Abstract
The largest gene cluster of human microRNAs (miRNAs), the chromosome 19 miRNA cluster (C19MC), is exclusively expressed in the placenta and in undifferentiated cells. The precise expression pattern and function of C19MC members are unknown. We sought to profile the relative expression of C19MC miRNAs in primary human trophoblast (PHT) cells and exosomes. Using high-throughput profiling, confirmed by PCR, we found that C19MC miRNAs are among the most abundant miRNAs in term human trophoblasts. Hypoxic stress selectively reduced miR-520c-3p expression at certain time-points with no effect on other C19MC miRNAs. Similarly, differentiation in vitro had a negligible effect on C19MC miRNAs. We found that C19MC miRNAs are the predominant miRNA species expressed in exosomes released from PHT, resembling the profile of trophoblastic cellular miRNA. Predictably, we detected the similar levels of circulating C19MC miRNAs in the serum of healthy pregnant women at term and in women with pregnancies complicated by fetal growth restriction. Our data define the relative expression levels of C19MC miRNAs in trophoblasts and exosomes, and suggest that C19MC miRNAs function in placental–maternal signaling.
Keywords: trophoblasts, microRNAs, C19MC, exosome, hypoxia
Introduction
The trophoblasts at the surface of human placental villi play a pivotal role in placental gas exchange, nutrition, waste removal, endocrine function and immunological support for the developing fetus. The terminally differentiated, post-mitotic syncytiotrophoblasts are directly bathed in maternal blood, and overlay a discontinuous layer of less-differentiated cytotrophoblasts. Whereas early placental development occurs in an environment of relative hypoxia, placental hypoxia beyond the first trimester is associated with villous trophoblast injury and with changes in gene expression that are implicated in fetal growth restriction (FGR) (Genbacev et al., 1997; Levy et al., 2000; Roh et al., 2005; Chen et al., 2006; Rimon et al., 2008; Simon and Keith, 2008; Pringle et al., 2010; Struwe et al., 2010).
MicroRNAs (miRNAs) are endogenous, 19–24 nucleotide, non-coding RNAs that post-transcriptionally regulate gene expression through sequence-specific base-pairing with target mRNAs. In the human placenta, miRNAs are synthesized and expressed primarily in trophoblasts, where they regulate cellular function and adaptation to stress (Barad et al., 2004; Bentwich et al., 2005; Donker et al., 2007; Luo et al., 2009; Leung and Sharp, 2010; Mouillet et al., 2010b). In addition to their intracellular function, secreted miRNAs may play an important role in intercellular communication by repressing mRNA targets in neighboring or distant cells (Pegtel et al., 2010; Zhang et al., 2010). Although the precise mechanisms of miRNA release is poorly understood, the miRNA effector protein Ago2 and high-density lipoprotein cholesterol, as well as microvesicles were identified as carriers of circulating miRNAs, and likely contribute to miRNA stability in the circulation (Valadi et al., 2007; Arroyo et al., 2011; Vickers et al., 2011). Trophoblast cell lines and first trimester primary human trophoblasts (PHTs) are capable of secreting the exosome nanovesicles (Frangsmyr et al., 2005; Redman and Sargent, 2007; Valadi et al., 2007; Thery et al., 2009; Mincheva-Nilsson and Baranov, 2010; Atay et al., 2011), which are known to contain miRNAs (Valadi et al., 2007; Luo et al., 2009). The production of exosomes by third trimester trophoblasts and their miRNA content have not been previously examined.
The chromosome 19 miRNA cluster (C19MC) is the largest human miRNA gene cluster, comprising roughly 8% of all known human miRNA genes, and is exclusively expressed in the placenta and in undifferentiated cells (Bentwich et al., 2005; Landgraf et al., 2007; Bortolin-Cavaille et al., 2009; Lin et al., 2010). This primate-specific miRNA cluster spans ∼100 kb at human chromosome 19q13.41, and spans 46 miRNA genes (Bortolin-Cavaille et al., 2009; Lin et al., 2010). In the human placenta, the C19MC cluster is imprinted, and is exclusively expressed from the paternally inherited allele, likely governed by an upstream CG nucleotide rich promotor region (Noguer-Dance et al., 2010). C19MC miRNAs are intron-encoded, and processed by the DGCR8-Drosha complex from a non-protein coding Pol-II transcript (Bortolin-Cavaille et al., 2009), rather than RNA Pol-III as initially thought (Noguer-Dance et al., 2010). C19MC miRNAs include two subfamilies, defined by sequence homology (Lin et al., 2010). Aberrant expression of C19MC miRNAs is observed in specific human malignancies, where they may contribute to tumor invasiveness (Li et al., 2009; Rippe et al., 2010).
We sought to interrogate the expression of C19MC miRNAs in primary term human trophoblasts and provide an extensive characterization of C19MC miRNAs profiles in PHT cells and exosomes.
Materials and Methods
Participants
The study was approved by the University of Pittsburgh's Institutional Review Board. Placentas were obtained after term singleton delivery. Plasma samples were collected from non-pregnant women, pregnant women with a singleton healthy pregnancy or pregnancy complicated by FGR, as described previously (Mouillet et al., 2010b). FGR was defined as birthweight <10th percentile after exclusion of pregnancies complicated by systemic maternal diseases that might be associated with placental abnormalities. Clinical characteristics of participants are listed in Table I. All newborns were delivered by vaginal delivery.
Table I.
