Abstract
Ceramidase plays an important role in regulating the metabolism of sphingolipids, such as ceramide, sphingosine (SPH), and sphingosine-1-phosphate (S1P), by controlling the hydrolysis of ceramide. Here we report the cloning and biochemical characterization of a neutral ceramidase from the red flour beetle Tribolium castaneum which is an important storage pest. The Tribolium castaneum neutral ceramidase (Tncer) is a protein of 696 amino acids. It shares a high degree of similarity in protein sequence to neutral ceramidases from various species. Tncer mRNA levels are higher in the adult stage than in pre-adult stages, and they are higher in the reproductive organs than in head, thorax, and midgut. The mature ovary has higher mRNA levels than the immature ovary. Tncer is localized to the plasma membrane. It uses various ceramides (D-erythro-C6, C12, C16, C18:1, and C24:1-ceramide) as substrates and has an abroad pH optimum for its in vitro activity. Tncer has an optimal temperature of 37 °C for its in vitro activity. Its activity is inhibited by Fe2+. These results suggest that Tncer has distinct biochemical properties from neutral ceramidases from other species.
Keywords: ceramidase, Tncer, activity, biochemical properties, plasma membrane, mRNA levels
1. Introduction
Sphingolipids are important lipid components of membranes of eukaryotic cells, and some of their metabolites, such as ceramide, sphingosine (SPH), and sphingosine-1-phosphate (S1P) also act as bioactive molecules that mediate various biological responses. It is generally believed that ceramide and SPH are anti-proliferative and pro-apoptotic whereas S1P is pro-proliferative and pro-survival. Because of the opposing roles of ceramide and SPH versus S1P in cellular responses, the relative cellular levels of ceramide and SPH versus S1P may determine a cell’s fate, to proliferate and survive or to undergo growth arrest and die.
Ceramidases (EC 3.5.1.23) are a group of enzymes that catalyze the hydrolysis of ceramide to generate SPH, which is further phosphorylated by sphingosine kinase to form S1P. Therefore, ceramidases may play an important role in regulating the levels of ceramide, SPH, and S1P and thereby cellular responses mediated by these bioactive lipids.
To date, five human ceramidase genes have been cloned in humans and their protein products are classified as the acid, neutral, and alkaline types according to their pH optima for in vitro activities [1]. The acid ceramidase is located in lysosomes and its genetic deficiency causes Farber disease. The neutral ceramidase is located to the plasma membrane or is secreted extracellularly. The alkaline ceramidases are localized to the endoplasmic reticulum (ER) and/or the Golgi complex.
Like mammals, insects also express ceramidases. We recently identified the Drosophila melanogaster alkaline ceramidase (Dacer), which is homologous to the human or mouse alkaline ceramidases [2]. We demonstrated that Dacer inactivation increases the levels of various ceramide species in Drosophila melanogaster, thus, leading to an increase in the Drosophila pre-adult development time, lifespan, or anti-oxidative stress capacity, suggesting that Dacer plays an important role in the Drosophila development and longevity by controlling the metabolism of ceramides. Ito et al. are the first group to clone the Drosophila neutral ceramidase [3], which plays a key role in photoreceptor homeostasis in Drosophila by controlling the endocytic turnover of the arrestin-rhodopsin complex in photoreceptors [4, 5]. These results suggest that the catabolism of ceramides is conserved between mammals and insects.
Here we report the cloning and biochemical characterization of a neutral ceramidase (Tncer) in the red flour beetle, Tribolium castaneum that is the most ubiquitous beetle species harmful to grains and cereals. We demonstrate that although Tncer shares a high degree of similarity in protein sequence to neutral ceramidases from other species, it has distinct biochemical properties.
2. Materials and methods
2.1 Material
Tribolium castaneum was raised with whole wheat flour plus 5% dried yeast extracts at 30 °C [6]. High Five cells and pFastBac HTB vector were a kind gift of Dr. Chuanxi Zhang (Zhejiang University). Lipofectamine 2000 and fetal bovine serum (FBS) were purchased from Invitrogen (Carlsbad, USA). ECL Plus kit was obtained from GE Healthcare (Buckinghamshire, UK). Various ceramides, D-e-sphingosine (C18SPH) with 18 carbons, and C17SPH were purchased from Avanti Polar Lipids, Inc. (Alabaster, USA). Cell culture medium was purchased from Sigma-Aldrich (St. Louis, USA).
2.2 Tncer cDNA cloning
Total RNA was isolated from 6 adult Tribolium castaneum with Trizol (Invitrogen) according to manufacturer’s instruction. cDNA was synthesized by PrimeScript™ 1st Strand cDNA Synthesis Kit (Takara). The full length Tncer gene was generated by PCR with sense primer 5′-GTGACCAAGGAGTGTAAA-3′ and anti-sense primer 5′-ACATTTTAACTAGAGTACTTGT-3′ under the conditions of one cycle of 94 °C for 3 min, 35 cycles of 94 °C for 30 s, 55 °C for 30 s and 72 °C for 2 min 30 s. Amplified PCR product was cloned into the pGEM T-easy vector (Promega), and the resulting cDNA plasmid was confirmed by sequencing.
