Abstract
The H19 large intergenic noncoding RNA (lincRNA) is one of the most highly abundant and conserved transcripts in mammalian development, being expressed in both embryonic and extraembryonic cell lineages, yet its physiological function is unknown. Here we show that miR-675, a microRNA (miRNA) embedded within H19’s first exon, is expressed exclusively in the placenta from the gestational time point when placental growth normally ceases, and placentas that lack H19 continue to grow. Overexpression of miR-675 in a range of embryonic and extraembryonic cell lines results in their reduced proliferation; targets of the miRNA are upregulated in the H19 null placenta, including the growth promoting Insulin-like growth factor 1 receptor (Igf1r). Moreover, the excision of miR-675 from H19 is dynamically regulated by the stress response RNA binding protein HuR. These results suggest that H19’s main physiological role is in limiting growth of the placenta prior to birth, by regulated processing of miR-675. The controlled release of miR-675 from H19 may also allow rapid inhibition of cell proliferation in response to cellular stress or oncogenic signals.
The H19 locus expresses high levels of a 2.5 kb RNA Polymerase II dependent transcript, which is spliced, capped, polyadenylated, and exported into the cytoplasm1, 2. Indeed, H19 is the second most abundant transcript in placenta (Supplementary Fig. S6b) and levels are higher still in fetal liver (Fig. 1c). H19 is imprinted with maternal expression, and has been implicated in tumour suppression, but its physiological function is currently unknown3-6. The H19 transcript contains only short open reading frames, which are poorly conserved between mice and human and thus appears to be a noncoding RNA, one of the first lincRNAs to be discovered1, 7. The H19 gene is located downstream of the growth promoting insulin like growth factor 2 (Igf2) gene on chromosome 7 in mouse and 11p15.5 in humans. Expressed from opposite parental alleles but co-regulated, H19 and Igf2 share a common imprinting mechanism and are found to be deregulated in many cancers and fetal growth syndromes in humans (reviewed in4). The H19 RNA itself does not appear to have a role in the imprinting mechanism, consistent with its cytoplasmic localization1, 2. Instead, the lincRNA has a tumour suppressive effect both in cell culture experiments5 and in in vivo mouse models6, and also has the potential of regulating a network of imprinted genes in trans8. How the RNA exerts these functions and what its physiological role is remains unknown.
Figure 1.
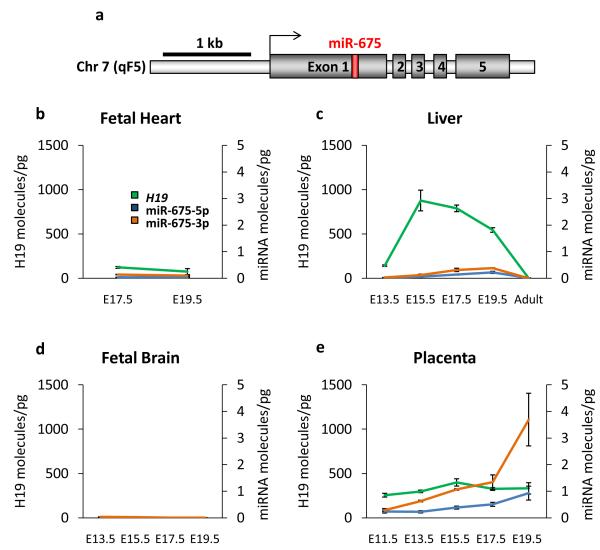
miR-675 is expressed in the late gestation placenta but suppressed in the embryo. (a) Schematic representation of the H19 transcriptional unit with miR-675 shown in red embedded within H19 exon 1. The black arrow indicates the transcriptional start site. (b,c,d,e) Expression determined by qRT-PCR for miR-675-5p (blue), miR-675-3p (orange) and H19 (green) in fetal heart (b), liver (c), fetal brain (d) and placenta (e). Expression levels are expressed as the number of molecules per picogram of total RNA with H19 levels on the left y-axis and miR-675 levels on the right. n ≥ 3. Error bars indicate the s.e.m.
One way by which lincRNAs may acquire functionality is by acting as the precursor to small RNAs capable of regulatory function, such as microRNAs (miRNAs)9-11. Indeed, exon 1 of H19 harbours a miRNA containing hairpin which has been found to serve as the template for two distinct miRNAs, miR-675-5p and miR-675-3p12, and it has been suggested that it may be these miRNAs that confer functionality on H194, 12, 13 (Fig. 1a). Moreover, the miR-675 stem loop was shown to be one of the most highly conserved features of the H19 RNA during mammalian evolution, indicating that selective pressure may be higher on the miRNAs than on H19 as a whole7.
RESULTS
miR-675 is expressed in placenta but processing is strongly inhibited
To investigate the possibility that H19 derives its functionality from miR-675, we determined the expression profile of the miRNA. As H19 is the primary miRNA (pri-miRNA) template for miR-675 we decided to examine its expression in tissues where H19 is highly transcribed. Unexpectedly, both species of miR-675 were barely detectable in fetal liver (endoderm derived tissue) and fetal heart (mesoderm derived tissue) at all stages of embryonic development tested, despite the vast levels of H19 (Fig. 1b, c), suggesting that processing of the miRNA from H19 is inhibited. Indeed, large scale miRNA profiling studies have also found only low levels of miR-675 in the embryo despite abundant H1914, 15. Low levels of miR-675 were also identified in human cell lines16, implying that the inhibition of miR-675 processing is evolutionarily conserved. In contrast to fetal tissues however, we observed miR-675 to be expressed in the placenta exclusively from the maternal allele and with increasing abundance from E11.5 until term, despite there being no change in the abundance of H19 (Fig. 1e, 3a and Supplementary Fig. S1a). This suggests that not only is miR-675 processing inhibited but that this inhibition is a dynamic process that can be relaxed to allow expression of the miRNA. No miR-675 expression was detected in fetal brain, where H19 is absent (Fig. 1d).
Figure 3.
