Abstract
A 10-year-old speechless, mentally deficient male, with low arylsulfatase A (ARSA) activity, and presumably, methachromatic leukodystrophy, underwent genetic evaluation. As the clinical picture was not compatible with this diagnosisan ARSA gene and chromosome analysis were performed, showing the presence of a pseudodeficiency ARSA allele and a de novo apparently balanced t(16;22)(p11.2;q13) translocation. A deletion on the long arm of chromosome 22 encompassing the ARSA gene, as shown by FISH and array-CGH, indicated a 22q13 deletion syndrome. This case illustrates the importance of detailed cytogenetic investigation in patients presenting low arylsulfatase A activity and atypical/unspecific clinical features.
Keywords: 22q13 deletion, apparently balanced translocation, ARSA gene, arylsulfatase A pseudodeficiency, metachromatic leukodystrophy
Very often genetic syndromes are underdiagnosed, their true incidence remaining unknown through inadequate clinical recognition or incomplete laboratory investigation. In Brazil, this mainly derives from structural deficiencies in the public health system, inappropriate for diagnosing rare diseases (Schwartz et al., 2007). The present report illustrates this, on dealing with the Phelan-McDermid syndrome (22q13 deletion syndrome), with a relatively nonspecific phenotype (Phelan, 2008), and meta-chromatic leukodystrophy (MLD), all of which requiring careful laboratory investigation to so avoid inaccurate diagnosis (Von Figura et al., 2001; Artigalás et al., 2010).
Assessment was directed to a 10-year-old boy presenting low arylsulftase A (ARSA) activity in leukocytes (1.6 nmol/h/mg prot, RV: 5–20) and diagnosed as MLD. Nevertheless, his clinical picture was atypical, since, after clinical and radiological evaluation, the prevailing neurological conditions were found to be stable, without signs of neurodegeneration or white matter disorder. The reason for initially determining ARSA activity was not apparent. He was the first son of a healthy, young, non-consanguineous couple, without a family history of genetic diseases. Furthermore, his younger brother was clinically normal. The boy, born through vaginal delivery after a 35-week pregnancy associated with oligohydramniosis had, received 7/8 APGAR scores. At birth, his weight was 2,630 g, length 46 cm, and head circumference 32 cm. He held up his head at 7 months, sat without support at 18, and walked without support at 24. Even so, he was unable to develop verbal language. Sphincter control occurred only at 8-years. At 3-years, he suffered from absence-type epileptic crises, which progressed to frequent-atonic crises; full control of these was achieved only after 18 months of treatment with phenobarbital, carbamazepine, and valproic acid. Autistic-like behavior was manifest in the form of restricted intuitive social interaction, delayed communication and inflexibility of interests. Physical examination revealed an elongated face, arched eyebrows, long eyelashes, diastasis of the upper central incisors, drooping feet, normal and symmetric deep tendinous reflexes, hypertonia of the lower limbs, and choreoathetosis. Results from subsequent investigations were normal. These consisted of CT brain scan and MRI, the molecular fragile-X syndrome test, blood amino acid thin-layer chromatography, dosage of very-long-chain fatty acids and A and B hexosaminidase activitiy, as well as chitotriosidase in plasma, urinary thin-layer chromatography of amino acids, oligosaccharides and sialosaccharides, and gas chromatography of urinary organic acids.
We determined ARSA activity three times in leukocytes (0.2, 2.1 and 1.2 nmol/h/mg prot, respectively), and twice in fibroblasts (0.76 and 0.31 nmol/h/mg prot – RV: 20–50 nmol/h/mg prot). All were below the normal range. As apparently sulfatide levels in urine (three different samples) were also low, the presence of the most common ARSA pseudodeficiency allele [p.N350S; c.1524+95A >G] was investigated, with positive results. Although the parents had been diagnosed as heterozygous for the PD-ARSA allele, the patient, on the contrary, was initially misinterpreted as being homozygous for PD-ARSA.