Maternal and neonatal characteristicsa.
| Normal pregnancy (n = 14) | FGR (n = 14) | P-value | |
|---|---|---|---|
| Maternal age (years) (mean ± SD) | 26 ± 5.8 | 24.3 ± 5.3 | NS |
| African-Americanb (n, %) | 5 (36%) | 5 (36%) | NS |
| Pre-pregnancy BMI (kg/m2) (mean ± SD) | 24.8 ± 4.7 | 23.8 ± 6.4 | NS |
| Gestational age at delivery (weeks) (mean ± SD) | 39.9 ± 1.0 | 39.8 ± 2.0 | NS |
| Neonatal weight (g) (mean ± SD) | 3452 ± 456 | 2582 ± 131 | P < 0.01 |
| Neonatal weight (percentile) (mean ± SD) | 58.4 ± 19.9 | 3.4 ± 2.7 | P < 0.01 |
| Apgar score <7 at 5 min | 0 | 0 | NS |
| Neontal sex (female) (n, %) | 7 (50%) | 8 (57%) | NS |
| Smoking during pregnancy (n, %) | 0 (0%) | 1 (7%) | NS |
| Alcohol use during pregnancy (n, %) | 1 (7%) | 1 (7%) | NS |
| No alcohol use one year prior to pregnancyc (n, %) | 5 (36%) | 4 (29%) | NS |
| No post-secondary education (n, %) | 3 (21%) | 4 (28%) | NS |
| Receiving public assistance (n, %) | 1 (7%) | 3 (21%) | NS |
aThe mean age of women in the non-pregnant control group was 26.1 ± 3.0 years (n = 7).
bAll other participants were white, except for one participant with FGR who was of Pacific Islander origin.
cNone of the participants was alcoholic. Several reported occasional alcohol use before pregnancy, but none exceeded three drinks per week. All drug users were excluded from the study.
Cell culture
PHT cells were prepared from normal term placentas using the trypsin-deoxyribonuclease-dispase/Percoll method as described by Kliman et al. (1986), with previously published modifications (Nelson et al., 1999). PHT, human placental choriocarcinoma cells JEG-3 and BeWo were grown and maintained as described previously (Donker et al., 2007; Mouillet et al., 2010b). After a 4-h period, designed to allow cell attachment, the culture plates in some of the paradigms were allocated to either standard (O2 = 20%) or hypoxic (O2 <1%) environment (Mouillet et al., 2010b). Differentiation of PHT cells in culture was routinely monitored by medium hCG levels, showing a characteristic increase in medium hCG as cytotrophoblasts differentiate into syncytiotrophoblasts, with attenuation of this process in hypoxic cells (Nelson et al., 1999; Chen et al., 2006).
RNA isolation and miRNA real-time RT-qPCR
Total cellular RNA was purified from cells, exosomes, culture supernatants (300 μl) or serum samples (600 μl) using miRNeasy Mini Kit (Qiagen, Valencia, CA, USA), according to the manufacturer's instructions. Exosomes were treated with 400 ng/ml RNase at 37°C for 15 min to confirm that only exosomal RNA and not medium RNA is extracted prior to RNA isolation from 25 to 40 μg of purified exosomes (Valadi et al., 2007).
For miRNA analysis, reverse transcription and quantitative PCR (RT-qPCR) of duplicate samples were performed using the miScript PCR system (Qiagen), following the manufacturer's instructions. miScript primers were used to detect expression of miR-512-5p, miR-517a, miR-518a-5p, miR-518b, miR-518c*, miR-518e, miR-519d, miR-520c-3p, miR-525-5p and RNU6B (Qiagen: MS00007007, MS00004459, MS00009961, MS00004466, MS00007070, MS00004487, MS00004508, MS00007413, MS00004557, MS00014000). Dissociation curves were run on all reactions to ensure amplification of a single product. Control samples of H2O were included for both RT and PCR in each experiment. Total RNA input was normalized using RNU6B RNA as an endogenous control, except for miRNA measurements in serum. The fold increase relative to control samples was determined by the 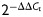 method (Livak and Schmittgen, 2001).
method (Livak and Schmittgen, 2001).
MiRNA microarray and NanoString miRNA expression analysis
Total cellular RNA was purified from cells using miRNeasy Mini Kit (Qiagen) and total cellular RNA (100 ng) was used for human miRNA microarray analysis (Agilent Technologies, Santa Clara, CA, USA), both according to the manufacturer's instructions. Because this preparation of RNA includes 3.9 ng of miRNA (range 10–40 nucleotides BioAnalyzer 2100, Agilent), we used the same amount of exosomal miRNA as input for miRNA microarray. Array data were extracted on a High-Resolution C scanner (Agilent Technologies). We also extracted total RNA as described above for human miRNA expression analysis, which was performed by NanoString Technologies (Seattle, WA, USA).
Exosome purification
Cells were maintained in Dulbecco's modified Eagle's medium (Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10% exosome-free fetal bovine serum (FBS, Hyclone, Logan, UT, USA) and antibiotics. FBS was depleted of exosomes by overnight ultracentrifugation (100 000g). Exosomes were isolated as described previously (Montecalvo et al., 2008; Rani et al., 2011). Briefly, supernatants from 200 to 250 × 106 PHT cells were pooled and subsequently centrifuged at 300g for 5 min, 1200g for 10 min and 10 000g for 30 min. Exosomes were concentrated by centrifugation at 2500g for 25 min using a Vivacell 100 filter (BioExpress, Kaysville, UT, USA). The filtered supernatant was ultracentrifuged at 100 000g for 1 h, and the exosome pellet was subsequently ultracentrifuged on top of a 30% sucrose/D2O density cushion at 100 000g for 1 h (Lamparski et al., 2002). The phase containing the exosomes was collected and resuspended in phosphate-buffered saline, and then ultracentrifuged at 100 000g for 1 h. The amount of exosome protein was measured with a NanoDrop 1000 (Thermo Scientific, Lafayette, CO, USA).