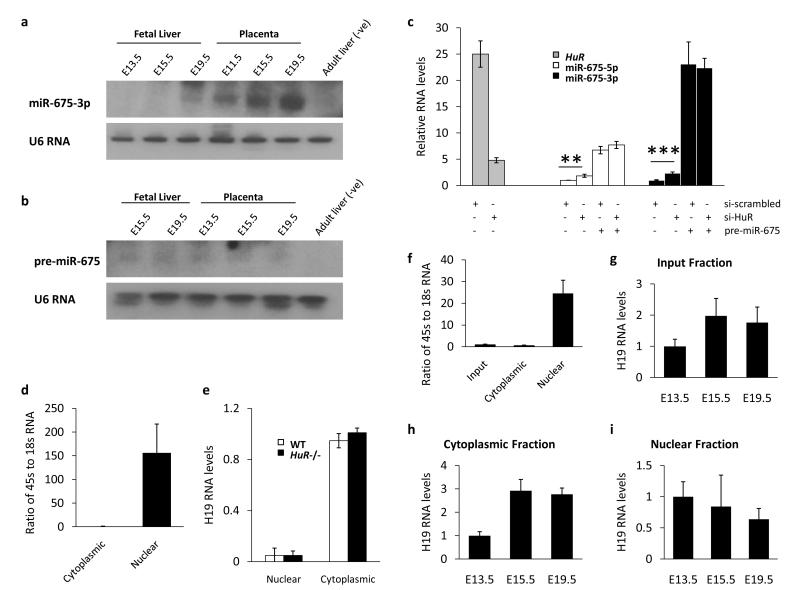
Evidence that HuR inhibits miR-675 at the Drosha processing step. (a, b) Northern blots using probes specific to either miR-675-3p (a) or pre-miR-675 (miRBase accession MI0004123) (b), against total RNA from fetal liver and placenta. Total RNA from adult liver is included as a negative control. (a) The expression pattern here agrees with the qRT-PCR results from Fig. 1e. (b) The image shown was exposed for 72 hours and shows the very low abundance of pre-miR-675 in these tissues. Full scans are shown in Supplementary Fig S8. (c) Expression of HuR, miR-675-5p and miR-675-3p following transfection of synthetic pre-miR-675 and either an siRNA against HuR (si-HuR) or a scrambled control (si-scrambled) in MEF cells, showing that HuR has no effect on Dicer processing of pre-miR-675. n = 3. Statistical significance was determined by Student’s t-test. ** P < 0.01, *** P < 0.001. (d, e) H19 expression in nuclear and cytoplasmic cell fractions from both wild type (WT) and HuR null (HuR-/-) MEF cells, measured by qRT-PCR. To assess the quality of fractionation the nuclear 45s transcript and the cytoplasmic 18s transcript were measured (d). H19 expression was measured in the different fractions (e). n = 6. (f, g, h, i) Nuclear and cytoplasmic fractionation of placental tissue was performed from different developmental time points. Quality of fractionation was assessed by measuring the 45s and 18s transcripts by qRT-PCR (f). H19 expression was measured in the input (g), cytoplasmic (h) and nuclear (i) fractions. n = 3. All error bars show the s.e.m.
By comparison to a miRNA universal standard, which contains known concentrations of most miRNAs, the number of molecules of miR-675 per cell were found to be very low in embryonic tissue (approximately 40 copies in fetal heart and 70 in fetal liver), indicating that miR-675 is most likely non-functional in these tissues (Fig. 1 b - e). When miR-675 reaches its peak of expression in the E19.5 placenta however, there are approximately 300 copies of miR-675-5p and 1000 copies of miR-675-3p per placental cell. The previously characterized miRNA let-7 is known to be functional in HeLa cells at a similar concentration17, 18. Despite this potentially functional abundance, miR-675 levels are only 1 % of H19 levels, demonstrating both the very high abundance of H19 and also an unusually tight inhibition of miRNA processing.
miR-675 processing is inhibited by the RNA binding protein HuR
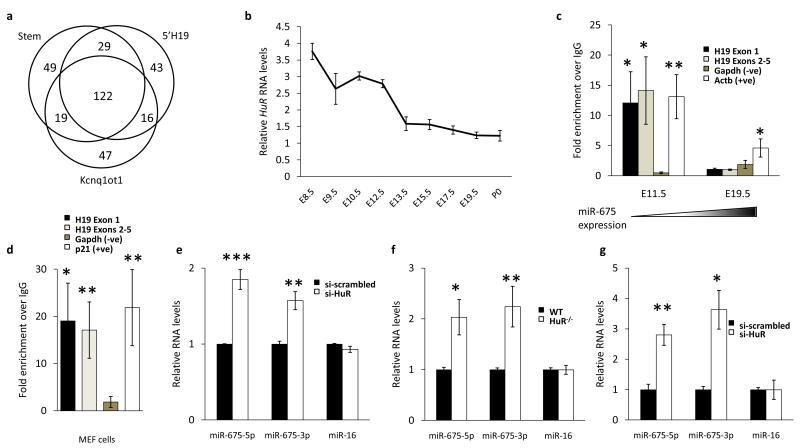
As inhibition appears to be a dynamic process in the placenta, we hypothesized that RNA binding proteins might contribute to miR-675 regulation and hence performed RNA affinity assays followed by mass spectrometry to identify proteins that bind H19 in the region of the miR-675 stem loop (see Methods). We used partially differentiated trophoblast stem (TS) cell lysate as a cell culture approximation of early gestation placenta as these cells do not express miR-675 despite robust H19 levels (Supplementary Fig. S1b, c). Using this approach we identified 49 proteins that were bound selectively to H19 in the region of the miR-675 stem loop (Fig. 2a). Notably, these included a number of proteins that are implicated in RNA metabolism and small RNA regulation, such as Upf1, Zcchc11, Luc7l and HuR. Amongst these, the RNA binding protein HuR stood out as a candidate for regulating miR-675 processing, for a number of reasons: first, there is a putative HuR binding site in H19 55 bp upstream from the miR-675 stem loop (AUUUUA), second, expression of HuR in the placenta declines during development as that of miR-675 increases19 (Fig. 2b), and third, HuR is known to afford mRNAs protection from endonucleases (the miRNA processing enzymes Drosha and Dicer are both RNAse III class endonucleases)20. Finally, knockout of HuR results in specific defects of the placenta21. We also found HuR expression to be higher in the liver, where miR-675 is silent, than it is in the late gestation placenta (data not shown).
Figure 2.
HuR binds to full length H19 and inhibits processing of miR-675. (a) Venn diagram indicating the numbers of proteins that were identified by RNA affinity assay (see methods) as binding to H19 in the region of the miR-675 stem loop (Stem) and to control segments including H19 upstream of the stem loop (5′ H19) and of the Kcnq1ot1 RNA (Kcnq1ot1). (b) Microarray data from a published study of transcription during placental development19 were reanalysed and the kinetics of HuR expression are shown from E8.5 until birth (P0). Note that HuR expression is inverse to that of miR-675 during placental development. Data is from at least 2 biological and two technical replicates. (c) RNA immunoprecipitation with an antibody to HuR indicates binding to H19 in the placenta at a gestational timepoint when miR-675 is suppressed (E11.5) but not when it is expressed (E19.5). The enrichment of RNA over a random IgG control is shown following normalisation to the 18s RNA. Levels of Actb and Gapdh are included as positive and negative controls respectively. n=9. (d) RNA immunoprecipitation with an antibody to HuR indicates binding to H19 in MEF cells. The enrichment of RNA over a random IgG control is again shown following normalisation to the 18s RNA. Levels of p21 and Gapdh are included as positive and negative controls respectively. n = 11. (e, g) The expression of miR-675-5p, miR-675-3p or miR-16 in MEF cells (e) and C2C12 cells (g) following treatment with either an siRNA against HuR (si-HuR) or a non-targeting scrambled control (si-scrambled) as determined by qRT-PCR. n = 4. (f) The expression of miR-675-5p, miR-675-3p or miR-16 in MEF cells that genetically lack HuR (HuR-/-). n = 5. All error bars indicate the s.e.m. p-values were determined by Student’s t-test. * p < 0.05, ** p < 0.01, *** p < 0.001.