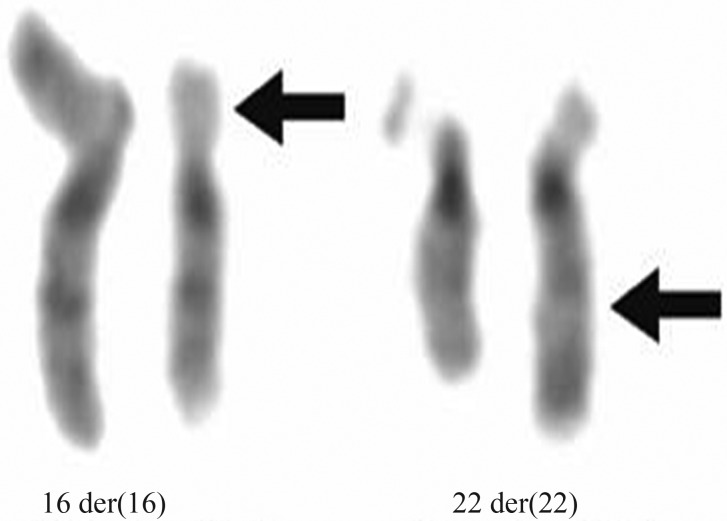
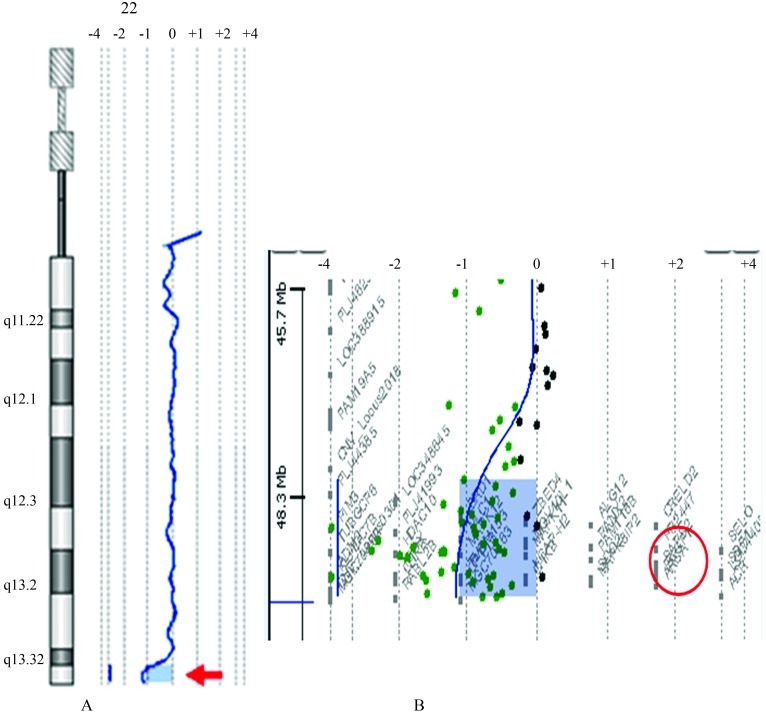
On considering mental deficiency through an unknown cause, we performed a GTG chromosome analysis of peripheral blood leucocytes that revealed an apparently balanced translocation between the short arm of chromosome 16 and the long arm of chromosome 22 [46,XY,t(16;22)(p11.2;q13)] in all the 30 metaphases analyzed (Figure 1). Incidentally, the parents presented normal karyotypes. FISH was performed with subtelomeric probes for the short arm of chromosome 16 and the long arm of chromosome 22, together with the DiGeorge/VCFS Probe TUPLE1(22q11.2)/ARSA control probe (22q13.3) (Vysis, Abbott Molecular Inc.). The ARSA (control) probe (22q13.3) and the 22q subtelomeric probe did not hybridize to either der(22) or der(16), whereas theDiGeorge/VCFS Probe TUPLE1 (22q11.2) and the 16p subtelomeric probe were detected on der(22). Array CGH was carried out on propositus DNA, by using an oligonucleotide-based micro-array containing about 44,000 60-mer probes (Agilent Human Genome Microarray, customer array design AMADID number 017457). Arrays were analyzed with the AGILENT DNA Microarray Scanner and AGILENT Feature Extraction software (v9.5.3). This revealed a ∼1.4 Mb deletion on the long arm of chromosome 22 involving the ARSA and SHANK3 genes (Figure 2). No deletion was detected on chromosome 16. These results were compatible with diagnosis of the 22q13 deletion syndrome. The low ARSA activity, seen in this case, was due to a de novo deletion of the ARSA gene, concomitant with an inherited pseudodeficiency allele.