Electron microscopy
Exosomes were loaded on Formvar/carbon-coated grids, negatively stained with 1% uranyl acetate and examined with a JEM-1011 transmission electron microscope (JEOL, Peabody, MA, USA) fitted with an AMT digital camera (Danvers, MA, USA).
Western immunoblotting
Cells were lysed in buffer containing 0.1% SDS, 0.5% sodium deoxycholate, 1% Triton X-100, 1 mM phenylmethylsulphonyl fluoride and Halt protease inhibitor cocktail (Thermo Scientific). Cellular protein concentration was determined by bicinchoninic acid protein assay (Pierce, Thermo Scientific), according to the manufacturer's instructions. Proteins (30–40 µg) were resolved using 10% SDS–PAGE. The proteins were transferred to polyvinylidene difluoride membranes (Immobilon-P, Millipore, Bedford, MA, USA), and membranes were blocked by incubation in 10% non-fat dry milk in tris-buffered saline with Tween-20 (TBST) (0.01 M TBS with 0.05% Tween-20). Next, the membranes were incubated with mouse anti-human tumor susceptibility gene 101 (TSG101) antibody (1 μg/ml, Santa Cruz Biotechnology, Santa Cruz, CA, USA), followed by horse-radish peroxidase-conjugated goat anti-mouse immunoglobulin G (Jackson ImmunoResearch Laboratories, West Grove, PA, USA). The blots were washed and processed for chemiluminescence using SuperSignal West Pico (Thermo Scientific), and analyzed using Epichemi-3 software (UVP BioImaging, Upland, CA, USA).
Statistics
All experiments were repeated at least three times. Data were expressed as mean ± SD. qPCR data were computed as fold using geometric means and normalized to the mean value of the control sample in each paradigm, defined as one. The data were analyzed using analysis of variance or Student's t-test (Stata, release 11, StataCorp LP, College Station, TX, USA), where appropriate. Significance was defined as P < 0.05. The data in Table I were analyzed using Student's t-test or χ2 test (Stata), where appropriate.
For microarray analysis, the RMA (robust multi-array average) method (Irizarry et al., 2003), as implemented in R package AgiMicroRna (Lopez-Romero, 2011), was used to obtain the summarized and normalized miRNA expression level. The R package Limma (Linear Models for Microarray data), which implements a moderated t-test, was used to identify differentially expressed miRNAs (Smyth, 2004). The Storey's q-value method (Storey and Tibshirani, 2003), as implemented in R package q-value, was used to calculate the adjusted P-values in order to control the false discovery rate.
Results
C19MC miRNAs comprise a major part of miRNAs in PHTs
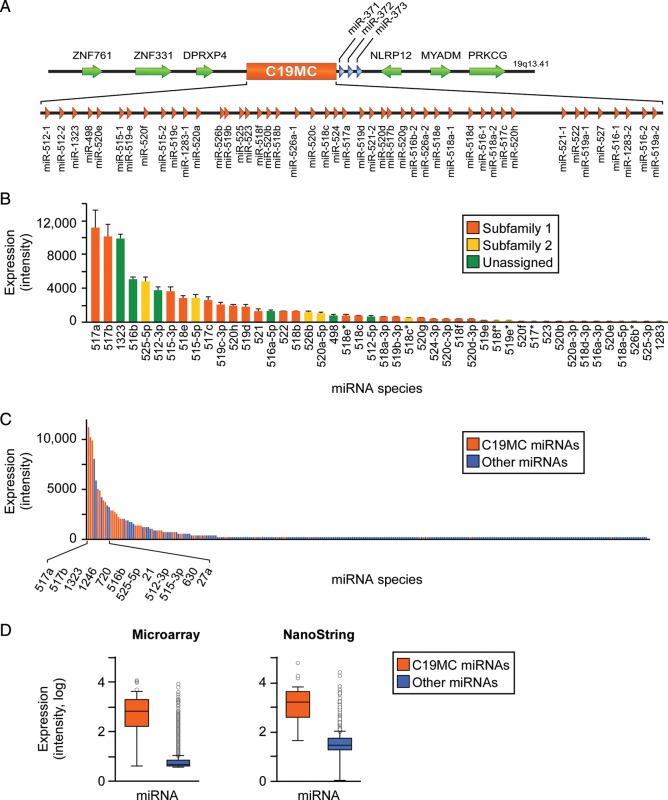
The human C19MC miRNA cluster is the largest known, comprising 46 miRNA genes (Fig. 1A) (Bentwich et al., 2005; Landgraf et al., 2007; Bortolin-Cavaille et al., 2009; Lin et al., 2010). Using high-throughput miRNA microarray profiling of PHT cells, we detected the expression of 41 of the 46 miRNA genes (Fig. 1B), which stratified by subfamilies as described by Lin et al. (2010). Furthermore, using microarrays and NanoString analyses and we found that C19MC miRNAs comprise the majority of miRNAs expressed in PHT cells (Fig. 1C and D). Using miRNA RT-qPCR, we also identified the expression of many C19MC miRNA species in the trophoblast-derived cell lines JEG-3 and BeWo, but not in trophoblast-derived HTR-8/SVneo cells (not shown). Interestingly, using NanoString analysis, we found that the proportion of C19MC miRNAs among all miRNA species was mildly reduced (56–52%) when PHT cells differentiate over 48 h in culture (P < 0.05, not shown).