We performed RNA immunoprecipitation with an antibody to HuR and found that it bound to full length H19 in placenta at E11.5, when miR-675 is suppressed and HuR levels are high, but not at E19.5 when miR-675 is expressed and HuR levels are lower (Fig. 2c). We sought to identify a cell culture model in which to perform functional studies. miR-675 is suppressed in primary MEF cells despite robust expression of H19 (Supplementary Fig. S1d), suggesting that inhibition of processing occurs in these cells; RNA immunoprecipitation experiments indeed confirmed HuR binding to H19 (Fig. 2d). To assess the role of HuR in the suppression of miR-675 processing we performed siRNA mediated knockdown in MEF cells and found that levels of both miR-675-5p and miR-675-3p were increased approximately 2- and 1.5-fold respectively in the absence of HuR (Fig. 2e and Supplementary Fig. S2a, b). A similar effect was observed in MEF cells genetically deficient for HuR (HuR-/-) where both miR-675-5p and miR-675-3p were increased by approximately 2- and 2.2-fold respectively relative to wild type controls (Fig. 2f). Finally, we ablated HuR in a myoblast cell line (C2C12) by siRNA knockdown and again found that miR-675-5p and miR-675-3p levels were increased in the absence of HuR, by 2.8- and 3.5 fold respectively (Fig. 2g and Supplementary Fig. S2c, d). In all cell culture models of HuR deficiency, levels of miR-16 were unchanged relative to controls suggesting that the regulatory effect of HuR ablation is specific to miR-675 as opposed to the broader miRNA processing pathway. Thus HuR negatively regulates processing of both species of miR-675. Luc7l knockdown also resulted in increased miR-675 levels suggesting that HuR is not the only negative regulator of processing (Supplementary Fig. S2b).
HuR does not block miR-675 processing at the Dicer step and likely blocks Drosha
We next sought to identify where in the miR-675 processing pathway HuR acts to inhibit processing. miRNAs are processed sequentially from longer hairpin containing RNAs, first in the nucleus from the pri-miRNA by Drosha to create the pre-miRNA and second in the cytoplasm by Dicer to create the mature miRNA22. The abundance of H19 (the pri-miRNA) suggests that processing of miR-675 is inhibited at the Drosha stage. To investigate this further we performed Northern blot experiments on fetal liver and placental RNA and found only low levels of pre-miR-675, the product of Drosha cleavage of H19 (Fig. 3a, b). Moreover when we transfected synthetic pre-miR-675 into MEF cells we observed an increase in mature miR-675 and this increase was not altered by the presence or absence of HuR, showing that processing by Dicer was not affected by HuR (Fig. 3c). Finally, our RNA immunoprecipitation experiments showed that HuR binds to the full length H19 RNA (Fig. 2c, d). Taken together these data suggest that HuR binds the full length H19 RNA and inhibits processing of miR-675 at the Drosha step. Nevertheless, HuR ablation does not result in complete processing of H19, suggesting that other mechanisms exist to protect H19 from Drosha cleavage, possibly including the other H19 RNA binding proteins we identified. Drosha localizes exclusively to the nucleus while the H19 RNA is known to localize predominantly to the cytoplasm1, 2 and thus it is likely that HuR (and perhaps other RNA binding proteins) protects H19 from processing by Drosha while it is localized to the nucleus, but that another level of protection is afforded by nuclear export of H19. A family of Igf2 mRNA binding proteins have been implicated in the cytoplasmic localization of H1923 and these proteins are thus additional candidates for conferring inhibition of miR-675 processing. To determine whether HuR affects the nuclear export of H19, we performed nuclear and cytoplasmic RNA fractionations from wildtype and HuR-/- MEF cells and found no change in the abundance of nuclear H19 (Fig. 3d, e). Moreover, fractionation of placental RNA revealed that the increasing miR-675 expression observed in that organ is not due to increased nuclear H19 levels (Fig. 3f - i).
miR-675 slows cell proliferation
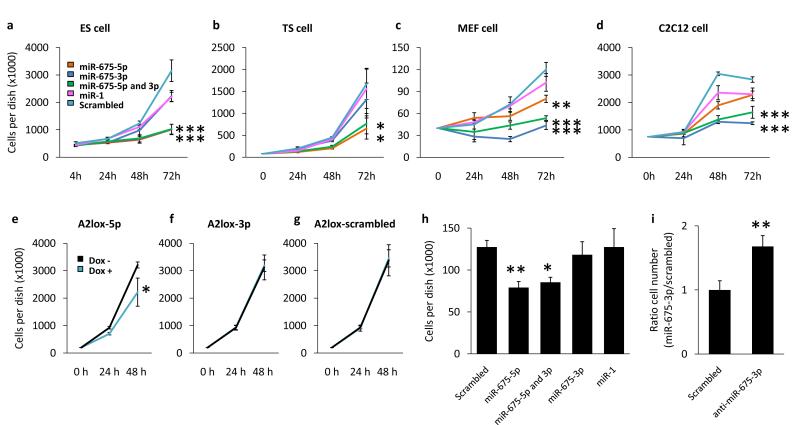
The fact that miR-675 has been conserved during evolution, despite suppression of its processing, suggests a function for the miRNA as well as a need for precise regulation of its dosage. To examine its function we transiently transfected miR-675-5p and miR-675-3p mimics into MEF, C2C12, TS and ES cell lines and found that the proliferation rate of all cell lines was reduced by at least 50% in the presence of miR-675 compared to a scrambled control (Fig. 4a - d). TUNEL staining showed this effect was not due to increased apoptosis (Supplementary Fig. S3a), rather we noticed that the negative regulator of cell cycle Rb1 was upregulated by miR-675, indicating a possible reduction of cell cycle rate (Supplementary Fig. S3b). Interestingly, it was not a single species of miR-675 that was responsible for the reduced proliferation, but rather miR-675-5p slowed the rate of proliferation of ES, TS and to a lesser extent MEF cells, while miR-675-3p slowed that of MEF and C2C12 cells. To confirm these results we utilized the A2lox.cre ES cell line24 to create cells capable of doxycycline induced expression of either miR-675-5p (A2lox-5p), miR-675-3p (A2lox-3p) or a scrambled control (A2lox-scrambled). To avoid inhibition of processing we placed these miRNAs in the miR-30 hairpin context and observed selective upregulation of either species of miR-675 upon the addition of doxycycline (Supplementary Fig. S4). Cell proliferation assays confirmed the results of the transient miR-675 mimic experiments in ES cells in that proliferation of the A2lox-5p cell line was slowed by the addition of doxycycline, while no effect was observed on the A2lox-3p or A2lox-scrambled cell lines (Fig. 4e - g).
