Abstract
Cannabinoids, endocannabinoids and marijuana activate two well-characterized cannabinoid receptors (CBRs), CB1-Rs and CB2-Rs. The expression of CB1-Rs in the brain and periphery has been well studied but neuronal CB2-Rs have received much less attention than CB1-Rs. Many studies have now identified and characterized functional glial and neuronal CB2-Rs in the central nervous system. However, many features of CB2-R gene structure, regulation and variation remain poorly characterized in comparison to the CB1-R. Here, we report on the discovery of a novel human CB2 gene promoter encoding testis (CB2A) isoform with starting exon located ca 45 kb upstream from the previously identified promoter encoding the spleen isoform (CB2B). The 5′ exons of both CB2 isoforms are untranslated 5′UTRs and alternatively spliced to the major protein coding exon of the CB2 gene. CB2A is expressed higher in testis and brain than CB2B that is expressed higher in other peripheral tissues than CB2A. Species comparison found that the CB2 gene of human, rat and mouse genomes deviated in their gene structures and isoform expression patterns. mCB2A expression was increased significantly in the cerebellum of mice treated with the CB-R mixed agonist, WIN55212-2. These results provide much improved information about CB2 gene structure and its human and rodent variants that should be considered in developing CB2-R-based therapeutic agents.
Keywords: CB2 cannabinoid receptors, CB2A, CB2B, testis, spleen, brain
Introduction
Marijuana remains one of the most widely used drugs and much progress in cannabinoid research has revealed the existence of an elaborate endocannabinoid physiological control system (EPCS) in animals and humans (Mackie 2008; Onaivi et al. 2008), with increasing new knowledge on the scientific and therapeutic bases for the use of cannabinoids (CBs) as medicine. The EPCS consists of (1) cannabinoid receptors (CB-Rs), (2) endogenous compounds termed endocannabinoids (eCBs) that activate these receptors and (3) enzymes that synthesize and degrade the eCBs. The study of the distribution and function of various components of the EPCS in the brain has provided the basis for the established effects of marijuana and eCBs. Thus, the use of cannabis or administration of phytocannabinoids or synthetic CBs interacts with the EPCS to modulate synaptic transmission. This modulatory action on synaptic transmission has significant functional implications on the interactions between exogenous cannabinoids such as smoking marijuana or administration of Δ9-THC and eCBs mediated synaptic plasticity.
The expression of CB1 cannabinoid receptors (CB1-Rs) in the brain and periphery has been well studied but CB2-Rs have received much less attention than have CB1-Rs (Mackie 2008; Onaivi et al. 2006). Both CB1-Rs and CB2-Rs are widely distributed in peripheral tissues with CB2-Rs particularly enriched in immune tissues and cells such as spleen, thymus, and leukocytes (Sugiura & Waku 2002). The functional neuronal expression of CB2-Rs in the brain had therefore been ambiguous and controversial (Onaivi et al. 2006; Van Sickle et al. 2005). The lack of CB2 information on brain expression is because CB2-Rs were thought previously to be expressed primarily in some peripheral tissues like the spleen and in other immune cells and has therefore been traditionally referred to as the peripheral CB-Rs (Gerard et al. 1990; Griffin et al. 1999; Munro et al. 1993). Several studies have now identified and characterized glial and neuronal CB2-Rs in the central nervous system. However, many features of CB2-R gene structure, regulation, and variation remain poorly characterized (Benito et al. 2008; Gong et al. 2006; Nunez et al. 2008; Onaivi et al. 2006; Van Sickle et al. 2005). This poor characterization of CB2-R gene structure and polymorphisms hampers progress in the determination of the functional role of CB2-Rs, particularly in neurodegenerative and neuropsychiatric disorders. Furthermore, the neurobiological basis for CB2-R physiological activity and its putative interaction with or without CB1-Rs remains to be determined.
Our recent studies indicate that genetic variants of CB2 gene expression, with a high incidence of Q63R but not the H316Y polymorphism, is involved in eating disorders, substance abuse, and depression in a human population (Onaivi et al. 2008). Other studies in human tissue samples from Alzheimer's disease subjects revealed that CB2-Rs are selectively over expressed in the microglial cells that are associated with Aβ-enriched neuritic plaques (Pazos et al. 2004). Data obtained in vitro and from animal models demonstrated the inducible nature of CB2-Rs under neuroinflammatory conditions and suggests that the up regulation of CB2-Rs is a common pattern of response against different types of chronic human brain neuropathology (Benito et al. 2008). Accruing evidence shows that CB2-Rs and its gene variants may play possible roles in neuroinflammation occurring in multiple sclerosis, traumatic brain injury, HIV-induced encephalitis, Alzheimer's, Parkinson's, and Huntington's diseases (Benito et al. 2008). It would be important to understand the role of CB2-Rs and its gene variants in disturbances of the brain involving neuroinflammation. Therefore, improved information about the CB2 gene and its human variants might add to our understanding of the role of CB2-Rs during neuroinflammatory conditions in the CNS and beyond immunocannabinoid activity.
To date, the complete gene structure, 5′- and 3′-UTR, and transcription initiation sites of human CB2 receptor have not been fully characterized (Abood 2005; Onaivi et al. 2006). Since the identification of mouse CB2 expression in brain regions was reported (Gong et al. 2006; Van Sickle et al. 2005), the specific expression of human or mouse CB2 isoforms in brain regions was not known. However, the published evidence showed significant species differences of CB2-Rs in human, mouse, and rat in terms of peptides, mRNA sizes, gene structure, and pharmacology (Brown et al. 2002; Munro et al. 1993; Shire et al. 1996). We thereafter name human, mouse, and rat CB2 genes as hCB2, mCB2, and rCB2, respectively. For instance, hCB2 mRNA was detected in the HL60 cell line (human acute promyelocytic leukemia cells) by Northern blot as 2.5 kb and 5.0 kb mRNA bands that were elevated by TPA- (12-O-tetradecanoylphorbol-13-acetate) and DMF- (N,N-dimethylformamide) induced differentiation to myeloid and granulocyte, respectively (Munro et al. 1993). The mRNA 5.0 kb and 2.5 kb bands of hCB2 were also observed in the Jurkat E6-1 cell line (human T-cell leukemia cells); however, only the 2.5 kb band was observed in HPB-ALL cells (human peripheral blood acute leukemia cell line) (Schatz et al. 1997). In contrast, mCB2 mRNA were shown to be predominantly expressed as a single band of 4.0 kb in spleen (Schatz et al. 1997). Different rCB2 mRNA species were observed as a 2.4 kb band (Schatz et al. 1997) and a 3.9 kb band in rat spleen, thymus, testis, and leukocytes (Brown et al. 2002). The discrepancies on CB2 mRNA sizes in the literature indicated incomplete gene structure of CB2 gene in different species or polymorphism in the same species. We now report on the discovery of a novel CB2 gene promoter that differentially encodes tissue-specific CB2 gene expression in human tissues. Intriguingly, the testis-specific isoform hCB2A expression level was much higher in brain regions than the spleen-dominant hCB2B isoform that is expressed at higher levels in spleen and other peripheral tissues except testis. Our results demonstrated tissue-specific isoforms of CNR2 gene that may provide tissue-specific targeting of cannabinoids in inflammatory and neuropathic pain disorders associated with CB2-Rs in the brain and peripheral tissues.
Materials and Methods
RNA isolation, cDNA synthesis, and real-time PCR quantification
Human total RNA from brain and peripheral tissues were obtained from Clontech (Mountain View, CA) and human brain tissues were obtained from the NICHD Brain and Tissue Bank for Developmental Disorders at the University of Maryland. Mouse total RNAs were extracted from C57BL/6J, C57BL/10J and the BTBR T+tF/J mouse peripheral tissues and brain regions using RNAzolB protocol (Tel-Test, Inc. Friendwood, TX). Single strand cDNA was synthesized from total RNA using SuperScript™ III One-Step RT-PCR System (Invitrogen, Carlsbad, CA). Novel hCB2A exons (exon 1a and 1b) were identified by aligning human genomic sequences with EST clone AL527378 isolated from normalized neuroblastoma cDNA library. For quantitative real time PCR assays, the exon-specific primers and fluorescent Fam-labeled and minor grove binder (MGB) conjugated probes of hCB2 and mCB2 genes were designed (Table 1) using Primer Express (Applied Biosystems, Foster City, CA). For hCB2 expression study, a CB2A-specific probe was designed to span exon 1a and 1b and a hCB2B-specific probe was designed to span exon 2 and 3. For the mCB2 expression study, an mCB2A-specific probe was designed to span exon 1 and 3 and an mCB2B-specific probe was designed to span exon 2 and 3. For the rCB2 expression study, specific probes (Table 1) were designed to span exon 1 and 3 and to exon 2 and 3 to detect rCB2A and rCB2B expression, respectively. The rCB2 primers were also designed (Table 1) to detect retroposon B2 insertion into 3′UTR of rCB2.