Figure 1.
C19MC miRNAs landscape in term PHT cells. (A) Genomic organization of the C19MC miRNA cluster. (B) The expression of C19MC miRNAs species in PHT cells, as determined by miRNA microarrays (t= 48 h, n= 4). The expression represents the mean of normalized signal intensities, which are stratified by miRNA family as described in the text. (C) C19MC miRNAs comprise a major part of PHT miRNAs, as determined by miRNA microarrays (t = 48 h, n = 4). The expression represents the mean of normalized signal intensities. (D) The expression level of all C19MC miRNAs versus all other miRNAs in PHT cells, as measured by microarray or NanoString (t = 48 h, n = 4 for both analyses). Boxplots represent mean, 25th and 75th percentile, and outliers (differences were significant at P < 0.05). All error bars represent SD.
The effect of hypoxia or in vitro differentiation on the expression of C19MC miRNAs
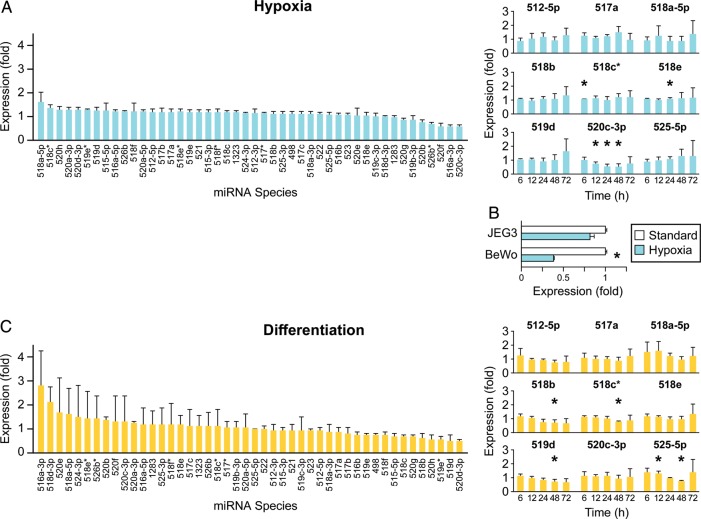
Placental hypoxia is associated with villous trophoblast injury (Levy et al., 2000; Pringle et al., 2010) and is known to modulate diverse trophoblast functions (Levy et al., 2000; Chen et al., 2006; Rimon et al., 2008; Simon and Keith, 2008). To investigate the effect of hypoxia on C19MC miRNA expression, we exposed PHT cells to hypoxia (O2 <1%) for 6, 12, 24, 48 or 72 h. Based on microarray analysis, hypoxia had an insignificant influence on the expression of C19MC miRNAs at all time points (Fig. 2A left panel, showing results for the 48 h time-point). We selected a subset of 9 out of 41 (22%) C19MC miRNAs for subsequent RT-qPCR analysis. The selection of miRNAs was designed to represent diverse array signal intensity, genomic loci within the C19MC cluster, miRNA subfamily (Fig. 1B) and a trend toward altered expression based on our microarray results (Fig. 2A, left panel). Except for sporadic changes of several species, we found that miR-520c-3p was consistently and significantly down-regulated after 12, 24 and 48 h exposure to hypoxia (Fig. 2A, right panel). Exposure of the trophoblast cell line BeWo to hypoxia resulted in a similar decrease in miR-520c-3p expression level (Fig. 2B). In contrast, we found that serum deprivation had an insignificant effect on the expression of the selected subset of miRNA species (not shown).
Figure 2.
The effect of hypoxia or in vitro differentiation on the expression of C19MC miRNAs. (A) Hypoxia does not affect C19MC miRNA expression, except for down-regulation of miR-520c-3p at 12, 24 and 48 h of exposure to hypoxia, as determined by high-throughput microarray profiling (left panel, t = 48 h, n = 4) or RT-qPCR (right panel, t = 48 h, n = 4). Fold expression represents mean of fold change in hypoxia (O2 <1%) over standard conditions (O2= 21%). (B) The expression of miR-520c-3p in placental cell lines after 36–48 h of exposure to hypoxia, as determined by RT-qPCR (representative experiment of n = 2 for BeWo, and n= 3 for JEG-3 cells). Fold expression represents mean of fold change in hypoxia (O2 <1%) over control (O2= 21%). *Denotes P < 0.05. (C) C19MC miRNA expression is not affected by PHT differentiation in vitro, as determined by high-throughput microarray profiling (left panel, t = 48 h, n = 4) or RT-qPCR (right panel, t = 48 h, n = 4). Fold expression represents the mean of fold change at t = 48 h over t = 0 h. *Denotes P < 0.05. All error bars represent SD.
Because PHT cells differentiate from cytotrophoblast into multinucleated syncytia over a 72 h time period (Nelson et al., 1999), we determined miRNA expression changes during that period. Using microarrays, we detected insignificant changes in miRNA expression (Fig. 2C left panel, showing results for the 48 h time-point). Although several of the miRNAs we selected for RT-qPCR analysis exhibited sporadic expression change (Fig. 2C, right panel), these miRNAs were characteristically expressed at a low level (Fig. 1B), rendering those measurements less reliable.