2.3 Tncer expression constructs
The Tncer coding sequence or open reading frame (ORF) was amplified from the Tncer cDNA plasmid by PCR with sense primer 5′-CGCGGATCCATGCGAAAAAAGCTCTTAG-3′ and anti-sense primer 5′-CCCAAGCTTTCACTTAGTGACTTGGAAAG-3′. The PCR conditions were one cycle of 94 °C for 3 min, 35 cycles of 94 °C for 30 s, 59 °C for 30 s and 72 °C for 2 min 30 s. The 2109 base pairs PCR product was cloned into the vector pFastBac HTB at the BamHI and HindIII sites and confirmed by sequencing. The resulting Tncer expression construct (pFastBac HTB/Tncer) that directed the expression His-tagged Tncer was transformed into E. coli DH10Bac according to the manufacturer’s instructions (Invitrogen).
2.4 Tncer expression in High Five cells
High Five cells (Tn5B-1-4) were cultured in TNM-FH Insect Medium (Sigma-Aldrich) supplemented with 10% fetal bovine serum (Invitrogen). High Five cells (105/well) were seeded in a 6-well plate one day before transfection. The bacmid DNA of pFastBac HTB/Tncer or pFastBac HTB was isolated from E. coli DH10Bac according to the manufacturer’s instruction (Invitrogen). These bacmids were transfected by Lipofectamine 2000 (Invitrogen) into High Five cells to yield the Autographa californica nuclear polyhedrosis viruses (AcNPV) carrying the Tncer ORF and control bacmid respectively, according to the manufacturer’s instructions. The recombinant baculoviruses were harvested 72-h post transfection, and used to infect High Five cells at a multiplicity of infection (MOI) of 0.1. After being amplified for 5 generations, recombinant baculoviruses were used to infect High-Five cells seeded in 75 cm2 culture flasks for 48 h to generate the His-tagged Tncer for ceramidase activity assay.
2.5 Microsomes preparation
High Five cells were harvested by centrifugation at 1200 g for 10 min, washed twice by PBS, and resuspended in a lysis buffer (20 mM Tris-HCl, 0.25 M Sucrose, pH 7.4) with 20 mg/ml protease inhibitor (Roche Applied Science). The cells were sonicated at a power level of 35% on a microtip-equipped Sonic Dismembrator for 4 – 5 times, 5 s each. After sonication, the cell lysates were centrifuged at 1,000 g for 10 min to remove nuclei and cell debris. The resulting supernatants were then centrifuged at 100,000 g for 1 h to sediment microsomes (the Mr = 100,000 membrane fraction).
2.6 Protein concentration determination
Protein concentration was determined using a bicinchoninic acid (BCA) protein determination kit (Beyotime) according to the manufacturer’s instructions.
2.7 Western Blot
Micrcosomes (40 μg proteins per lane) were resolved on a 10% SDS-polyacrylamide gel and transferred to a nitrocellulose membrane, which was probed with an anti-His×6 antibody at a dilution of 1:1000, followed by an anti-mouse secondary antibody at a dilution of 1:3000. The His-tagged Tncer was detected with an ECL Plus detection kit (GE Healthcare) according to manufacturer’s instruction.
2.8 Ceramidase activity assays
Ceramidase activity was assayed exactly according to our previous study [2].
2.9 Subcelluar localization study High Five cells
The Tncer ORF was cloned both downstream and upstream of the enhanced green fluorescent protein (eGFP) coding sequence of the vectors pFastBac HTB-GFPC and pFastBac HTB-GFPN to generate expression constructs (pFastBac HTB-GFPC/Tncer and pFastBac HTB-GFPN/Tncer) that directed the expression the eGFP-tagged Tncer, eGFP-Tncer. The Bacmid containing pFastBac HTB-GFPC/Tncer or pFastBac HTB-GFPN/Tncer was transfected into High Five cells as described earlier. After 24-h infection, the cells were fixed and stained with the nuclear dye 4′, 6-diamidino-2-phenylindole (DAPI) before being examined under a confocal fluorescence microscope (Leica TCS SP5, Germany).
2.10 Quantitative real time PCR (qPCR)
qPCR was performed with the cDNA reversely transcribed from total RNA isolated from different developmental stages of Tribolium castaneum or total RNA isolated from different organs of the adult Tribolium castaneum. cDNA was obtained using PrimeScript® RT reagent Kit according to the manufacture’s instruction (Takara). The life stages included egg (approximately 100 eggs), early instar larvae (20 mixed-sex larvae), last instar larvae (4 mixed-sex larvae), pupae (4 pupae) and adult (4 mixed-sex adults). And the organs included head, thorax, midgut, ovary and spermary. Approximately 50 female and male adults were dissected on ice respectively. The variety of Tncer expression in ovary and spermary was also analyzed using female and male adults of 1st day, 7th day and 14th day respectively. Approximately 50 females and 50 males were dissected for ovary and spermary on ice respectively. The qPCR primers were 5′-AAGGCACCAGATCGACAAAC-3′ (sense) and 5′-CTTCACCCACTTGGACGATT-3′ (anti-sense). The ribosome protein S6 gene was used as an internal standard. Its qPCR primers were 5′-CGATGAAGCAGGGTGTTCTC-3′ (sense) and 5′-ATCCAGGGATTTCCTGTTCG-3′ (anti-sense). Each qPCR reaction (50 μl) was assembled in a 0.2-ml microtube by adding a SYBR EX Taq mixture (25 μl) (Takara), a sense primer (10 μM), an antisense primer (10 μM), ROX reference dyeII (1 μl) (Takara), a cDNA template (4 μl), and ddH2O (18 μl). qPCR was run on an ABI 7500 Real-Time PCR System for one cycle of 95 °C for 30 s, 40 cycles of 95 °C for 5 s, 60 °C for 34 s.