Figure 4.
miR-675 overexpression decreases the proliferation rate of a number of cultured cell lines. (a,b,c,d) Cell lines were transfected with miRNA mimics of either miR-675-5p (orange), miR-675-3p (dark blue), both miR-675 species in combination (green), miR-1 (pink) or a scrambled control (light blue) and cell numbers were counted at timepoints post transfection. This procedure was performed in ES (a), TS (b), MEF (c) and C2C12 (d) cells. (e,f,g) Using the A2lox-cre system (see methods) ES cell lines were created which upon the addition of doxycycline selectively express miR-675-5p (A2lox-5p) (e), miR-675-3p (A2lox-3p) (f) or a non-targeting scrambled control (A2lox-scrambled) (g). Cell numbers were counted at timepoints following the addition of doxycycline to the growth media (dox+, blue line) or in the absence of doxycycline (dox−, black line). n = 4. p-values were determined by two-way ANOVA followed by post hoc Tukey’s Test. (h) G401 cells were treated with the indicated miRNA mimics and cell numbers were counted 72 hours post transfection. n = 4. p-values were determined by Student’s t-test. (i) C2C12 cells were treated with either an inhibitor of miR-675-3p or a scrambled control. miR-675 expression was induced by differentiation and the cells counted following 96 hours of differentiation. n = 9. p-values were determined by Student’s t-test. All error bars indicate the s.e.m. * p < 0.05, ** p < 0.01, *** p < 0.001.
H19 was first shown to have tumor suppressor properties by overexpression in the G401 metastatic rhabdoid tumor cell line5. We investigated whether miR-675, rather than H19 itself, may have been responsible for this previously reported effect by overexpressing the smaller RNA in these cells. Indeed we observed reduced proliferation of G401 cells when treated with miR-675-5p as opposed to a scrambled control (Fig. 4h). Finally, we took advantage of the fact that both H19 and miR-675 become expressed in differentiating C2C12 cells (Supplementary Fig. S5) to perform antagomiR mediated loss of function experiments and found that proliferation of these cells increased when miR-675-3p was inhibited (Fig. 4i). Taken together these results show that miR-675 is a functional component of the H19 RNA, capable of suppressing cell proliferation.
Deletion of both H19 and miR-675 results in placental overgrowth
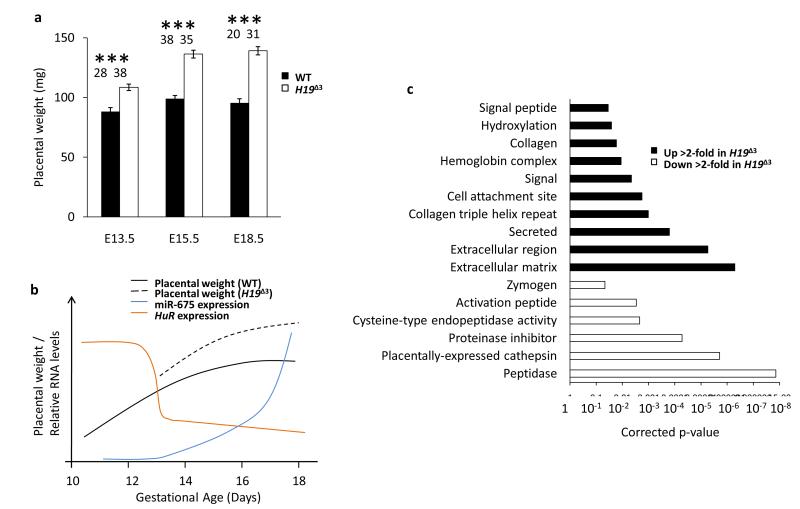
Given the growth limiting effect of miR-675 it was interesting to note that expression of the miRNA in the placenta is concomitant with the natural cessation of the growth of that organ25. Moreover mice that carry a 13 kb deletion that includes H19, miR-675 and 10 kb of regulatory sequences upstream of H19 show an overgrowth phenotype that is more severe in the placenta (145 % WT) than the embryo (123 % WT)26-28. The interpretation of this phenotype however is complicated by the fact that the deletion (of the imprinting control region upstream of H19) also results in overexpression of the linked Igf2 gene29. To examine the possibility that the H19 RNA itself regulates placental growth we studied an alternative H19 mouse model (H19 Δ3) which carries a 3 kb deletion of just the H19 transcription unit, including miR-67530. This deletion specifically ablates the H19 RNA without affecting expression of Igf2 in the placenta (Supplementary Fig. S6a)8. Notably, placentas carrying the H19Δ3 allele were 32% larger than wild types at E18.5, while the mutant embryos themselves were only 8% larger than wild type, in agreement with previous results30 (Fig. 5a and Supplementary Fig. S6c). Expression of miR-675 in the placenta correlates with a rapid drop in the expression of HuR and the natural cessation of placental growth, which does not occur in the absence of H19 and miR-675 (Fig. 5b), implying that miR-675 is a negative regulator of placental size.
Figure 5.
The phenotype and transcriptome of the H19Δ3 placenta implies that miR-675 is a negative growth regulator in this tissue. (a) Placental weight was measured for homozygous H19Δ3 crosses and compared to wild type revealing an overgrowth phenotype for the knockout that is more severe in the placenta than the embryo. The number of placentas measured for each data point is indicated above each bar. Error bars indicate the s.e.m. p-values were determined by Student’s t-test. ** p < 0.01, *** p < 0.001. (b) Representation based on real data showing the correlation of mir-675 (blue) and HuR19 (orange) expression in the placenta with the developmental weight of that organ (black). The weight of the H19Δ3 placenta is also indicated (slotted line). At E13.5 there is a rapid drop in HuR expression concomitant with the induction of miR-675. At approximately E15.5 the wild type placenta ceases to increase in mass, however this does not occur in the H19Δ3 placenta25. (c) Enriched gene ontology terms in RNA-seq data from the labyrinth layer of a day E18.5 wild type and H19Δ3 placenta. Categories for genes upregulated (black) and downregulated (white) in the H19Δ3 placenta are shown.