Table 1.
TaqMan probes, and flanking forward and reverse primer sequences for human, mouse, and rat CB2 genes and PCR primers to amplify rat B2 insertion site.
| Isoforms | Exons | MGB TaqMan probes | Forward primer | Reverse primer |
|---|---|---|---|---|
| hCB2A | Exon 1a, 1b | CCAGATGCAGCCGC | GGAAGAAAGAGAATATTGTTCAGTTGATT | GCTGGCCTTGGAGAGTGACA |
| hCB2B | Exon 2, 3 | ACAACACAACCCAAAGC | TGGGAGAGGACAGAAAACAACTG | TGGGCCCTTCAGATTCCA |
| rCB2A | Exon 1, 3 | CTGACAAATGACACCCAGTC | CAGGACAAGGCTCCACAAGAC | GATGGGCTTTGGCTTCTTCTAC |
| rCB2B | Exon 1, 3 | TGGGCCCAGTCTT | GCCACCCAGCAAACATCTCT | GATGGGCTTTGGCTTCTTCTAC |
| rB2 insert | Exon 3 | CCAGGCTGCTCCAATTGCT | TCTGACTCGGACTGCTTCCA | |
| mCB2A | Exon 1, 3 | CTGACAAATGACTCCCAGTC | CAGGACAAGGCTTCACAAGAC | GACAGGCTTTGGCTGCTTCTAC |
| mCB2B | Exon 2, 3 | TGGGCCCAGTCCT | GCCACCCAGCAAACATCTAT | GACAGGCTTTGGCTGCTTCTAC |
| mCB2-KO | Exon 3 | ATGCTGGTTCCCTGCAC | AGCTCGGATGCGGCTAGAC | AGGCTGTGGCCCATGAGA |
Endogenous controls were provided by pre-developed human GAPDH or mouse and β-actin with fluorescent FAM-labeling (Applied Biosystems, Foster City, CA). ABI 7900HT Sequence Detection System default program was used for real time PCR with 40 cycles, as well as rCB2 B2 insertion detection.
The novel isoforms were confirmed by agarose gel analysis of PCR fragments that were in the expected sizes and by dideoxynucleotide sequencing. To confirm hCB2A testis-specific expression, TaqMan assays were carried out independently by different individuals. Mouse mCB2 gene expression and regulation by treatment with cannabinoids was determined in mouse brain regions and other peripheral tissues after sub-acute treatment with CB-R ligands in comparison to vehicle-treated control mice. Sprague-Dawley and Long Evans rats were decapitated and the brain and peripheral tissues were dissected for RNA isolation and cDNA synthesis. The different brain regions included striatum, brain stem, cerebellum, hypothalamus and frontal cortex, and peripheral tissues included spleen and testis.
Animal subjects and cannabinoid receptor ligand treatmens
The selected mouse strain C57BL/6Js were treated daily for seven days with the mixed cannabinoid agonist, WIN55212-2 (2 mg/kg). On test days, animals were treated with the test drug for 30 mins before assessment of the performance of the mice in FST for 5-mins. In this study, the effect of WIN55212-2 acutely and on day six in the FST was determined and compared to control mice following seven days of daily administration. Six mice per group were sacrificed and non-injected BTBR mice tissue parts included for mCB2 gene expression analysis as follows: Vehicle = (V1-V6), WIN55212-2 (2 mg/kg) = (W1-W6) and untreated BTBR = (B1-B6). We also analyzed mCB2 gene expression in chronically treated C57BL/10Js treated with CB1-R antagonist (AM 251, 3 mg/kg) or CB2-R antagonist (AM 630, 10 mg/kg) in comparison to vehicle treated control mice.
The mCB2 knockout mice were bred at NIDA-IRP from two CB2+ ⁄ – breeding pairs, generously donated by Dr. George Kunos at the NIAAA. Genotyping was performed by the Charles River National Laboratories. We analyzed mCB2 expression in brain regions, testis, and spleen using the three TaqMan probes against two promoters of mouse mCB2 gene and the deleted part of mCB2 gene (Table 1).
Statistical analysis
Prism-3 program (GraphPad Software, Inc, San Diego, CA) was used for graphical and statistical analyses including t-tests and one-way analyses of variance. The accepted level of significance is p<0.05.
Results
Human CB2 genomic structure (1p36.1) and discovery of a novel hCB2A promoter
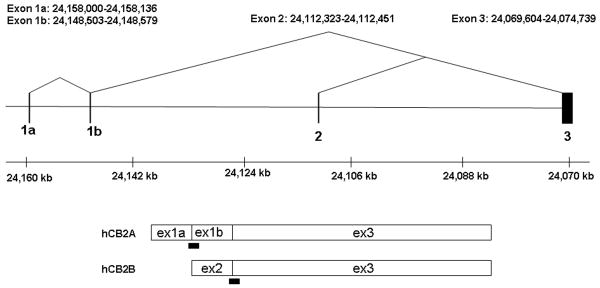
With the published evidence showing clear species differences of CB2-Rs in human, mouse, and rat in terms of mRNA sizes in tissues and cell lines (Brown et al. 2002; Munro et al. 1993; Shire et al. 1996), we set out to identify unidentified human hCB2 isoforms by first aligning the EST sequences with hCB2 genomic sequences and then by PCR amplification. We found that an EST clone (AL527378) derived from a normalized neuroblastoma cDNA library aligned with two upstream human hCB2 exons (exon 1a and 1b) and down stream major coding exon (exon 3) with two conserved exon-intron (AG-GT) junctions (Figure 1A). We then sequenced the PCR fragments that were amplified using primers designed according to the novel exons and we deposited hCB2A cDNA sequence in GenBank (EU517121). The novel exon 1a and 1b of hCB2 are located 85,478 bp and 75,971 bp, respectively, upstream from the major coding exon 3. Both of the novel exon 1a and 1b are 5′UTR sequences because they do not contain open reading frame in frame with the initiation methionine of hCB2 encoded by the exon 3. In contrast, the previously identified upstream 5′UTR exon 2 is located 39,769 bp upstream from the major coding exon 3 (Munro et al. 1993). Therefore, the size of the newly identified hCB2A isoform spans a genomic region of 90.8 kb, about twice as large as the previously identified human hCB2B gene of 45.7 kb (Figure 1A).
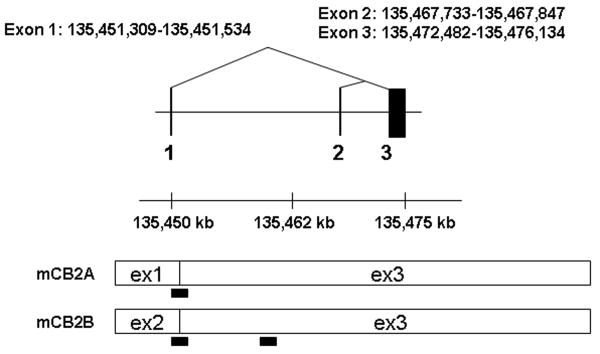
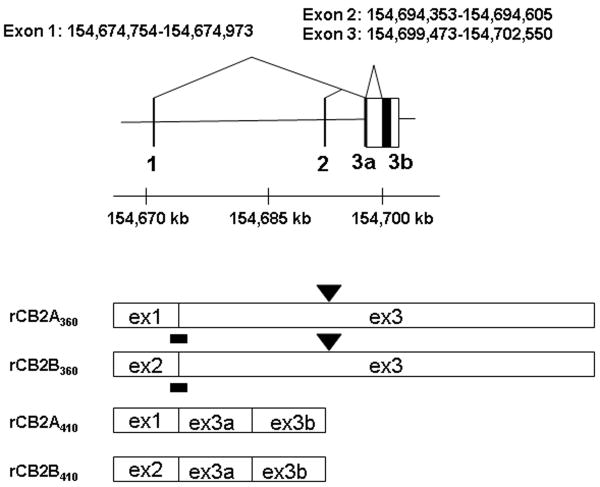
Figure 1.
A: Human hCB2 genomic structure and transcripts; B: Mouse mCB2 genomic structure and transcripts; C: Rat rCB2 genomic structure and transcripts. Horizontal line represents NCBI MapViewer's Homo sapians chromosome1p36.1 position Build 36.3, mus musculus chromosome 4D3 position Build 37.1, and Rattus norvegicus chromosome 5q36 position, Build RGSC 34.1. Vertical bars represent exons. Triangular lines represent splicing events. Open boxes represent spliced exons in mRNA transcripts. Black bars represent the location of TaqMan probes. Black boxes represent intraexonal splicing of rat rCB2 major coding exon and black triangles represent rat B2 retroposon insertions sites.