The C19MC miRNA landscape in the cellular and exosomal fraction of cultured PHT cells
We assessed whether term PHT cells secrete exosomes that contain C19MC miRNAs. Using ultracentrifugation on a sucrose gradient, we isolated a population of small microvesicles with a diameter range of 30–250 nm, as determined by electron microscopy (Fig. 3A). The microvesicle population was enriched for the exosomal marker TSG101 (Fig. 3B). The level of secreted PHT exosomes, determined by total exosomal protein, was insignificantly influenced by normoxic or hypoxic culture conditions (not shown). Interestingly, we found that TSG101 expression level was reduced by 1.6- and 2.0-fold in hypoxic PHT cells and exosomes, respectively (Fig. 3B). Using microarray analysis of PHT exosomal miRNAs, we identified the expression of C19MC and non-C19MC miRNAs in exosomes from PHT cells cultured for 48 h, as described in ‘Materials and Methods’ section (Fig. 3C). Akin to the landscape of C19MC miRNAs in PHT cells, C19MC miRNAs comprised the majority of exosomal miRNAs (Fig. 3C), and the pattern of expression of selected C19MC miRNAs during hypoxia was maintained in PHT cells and in exosomes released from these cells (Fig. 3D).
Figure 3.
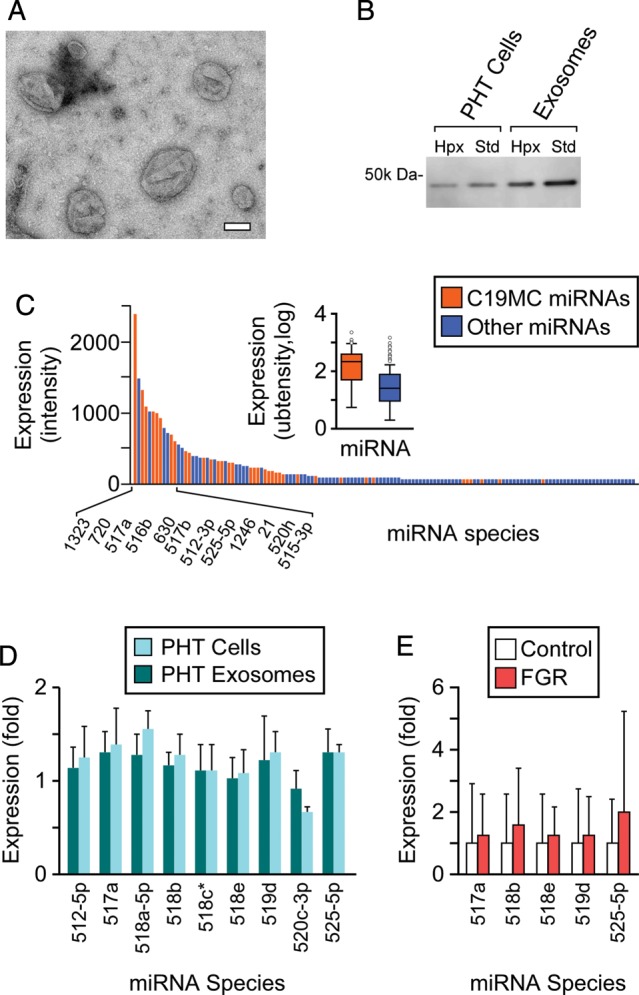
The profile of C19MC miRNAs in PHT exosomes resembles that of PHT cells. (A) PHT cells release exosomes and microvesicles of diverse sizes (see ‘Materials and Methods’ section for details of electron microscopy measurements). Bar = 100 nm. (B) PHT microvesicle population is enriched for the exosome marker TSG101, in standard (SD) and hypoxic conditions. Expression determined by western immunoblotting. (C) C19MC miRNAs comprise the majority of miRNAs in PHT exosomes, as determined by miRNA microarray without replicates (t = 48 h). Expression represents signal intensity. (D) The relative expression level of C19MC miRNA in PHT exosomes resembles the expression level detected in PHT cells cultured in hypoxic conditions, as determined by RT-qPCR (t = 48 h, n = 3). The fold expression represents mean fold change in hypoxia (O2 <1%) over normoxia (O2= 21%). (E) Circulating C19MC miRNA species are detected in the plasma of normal human pregnancy or pregnancy complicated by FGR as described in ‘Materials and Methods’ section. MiRNAs were detected using RT-qPCR (n = 14 for each group), with fold expression in normal pregnancy defined as 1. All error bars represent SD.
Lastly, we sought to corroborate our findings regarding the level of released C19MC miRNAs in vivo. As expected, the level of several C19MC miRNA species in the serum from non-pregnant women was either non-detectable or near the detection limit by RT-qPCR (not shown). In contrast, several C19MC miRNA species that are relatively highly expressed in PHT cells (Fig. 1B) were detected in normal pregnancy and pregnancy complicated by FGR. As suggested by our in vitro data using isolated exosomes, we did not observe any significant differences in C19MC miRNA levels in un-fractionated plasma pregnancies complicated by FGR (Fig. 3E).
Discussion
C19MC is the largest human miRNA cluster, and is exclusively expressed in the primate placenta and, to some degree, in other undifferentiated cells (Bentwich et al., 2005; Landgraf et al., 2007; Bortolin-Cavaille et al., 2009; Lin et al., 2010). As observed in other tissues and cell types (Landgraf et al., 2007), we found the miRNA content in PHT cells largely comprises only a limited number of miRNA species, many derived from the C19MC. We identified considerable variability in C19MC miRNA expression levels (Fig. 1B), suggesting post-transcriptional regulation of C19MC miRNA transcripts, or individual differences in miRNA stability.