The H19Δ3 placental transcriptome reveals targets of miR-675
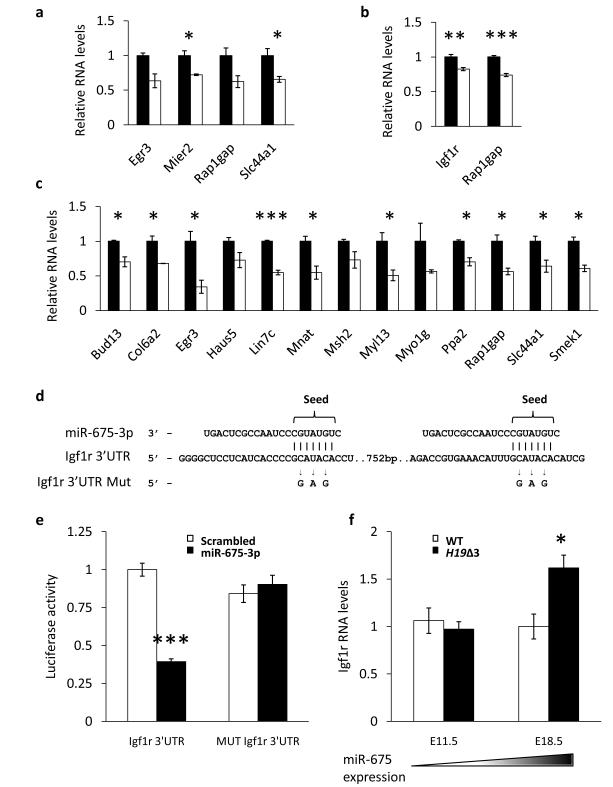
miR-675 is expressed most highly in the labyrinthine zone of the placenta as compared to the junctional zone (Supplementary Fig. S7a - d); to determine the effect of miR-675 on the H19Δ3 transcriptome we performed RNA sequencing (RNA-Seq) on RNA from the labyrinthine zone in the E18.5 H19Δ3 placenta. We found 285 genes upregulated and 59 downregulated by greater than 2-fold in the mutant versus the control labyrinth (Supplementary Fig. S7e). Gene ontology categories enriched in the H19Δ3 transcriptome were consistent with growth and morphogenesis of the placenta (Fig. 5c). H19 has previously been reported to regulate a network of imprinted genes in fetal muscle but not placenta8. Indeed we observed no specific deregulation of imprinted genes in the H19Δ3 placenta. We found computationally predicted targets of miR-675 enriched in the genes upregulated in the H19Δ3 placenta (p = 0.0254). We co-transfected miR-675-5p and miR-675-3p mimics into ES, C2C12 and MEF cells and monitored the effect on 35 predicted targets by qRT-PCR (Fig. 6a–c), many of which (45 %) were found to be downregulated by miR-675. However, genes that appeared to be targets in one cell line were not necessarily targets in another. This suggests that the target network by which miR-675 slows cell proliferation is of a complex nature and may be distinct in different cell types.
Figure 6.
Identification of miR-675 targets. (a, b, c) Expression of predicted miR-675 targets, detected by qRT-PCR, following the co-transfection of both miR-675 species mimics or a scrambled control in C2C12 (a), ES (b) and MEF (c) cells. n = 3. Note that only targets which were affected by miR-675 transfection are shown. (d) Schematic representation of miR-675-3p binding to two potential sites in the Igf1r 3′UTR. The bases mutated to make the mutant Igf1r 3′UTR are indicated. (e) Effect of miR-675-3p transfection on the expression of a Renilla luciferase reporter fused to either the wild type or mutant Igf1r 3′UTR. (f) Igf1r expression, determined by qRT-PCR, in the labyrinth layer of wild type and H19Δ3 placentas at either E11.5 or E18.5. Endogenous miR-675 expression in the placenta is indicated by the shaded triangle. n = 4. All error bars indicate the s.e.m. p-values were determined by Student’s t-test. * p < 0.05, *** p < 0.001.
Igf1r is a potential target of miR-675
Among the targets validated in cell lines, Igf1r was of particular interest since it is the key receptor through which Igf2 exerts its growth promoting effect during fetal development. Igf1r is a predicted target of miR-675-3p and contains two 7-mer seed matches in its 3′UTR (Fig. 6d). We cloned the Igf1r 3′UTR downstream of a luciferase reporter construct and co-transfected it into cells alongside either a miR-675-3p mimic or a scrambled control. We found that luciferase levels were reduced by more than 60 % in the presence of miR-675-3p (Fig. 6e). This effect was not observed when the miR-675-3p binding sites were mutated, thus Igf1r is a potential target of miR-675-3p. Moreover Igf1r was found to be upregulated in H19Δ3 placentas only at the time when miR-675 is expressed (E18.5) and not when it is silent (E11.5) (Fig. 6f), suggesting it is a target of the microRNA in vivo.
DISCUSSION
Our results suggest that a physiological role of H19 is to slow down growth of the placenta in the second half of gestation, in preparation for parturition. This appears to be achieved at least in part by downregulation of the RNA binding protein HuR during gestation which normally blocks processing of miR-675 at the Drosha stage. Increased levels of miR-675 in the placenta are concomitant with downregulation of Igf1r, amongst other targets which may also contribute to reduced growth. Igf1r has been shown to be an important regulator of growth31, 32, with its main ligand being Igf2. Remarkably, the H19 locus therefore regulates the abundance of Igf2 (through imprinting) and that of its receptor, Igf1r, through miR-675.
While data presented here applies to growth regulation in the placenta under physiological conditions, it is notable that in embryonic tissues miR-675 remains tightly repressed despite the high levels of H19. We speculate that the enormous reservoir of the growth suppressing miR-675 could be rapidly mobilized in response to cellular stress or abnormal proliferation. Indeed, HuR is known to relocate from the nucleus to the cytoplasm in stress conditions33-36 and upon pharmacologically induced neoplastic transformation37. This would expose H19 to processing by Drosha, thus liberating miR-675. HuR was recently shown to suppress processing of human miR-7 from the highly expressed HNRNPK transcript38. That miR-7 is also a known tumor suppressor39-41 that functions partly through Igf1r targeting41, may suggest a wider miRNA repertoire for a HuR mediated response to aberrant cell proliferation. The H19 RNA being a regulatable reservoir of the proliferation suppressing miR-675 may thus explain the lincRNAs tumour suppressive role, especially in childhood tumours42. The mechanisms of processing and function of miR-675 are likely to be relevant to the molecular pathology of fetal growth and cancer syndromes.
Materials and Methods
Cell Culture
ES cells were the J1 line and were maintained on gelatinized dishes in ES media (Dulbecco’s modified Eagle’s medium containing sodium pyruvate and l-glutamine (DMEM), supplemented with 15% fetal bovine serum (Invitrogen), 1% penicillin / streptomycin, 1x non-essential amino acids, 0.05 mM 2-mercaptoethanol and leukaemia inhibitory factor (LIF) (1000 units / ml) (Millipore)).
TS cells were the TS-rs26 line derived in the Rossant Laboratory, cultured in standard conditions (RPMI 1640 supplemented with 20% fetal bovine serum (Invitrogen), 1% antibiotic / antimycotic solution, 1x sodium pyruvate and 0.05 mM 2-mercaptoethanol, 20 ng/ml b-fetal growth factor (bFGF) (Sigma) and 1 μg / ml heparin (Sigma) with 70% of the media preconditioned with embryonic feeder cells43). Differentiation was achieved by culturing without bFGF or heparin in unconditioned media.