There are four probable poly A signals and poly A sites in the extended 3′UTR sequence of hCB2 cDNA. The first poly A site is the previously identified cDNA clone (HCX5.36) with imperfect polyadenylation signal sequence of GATAAA (Munro et al. 1993). There are three additional poly A sites with perfect polyadenylation signal sequence AATAAA; the second poly A site is located at 2,799 bp of hCB2A (EU517121) with polyA containing 3′EST alignments (AW515513, AI864752, and AI475798). The third is located at 4,632 bp with polyA containing 3′EST alignments (AW967543, AW966929, AV715015, and AA250845) and the fourth is located on 5,349 bp with polyA containing 3′EST alignments (AW405600, AW294847, AA909480, AI218261, and AI424994). All these 3′EST clones were derived from cDNA libraries of B-cells or lymphocytes. The additional evidence of hCB2 mRNA that contains extended 3′UTR sequence is another hCB2 cDNA clone (HCX5.1) with extended sequence beyond the first polyA site (Munro et al. 1993). Therefore, human CNR2 gene contains two separate promoters and several alternative polyA sites, generating multiple mRNA sizes as previously observed in Northern blot of human HL-60 and Jurkat E6-1 cell lines (Munro et al. 1993; Schatz et al. 1997). The first promoter initiates from 5′UTR exon 1a and is spliced to exon 1b and then to the major coding exon 3 (Figure 1). The second promoter initiates from exon 2 that is also spliced to the major coding exon 3 (Figure 1). We named the novel hCB2 isoform that is transcribed from the first exon as hCB2A and the second promoter as hCB2B.
Human hCB2A and hCB2B isoform tissue expression patterns
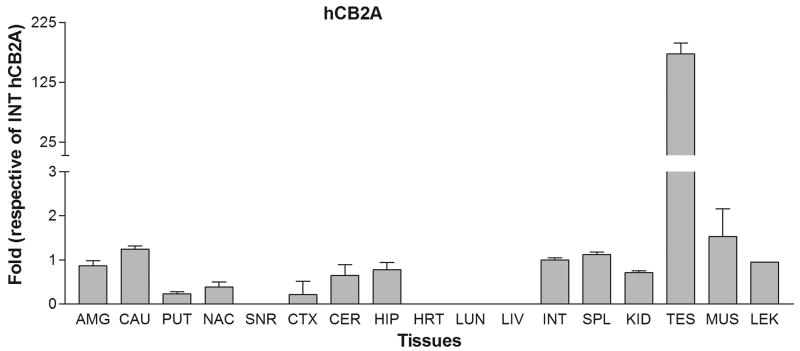
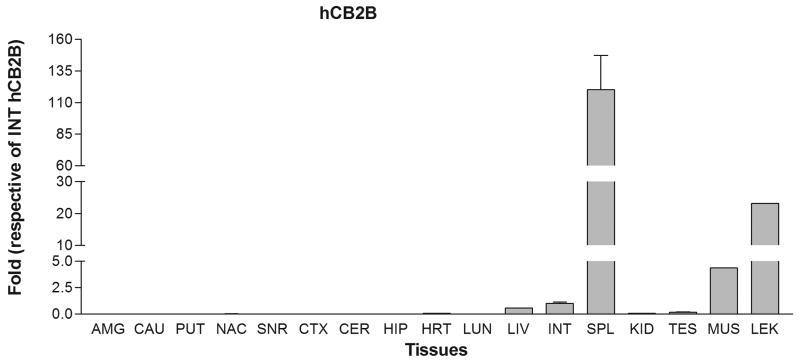
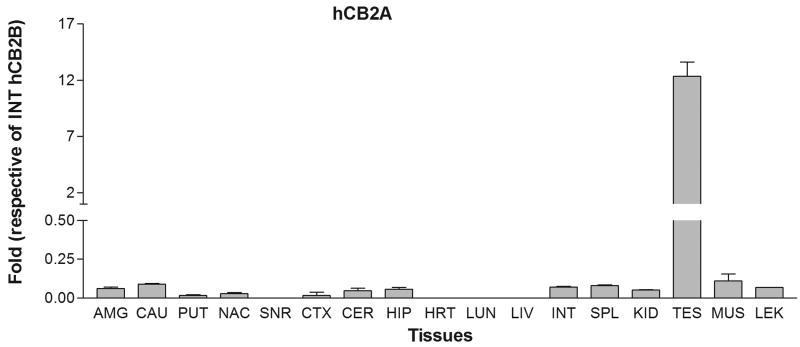
We next tested the tissue expression patterns of the newly identified hCB2A isoform in comparison with the previously identified hCB2B isoform in various human peripheral tissues and brain regions. The tissue cDNA templates were synthesized from human RNA preparations that consist of intact RNA with no genomic DNA contamination (Clontech, Mountain View, CA). The TaqMan probes were designed across exon 1a/1b and exon 2/3, respectively, for detecting specific hCB2A and hCB2B isoform expression and avoiding the detection of genomic contamination (Table 1 and Figure 1A). Strikingly, hCB2A expression was observed predominantly in testis i.e. more than 100 fold than that of spleen and leukocytes. The hCB2A expression was also observed in various brain regions at levels, i.e. 0-2 fold using intestine hCB2A as a reference (Figure 2A). As expected, hCB2B expression was observed predominantly in spleen and leukocytes. Spleen and leukocytes hCB2B mRNA levels were 120 and 30 fold higher than that of intestine reference of hCB2B (Figure 2B), respectively. However, there was little expression of hCB2B in brain regions. In comparison with intestine hCB2B reference, hCB2A expression levels were significantly lower in peripheral tissues except in testis; however, hCB2A expression in brain regions was observed in amygdala, caudate, putaman, nucleus accumbens, cortex, hippocampus and cerebellum at low levels. The caudate hCB2A mRNA level was about 0.07% of the spleen hCB2B expression level (Figure 2C). This is the first observation of the isoforms-specific expression of human hCB2A at significant, albeit low levels (<1% of hCB2A testis expression and <0.1% of CB2B spleen expression) in human brain regions (Figure 2C).
Figure 2.
Tissue expression patterns of human hCB2A and hCB2B isoforms: A. human hCB2A isoform expression in different brain regions and peripheral tissues using human intestine hCB2A as a reference; B. human hCB2B isoform expression in different brain regions and peripheral tissues using human intestine hCB2B as a reference; C. human hCB2A isoform expression patterns using human intestine hCB2B as a reference. Error bars were derived from standard deviation (SD) of triplicate TaqMan assay. The abbreviations for brain regions and peripheral tissues are AMG: amygdala, CAU: caudate, PUT: putaman, NAC: nucleus acumbens, SNR: substantia nigra, CTX: cortex, CER: cerebellum, HIP: hippocampus, HRT: heart, LUN: lung, LIV: liver, INT: intestine, SPL: spleen, KID: kidney, TES: testis, MUS, muscle, and LEU: leukocytes.
