F, Satoh Y, Korenaga M, et al. Hepatic expression of type I interferon receptor for predicting response to interferon therapy in chronic hepatitis C patients: a comparison of immunohistochemical method vs. competitive polymerase chain reaction assay. Hepatol Res. 2003;25(4):377–84. doi: 10.1016/S1386-6346(02)00312-1. [DOI] [PubMed] [Google Scholar]
- 26.Lok AS, McMahon BJ. Chronic hepatitis B. Hepatology. 2007;45(2):507–39. doi: 10.1002/hep.21513. [DOI] [PubMed] [Google Scholar]
- 27.Lampertico P, Del Ninno E, Vigano M, Romeo R, Donato MF, Sablon E, et al. Long-term suppression of hepatitis B e antigen-negative chronic hepatitis B by 24-month interferon therapy. Hepatology. 2003;37(4):756–63. doi: 10.1053/jhep.2003.50148. [DOI] [PubMed] [Google Scholar]