In vitro differentiation of PHT cells was associated with a small reduction in the fraction of C19MC miRNAs among the total cellular miRNA, suggesting that C19MC miRNAs are likely functional in undifferentiated and differentiated trophoblasts. Although hypoxic stress modulates the trophoblast transcriptome (Roh et al., 2005; Struwe et al., 2010) and consequently, cellular functions (Levy et al., 2000; Chen et al., 2006; Rimon et al., 2008; Simon and Keith, 2008), we found that hypoxic stress does not affect C19MC miRNA expression, except for down-regulation of miR-520c-3p. These findings suggest that C19MC miRNAs do not contribute to trophoblast response to hypoxic stress, although we cannot exclude hypoxia-specific regulation of C19MC miRNA function (Leung and Sharp, 2010). The role of trophoblastic miR-520c-3p remains unclear. Interestingly, aberrant expression of miR-520c-3p in breast cancer promotes an invasive phenotype (Huang et al., 2008). Identification of miR-520c-3p targets in PHT cells may clarify its function in placental response to hypoxic stress. We also found that the expression of miR-525-5p is reduced during differentiation, but not in hypoxia. This C19MC miRNA is expressed in the placenta and released to the maternal circulation (Kotlabova et al., 2011), yet its function remains to be determined.
Secreted miRNAs have recently emerged as a potential mechanism of intercellular communication (Valadi et al., 2007; Pegtel et al., 2010; Zhang et al., 2010). To our knowledge, isolation of exosomes from primary term human trophoblasts and demonstration of their miRNA content has not been hitherto described. Our data show that the C19MC miRNA landscape of PHT exosomes resembles that of C19MC miRNA in PHT cells. Because we profiled exosomal miRNAs at 48 h of culture, when the majority of the cells are differentiated into multinucleated syncytiotrophoblasts, we cannot rule out a different pattern of exosomal miRNA expression in less or more differentiated PHT cells. Our data also indicate that unlike observations in other cell types (Zhang et al., 2010; Mittelbrunn et al., 2011), hypoxia does not cause selective packaging of specific C19MC miRNA species in secreted PHT exosomes. Although we previously detected an increase in the overall levels of miRNA species in the plasma of women with pregnancies complicated by FGR (Mouillet et al., 2010a), we note that plasma or cell culture supernatant may include miRNAs that were released by dead cells (Wang et al., 2010; Turchinovich et al., 2011), particularly in conditions of cell injury that are associated with apoptosis (Levy et al., 2000). We also highlight that our analyses were performed in vitro using PHT cells derived from healthy participants. It might have been interesting to examine C19MC expression and release in cultured cells that represent FGR. However, at the present time, it is not clear that cultured PHT cells, even if derived from women with FGR, retain their in vivo phenotype (or that of other clinical entities, such as the presence or absence of labor) (Oh et al., 2011). Lastly, although we used the exosomal marker TSG101 merely to validate the presence of exosomes among our isolated multivesicular bodies, we noted that TSG101 levels were reduced in hypoxic cells or exosomes. This observation likely reflects cell stress, known to influence exosomal phenotype (Yu et al., 2006; Eldh et al., 2010), but not a general effect on exosomal release, because total exosomal protein level as well as exosomal miRNA levels were unchanged in hypoxia.
The unique C19MC miRNA landscape suggests that these miRNAs contribute to the establishment or maintenance of a trophoblast-specific mRNA and protein profile (Roh et al., 2005; Sood et al., 2006; Guo et al., 2010), or to trophoblast adaptation to homeostatic perturbations. Secreted trophoblast-specific C19MC miRNAs, in exosomes or protein-bound in the plasma, may play an important role in placental-maternal communication, possibly directing maternal adaptation to pregnancy. Future work may also elucidate the mechanisms of biosynthesis, stability and degradation of C19MC miRNA, and shed light on the expression changes in C19MC miRNAs from early pregnancy until delivery.
Authors’ roles
R.B.D., J.F.M. and T.C. conceived the manuscript; designed the study; performed data acquisition, analysis and interpretation and provided final approval. C.A.H. contributed to the study design, performed data acquisition, analysis and interpretation and provided final approval. D.B.S. and A.E.M. contributed to the study design, performed data acquisition and provided final approval. Y.S. conceived the manuscript, designed the study, conducted analysis and interpretation and provided final approval. R.B.D. and Y.S. wrote the manuscript.
Funding
R.B.D. is supported by a postdoctoral research fellowship from Magee-Womens Research Institute at the University of Pittsburgh, PA, USA. The project was also supported by the Pennsylvania Department of Health Research Formula Funds (to J.F.M. and T.J.C.), NIH P01-HD030367 (to C.A.H.) and NIH R21-HD053878 and R01-HD065893 (to Y.S.).
Conflict of interest
None declared.
Acknowledgements
The authors thank Dr C.H. Graham (Queen's University, Kingston, ON, Canada) for the HTR-8/SVneo cells. The authors thank Magdalena Jennings, Elena Sadovsky, Ming Sun and William Shufesky for technical assistance, and Lori Rideout for assistance during preparation of the manuscript.