Figure 1.
t(16;22)(p11.2;q13), GTG-banded der(16), der (22) and their normal homologues. Arrows point to breakpoints.
Figure 2.
Chromosome 22 array-CGH profile of the propositus (normal male as a reference DNA). (A) The probe log2 ratios were plotted according to genomic coordinates (based on the UCSC Genome Browser, February 2009, NCBI Build 37 reference sequence). A deletion (copy number loss) was detected on the distal 22q region (red arrow) in a genomic segment with median log2 ratioshifted to <−0.8. (B) Detail of the 22q subtelomeric region showing the deletion of ∼1.4 Mb segment which includes the ARSA gene (red circle). The minimum deleted segment was mapped between chr22:48138838 (first aberrant probe) and chr22:49525130 (last aberrant probe).
In spite of variation in expression and severity, patients with the 22q13 deletion syndrome (OMIM #606232) generally show global developmental delay, generalized hypotonia, autist-like behavior, absence of, or severely retarded speech, normal to accelerated growth, and other minor dysmorphic anomalies (Phelan, 2008). Brain MRI is usually normal or with a thin or morphologically atypical corpus callosum (Philippe et al., 2008). Liver dysfunction has also been observed (Bartsch et al., 2010). Chromosome alterations involving the 22q13 region have been described in over 100 cases (Bisgaard et al. 2009). The most frequent abnormality is a simple terminal deletion (Bonaglia et al., 2011). Nevertheless, in over 30% of the cases with a 22q13 deletion syndrome two or more chromosome studies were required to cytogenetically detect the deletion. Moreover, due to clinical features being unrecognizable to, very subtle and unspecific, in many patients the cytogenetic investigation was not sufficiently profound, thus leading to faulty diagnosis (Phelan, 2008). The SHANK3 gene mapped at 22q13.3 encodes a structural protein found in the postsynaptic density that connects ion channels and receptors on the postsynaptic membrane to the cytoskeleton membrane, in the signal transduction pathway (Durand et al., 2007). Thus, SHANK3 haploinsufficiency appears to be responsible for the main neurological manifestations of the 22q13 deletion syndrome (Wilson et al. 2008; Bonaglia et al., 2011; Waga et al., 2011).
On contemplating differential diagnosis of low ARSA activity, six conditions should be considered, namely: 1) MLD, 2) ARSA pseudodeficiency, 3) multiple sulfatase deficiency, 4) saposin B deficiency (associated with ARSA deficiency in vivo only) (Von Figura et al., 2001), 5) compound heterozygosity for a null and pseudo-deficiency alleles of the ARSA gene (without white matter disease), and 6) the 22q13 deletion syndrome. This syndrome can be associated with reduced ARSA activity, even if the remaining allele is normal (Phelan et al., 1992). If, on the other hand, the remaining allele bears a pathogenic mutation, the patient will present LDM and 22q13 syndrome features (Bisgaard et al., 2009). Defining the cause of low ARSA activity is thus essential for genetic counseling, since the risk of recurrence is negligible in the case of a de novo 22q13 deletion. Moreover, the therapeutic management of low ARSA activity depends on a definitive etiologic diagnosis and may include, as in the case of MLD, bone marrow transplantation and enzyme replacement (Biffi et al., 2008). In the case of the 22q13 deletion syndrome, the use of intranasal insulin is being evaluated (Schmidt et al., 2008).
The case herein reported reinforces the importance of a detailed cytogenetic investigation in patients presenting low arylsulfatase A activity, in association with atypical/unspecific clinical features.
Acknowledgments
The authors thank Drs. Roberto Giugliani, Louise Lapagesse Pinto, Cristina B. Netto, and Mercedes Villanueva for their help with this study. This study was conducted with the support of FIPE/HCPA, CNPq/Brazil (RedeBRIM Nr. 402012/2010-0) and the Swiss National Foundation (n. 320000-113635).