C2C12 cells were cultured in growth media (Dulbecco’s modified Eagle’s media (DMEM) supplemented with 200 mM l-glutamine and 10% fetal bovine serum (PA)). Differentiation was induced by culturing in differentiation media (DMEM supplemented with 200 mM l-glutamine and 2% horse serum (Invitrogen)).
MEF cells were derived from C57/BL6J x CBA mice and cultured in ES cell media without LIF. HuR-/- MEFs have been previously described21.
RNA was isolated from cells using the Qiazol method (Qiagen).
Animal studies
Mice used for the expression studies were the C57/BL6 strain. The H19Δ3 transgenic line has been previously described30. All experiments were performed under Home Office (UK) licence and in accord with the Animals (Scientific Procedures) Act of 1986. To define developmental stage, the day of conception is considered to be day 0.5 of pregnancy.
RNA was isolated from animal tissue using the Qiazol method (Qiagen).
Generation of A2lox-miR-675 cell lines
Hairpins containing the microRNA required for expression, miR-30 flanking sequences and overhangs matching the restriction site fragments, XhoI and EcoRI, were annealed to their corresponding complementary oligomer (Supplementary Table S1) and cloned into the PSM2 vector (Open Biosystems) using the restriction site overhangs. Constructs were PCR amplified using primers containing artificial NotI and HindIII restriction sites (ATCAAGCTTCAGGGTAATTGTTTGAATGAGGC and AGCGGCCGCGTCTTCCAATTGAAAAAAGTGA) and cloned into the p2lox vector followed by transfection into the A2lox.cre ES cell line24. The day prior to transfection Cre was induced by the addition of doxycycline (0.5 μg / ml). Cells were transfected with 2 μg of p2lox plasmid using Lipofectamine 2000 (Invitrogen) and selection for stable integration with geneticin (300 μg/ml, Melford) for 10 days. Single colonies were picked and expanded. Expression of hairpin constructs was achieved by the addition of doxycycline (2 μg/ml) to the ES media.
RNAi knockdown
RNAi mediated knockdown of mRNAs was achieved in all cell types by Stealth™ RNAi oligos (Invitrogen) against HuR (Catalogue no. mss205313), Serbp1 (Catalogue no. mss288826), Upf1 (Catalogue no. mss208598), Zcchc11 (Catalogue no. mss279478), Ptbp2 (Catalogue no. mss225938), Luc7l (Catalogue no. mss250251) or a non-targeting control (Catalogue no. 12935-300), at a final concentration of 50 pmol / ml. Transfection of RNAi oligos into cell lines achieved using Lipofectamine 2000 (Invitrogen). RNA was extracted 48 hours later.
Cell proliferation assays
Pre-miR™ miRNA precursor molecules for miR-675-5p, miR-675-3p, has-miR-1 or Negative Control #1 (Ambion) were transfected into all cell lines at a final concentration of 24 nM using Lipofectamine 2000 (Invitrogen). Cells were harvested using Trypsin (0.05%) and then counted using a Nikon TMS microscope and haemacytometer (Assistent).
C2C12 cells maintained in growth media (described above) were transfected with either antagomiR molecules against miR-675-3p or a scrambled control (Invitrogen) to a final concentration of 30 nM using Lipofectamine 2000 (Invitrogen) for 6 hours, before inducing differentiation by the addition of differentiation media (described above). Cells were maintained in differentiation media for 2 days before a second round of antagomiR transfection. Cells were maintained for a further 3 days before being harvested with Trypsin (0.05%) and counted using a Nikon TMS microscope and haemacytometer (Assistent).
Quantitative PCR
Quantitative reverse transcription PCR (qRT-PCR) was performed using primers from Supplementary Table S2.
MicroRNA qRT-PCR was performed with TaqMan® MicroRNA assays (Applied Biosystems) and normalized to the U6 RNA according the manufacturers guidelines. For absolute quantification of miRNA levels, ct values were compared to the ct values of a standard curve produced by serial dilution and Taqman® MicroRNA amplification of the miRXplore™ Universal Reference (Miltenyi Biotec).
For absolute quantification of H19 levels, part of H19 was amplified using the H19 RT primers (see Supplementary tables) and cloned into the pGEM®T-easy vector (Promega). Standard curves were created by qRT-PCR using a dilution series of this vector which was used to measure absolute H19 levels in tissue samples.
Northern blotting
To detect pre-miR-675 probes were PCR amplified from H19 containing plasmid DNA using a forward (TGCGGCCCAGGGACTGGT) and reverse primer harbouring the T7 promoter (GGATCCTAATACGACTCACTATAGGGAGAGGAGCCAGACCCAGGGACTGA). PCR products were reverse transcribed and radioactively labelled using [α-32P] rUTP. Total RNA was separated on a 15% TBE-Urea denaturing gel (Invitrogen). RNA was transferred to a GeneScreen Plus® Hybridization Transfer membrane (Perkin Elmer) and incubated with labelled probe or labelled U6 RNA oligo (TTGCGTGTCATCCTTGCGCAGG) for approximately 16 hours. Membranes were exposed to X-ray film (Fujifilm).
For detection of mature miR-675, total RNA was separated on a 15% TBE-Urea denaturing gel (Invitrogen). RNA was transferred to a GeneScreen Plus® Hybridization Transfer membrane (Perkin Elmer) and UV cross-linked. Probes specific to miR-675-3p (Exiqon) or U6 RNA were labelled with ATP[γ-32P] and incubated with membrane for approximately 16 hours before exposure to X-ray film (Fujifilm).
mRNA library preparation (RNAseq)
RNAseq libraries were created, sequenced and mapped using an in house protocol which has already been described44. Mapped sequence reads were analysed with the aid of SeqMonk Mapped Sequence Analysis Tool (http://www.bioinformatics.bbsrc.ac.uk/projects/seqmonk/). These data are available on the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress/) with accession number E-MTAB-895.