Mouse mCB2 genomic structure (4D3) and tissue expression patterns
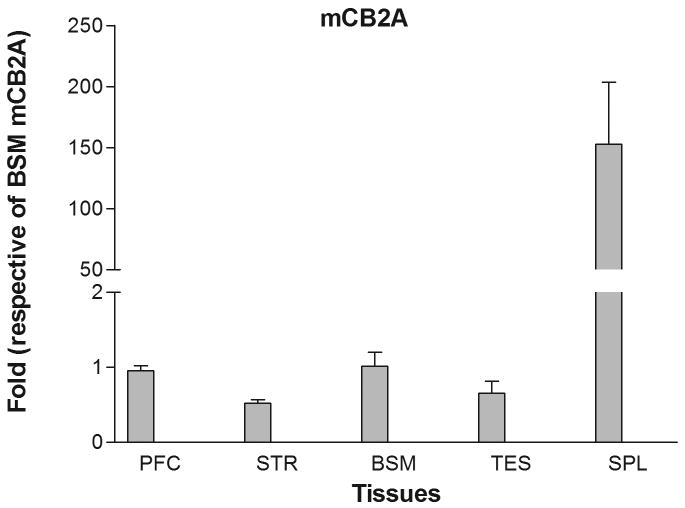
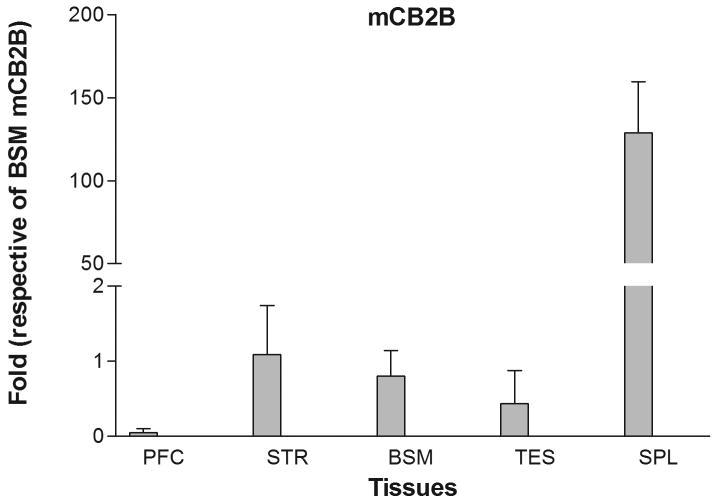
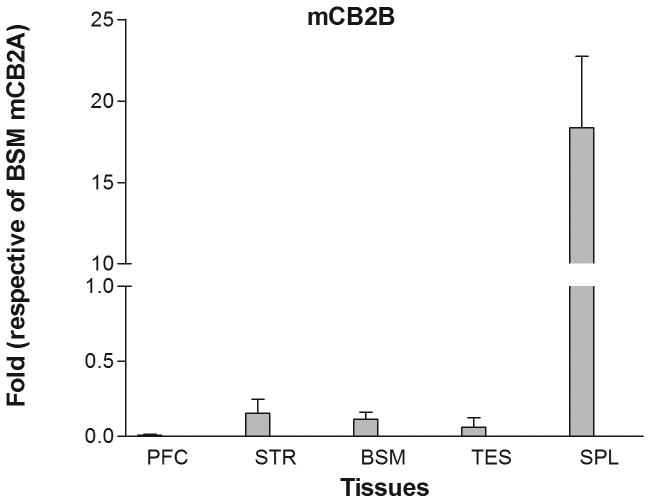
Mouse mCB2 gene contains three exons with two separate promoters (Figure 1B) (Onaivi et al. 2006). Transcription of the first cloned mouse mCB2A cDNA (X86405) was initiated from the first exon (Shire et al. 1996) and that of the mouse mCB2B cDNA clones from FANTOM Consortium and Mammalian Gene Collection (AK036658 and BC024052, respectively) was initiated from exon 2 (Figure 1B) (Okazaki et al. 2002; Strausberg et al. 2002). However, the tissue expression patterns of the specific mCB2A and mCB2B isoforms were not known. We designed the isoform specific TaqMan probes across exon 1/3 and exon 2/3 (Table 1 and Figure 1B) to determine mCB2A and mCB2B tissue expression patterns, respectively. Mouse mCB2A and mCB2B were expressed predominantly in spleen although the mCB2A and mCB2B isoform transcripts were also detected in brain regions such as frontal cortex, striatum, and brain stem at about 1% of the spleen expression (Figure 3A and 3B). Although mCB2A and mCB2B tissue expression patterns were similar, mCB2A mRNA levels were generally about five fold higher than mCB2B using brain stem CB2A as a reference (Figure 3C); therefore, mouse mCB2A isoform is expressed at higher levels in brain regions and other peripheral tissues.
Figure 3.
Tissue expression patterns of mouse mCB2A and mCB2B isoforms: Error bars were derived from standard error of means (SEM) from six mice. A. mouse mCB2A isoform expression in different brain regions and peripheral tissues using mouse brain stem mCB2A as a reference; B. mouse mCB2B isoform expression in different brain regions and peripheral tissues using mouse brain stem mCB2B as a reference; C. mCB2A isoform expression patterns using mouse brain stem mCB2A as a reference. The abbreviations for brain regions and peripheral tissues are PFC: prefrontal cortex, STR: striatum, BSM: brain stem, TES: testis, and SPL: spleen.
Up-regulation of undeleted portion of mCB2 in the C-terminal knockout mouse tissues
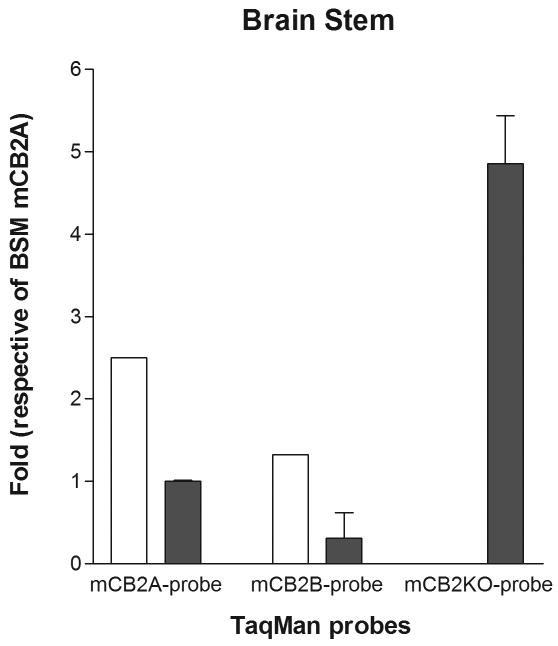
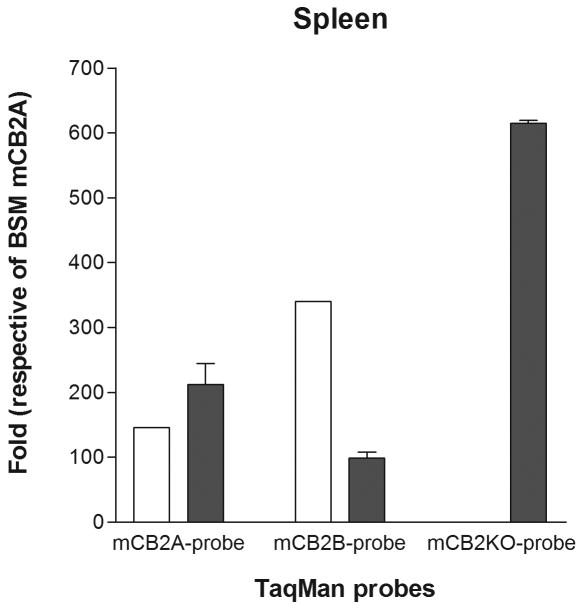
We further investigated the promoter activities of mCB2A and mCB2B in the knockout mouse brain and peripheral tissues, because the knockout strain deleted the C-terminal 131 amino acid including last two transmembrane and intracellular domains (Buckley 2008; Buckley et al. 2000). We found that there was a compensatory effect of mCB2A and mCB2B promoter activities that up-regulate mCB2A and mCB2B isoforms in the brain stem of mCB2 knockout mouse (Figure 4A) using wild-type brain stem mCB2A as a reference. The expression level of mCB2B was also up-regulated, although mCB2A expression was reduced in the spleen tissue of mCB2 knockout mice (Figure 4B). We further designed the TaqMan probe mCB2-KO (Table 1) that hybridized to the deleted C-terminus of mCB2 knockout mouse (Buckley et al. 2000) to validate the mCB2 expression in the mCB2 knockout mouse. We found that the knockout mouse did not express the deleted portion of mCB2 in the brain stem and spleen, however, the mCB2-KO TaqMan probe could readily detect the C-terminal expression in the brain stem and spleen tissue of the wild-type mouse (Figure 4A, and 4B).
Figure 4.
Detection of mCB2 tissue expression in the C-terminus deletion of mCB2 knock out mouse (open bars) and wild type (filled bars) by mCB2A, mCB2B, and mCB2-KO TaqMan probes. Figure 4A, and B represent brain stem (BSM) and spleen (SPL), respectively.
Rat rCB2 genomic structure (5q36) and novel isoforms without retroposon B2 insertion at 3′UTR
Rat rCB2 gene was reported previously to contain three coding exons that encode two different intracellular C-terminal peptides with one homologous (Griffin et al. 2000) and the other non-homologous with hCB2 and mCB2 C-terminal peptides (Brown et al. 2002). The upstream promoters of rCB2 gene were not reported in the literature. In order to identify rCB2 isoforms by PCR cloning, we used homologous region of mouse exon 1 and 2 for forward primers and used downstream exon 3 sequence for the reverse primers (Table 1 and Figure 1C). We found that rCB2 gene also contains isoform rCB2A and rCB2B that were transcribed from upstream promoters of exon 1 and 2, respectively, similar to mCB2 gene (Figure 1C). The rCB2 genomic region of 29 kb is also significantly shorter than that of the hCB2 gene of 90 kb. Surprisingly, we found only one coding exon instead of three coding exons (Brown et al. 2002) in Sprague-Dawley and Long Evans rat cDNA templates synthesized from spleen and brain regions (Figure 1C). The uninterrupted coding exon encodes rCB2 of 360 amino acids with shorter intracellular C-terminal peptide (Figure 5) as Griffin et al. reported (Griffin et al. 2000) than that of previously cloned rCB2 of 410 amino acids (Brown et al. 2002). We named the rat C-terminal isoform as rCB2360 and rCB2410 for rCB2 isoforms containing 360 and 410 amino acids, respectively. Interestingly, rCB2360 isoform contains the same size of hCB2 intracellular C-terminal peptide with 67% identity and is 13 amino acids longer than that of mCB2 intra-cellular C-terminal region with 72% identity (Figure 5). We could not detect rCB2410 in the rat strains of Sprague-Dawley and Long Evans that were used for the experiment; therefore, we could not compare the expression levels of rCB2360 and rCB2410 isoforms. The tissue expression patterns of rCB2A and rCB2B were very similar with mCB2A and mCB2B (data not shown, primer sequences are listed in Table 1).