References
- Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atay S, Gercel-Taylor C, Kesimer M, Taylor DD. Morphologic and proteomic characterization of exosomes released by cultured extravillous trophoblast cells. Exp Cell Res. 2011;317:1192–1202. doi: 10.1016/j.yexcr.2011.01.014. [DOI] [PubMed] [Google Scholar]
- Barad O, Meiri E, Avniel A, Aharonov R, Barzilai A, Bentwich I, Einav U, Gilad S, Hurban P, Karov Y, et al. MicroRNA expression detected by oligonucleotide microarrays: system establishment and expression profiling in human tissues. Genome Res. 2004;14:2486–2494. doi: 10.1101/gr.2845604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- Bortolin-Cavaille ML, Dance M, Weber M, Cavaille J. C19MC microRNAs are processed from introns of large Pol-II, non-protein-coding transcripts. Nucleic Acids Res. 2009;37:3464–3473. doi: 10.1093/nar/gkp205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Nelson DM, Sadovsky Y. N-myc down-regulated gene 1 modulates the response of term human trophoblasts to hypoxic injury. J Biol Chem. 2006;281:2764–2772. doi: 10.1074/jbc.M507330200. [DOI] [PubMed] [Google Scholar]
- Donker RB, Mouillet JF, Nelson DM, Sadovsky Y. The expression of argonaute2 and related microRNA biogenesis proteins in normal and hypoxic trophoblasts. Mol Hum Reprod. 2007;13:273–279. doi: 10.1093/molehr/gam006. [DOI] [PubMed] [Google Scholar]
- Eldh M, Ekstrom K, Valadi H, Sjostrand M, Olsson B, Jernas M, Lotvall J. Exosomes communicate protective messages during oxidative stress; possible role of exosomal shuttle RNA. PLoS ONE. 2010;5:e15353. doi: 10.1371/journal.pone.0015353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangsmyr L, Baranov V, Nagaeva O, Stendahl U, Kjellberg L, Mincheva-Nilsson L. Cytoplasmic microvesicular form of fas ligand in human early placenta: switching the tissue immune privilege hypothesis from cellular to vesicular level. Mol Hum Reprod. 2005;11:35–41. doi: 10.1093/molehr/gah129. [DOI] [PubMed] [Google Scholar]
- Genbacev O, Zhou Y, Ludlow JW, Fisher SJ. Regulation of human placental development by oxygen tension. Science. 1997;277:1669–1672. doi: 10.1126/science.277.5332.1669. [DOI] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q, Gumireddy K, Schrier M, le Sage C, Nagel R, Nair S, Egan DA, Li A, Huang G, Klein-Szanto AJ, et al. The microRNAs miR-373 and miR-52°c promote tumour invasion and metastasis. Nat Cell Biol. 2008;10:202–210. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Kliman HJ, Nestler JE, Sermasi E, Sanger JM, Strauss JF., III Purification, characterization, and in vitro differentiation of cytotrophoblasts from human term placentae. Endocrinology. 1986;118:1567–1582. doi: 10.1210/endo-118-4-1567. [DOI] [PubMed] [Google Scholar]
- Kotlabova K, Doucha J, Hromadnikova I. Placental-specific microRNA in maternal circulation—identification of appropriate pregnancy-associated microRNAs with diagnostic potential. J Reprod Immunol. 2011;89:185–191. doi: 10.1016/j.jri.2011.02.006. [DOI] [PubMed] [Google Scholar]
- Lamparski HG, Metha-Damani A, Yao JY, Patel S, Hsu DH, Ruegg C, Le Pecq JB. Production and characterization of clinical grade exosomes derived from dendritic cells. J Immunol Methods. 2002;270:211–226. doi: 10.1016/s0022-1759(02)00330-7. [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK, Sharp PA. MicroRNA functions in stress responses. Mol Cell. 2010;40:205–215. doi: 10.1016/j.molcel.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy R, Smith SD, Chandler K, Sadovsky Y, Nelson DM. Apoptosis in human cultured trophoblasts is enhanced by hypoxia and diminished by epidermal growth factor. Am J Physiol Cell Physiol. 2000;278:C982–C988. doi: 10.1152/ajpcell.2000.278.5.C982. [DOI] [PubMed] [Google Scholar]
- Li M, Lee KF, Lu Y, Clarke I, Shih D, Eberhart C, Collins VP, Van Meter T, Picard D, Zhou L, et al. Frequent amplification of a chr19q13.41 microRNA polycistron in aggressive primitive neuroectodermal brain tumors. Cancer Cell. 2009;16:533–546. doi: 10.1016/j.ccr.2009.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Cheung WK, Chen S, Lu G, Wang Z, Xie D, Li K, Lin MC, Kung HF. Computational identification and characterization of primate-specific microRNAs in human genome. Comput Biol Chem. 2010;34:232–241. doi: 10.1016/j.compbiolchem.2010.08.001. [DOI] [PubMed] [Google Scholar]
-
Livak K, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the
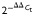 method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar] - Lopez-Romero P. Pre-processing and differential expression analysis of agilent microRNA arrays using the agi microRNA bioconductor library. BMC Genomics. 2011;12:64. doi: 10.1186/1471-2164-12-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo SS, Ishibashi O, Ishikawa G, Ishikawa T, Katayama A, Mishima T, Takizawa T, Shigihara T, Goto T, Izumi A, et al. Human villous trophoblasts express and secrete placenta-specific microRNAs into maternal circulation via exosomes. Biol Reprod. 2009;81:717–729. doi: 10.1095/biolreprod.108.075481. [DOI] [PubMed] [Google Scholar]
- Mincheva-Nilsson L, Baranov V. The role of placental exosomes in reproduction. Am J Reprod Immunol. 2010;63:520–533. doi: 10.1111/j.1600-0897.2010.00822.x. [DOI] [PubMed] [Google Scholar]