Footnotes
Associate Editor: Angela M. Vianna-Morgante
References
- Artigalás O, Lagranha VL, Saraiva-Pereira ML, Burin MG, Lourenço CM, van der Linden H, Jr, Santos ML, Rosemberg S, Steiner CE, Kok F, et al. Clinical and biochemical study of 29 Brazilian patients with metachromatic leukodystrophy. J Inherit Metab Dis. 2010 doi: 10.1007/s10545-010-9140-4. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- Bartsch O, Schneider E, Damatova N, Weis R, Tufano M, Iorio R, Ahmed A, Beyer V, Zechner U, Haaf T. Fulminant hepatic failure requiring liver transplantation in 22q13.3 deletion syndrome. Am J Med Genet. 2010;152:2099–2102. doi: 10.1002/ajmg.a.33542. [DOI] [PubMed] [Google Scholar]
- Biffi A, Lucchini G, Rovelli A, Sessa M. Metachromatic leukodystrophy: An overview of current and prospective treatments. Bone Marrow Transplant. 2008;42:S2–S6. doi: 10.1038/bmt.2008.275. [DOI] [PubMed] [Google Scholar]
- Bisgaard AM, Kirchhoff M, Nielsen JE, Kibaek M, Lund A, Schwartz M, Christensen E. Chromosomal deletion unmasking a recessive disease: 22q13 deletion syndrome and metachromatic leukodystrophy. Clin Genet. 2009;75:175–179. doi: 10.1111/j.1399-0004.2008.01113.x. [DOI] [PubMed] [Google Scholar]
- Bonaglia MC, Giorda R, Beri S, De Agostini C, Novara F, Fichera M, Grillo L, Galesi O, Vetro A, Ciccone R, et al. Molecular mechanisms generating and stabilizing terminal 22q13 deletions in 44 subjects with Phelan/McDermid Syndrome. PLoS Genet. 2011;7:e1002173. doi: 10.1371/journal.pgen.1002173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F, Nygren G, Rastam M, Gillberg IC, Anckarsäter H, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39:25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan MC. Deletion 22q13.3 syndrome. Orphanet J Rare Dis. 2008;3:14–19. doi: 10.1186/1750-1172-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan MC, Thomas GR, Saul RA, Rogers RC, Taylor HA, Wenger DA, McDermid HE. Cytogenetic, biochemical, and molecular analyses of a 22q13 deletion. Am J Med Genet. 1992;43:872–876. doi: 10.1002/ajmg.1320430524. [DOI] [PubMed] [Google Scholar]
- Philippe A, Boddaert N, Vaivre-Douret L, Robel L, Danon-Boileau L, Malan V, de Blois MC, Heron D, Colleaux L, Golse B, et al. Neurobehavioral profile and brain imaging study of the 22q13.3 deletion syndrome in childhood. Pediatrics. 2008;122:376–382. doi: 10.1542/peds.2007-2584. [DOI] [PubMed] [Google Scholar]
- Schmidt H, Kern W, Giese R, Hallschmid M, Enders A. Intranasal insulin to improve the developmental delay in children with 22q13 deletion syndrome: An exploratory clinical trial. J Med Genet. 2008;46:217–222. doi: 10.1136/jmg.2008.062141. [DOI] [PubMed] [Google Scholar]
- Schwartz IV, Ribeiro MG, Mota JG, Toralles MB, Correia P, Horovitz D, Santos ES, Monlleo IL, Fett-Conte AC, Sobrinho RP, et al. A clinical study of 77 patients with mucopolysaccharidosis type II. Acta Paediatr Suppl. 2007;96:63–70. doi: 10.1111/j.1651-2227.2007.00212.x. [DOI] [PubMed] [Google Scholar]
- Von Figura K, Gieselmann V, Jaeken J. Methacromatic leukodystrophy. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. The Metabolic and Molecular Basis of Inherited Disease. 7th edition. McGraw-Hill; New York: 2001. pp. 3695–3724. [Google Scholar]
- Waga C, Okamoto N, Ondo Y, Fukumura-Kato R, Goto Y, Kohsaka S, Uchino S. Novel variants of the SHANK3 gene in Japanese autistic patients with severe delayed speech development. Psychiatr Genet. 2011;21:208–211. doi: 10.1097/YPG.0b013e328341e069. [DOI] [PubMed] [Google Scholar]
- Wilson HL, Crolla JA, Walker D, Artifoni L, Dallapiccola B, Takano T, Vasudevan P, Huang S, Maloney V, Yobb T, et al. Interstitial 22q13 deletions: Genes other than SHANK3 have major effects on cognitive and language development. Eur J Hum Genet. 2008;16:1301–1310. doi: 10.1038/ejhg.2008.107. [DOI] [PubMed] [Google Scholar]