3. Results
3.1 Cloning
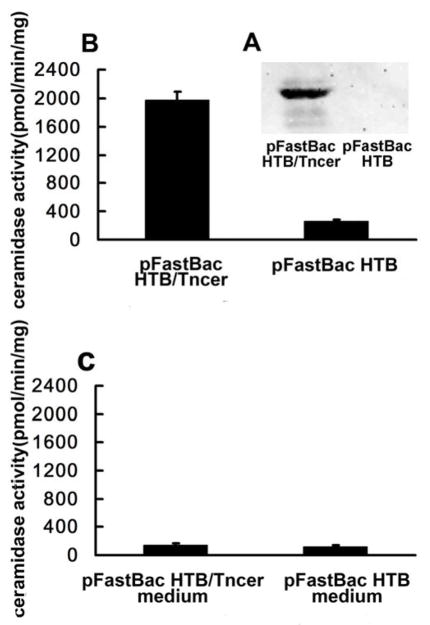
A BLAST search of the GenBank™ using Drosophila melanogaster neutral ceramidase as queries revealed three putative Tribolium castaneum neutral ceramidase genes with the GeneBank Gene ID 657468, 657392, and 657315, respectively. All of these putative genes are neighborhoods on chromosome 10 and have a pfam04734 ceramidase_alk domain conserved among neutral ceramidases. We cloned the ORF of each of these putative genes and expressed them in High Five cells using the Bac-to-Bac baculovirus expression system. The expression constructs were confirmed by sequencing. Western Blot showed the expression of 657315 (Figure 1A). We referred to the product of this gene as the Tribolium castaneum neutral ceramidase, Tncer. But Western Blot using the concentrated cell culture medium and whole cell lysates showed no other genes gave rise to a polypeptide with an estimated Mr. of 80.1 kDa in High Five cells.
Fig. 1. Expression of Tncer in High Five cells and the Tncer activity assay.
(A) Microsomes were prepared from pFastBac HTB/Tncer and pFastBac HTB cells and were resolved on a SDS-PAGE gel. A portion of the microsomes was subjected to Western Blot analysis with the anti-His×6 antibody (1:1,000). The relative Mr. (*80.1 kDa) was estimated according to protein standards. (B) Another portion of the microsomes was assayed for ceramidase activity at pH 7.0. (C) The medium of pFastBac HTB/Tncer or pFastBac HTB transfected cells was assayed for ceramidase activity at pH 7.0.
To determine whether Tncer has ceramidase activity, we measured ceramidase activity in microsomes isolated from High-Five cells transfected with empty vector (pFastBac HTB) or vector containing the full-length Tncer (pFastBac HTB/Tncer). Transfection with pFastBac HTB/Tncer increased microsomal ceramidase activity significantly compared to transfection with the empty vector (Figure 1B), suggesting that Tncer has ceramidase activity.
3.2 Sequence comparison
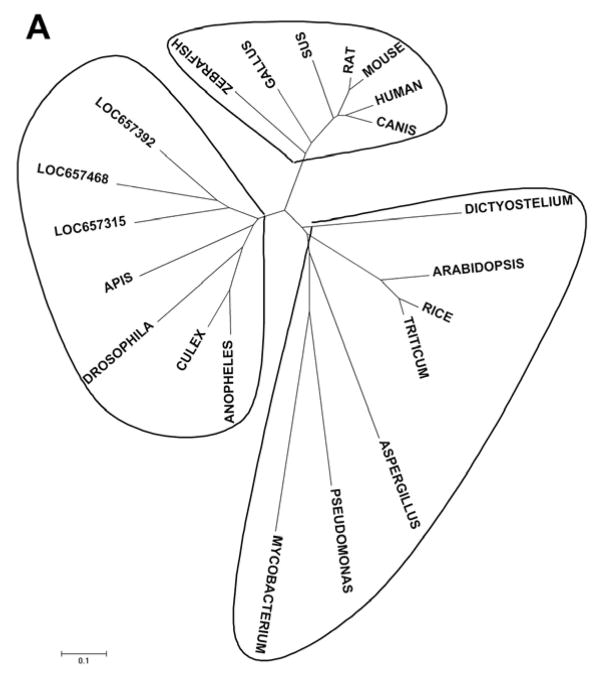
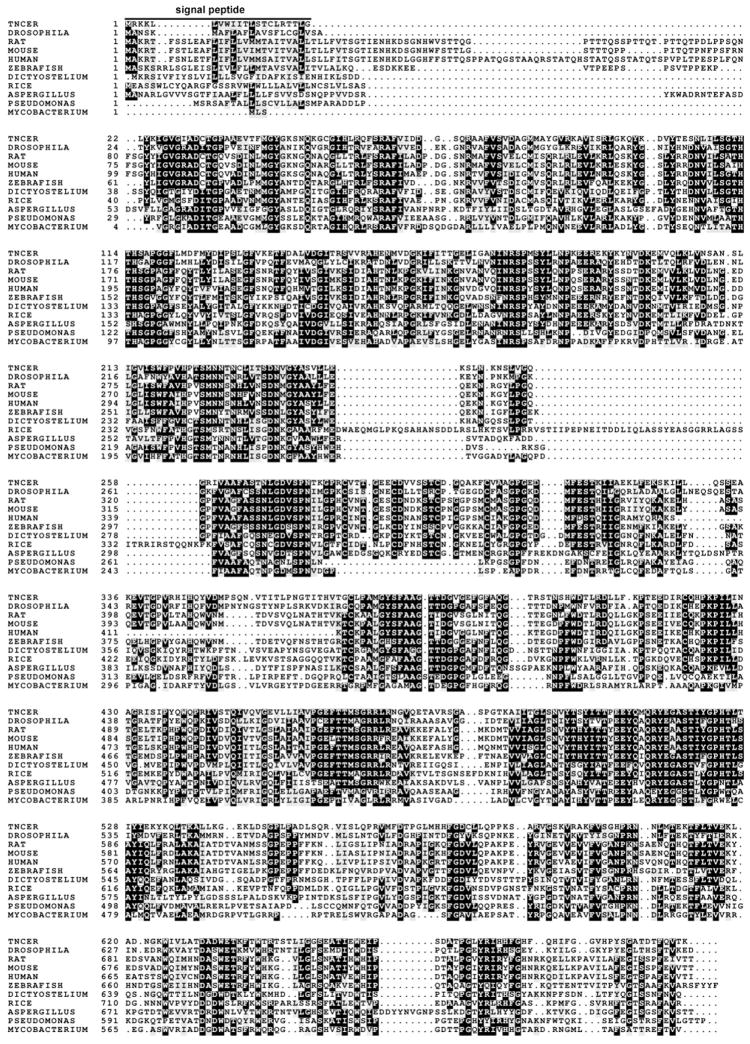