- Mittelbrunn M, Gutierrez-Vazquez C, Villarroya-Beltri C, Gonzalez S, Sanchez-Cabo F, Gonzalez MA, Bernad A, Sanchez-Madrid F. Unidirectional transfer of microRNA-loaded exosomes from t cells to antigen-presenting cells. Nat Commun. 2011;2:282. doi: 10.1038/ncomms1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montecalvo A, Shufesky WJ, Stolz DB, Sullivan MG, Wang Z, Divito SJ, Papworth GD, Watkins SC, Robbins PD, Larregina AT, et al. Exosomes as a short-range mechanism to spread alloantigen between dendritic cells during t cell allorecognition. J Immunol. 2008;180:3081–3090. doi: 10.4049/jimmunol.180.5.3081. [DOI] [PubMed] [Google Scholar]
- Mouillet JF, Chu T, Hubel CA, Nelson DM, Parks WT, Sadovsky Y. The levels of hypoxia-regulated microRNAs in plasma of pregnant women with fetal growth restriction. Placenta. 2010a;31:781–784. doi: 10.1016/j.placenta.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouillet JF, Chu T, Nelson DM, Mishima T, Sadovsky Y. Mir-205 silences med1 in hypoxic primary human trophoblasts. FASEB J. 2010b;24:2030–2039. doi: 10.1096/fj.09-149724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DM, Johnson RD, Smith SD, Anteby EY, Sadovsky Y. Hypoxia limits differentiation and up-regulates expression and activity of prostaglandin h synthase 2 in cultured trophoblast from term human placenta. Am J Obstet Gynecol. 1999;180:896–902. doi: 10.1016/s0002-9378(99)70661-7. [DOI] [PubMed] [Google Scholar]
- Noguer-Dance M, Abu-Amero S, Al-Khtib M, Lefevre A, Coullin P, Moore GE, Cavaille J. The primate-specific microRNA gene cluster (CI9MC) is imprinted in the placenta. Hum Mol Genet. 2010;19:3566–3582. doi: 10.1093/hmg/ddq272. [DOI] [PubMed] [Google Scholar]
- Oh SY, Chu T, Sadovsky Y. The timing and duration of hypoxia determine gene expression patterns in cultured human trophoblasts. Placenta. 2011;32:1004–1009. doi: 10.1016/j.placenta.2011.09.010. [DOI] [PubMed] [Google Scholar]
- Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL, de Gruijl TD, Wurdinger T, Middeldorp JM. Functional delivery of viral microRNA via exosomes. Proc Natl Acad Sci USA. 2010;107:6328–6333. doi: 10.1073/pnas.0914843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle KG, Kind KL, Sferruzzi-Perri AN, Thompson JG, Roberts CT. Beyond oxygen: complex regulation and activity of hypoxia inducible factors in pregnancy. Hum Reprod Update. 2010;16:415–431. doi: 10.1093/humupd/dmp046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rani S, O'Brien K, Kelleher FC, Corcoran C, Germano S, Radomski MW, Crown J, O'Driscoll L. Isolation of exosomes for subsequent mRNA, microRNA, and protein profiling. Methods Mol Biol. 2011;784:181–195. doi: 10.1007/978-1-61779-289-2_13. [DOI] [PubMed] [Google Scholar]
- Redman CW, Sargent IL. Microparticles and immunomodulation in pregnancy and pre-eclampsia. J Reprod Immunol. 2007;76:61–67. doi: 10.1016/j.jri.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Rimon E, Chen B, Shanks AL, Nelson DM, Sadovsky Y. Hypoxia in human trophoblasts stimulates the expression and secretion of connective tissue growth factor. Endocrinology. 2008;149:2952–2958. doi: 10.1210/en.2007-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippe V, Dittberner L, Lorenz VN, Drieschner N, Nimzyk R, Sendt W, Junker K, Belge G, Bullerdiek J. The two stem cell microRNA gene clusters C19MC and miR-371-3 are activated by specific chromosomal rearrangements in a subgroup of thyroid adenomas. PLoS ONE. 2010;5:e9485. doi: 10.1371/journal.pone.0009485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh CR, Budhraja V, Kim HS, Nelson DM, Sadovsky Y. Microarray-based identification of differentially expressed genes in hypoxic term human trophoblasts and in placental villi of pregnancies with growth restricted fetuses. Placenta. 2005;26:319–328. doi: 10.1016/j.placenta.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Simon MC, Keith B. The role of oxygen availability in embryonic development and stem cell function. Nat Rev Mol Cell Biol. 2008;9:285–296. doi: 10.1038/nrm2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:1–25. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Sood P, Krek A, Zavolan M, Macino G, Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc Natl Acad Sci USA. 2006;103:2746–2751. doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struwe E, Berzl G, Schild R, Blessing H, Drexel L, Hauck B, Tzschoppe A, Weidinger M, Sachs M, Scheler C, et al. Microarray analysis of placental tissue in intrauterine growth restriction. Clin Endocrinol (Oxf) 2010;72:241–247. doi: 10.1111/j.1365-2265.2009.03659.x. [DOI] [PubMed] [Google Scholar]
- Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39:7223–7233. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. microRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Zhang S, Weber J, Baxter D, Galas DJ. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010;38:7248–7259. doi: 10.1093/nar/gkq601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Harris SL, Levine AJ. The regulation of exosome secretion: a novel function of the p53 protein. Cancer Res. 2006;66:4795–4801. doi: 10.1158/0008-5472.CAN-05-4579. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu D, Chen X, Li J, Li L, Bian Z, Sun F, Lu J, Yin Y, Cai X, et al. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol Cell. 2010;39:133–144. doi: 10.1016/j.molcel.2010.06.010. [DOI] [PubMed] [Google Scholar]