RNA affinity assay
Bait regions were PCR amplified from genomic DNA using appropriate primers (miR-675 stem: AATGGAAAAGAAGGGCAGTG and CCCAGCTACTCGCTCTACCT, 5′ H19: AGACCTGGGCAGTGAAGGTA and GCCACTGTCTCCAAGGACTC, and Kcnq1ot1: TGGGTGGCCTCTAATACTGG and CTGCCCCTTCTCTATTGCAG) to produce amplicons of 343, 363 and 411 bp respectively. Note that the miR-675 stem fragment includes the putative HuR binding site adjacent to the miR-675 stem loop. PCR products were cloned into the pGEM®T-easy vector (Promega) and digested with ScaI (New England Biolabs) to define the 3′ end before in vitro transcription. RNA binding proteins were then isolated using previously described methods45. Proteins were identified by mass spectrometry. Briefly, coomassie-stained gel lanes were excised, destained, reduced, carbamidomethylated and digested overnight with 10 ng / ml modified trypsin (Promega) in 25mM ammonium bicarbonate at 30 °C. The resulting peptide mixtures were separated by reverse-phase liquid chromatography (column: 0.05 × 100 mm, Vydac C18, 5 mm particle size), with an acetonitrile gradient (5 – 40 % over 30 min) containing 0.1 % formic acid, at a flow rate of 150 nL / min. The column was coupled to a nanospray ion source (Protana Engineering) fitted to a quadrupole-TOF mass spectrometer (Qstar Pulsar i; Applied Biosystems / MDS Sciex), operating in information dependent acquisition mode.
RNA immunoprecipitation
Cell lysate was produced from either cultured MEF cells or mechanically homogenised whole placentas by incubation in Lysis Buffer (100 mM KCl, 5 mM MgCl2, 10 mM Hepes, pH 7, 0.5% NP-40, 1x Complete mini Protease Inhibitor Cocktail tablets (Roche)). RNA immunoprecipitation was performed using 4 μg of antibody against HuR (3A2, Santa Cruz) or non-specific mouse IgG (Santa Cruz) and a previously described method46.
Luciferase assay
The miR-675 target sites within the Igf1r were PCR amplified using primers containing artificial XhoI and NotI restriction sites and cloned into the Psicheck2 vector (Promega) directly downstream of a luciferase reporter gene. miRNA binding sites were mutated by PCR amplifying the Psicheck2-Igf1r vector with primers containing the required point mutations followed by removal of un-mutated plasmid by DpnI digestion. Plasmids (200 μg) were co-transfected into ES cells with miR-675 mimic (24 nM, Ambion) and the luciferase activity measured 24 hours later using the Dual Glo luciferase assay (Promega).
TUNEL staining
Cells were treated with Pre-miR™ miRNA precursor molecules (Ambion), as described above, and maintained for 48 hours before being harvested and cytospun on to poly-L-lysine coated slides. Apoptotic cells were detected using the DeadEnd™ Fluorometric TUNEL system (Promega). Positive controls were made by treatment with DNaseI (Roche) according to instructions supplied with the TUNEL staining kit. Cell nuclei were stained with Vectashield® with DAPI (Vector Laboratories) and visualized on an Olympus BX41 fluorescence microscope.
Nuclear cytoplasmic RNA fractionation
Nuclear and cytoplasmic RNA was extracted from MEF cells with a PARIS™ kit (Ambion). Fractionated placental RNA was obtained by mechanical homogenization of placentas in lysis buffer (0.32M sucrose, 5mM CaCl2, 5mM EDTA, 3mM MgAc, 10 mM Tris-HCl pH 8 and 40 U/ml RNAsin (Promega)) and filtration. Placental lysate was then centrifuged at 700 g for 10 min and the supernatant retained as the cytoplasmic fraction for RNA extraction. Nuclei were resuspended in centrifuge buffer (2 M sucrose, 3mM MgAc and 10 mM Tris-HCl pH 8) and overlaid on a cushion of further centrifuge buffer before centrifugation at 26,000 g for 1 hour and RNA extraction.
Statistical analysis
Data were analysed either by two-way analysis of variance (ANOVA) followed by post hoc Tukey’s test, or two tailed Student’s t-test. p < 0.05 was considered as significant.
Supplementary Material
Acknowledgements
We thank Dimitris L. Kontoyiannis (Alexander Fleming Biomedical Sciences Research Center) for supplying HuR-/- MEF cells. We also thank Helene Jammes and Anne Gabory for assisting with collection of the H19Δ3 phenotypic data, Judith Webster for preparing samples for mass spectrometry and Wendy Dean for tissue collections. The authors would also like to thank Kristina Tabbada, Anne Segonds-Pichon and Simon Andrews for RNA-seq, statistical and bioinformatic assistance respectively. We thank Michelina Iacovino for creating the A2lox-cre ES cell line. We would also like to thank Tim Hore, Christel Krueger and Jonathan Houseley for critical reading of the manuscript and all members of the labs of Wolf Reik, Myriam Hemberger, Elena Vigorito and Jonathan Houseley for helpful discussions. This work was supported by BBSRC, the Wellcome Trust, MRC, EU NoE EpiGeneSys, EU BLUEPRINT, NIH/NHLBI (U01HL100407) and the Cambridge Commonwealth Trust.
Footnotes
Author Contributions AK designed and carried out experiments and interpreted results. DO performed mass spectrometry. PM collected and dissected placentas for RNA-seq. MK made the A2lox.cre ES cell line. LD, GS and WR designed and supervised this work and interpreted results. AK and WR wrote the manuscript.
Competing Financial Interests The authors have no competing financial interests to declare.