To determine the evolutionary relationship between Tncer and neutral ceramidases from other species, we performed phylogenetic analyses. Neutral ceramidases were clustered into three subclasses: insect, mammalian, and fungi, and plant neutral ceramidases were clustered into the fungal subclass (Figure 2A). The amino acid sequence of Tncer has a 49.1%, 38.1%, 43.4%, 36.8%, and 43.4% sequence similarity to the neutral ceramidase of Drosophila melanogaster, rice, humans, and Pseudomonas aeruginosa, respectively. Tncer shared several conserved domains with other neutral ceramidases (Figure 2B).
Fig. 2. Phylogenetic analysis of neutral ceramidase homologue.
(A) Classification of insect, mammalian and fungi neutral ceramidase based on amino acid sequence similarities. Neighbor-joining tree was generated using MAGE 4 software. (B) Alignment of deduced amino acid sequences of neutral ceramidase from different species. The neutral ceramidases from Tribolium castaneum (Gene ID 657315), rat, mouse, human, zebrafish, Drosophila melanogaster, rice, Dictyostelium discoideum, Pseudomonas aeruginosa, Mycobacterium tuberculosis, and Aspergillus oryza, respectively, were aligned. Conserved residues were shaded. Putative signal peptide was highlight. APIS: Apis mellifera; DROSOPHILA: Drosophila melanogaster; CULEX: Culex pipiens; ANOPHELES: Anopheles gambiae; MYCOBACTERIUM: Mycobacterium tuberculosis; PSEUDOMONAS: Pseudomonas aeruginosa; ASPERGILLUS: Aspergillus oryza; TRITICUM: Triticum aestivum; ARABIDOPSIS: Arabidopsis thaliana; DICTYOSTELIUM: Dictyostelium discoideum; CANIS: Canis familiaris; SUS: Sus scrofa; GALLUS:Gallus gallus.
3.3 Development-specific and tissue-specific expression of Tncer
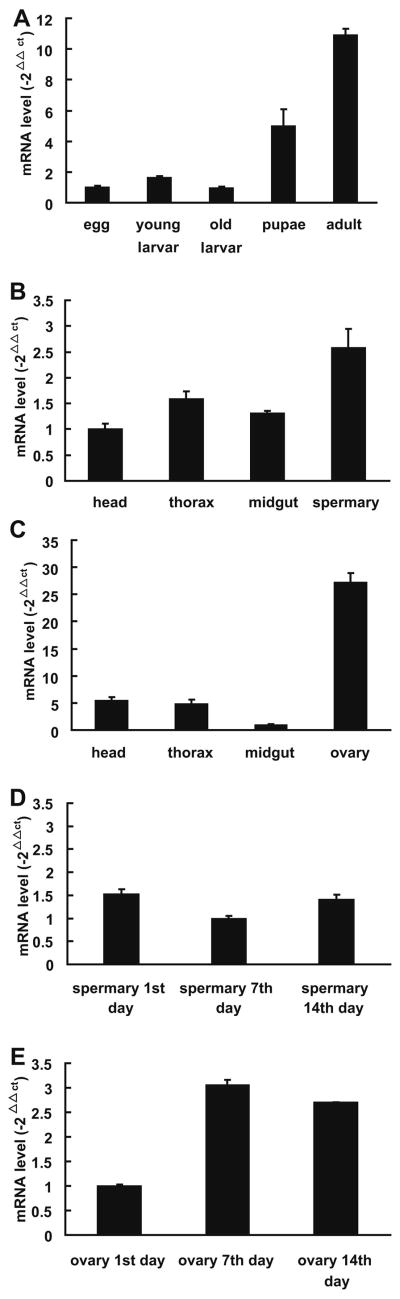
To better understand the physiological roles of Tncer, qPCR was performed to quantify the temporal and spatial expression of Tncer. qPCR showed that the mRNA levels of Tncer were significant higher in the adult Tribolium castaneum than in eggs, early instar larvae, last instar larvae, or pupae stage of life cycle (Figure 3A), suggesting that Tncer was up-regulated at the adult stage during the development of Tribolium castaneum. And in the female Tribolium castaneum, the Tncer mRNA levels were much higher in the ovary than in other organs we examined (Figure 3C), suggesting that Tncer was highly expressed in the female reproductive organ. However, this difference was not significant in the male Tribolium castaneum (Figure 3B).