Figure 5.
Alignment of the C-terminal intracellular peptide sequences of CB2 receptors from human, mouse, rat rCB2360, and rCB2410 isoforms. The numbers represent the amino acid numbers in each of the CB2 receptors from different species and isoforms. Asterisks represent identical amino acids; colons and dots represent similar amino acids. Bold and underlined S represents predicted PKA phosphorylation and PKC phosphorylation sites (Score> 0.65, NetPhosK 1.0: http://www.cbs.dtu.dk/services/NetPhosK/).
There is a rat B2 retroposon insertion (flanked by direct repeat) into the 3′ UTR region of the major coding exon in the reported rat rCB2 cDNA clone, (Brown et al. 2002) as well as the NCBI rat genomic sequence. Using primers that flank the B2 retroposon insertion site (Table 1), we demonstrated that the Sprague-Dawley and Long Evans rats that we used did not contain B2 retroposon insertion (data not shown). Indeed, Celera rat genome sequence does not contain the B2 retroposon insertion of the major coding exon of rCB2 gene.
Species Differences of CB2 gene between human, mouse, and rat
Including the newly identified exons of hCB2A, the human CNR2 gene spans the 90 kb genomic region, which is about four times larger than the 23 kb mouse mCB2 gene and 29 kb of rat rCB2 gene (Figure 1A, 1B, and 1C), respectively. Using the default setting of NCBI Blast 2 program to align human and rodent exon sequences, the only region that aligns significantly is in the protein coding region and no significant alignments could be found in the 5′ UTR and 3′ UTR sequences between human and rodent rCB2 and mCB2 cDNA sequences. The full-length of human hCB2 3′UTR sequence is 2,500 bp and 3,000 bp longer than that of mouse and rat 3′UTR sequences, respectively. In contrast, mouse and rat 5′UTR and 3′UTR were well aligned with more than 85% identity. However, mCB2 contains longer 3′UTR (779 bp longer) than that of rCB2. Mouse mCB2 contains a stop codon, that precedes the C-terminal 13 amino acids of human hCB2 and rat rCBS isoforms, although the remaining 12 amino acids of mCB2 after stop codon are completely identical to rCB360 isoform. It is possible that a nonsense mutation occurred during mouse evolution because all other mammalian species such as rat, bovine, sheep, dog, rhesus monkey, and chimpanzee do not contain the premature stop codon as does mouse mCB2 (Figure 5).
Although human and mouse CB2 genes both contain two promoters, the 5′ flanking sequences of the transcription initiation exons share no significant homology and the promoter activities of hCB2 and mCB2 genes are quite different. Human hCB2A is expressed predominantly in testis and hCB2B predominantly in spleen while both rodent CB2A and CB2B are expressed predominantly in spleen. Human hCB2A promoter transcription is stronger in testis and brain regions while hCB2B promoter transcription is stronger in spleen, leukocytes, and other peripheral tissues (Figure 2A, 2B, and 2C). In contrast, mouse mCB2A promoter activity is about five times stronger in the expressed tissues than that of mCB2B (Figure 3A, 3B, and 3C). The common features of human and rodent CB2 tissue expression patterns are the isoform-specific predominant tissue expression such as human hCB2A in testis, human hCB2B in spleen, and rodent mCB2A and mCB2B in spleen (Figure 2 and 3). The detectable brain expression of hCB2A was at very low levels at 1% of that of hCB2A in testis and.0.07% of that of hCB2B in spleen; however, the detectable brain expression of mCB2A was measurable at 1% of that of mCB2A in spleen. Therefore, CB2A isoform expression is higher in mouse brain than in human brain. In general, human hCB2B expression levels were higher than hCB2A except in testis and brain regions. Mouse mCB2A expression levels were generally higher in both brain and peripheral tissues. The gene regulation mechanisms might differ in human and mouse CB2 gene.
Regulation of mCB2 isoform expression by CB-R ligands in mouse brain regions and autistic mouse model of BTBR T+tF/J strain
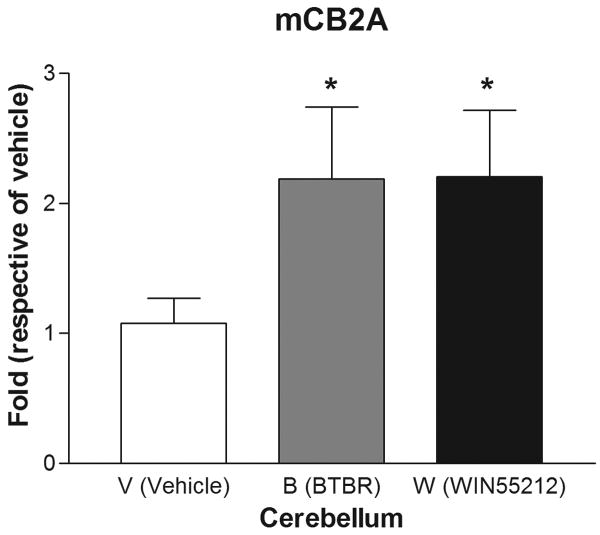
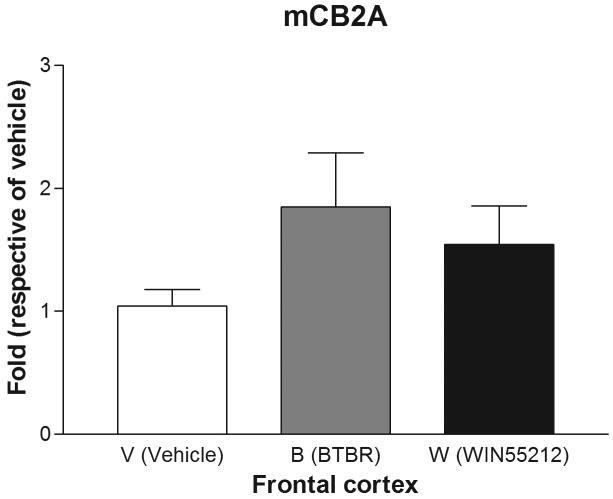
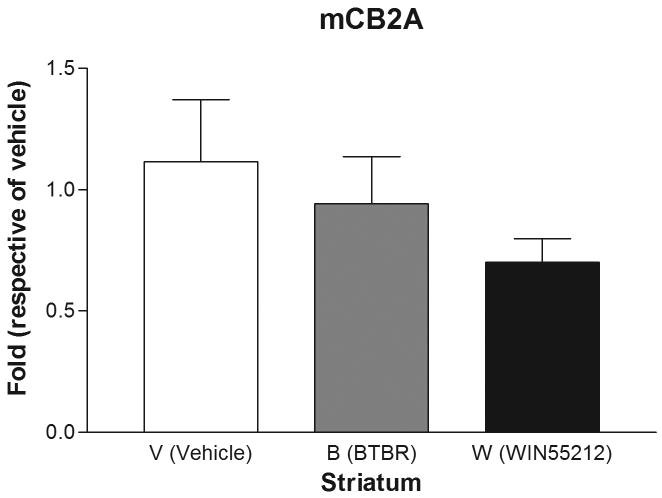
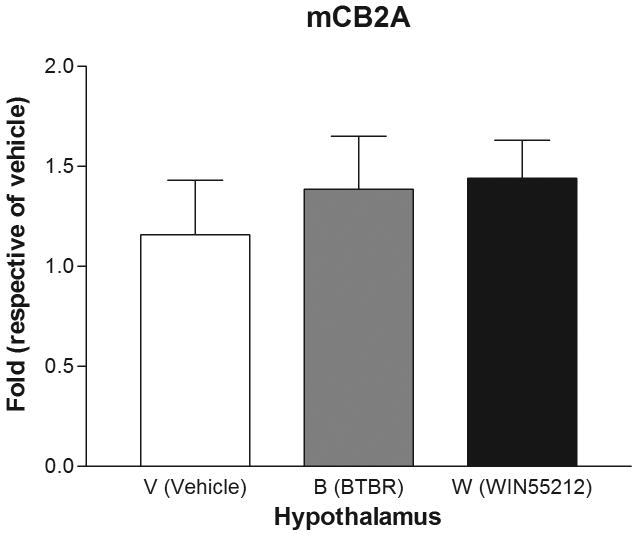
Are mice mCB2A and mCB2B receptor isoforms regulated differentially by treatment with the mixed cannabinoid agonist and CB2-R ligands? We therefore determined mCB2 gene expression in mice brain regions after sub-acute treatment with CB2-R ligands using CB1-R selective antagonist AM251 (3 mg/kg) and CB2-R selective antagonist AM 630 (10 mg/kg) and the mixed cannabinoid receptor agonist WIN55212-2 (2 mg/kg) after four weeks of treatment (Figure 6). We did not observe any significant changes of mCB2A and mCB2B in peripheral tissues such as spleen and testis (data not shown). We did not observe any significant expression changes in AM251 and AM630 treatment for CB1- and CB2-specific antagonist treatments, respectively, in brain regions (data not shown). However, there were mouse strain and brain regional specific mCB2 gene expression profiles following sub-acute administration with the mixed cannabinoid receptor agonist WIN55212-2 (2 mg/kg). In the cerebellum, for example, mCB2A was significantly (t test: t1=2.39, p=0.0438) upregulated by treatment of C57BL/6J mice with WIN55212-2 in comparison to vehicle controls. Interestingly, mCB2A was also increased significantly in the cerebellum of the BTBR T+tF/J strain of mouse, compared to C57BL/6J strain (Figure 6A). mCB2A isoform expression was increased about one fold in cerebellum in the mixed agonist WIN55212-2 treatment although no significant changes were observed in other brain regions such as frontal cortex, striatum, and hypothalamus (Figure 6B, C, and D). mCB2B regulation in mouse brain regions were not shown because mCB2B expression is several times lower than that of mCB2A and the mRNA levels could not be measured reliably by TaqMan assay.