References
- 1.Brannan CI, Dees EC, Ingram RS, Tilghman SM. The product of the H19 gene may function as an RNA. Mol Cell Biol. 1990;10:28–36. doi: 10.1128/mcb.10.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seidl CI, Stricker SH, Barlow DP. The imprinted Air ncRNA is an atypical RNAPII transcript that evades splicing and escapes nuclear export. Embo J. 2006;25:3565–3575. doi: 10.1038/sj.emboj.7601245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartolomei MS. Genomic imprinting: employing and avoiding epigenetic processes. Genes Dev. 2009;23:2124–2133. doi: 10.1101/gad.1841409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gabory A, Jammes H, Dandolo L. The H19 locus: role of an imprinted non-coding RNA in growth and development. Bioessays. 2010;32:473–480. doi: 10.1002/bies.200900170. [DOI] [PubMed] [Google Scholar]
- 5.Hao Y, Crenshaw T, Moulton T, Newcomb E, Tycko B. Tumour-suppressor activity of H19 RNA. Nature. 1993;365:764–767. doi: 10.1038/365764a0. [DOI] [PubMed] [Google Scholar]
- 6.Yoshimizu T, et al. The H19 locus acts in vivo as a tumor suppressor. Proc Natl Acad Sci U S A. 2008;105:12417–12422. doi: 10.1073/pnas.0801540105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smits G, et al. Conservation of the H19 noncoding RNA and H19-IGF2 imprinting mechanism in therians. Nat Genet. 2008;40:971–976. doi: 10.1038/ng.168. [DOI] [PubMed] [Google Scholar]
- 8.Gabory A, et al. H19 acts as a trans regulator of the imprinted gene network controlling growth in mice. Development. 2009;136:3413–3421. doi: 10.1242/dev.036061. [DOI] [PubMed] [Google Scholar]
- 9.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 12.Cai X, Cullen BR. The imprinted H19 noncoding RNA is a primary microRNA precursor. Rna. 2007;13:313–316. doi: 10.1261/rna.351707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dudek KA, Lafont JE, Martinez-Sanchez A, Murphy CL. Type II collagen expression is regulated by tissue-specific miR-675 in human articular chondrocytes. J Biol Chem. 2010;285:24381–24387. doi: 10.1074/jbc.M110.111328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiang HR, et al. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. 2010;24:992–1009. doi: 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mineno J, et al. The expression profile of microRNAs in mouse embryos. Nucleic Acids Res. 2006;34:1765–1771. doi: 10.1093/nar/gkl096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang JH, Shao P, Zhou H, Chen YQ, Qu LH. deepBase: a database for deeply annotating and mining deep sequencing data. Nucleic Acids Res. 2010;38:D123–130. doi: 10.1093/nar/gkp943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lim LP, et al. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knox K, Baker JC. Genomic evolution of the placenta using co-option and duplication and divergence. Genome Res. 2008;18:695–705. doi: 10.1101/gr.071407.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao Z, Chang FC, Furneaux HM. The identification of an endonuclease that cleaves within an HuR binding site in mRNA. Nucleic Acids Res. 2000;28:2695–2701. doi: 10.1093/nar/28.14.2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katsanou V, et al. The RNA-binding protein Elavl1/HuR is essential for placental branching morphogenesis and embryonic development. Mol Cell Biol. 2009;29:2762–2776. doi: 10.1128/MCB.01393-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 23.Runge S, et al. H19 RNA binds four molecules of insulin-like growth factor II mRNA-binding protein. J Biol Chem. 2000;275:29562–29569. doi: 10.1074/jbc.M001156200. [DOI] [PubMed] [Google Scholar]
- 24.Mallanna SK, et al. Proteomic analysis of Sox2-associated proteins during early stages of mouse embryonic stem cell differentiation identifies Sox21 as a novel regulator of stem cell fate. Stem Cells. 2010;28:1715–1727. doi: 10.1002/stem.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coan PM, Ferguson-Smith AC, Burton GJ. Developmental dynamics of the definitive mouse placenta assessed by stereology. Biol Reprod. 2004;70:1806–1813. doi: 10.1095/biolreprod.103.024166. [DOI] [PubMed] [Google Scholar]
- 26.Angiolini E, et al. Developmental adaptations to increased fetal nutrient demand in mouse genetic models of Igf2-mediated overgrowth. FASEB J. 2011;25:1737–1745. doi: 10.1096/fj.10-175273. [DOI] [PubMed] [Google Scholar]
- 27.Esquiliano DR, Guo W, Liang L, Dikkes P, Lopez MF. Placental glycogen stores are increased in mice with H19 null mutations but not in those with insulin or IGF type 1 receptor mutations. Placenta. 2009;30:693–699. doi: 10.1016/j.placenta.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leighton PA, Ingram RS, Eggenschwiler J, Efstratiadis A, Tilghman SM. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature. 1995;375:34–39. doi: 10.1038/375034a0. [DOI] [PubMed] [Google Scholar]
- 29.Thorvaldsen JL, Duran KL, Bartolomei MS. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 1998;12:3693–3702. doi: 10.1101/gad.12.23.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ripoche MA, Kress C, Poirier F, Dandolo L. Deletion of the H19 transcription unit reveals the existence of a putative imprinting control element. Genes Dev. 1997;11:1596–1604. doi: 10.1101/gad.11.12.1596. [DOI] [PubMed] [Google Scholar]
- 31.Liu JP, Baker J, Perkins AS, Robertson EJ, Efstratiadis A. Mice carrying null mutations of the genes encoding insulin-like growth factor I (Igf-1) and type 1 IGF receptor (Igf1r) Cell. 1993;75:59–72. [PubMed] [Google Scholar]
- 32.Baker J, Liu JP, Robertson EJ, Efstratiadis A. Role of insulin-like growth factors in embryonic and postnatal growth. Cell. 1993;75:73–82. [PubMed] [Google Scholar]
- 33.Jeyaraj SC, Dakhlallah D, Hill SR, Lee BS. Expression and distribution of HuR during ATP depletion and recovery in proximal tubule cells. Am J Physiol Renal Physiol. 2006;291:F1255–1263. doi: 10.1152/ajprenal.00440.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim HH, Abdelmohsen K, Gorospe M. Regulation of HuR by DNA Damage Response Kinases. J Nucleic Acids. 2010;2010 doi: 10.4061/2010/981487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pan YX, Chen H, Kilberg MS. Interaction of RNA-binding proteins HuR and AUF1 with the human ATF3 mRNA 3′-untranslated region regulates its amino acid limitation-induced stabilization. J Biol Chem. 2005;280:34609–34616. doi: 10.1074/jbc.M507802200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang W, et al. HuR regulates p21 mRNA stabilization by UV light. Mol Cell Biol. 2000;20:760–769. doi: 10.1128/mcb.20.3.760-769.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blaxall BC, et al. Differential expression and localization of the mRNA binding proteins, AU-rich element mRNA binding protein (AUF1) and Hu antigen R (HuR), in neoplastic lung tissue. Mol Carcinog. 2000;28:76–83. [PubMed] [Google Scholar]
- 38.Lebedeva S, et al. Transcriptome-wide Analysis of Regulatory Interactions of the RNA-Binding Protein HuR. Mol Cell. 2011 doi: 10.1016/j.molcel.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 39.Reddy SD, Ohshiro K, Rayala SK, Kumar R. MicroRNA-7, a homeobox D10 target, inhibits p21-activated kinase 1 and regulates its functions. Cancer Res. 2008;68:8195–8200. doi: 10.1158/0008-5472.CAN-08-2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saydam O, et al. miRNA-7 attenuation in Schwannoma tumors stimulates growth by upregulating three oncogenic signaling pathways. Cancer Res. 2011;71:852–861. doi: 10.1158/0008-5472.CAN-10-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang L, et al. MicroRNA-7 targets IGF1R (insulin-like growth factor 1 receptor) in tongue squamous cell carcinoma cells. Biochem J. 2010;432:199–205. doi: 10.1042/BJ20100859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lim DH, Maher ER. Genomic imprinting syndromes and cancer. Adv Genet. 2010;70:145–175. doi: 10.1016/B978-0-12-380866-0.60006-X. [DOI] [PubMed] [Google Scholar]
- 43.Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J. Promotion of trophoblast stem cell proliferation by FGF4. Science. 1998;282:2072–2075. doi: 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
- 44.Ficz G, et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 45.Caputi M, Mayeda A, Krainer AR, Zahler AM. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J. 1999;18:4060–4067. doi: 10.1093/emboj/18.14.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baroni TE, Chittur SV, George AD, Tenenbaum SA. Advances in RIP-chip analysis : RNA-binding protein immunoprecipitation-microarray profiling. Methods Mol Biol. 2008;419:93–108. doi: 10.1007/978-1-59745-033-1_6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.