Fig. 3. Stage and tissue specific expression of Tncer.
(A) mRNA levels of Tribolium castaneum neutral ceramidase at different life stages. (B) mRNA levels of male Tribolium castaneum neutral ceramidase of different organs. (C) mRNA levels of female Tribolium castaneum neutral ceramidase of different organs. (D) mRNA levels of Tncer in spermary of male adult Tribolium castaneum at different ages. (E) mRNA levels of Tncer in ovary of female adult Tribolium castaneum at different ages. Transcript abundance was calculated based on the difference in threshold cycle (Ct) values between Tncer and S6 transcripts based on the normalized relative quantification 2−ΔΔCt method. Data represent the mean value ± SE of three independent experiments performed in duplicate.
To investigate the temporal expression of Tncer in ovary and spermary, qPCR was performed with the ovary or spermary of adult Tribolium castaneum at different ages. The result showed that the Tncer mRNA levels in the ovary of the 7 days-age female were 3-fold higher than those of the 1 day-age female. The Tncer mRNA levels of the ovary were similar in females at the ages of 7 days to 14 days (Figure 3E). In contrast, no significant change was found in the Tncer mRNA levels in the spermary of the 1, 7, or 14-day-age male (Figure 3D). These results suggested that Tncer was up-regulated during ovary maturation.
3.4 Subcelluar localization of Tncer in High Five cells
Because neutral ceramidase has been found to be secreted from S2 cells [3], we investigated whether Tncer is also secreted from High Five cells. Western Blot did not show the present of Tncer in concentrated medium. And also we found that transfection with pFastBac HTB/Tncer did not increase ceramidase activity in medium compared to transfection with the empty vector (Figure 1C). These suggested that unlike Drosophila neutral ceramidase, Tncer may not be secreted to medium.
To determine the subcellular localization of Tncer, we cloned the Tncer ORF into the vectors pFastBac HTB-GFPC and pFastBac HTB-GFPN and expressed the eGFP-tagged Tncer (eGFP-Tncer) in High Five cells. Confocal microscopy demonstrated that the eGFP-Tncer protein was mostly localized to the plasma membrane (Figure 4), suggesting that Tncer had the same cellular localization as other neutral ceramidases.
Fig. 4. Tribolium castaneum neutral ceramidase was located mainly in plasma membrane.
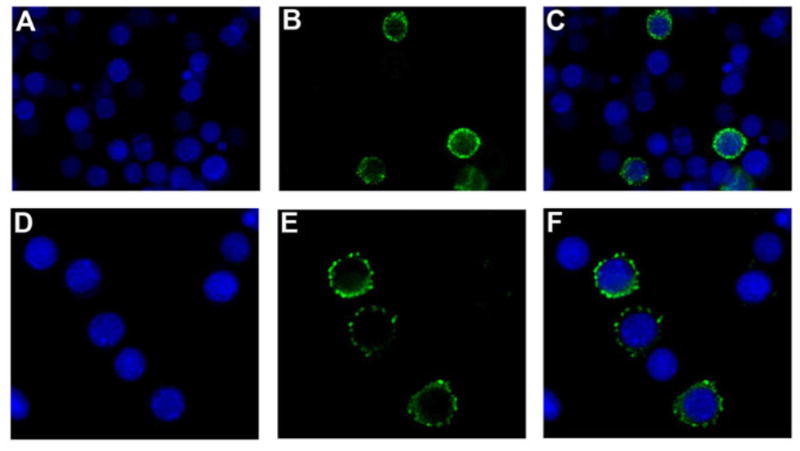
(A) Nucleolus stained by DAPI in blue. (B) eGFP-Tncer fusion protein expressed by pFastBac HTB-GFPC/Tncer observed in green under confocal fluorescence microscope. (C) Merged. (D) Nucleolus stained by DAPI in blue. (E) eGFP-Tncer fusion protein expressed by pFastBac HTB-GFPN/Tncer observed in green under confocal fluorescence microscope. (F) Merged.