Figure 6.
Regulation of mCB2A by the mixed CB-R agonist WIN55212-2 after seven days of sub-acute treatment in C57BL/6Js and in the brain regions of BTBR T+tF/J autism mouse model. Error bars were derived from standard error of means (SEM) from 6 mice. A: cerebellum, B: frontal cortex, C: striatum, and D: hypothalamus
Discussion
New data obtained from these studies shed some light on the ambiguity and sometimes confusing structure and expression patterns of CB2/CNR2 gene in different species (Brown et al. 2002; Munro et al. 1993; Shire et al. 1996). Using isoform-specific TaqMan probes, we have identified a novel human hCB2A isoform that is expressed predominantly in testis and at lower levels in brain regions of reward system in contrast to hCB2B that is expressed predominantly in spleen with very low expression in brain. We showed that the different sizes (2.5 and 5.0 kb) of hCB2 mRNA observed in Northern blot (Munro et al. 1993; Schatz et al. 1997) were due to alternative polyadenylation of the extended 3′UTR of hCB2 mRNA. Mouse mCB2A and mCB2B are both expressed predominantly in spleen with mCB2A expression about five times higher in the expressed tissues. mCB2 knock-out strain with the C-terminal deletion (Buckley et al. 2000) showed enhanced mCB2A and mCB2B promoter activities that might reflect increased expression of the truncated mCB2 receptor. The N-terminal specific antibody against mCB2 shall be used in the future experiment to determine whether the truncated mCB2 protein is indeed up-regulated. The truncated mCB2 receptor might introduce gain of function and influence interpretation of pharmacological and behavioral experiment results on mCB2 knock-out mice.
We also identified a rat rCB2360 short isoform (Griffin et al. 2000) that contains a shorter intracellular C-terminus peptide than previously identified rat rCB2410 long isoform (Brown et al. 2002). The rCB2360 C-terminus intracellular region is more homologous to human and mouse mCB2 C-terminal peptide than that of rCB2410 isoform. It is possible that the B2 retroposon insertion into the 3′ UTR of rCB2 alters the splicing pattern and created two cryptic alternative splicing sites that caused a frame shift and generated different intracellular C-terminal sequence of rCB2 protein, which also was shown by different sizes of rCB2 mRNA on Northern blot (Brown et al. 2002). Subsequently, Brown et. al., noted a G-to-T transversion following codon 343 of rCB2 might introduce a putative GT splice donor site that initiates an intron (Brown et al. 2002). However, Ashton et al., found that the 54 nucleotides following the G-to-T transversion share high identity with homologous hCB2 cDNA sequence including the stop codon (Ashton et al. 2008). On the other hand, another cDNA sequence (AF286722) from the same laboratory (Brown et al. 2002) contains upstream exon 1 spliced to the uninterrupted exon 3 of the rCB2 with B2 retroposon insertion sequence. Translation of this uninterrupted rCB2A sequence produced the same C-terminal intracellular peptide sequence as did rCB2360. It is very likely that the major rCB2 isoform is rCB2360. We could not find G-toT transversion or the B2 insertion in the rat strains that we used for the experiment. The expression levels of rCB2360 and rCB2410 isoforms, and the 3′UTR with or without B2 insertion, therefore, could not be measured. It would be interesting to use proteomic approach to identify rCB2360 and rCB2410 isoforms once the G-to-T transversion and B2 insertion are identified in rat strains. The C-terminal intracellular domain of CB2 interacts with Gi/o protein that is negatively coupled to adenylyl cyclase (Bayewitch et al. 1995) and activates the MAP kinase signal transduction pathway (Wartmann et al. 1995). The differences in rat rCB2360 and rCB2410 isoform might influence the coupling of Gi/o with adenylyl cyclase and downstream signal transduction because rat rCB2410 contains additional PKA and PKC consensus sites (S393 and S399, respectively) besides the common PKA consensus site (S336) that is conserved in the CB2 isoforms of other mammalian species (Figure 5). It would be interesting to study the pharmacological and behavioral differences of rat strains that contain different C-terminal intracellular peptide sequences.
Retroposon B2 integration into 3′UTR of rCB2 might also affect rCB2 expression. Retroviral integration was also observed in mCB2 gene and mCB2 3′ UTR contains a frequent provirus insertion site (Evi 11) after Ca-Br-M murine leukemia virus (MuLV) inoculation of NIH/Swiss mice that developed malignancies (Joosten et al. 2000; Valk et al. 1997). The special 3′UTR sequences of rodent CB2 and the chromatin structure in this region might form an open DNA conformation to attract retropositions as observed in other genomic regions (Liu & Chan 1990). The B2 retroposon insertion into similar 3′ UTR of rat rCB2 gene might also change the expression of rCB2 and influence hematopoietic differentiation.
The specific human hCB2A isoform that is expressed predominantly in testis might indicate that human hCB2 has a potential function related to spermatogenesis and fertilization. Chronic administration of (-)-Δ9-tetrahydrocannabinol (THC) induces impotence, reduction of testosterone, and decrease of sperm viability (Hall & Solowij 1998; Murphy et al. 1994; Park et al. 2004). Mouse immature Sertoli cells of testis express mCB2 receptor on the plasma membrane with a CB2-specific agonist binding profile (Maccarrone et al. 2003). Higher expression of hCB2A isoform in human testis might render humans more vulnerable to male gametoxicity by cannabinoids. The detailed localization of hCB2A in human testis needs to be studied further by in situ hybridization and immunohistochemistry using human testis biopsy samples with specific nucleotide and antibody probes.
The species differences between the human and rodent CB2 gene are striking. A meta-analysis of previous studies showed significant differences in ligand-binding affinities between hCB2 and rCB2 (McPartland et al. 2007a). Human hCB2 gene is four times larger than that of rodents. If the transcription rates are similar between human and rodents, hCB2A isoform would take much longer time to be transcribed in testis and brain. This is unusual because other gene orthologs between humans and mice are usually within one fold differences in genomic sizes. The CB2A promoter distances to the spliced coding exon and the species-specific splicing mechanisms might reflect tissue-specific expression differences between humans and mice. CB2A isoform could be detected in human and mouse brain regions at such low levels of <0.1% and <1% of the highest expressed CB2 isoform of human and mouse spleen expression, respectively. However, a selective postsynaptic localization pattern has been observed in rat hippocampus (Brusco et al. 2008). CB2A expression level was higher in mouse brain than in human brain but such immunohistochemical signals of CB2 has yet to be found in human brain regions.
Such promoter-specific CB2-R isoform distribution may in part explain why CB2-Rs were previously undetectable in both human and rodent brains using PCR primers designed in the region of CB2 exon 3 and Northern blot analysis (Brown et al. 2002; Griffin et al. 1999; Munro et al. 1993). Our study of CB2 isoform expression was more specific because the TaqMan probes were designed to hybridize the promoter exons and the major coding exon. TaqMan assay is more sensitive than in situ hybridization that was unable to detect significant mCB2 signals in mouse brain (Griffin et al. 1999; Munro et al. 1993). The weak mRNA signals of CB2 in brain would possibly be overcome by in situ PCR on brain slides using CB2A-specific primers to determine cellular types that express CB2A. Therefore, recent detection of mCB2 brain expression in mice by immunohistochemistry (Baek et al. 2008; Brusco et al. 2008; Gong et al. 2006; Van Sickle et al. 2005) might not represent human CB2 brain expression patterns because of different promoter activities in terms of quantity and quality for tissue distributions between human and rodents. The CB2 isoform specific TaqMan probes could differentiate the isoform-specific expression and are more sensitive and specific than CB2 antibodies that are currently available (N-terminal epitopes, Cayman Chemical Company and Sigma-Aldrich or C-terminal epitope, Santa Cruz Biotechnology, Inc.) that could not detect CB2 isoforms. The controversial CB2 brain expression in the literature could be of low expression of CB2A isoform in brain regions and less specific CB2 commercial antibodies in immunohistochemistry studies, especially those studies using antibodies against human hCB2 epitopes for rodent brain immunostaining. The specificities of CB1 receptor antibodies were also controversial because they could not detect the native and transfected CB1 antigen although they recognized unspecific proteins in Western blot and immunohistochemistry (Grimsey et al. 2008). The mCB2 knockout strain with ablation of N-terminal peptide of 156 amino acids (Deltagen, Inc. San Mateo, CA) could clarify the specificity of the antibodies that were used against the N-terminal epitopes (human CB2 1-33 amino acids from Sigma and human10-33 amino acids from Cayman Chemical Company). Another CB2 knockout strain with ablation of C-terminal peptide of 131 amino acids (Buckley et al. 2000) could clarify the specificity of the antibodies that were used against C-terminal epitopes (human CB2 301-360 amino acids from Santa Cruz and rat 328-342 amino acids from Ken Mackie's lab) (Suarez et al. 2008).
CB2-specific agonists are under development for therapeutic treatment of inflammatory and neuropathic pain (Guindon & Hohmann 2008) with minimum psychoactive effects (Malan et al. 2003). Although cannabinoids significantly reduce chronic pain in animal models, they have less analgesic effects in human subjects (Malan et al. 2003). The identification of novel human hCB2A isoform that is expressed at low levels in different brain regions might provide targets for any psychoactive side effects for high doses of CB2 agonist-mediated anti-nociception treatment in humans that are not observed in mouse and rat models (Campbell et al. 2001). Another possible side effect that should be considered is associated with the reproductive system because hCB2A is expressed at a high level in human testis. The differential polyadenylation at 3′UTR that generates 2.5 and 5.0 kb human hCB2 mRNA species might target hCB2 to different cell types and even different sub-cellular localizations (An et al. 2008; Liu et al. 2005) that might also differ from human and rodent.
The differential promoter activities and non-homologous 5′UTR and 3′UTR sequences in human and rodents might explain differential anti-nociceptive effects of CB2 agonists in human and rodent models. CB2 gene is conserved from fish to human and only an ancestral CB-R sequence was identified in some invertebrates (McPartland et al. 2006). Human CNR2 gene structure and transcription mechanisms are quite different from those of rodent Cnr2 gene. It would be interesting to compare the human CNR2 gene with other primate Cnr2 genes, however, the Cnr2 genomic sequences are still unavailable from other primate species such as chimpanzee and rhesus monkey. The CB2 gene might evolve more rapidly in recent evolution time than CB1 (McPartland et al. 2007b) to generate such species differences of CB2 gene structure, tissue expression, and regulation that should be taken into account for developing analgesic drugs for different species. hCB2A gene is possibly still evolving because many new transcripts first appear in testes that possess the facilitated transcription mechanism. Some functional genes could then migrate out of testes to other tissues (Vinckenbosch et al. 2006).
One interesting result from our study is that the BTBR T+tF/J strain that has been reported to exhibit autism-like behavioral phenotypes demonstrated a higher basal wheel running activity and was significantly sensitive to a low dose of BML-190 that induced wheel running activity in the BTBR mice in comparison to vehicle-treated controls (data not shown). While recent data indicate increasing rates of autism in children, future studies should investigate the role of the cannabinoid system in the etiology of autism spectrum disorders. Some recent events have linked the anti-obesity drug Rimonabant, a CB1-R antagonist, an appetite suppressant with a higher risk of depression and suicide. However, our results showing an association of CNR2 gene and depression in a human population (Onaivi et al. 2008) and an animal model suggest that cannabinoid CB2-R may be involved in the endocannabinoid signaling mechanisms associated with the regulation of emotionality. Thus, more studies are required to determine if CB2-R ligands have the risk of depression or suicide that has led to the withdrawal of Rimonabant from use as an appetite suppressant in control of obesity. The studies here provide some of the first opportunities to elucidate candidate promoter regions for the hCB2 gene of continuing interest.
Acknowledgments
This work was supported in part by NIDA/IRP and ESO acknowledges the technical assistance and financial support from William Paterson University Center for Research, Biology department, the Dean's (Dr. De Young) student worker fund, and the Provost office for assigned release time. The CB2 knockout and their wild-type control mice were developed by Buckley et al., 2000, and obtained from NIAAA through Dr. Kunos. Part of human brain tissue was obtained from the NICHD Brain and Tissue Bank for Developmental Disorders at the University of Maryland, Baltimore, MD. The authors would also like to acknowledge the technical assistance of Dr. Xia Li from Chemical Biology Research Branch and English editing by Dr. Mary Pfeiffer, Editor and Writer NIDA, IRP. ALB and MPV acknowledges Red de Trastornos Adictivos (RD06/0001/1013). We thank Dr. John McPartland, for his excellent and in-depth review of the manuscript and his suggestions.
Footnotes
References
- Abood ME. Molecular biology of cannabinoid receptors. Handb Exp Pharmacol. 2005:81–115. doi: 10.1007/3-540-26573-2_3. [DOI] [PubMed] [Google Scholar]
- An JJ, Gharami K, Liao GY, Woo NH, Lau AG, Vanevski F, Torre ER, Jones KR, Feng Y, Lu B, Xu B. Distinct role of long 3′ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell. 2008;134:175–187. doi: 10.1016/j.cell.2008.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashton JC, Wright JL, McPartland JM, Tyndall JD. Cannabinoid CB1 and CB2 receptor ligand specificity and the development of CB2-selective agonists. Curr Med Chem. 2008;15:1428–1443. doi: 10.2174/092986708784567716. [DOI] [PubMed] [Google Scholar]
- Baek JH, Zheng Y, Darlington CL, Smith PF. Cannabinoid CB2 receptor expression in the rat brainstem cochlear and vestibular nuclei. Acta Otolaryngol. 2008;128:961–967. doi: 10.1080/00016480701796944. [DOI] [PubMed] [Google Scholar]
- Bayewitch M, Avidor-Reiss T, Levy R, Barg J, Mechoulam R, Vogel Z. The peripheral cannabinoid receptor: adenylate cyclase inhibition and G protein coupling. FEBS Lett. 1995;375:143–147. doi: 10.1016/0014-5793(95)01207-u. [DOI] [PubMed] [Google Scholar]
- Benito C, Tolon RM, Pazos MR, Nunez E, Castillo AI, Romero J. Cannabinoid CB2 receptors in human brain inflammation. Br J Pharmacol. 2008;153:277–285. doi: 10.1038/sj.bjp.0707505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SM, Wager-Miller J, Mackie K. Cloning and molecular characterization of the rat CB2 cannabinoid receptor. Biochim Biophys Acta. 2002;1576:255–264. doi: 10.1016/s0167-4781(02)00341-x. [DOI] [PubMed] [Google Scholar]
- Brusco A, Tagliaferro P, Saez T, Onaivi ES. Postsynaptic localization of CB2 cannabinoid receptors in the rat hippocampus. Synapse. 2008;62:944–949. doi: 10.1002/syn.20569. [DOI] [PubMed] [Google Scholar]
- Buckley NE. The peripheral cannabinoid receptor knockout mice: an update. Br J Pharmacol. 2008;153:309–318. doi: 10.1038/sj.bjp.0707527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley NE, McCoy KL, Mezey E, Bonner T, Zimmer A, Felder CC, Glass M, Zimmer A. Immunomodulation by cannabinoids is absent in mice deficient for the cannabinoid CB(2) receptor. Eur J Pharmacol. 2000;396:141–149. doi: 10.1016/s0014-2999(00)00211-9. [DOI] [PubMed] [Google Scholar]
- Campbell FA, Tramer MR, Carroll D, Reynolds DJ, Moore RA, McQuay HJ. Are cannabinoids an effective and safe treatment option in the management of pain? A qualitative systematic review. Bmj. 2001;323:13–16. doi: 10.1136/bmj.323.7303.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerard C, Mollereau C, Vassart G, Parmentier M. Nucleotide sequence of a human cannabinoid receptor cDNA. Nucleic Acids Res. 1990;18:7142. doi: 10.1093/nar/18.23.7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong JP, Onaivi ES, Ishiguro H, Liu QR, Tagliaferro PA, Brusco A, Uhl GR. Cannabinoid CB2 receptors: immunohistochemical localization in rat brain. Brain Res. 2006;1071:10–23. doi: 10.1016/j.brainres.2005.11.035. [DOI] [PubMed] [Google Scholar]