3.5 Biochemical properties of Tncer
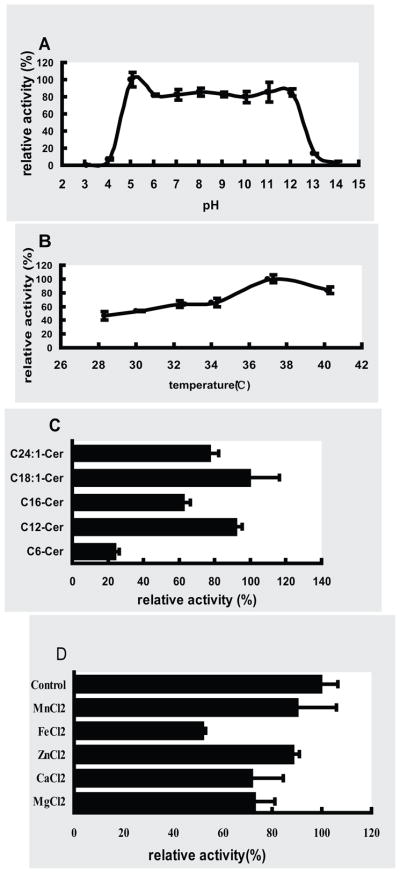
In order to better understand the physiological roles of Tncer, we studied its biochemical properties. First, its pH optimum was determined. Tncer ceramidase activity was assayed at pH from 3 to 14. We demonstrated that Tncer activity was highest between pH 5 and 12 and remained fairly constant within this range, suggesting that Tncer had a very broad pH optimum for its activity (Figure 5A). Second, its temperature optimum was determined. We found that the optimal temperature for Tncer activity was around 37 °C (Figure 5B). Third, the substrate specificity of Tncer was determined using different ceramides as substrates. Tncer efficiently hydrolyzed C12, C16, C18:1, or C24:1-ceramide with a similar rate, but it hydrolyzed C6-Cer with a much lower efficiency (Figure 5C), suggesting that Tncer prefers medium-chain or long-chain ceramides over short-chain ceramides as substrates. Finally, the cationic effects on Tncer activity were investigated. Tncer activity was determined in the presence of various cations at 5 mM or in the absence of any cation. We demonstrated that among the cations examined, only Fe2+ inhibited Tncer activity by 50% (Figure 5D).
Fig. 5. Biochemical properties of Tncer.
(A) The Tncer activity in microsomes of pFastBac HTB/Tncer and pFastBac HTB cells were assayed at different pH values. The pH was adjusted by adding the following buffer: acetate (pH 3–6), Tris (pH 7–8), Glycine (pH 9–13), 1M NaOH (pH 14). Ceramidase activity of the recombinant Tncer at each pH value was computed by subtracting ceramidase activity in pFastBac HTB microsomes from that in pFastBac HTB/Tncer microsomes. The Tncer ceramidase activity at pH 5 was highest and set as 100%, and ceramidase activity at other pH values was expressed as % of the maximal activity. (B) Temperature dependence of Tncer. The above microsomes were assayed for ceramidase activity at indicated temperature. Ceramidase activity of the recombinant Tncer at each temperature was computed as mentioned. The Tncer ceramidase activity at 37 °C was highest and set as 100%, and ceramidase activity at other temperature values was expressed as % of the maximal activity (C) Substrate specificity of Tncer. The microsomes of pFastBac HTB/Tncer and pFastBac HTB were assayed for ceramidase activity using indicated ceramide as substrate. Ceramidase activity was computed as mentioned. The Tncer ceramidase activity was highest with substrate of C18:1 ceramide and set as 100%, and ceramidase activity with other substrate values was expressed as % of the maximal activity (D) Tncer activity was inhibited by different cations. The microsomes of pFastBac HTB/Tncer and pFastBac HTB were assayed for ceramidase activity with adding indicated cation directly in reaction buffer. The Tncer ceramidase activity with no cation added was highest and set as 100%, and ceramidase activity with different cations values was expressed as % of the maximal activity. All data represent the mean value of three independent experiments performed in duplicate.
4. Discussion
In this study, we report the cloning, expression, and biochemical characterization of the first ceramidase, Tncer, in an important storage pest, Tribolium castaneum. Other two genes were only found in transcript level. The similarity between these three genes was fairly high. In Human, there are also two putative neutral ceramidase only found in transcript level. The function of these putative genes in mRNA level needs further research.
We demonstrate that although Tncer is highly homologous to other neutral ceramidases, it exhibits unique biochemical properties.
First, it has a much broader pH optimum than any other known neutral ceramidases. It has a pH optimum of 5 to 12. The neutral ceramidases in Dictyostelium discoideum, Aspergillus oryzae and rice have pH optima of 3, 5.7 – 6.0, and 4.0 – 4.5, respectively. Drosophila neutral ceramidase has optimal pH between 6.5 and 7.5 [3]. The neutral ceramidases in Pseudomonas aeruginosa and Mycobacterium tuberculosis have slightly alkaline pH optima [7].