- Griffin G, Tao Q, Abood ME. Cloning and pharmacological characterization of the rat CB(2) cannabinoid receptor. J Pharmacol Exp Ther. 2000;292:886–894. [PubMed] [Google Scholar]
- Griffin G, Wray EJ, Tao Q, McAllister SD, Rorrer WK, Aung MM, Martin BR, Abood ME. Evaluation of the cannabinoid CB2 receptor-selective antagonist, SR144528: further evidence for cannabinoid CB2 receptor absence in the rat central nervous system. Eur J Pharmacol. 1999;377:117–125. doi: 10.1016/s0014-2999(99)00402-1. [DOI] [PubMed] [Google Scholar]
- Grimsey NL, Goodfellow CE, Scotter EL, Dowie MJ, Glass M, Graham ES. Specific detection of CB1 receptors; cannabinoid CB1 receptor antibodies are not all created equal! J Neurosci Methods. 2008;171:78–86. doi: 10.1016/j.jneumeth.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Guindon J, Hohmann AG. Cannabinoid CB2 receptors: a therapeutic target for the treatment of inflammatory and neuropathic pain. Br J Pharmacol. 2008;153:319–334. doi: 10.1038/sj.bjp.0707531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall W, Solowij N. Adverse effects of cannabis. Lancet. 1998;352:1611–1616. doi: 10.1016/S0140-6736(98)05021-1. [DOI] [PubMed] [Google Scholar]
- Liu QR, Chan PK. Identification of a long stretch of homopurine. homopyrimidine sequence in a cluster of retroposons in the human genome. J Mol Biol. 1990;212:453–459. doi: 10.1016/0022-2836(90)90324-F. [DOI] [PubMed] [Google Scholar]
- Liu QR, Walther D, Drgon T, Polesskaya O, Lesnick TG, Strain KJ, de Andrade M, Bower JH, Maraganore DM, Uhl GR. Human brain derived neurotrophic factor (BDNF) genes, splicing patterns, and assessments of associations with substance abuse and Parkinson's Disease. Am J Med Genet B Neuropsychiatr Genet. 2005;134B:93–103. doi: 10.1002/ajmg.b.30109. [DOI] [PubMed] [Google Scholar]
- Maccarrone M, Cecconi S, Rossi G, Battista N, Pauselli R, Finazzi-Agro A. Anandamide activity and degradation are regulated by early postnatal aging and follicle-stimulating hormone in mouse Sertoli cells. Endocrinology. 2003;144:20–28. doi: 10.1210/en.2002-220544. [DOI] [PubMed] [Google Scholar]
- Mackie K. Cannabinoid receptors: where they are and what they do. J Neuroendocrinol. 2008;20 1:10–14. doi: 10.1111/j.1365-2826.2008.01671.x. [DOI] [PubMed] [Google Scholar]
- Malan TP, Jr, Ibrahim MM, Lai J, Vanderah TW, Makriyannis A, Porreca F. CB2 cannabinoid receptor agonists: pain relief without psychoactive effects. Curr Opin Pharmacol. 2003;3:62–67. doi: 10.1016/s1471-4892(02)00004-8. [DOI] [PubMed] [Google Scholar]
- McPartland JM, Glass M, Pertwee RG. Meta-analysis of cannabinoid ligand binding affinity and receptor distribution: interspecies differences. Br J Pharmacol. 2007a;152:583–593. doi: 10.1038/sj.bjp.0707399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPartland JM, Matias I, Di Marzo V, Glass M. Evolutionary origins of the endocannabinoid system. Gene. 2006;370:64–74. doi: 10.1016/j.gene.2005.11.004. [DOI] [PubMed] [Google Scholar]
- McPartland JM, Norris RW, Kilpatrick CW. Tempo and mode in the endocannaboinoid system. J Mol Evol. 2007b;65:267–276. doi: 10.1007/s00239-007-9004-1. [DOI] [PubMed] [Google Scholar]
- Munro S, Thomas KL, Abu-Shaar M. Molecular characterization of a peripheral receptor for cannabinoids. Nature. 1993;365:61–65. doi: 10.1038/365061a0. [DOI] [PubMed] [Google Scholar]
- Murphy LL, Gher J, Steger RW, Bartke A. Effects of delta 9-tetrahydrocannabinol on copulatory behavior and neuroendocrine responses of male rats to female conspecifics. Pharmacol Biochem Behav. 1994;48:1011–1017. doi: 10.1016/0091-3057(94)90213-5. [DOI] [PubMed] [Google Scholar]
- Nunez E, Benito C, Tolon RM, Hillard CJ, Griffin WS, Romero J. Glial expression of cannabinoid CB(2) receptors and fatty acid amide hydrolase are beta amyloid-linked events in Down's syndrome. Neuroscience. 2008;151:104–110. doi: 10.1016/j.neuroscience.2007.10.029. [DOI] [PubMed] [Google Scholar]
- Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, Nikaido I, Osato N, Saito R, Suzuki H, Yamanaka I, Kiyosawa H, Yagi K, Tomaru Y, Hasegawa Y, Nogami A, Schonbach C, Gojobori T, Baldarelli R, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- Onaivi ES, Ishiguro H, Gong JP, Patel S, Meozzi PA, Myers L, Perchuk A, Mora Z, Tagliaferro PA, Gardner E, Brusco A, Akinshola BE, Hope B, Lujilde J, Inada T, Iwasaki S, Macharia D, Teasenfitz L, Arinami T, et al. Brain neuronal CB2 cannabinoid receptors in drug abuse and depression: from mice to human subjects. PLoS ONE. 2008;3:e1640. doi: 10.1371/journal.pone.0001640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onaivi ES, Ishiguro H, Gong JP, Patel S, Perchuk A, Meozzi PA, Myers L, Mora Z, Tagliaferro P, Gardner E, Brusco A, Akinshola BE, Liu QR, Hope B, Iwasaki S, Arinami T, Teasenfitz L, Uhl GR. Discovery of the presence and functional expression of cannabinoid CB2 receptors in brain. Ann N Y Acad Sci. 2006;1074:514–536. doi: 10.1196/annals.1369.052. [DOI] [PubMed] [Google Scholar]
- Park B, McPartland JM, Glass M. Cannabis, cannabinoids and reproduction. Prostaglandins Leukot Essent Fatty Acids. 2004;70:189–197. doi: 10.1016/j.plefa.2003.04.007. [DOI] [PubMed] [Google Scholar]
- Pazos MR, Nunez E, Benito C, Tolon RM, Romero J. Role of the endocannabinoid system in Alzheimer's disease: new perspectives. Life Sci. 2004;75:1907–1915. doi: 10.1016/j.lfs.2004.03.026. [DOI] [PubMed] [Google Scholar]
- Schatz AR, Lee M, Condie RB, Pulaski JT, Kaminski NE. Cannabinoid receptors CB1 and CB2: a characterization of expression and adenylate cyclase modulation within the immune system. Toxicol Appl Pharmacol. 1997;142:278–287. doi: 10.1006/taap.1996.8034. [DOI] [PubMed] [Google Scholar]
- Shire D, Calandra B, Rinaldi-Carmona M, Oustric D, Pessegue B, Bonnin-Cabanne O, Le Fur G, Caput D, Ferrara P. Molecular cloning, expression and function of the murine CB2 peripheral cannabinoid receptor. Biochim Biophys Acta. 1996;1307:132–136. doi: 10.1016/0167-4781(96)00047-4. [DOI] [PubMed] [Google Scholar]
- Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, Wagner L, Shenmen CM, Schuler GD, Altschul SF, Zeeberg B, Buetow KH, Schaefer CF, Bhat NK, Hopkins RF, Jordan H, Moore T, Max SI, Wang J, et al. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci U S A. 2002;99:16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez J, Llorente R, Romero-Zerbo SY, Mateos B, Bermudez-Silva FJ, de Fonseca FR, Viveros MP. Early maternal deprivation induces gender-dependent changes on the expression of hippocampal CB(1) and CB(2) cannabinoid receptors of neonatal rats. Hippocampus. 2008 doi: 10.1002/hipo.20537. [DOI] [PubMed] [Google Scholar]
- Sugiura T, Waku K. Cannabinoid receptors and their endogenous ligands. J Biochem. 2002;132:7–12. doi: 10.1093/oxfordjournals.jbchem.a003200. [DOI] [PubMed] [Google Scholar]
- Van Sickle MD, Duncan M, Kingsley PJ, Mouihate A, Urbani P, Mackie K, Stella N, Makriyannis A, Piomelli D, Davison JS, Marnett LJ, Di Marzo V, Pittman QJ, Patel KD, Sharkey KA. Identification and functional characterization of brainstem cannabinoid CB2 receptors. Science. 2005;310:329–332. doi: 10.1126/science.1115740. [DOI] [PubMed] [Google Scholar]
- Vinckenbosch N, Dupanloup I, Kaessmann H. Evolutionary fate of retroposed gene copies in the human genome. Proc Natl Acad Sci U S A. 2006;103:3220–3225. doi: 10.1073/pnas.0511307103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wartmann M, Campbell D, Subramanian A, Burstein SH, Davis RJ. The MAP kinase signal transduction pathway is activated by the endogenous cannabinoid anandamide. FEBS Lett. 1995;359:133–136. doi: 10.1016/0014-5793(95)00027-7. [DOI] [PubMed] [Google Scholar]