Second, there are unique cationic effects on Tncer activity. The neutral ceramidase of Pseudomonas aeruginosa is found to be activated by Ca2+ [8]. Inoue et al. recently defined the Ca2+/Mg2+ binding site might be important for the interaction of N-terminal domain and C-terminal domain [9]. In contrast, none of eukaryotic neutral ceramidases have been found to be activated by Ca2+ [10, 11]. We demonstrated that like those eukaryotic enzymes, Tncer is not activated by any cations, including Ca2+. We find that similar to the Drosophila neutral ceramidase [3], Tncer is inhibited by Fe2+. However, distinct from the Drosophila ceramidase, Tncer is not inhibited by Zn2+. Zn2+ also inhibits the neutral ceramidase activity in rat and human [12, 13]. But the rice neutral ceramidase is also not inhibited by Zn2+. These results suggest that neutral ceramidases from different species differ greatly in the cationic effect on their activity.
The acid ceramidase prefers medium-chain ceramides as substrates [1] whereas the alkaline ceramidases mainly use long-chain or very long-chain ceramides as substrates [1, 2]. El et al demonstrate that the rat neutral ceramidase hydrolyzes various long-chain and very long-chain ceramides efficiently [14]. We demonstrate that Tncer hydrolyzes medium-chain ceramides as efficiently as long-chain or very long-chain ceramides although it hydrolyzes the short-chain ceramide D-e-C6-ceramide less efficiently, suggesting that Tncer has a broader substrate specificity than mammalian ceramidases.
The neutral ceramidase is localized to the plasma membrane or secreted extracellularly. Mammalian neutral ceramidases, such as the human, mouse, rat ceramidsaes, are mainly localized to the plasma membrane [15, 16]. The rice neutral ceramidase is localized to the ER and the Golgi comple [17]. In contrast, Pseudomonas aeruginosa neutral ceramidase is released to the medium [7]. The Drosophila neutral ceramidase is also secreted [3]. We demonstrated that like human and mouse neutral ceramidases, Tncer is localized to the plasma membrane, but it is not secreted. We searched a putative transmembrane domain (TM) or (TMs) using an on-line program, TMHMM (http://ww.cbs.dtu.dk/services/TMHMM-2.0), no putative TM was found. Using the program SignaIP (http://www.cbs.dtu.dk/services/SignalP/), we found a putative signal peptide (MRKKLLVWIITLSTCLRTTLG) at the N-terminal of Tncer. This signal peptide is conserved among Tncer and the mammalian neutral ceramidases, which are also localized to the plasma membrane [15]. This signal peptide may anchor Tncer to the plasma membrane. This possibility is under investigation.
Tncer expression is both developmental stage and tissue-specific. Like mammalian neutral ceramidase, Tncer is ubiquitously expressed [10, 11, 13]. However, its tissue-specific expression is distinct from its homologues in other species. Tncer mRNA levels are highest in reproductive organs in Tribolium castaneum, whereas the mouse neutral ceramidase mRNA levels are much lower in mouse testis than in mouse kidney [10]. The mouse neutral ceramidase mRNA levels are in fact highest in kidney and liver. The rat neutral ceramidase levels are much higher in kidney and brain than in other tissues including liver [13]. Human neutral ceramidase is also highest in kidney [11]. We demonstrate that the Tncer mRNA levels are increased during the development of Tribolium castaneum from eggs to the adult, being the highest in the adult, suggesting that Tncer may play a role in the insect development. Although Tncer is ubiquitously expressed in Tribolium castaneum tissues, its expression is much higher in the ovary than other tissues. Moreover, its expression in the ovary is increased with ovarian maturation, suggesting that Tncer may play a key role in female reproduction. The role of Tncer in Tribolium castaneum development will be focused in our further study.
In summary, we found a kind of ceramidase in Tribolium castaneum. It is a plasma membrane located protein. Although it is highly conserved compared with other reported neutral ceramidase, but showing quite broad pH optimal for its high activity. The preferable substrates for Tncer, medium or long-chain ceramides, are the same as other neutral ceramidase. Its activity can be inhibited by Fe2+, which is also the same as other neutral ceramidase. Compared with other neutral ceramidase in cation effect, we found ceramidase in different species showed differently to certain cation. The mRNA level was found to be highest in the ovary of adult female compared with other organs suggesting its potential roles in the development of ovary. The physiological function of Tncer in the development of Tncer will be emphasis in our further research.
Research Highlights
Tribolium castaneum ceramidase (Tncer) is mainly localized to the plasma membrane.
Its mRNA levels are much higher in ovary.
Tncer has a much broader pH optimum than any other known neutral ceramidases.
Tncer’s substrate specificity is broader than mammalian ceramidases.
Acknowledgments
This work was supported by China’s National Basic Research (973 Programs 2009CB119203, 2010CB126200) (ZZ), NSFC grants (30528024) (CM), and MOE project 111 (CM), and the United States National Institutes of Health grant R01CA104834 (CM). We thank Zhong-Jun Gong, Gui-Juan Kang, and Wen-Juan Jiao for their technical assistance in cell culture and red flour beetle colony maintenance.
Abbreviations
- Tncer
Tribolium castaneum neutral ceramidase
- SPH
sphingosine
- S1P
sphingosine-1-phosphate
- qPCR
Quantitative real time PCR
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mao C, Obeid LM. Ceramidases: regulators of cellular responses mediated by ceramide, sphingosine, and sphingosine-1-phosphate. Biochimica et biophysica acta. 2008;1781:424–434. doi: 10.1016/j.bbalip.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang Q, Gong ZJ, Zhou Y, Yuan JQ, Cheng J, Tian L, Li S, Lin XD, Xu R, Zhu ZR, Mao C. Role of Drosophila alkaline ceramidase (Dacer) in Drosophila development and longevity. Cell Mol Life Sci. 2010;67:1477–1490. doi: 10.1007/s00018-010-0260-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoshimura Y, Okino N, Tani M, Ito M. Molecular cloning and characterization of a secretory neutral ceramidase of Drosophila melanogaster. Journal of biochemistry. 2002;132:229–236. doi: 10.1093/oxfordjournals.jbchem.a003215. [DOI] [PubMed] [Google Scholar]
- 4.Acharya JK, Dasgupta U, Rawat SS, Yuan C, Sanxaridis PD, Yonamine I, Karim P, Nagashima K, Brodsky MH, Tsunoda S, Acharya U. Cell-nonautonomous function of ceramidase in photoreceptor homeostasis. Neuron. 2008;57:69–79. doi: 10.1016/j.neuron.2007.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Acharya U, Mowen MB, Nagashima K, Acharya JK. Ceramidase expression facilitates membrane turnover and endocytosis of rhodopsin in photoreceptors. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:1922–1926. doi: 10.1073/pnas.0308693100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arakane Y, Li B, Muthukrishnan S, Beeman RW, Kramer KJ, Park Y. Functional analysis of four neuropeptides, EH, ETH, CCAP and bursicon, and their receptors in adult ecdysis behavior of the red flour beetle, Tribolium castaneum. Mech Dev. 2008;125:984–995. doi: 10.1016/j.mod.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 7.Okino N, Ichinose S, Omori A, Imayama S, Nakamura T, Ito M. Molecular cloning, sequencing, and expression of the gene encoding alkaline ceramidase from Pseudomonas aeruginosa. Cloning of a ceramidase homologue from Mycobacterium tuberculosis. The Journal of biological chemistry. 1999;274:36616–36622. doi: 10.1074/jbc.274.51.36616. [DOI] [PubMed] [Google Scholar]
- 8.Wu BX, Snook CF, Tani M, Bullesbach EE, Hannun YA. Large-scale purification and characterization of recombinant Pseudomonas ceramidase: regulation by calcium. Journal of lipid research. 2007;48:600–608. doi: 10.1194/jlr.M600423-JLR200. [DOI] [PubMed] [Google Scholar]
- 9.Inoue T, Okino N, Kakuta Y, Hijikata A, Okano H, Goda HM, Tani M, Sueyoshi N, Kambayashi K, Matsumura H, Kai Y, Ito M. Mechanistic insights into the hydrolysis and synthesis of ceramide by neutral ceramidase. The Journal of biological chemistry. 2009;284:9566–9577. doi: 10.1074/jbc.M808232200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tani M, Okino N, Mori K, Tanigawa T, Izu H, Ito M. Molecular cloning of the full-length cDNA encoding mouse neutral ceramidase. A novel but highly conserved gene family of neutral/alkaline ceramidases. The Journal of biological chemistry. 2000;275:11229–11234. doi: 10.1074/jbc.275.15.11229. [DOI] [PubMed] [Google Scholar]
- 11.El Bawab S, Roddy P, Qian T, Bielawska A, Lemasters JJ, Hannun YA. Molecular cloning and characterization of a human mitochondrial ceramidase. The Journal of biological chemistry. 2000;275:21508–21513. doi: 10.1074/jbc.M002522200. [DOI] [PubMed] [Google Scholar]
- 12.Ohlsson L, Palmberg C, Duan RD, Olsson M, Bergman T, Nilsson A. Purification and characterization of human intestinal neutral ceramidase. Biochimie. 2007;89:950–960. doi: 10.1016/j.biochi.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 13.Mitsutake S, Tani M, Okino N, Mori K, Ichinose S, Omori A, Iida H, Nakamura T, Ito M. Purification, characterization, molecular cloning, and subcellular distribution of neutral ceramidase of rat kidney. The Journal of biological chemistry. 2001;276:26249–26259. doi: 10.1074/jbc.M102233200. [DOI] [PubMed] [Google Scholar]
- 14.El Bawab S, Usta J, Roddy P, Szulc ZM, Bielawska A, Hannun YA. Substrate specificity of rat brain ceramidase. Journal of lipid research. 2002;43:141–148. [PubMed] [Google Scholar]
- 15.Tani M, Iida H, Ito M. O-glycosylation of mucin-like domain retains the neutral ceramidase on the plasma membranes as a type II integral membrane protein. The Journal of biological chemistry. 2003;278:10523–10530. doi: 10.1074/jbc.M207932200. [DOI] [PubMed] [Google Scholar]
- 16.Hwang YH, Tani M, Nakagawa T, Okino N, Ito M. Subcellular localization of human neutral ceramidase expressed in HEK293 cells. Biochemical and biophysical research communications. 2005;331:37–42. doi: 10.1016/j.bbrc.2005.03.134. [DOI] [PubMed] [Google Scholar]
- 17.Pata MO, Wu BX, Bielawski J, Xiong TC, Hannun YA, Ng CK. Molecular cloning and characterization of OsCDase, a ceramidase enzyme from rice. Plant J. 2008;55:1000–1009. doi: 10.1111/j.1365-313X.2008.03569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]