Abstract
Phosphoinositide-3-OH kinases (PI3K) are critical regulators of cell metabolism, growth, and survival. In a recent publication in Nature, Jia et al. (2008) identify specific functions of the p110β isoform of PI3K in glucose metabolism, cellular proliferation, and tumorigenesis.
Class IA PI3Ks are members of a conserved family of lipid kinases comprised of a p85 regulatory subunit and a p110 catalytic subunit. There are two ubiquitously expressed class IA catalytic isoforms, p110α and p110β. Class IA PI3Ks are activated by receptor tyrosine kinases (RTKs). The p110β catalytic subunit can also be activated through G protein-coupled receptors (GPCRs) (Hazeki et al., 1998). Activation of PI3K results in the conversion of PI(4,5)P2 to PIP3; the latter binds to pleckstrin homology (PH) domains of various signaling proteins, including the serine/threonine kinase Akt. Dysregulation of PI3K signaling is implicated in the pathogenesis of diabetes mellitus and cancer (Engelman et al., 2006). The p110α isoform of PI3K has been the most extensively studied to date, and the gene encoding this enzyme is frequently mutated in human cancers. However, in a recent issue of Nature, Jia et al. (2008) identify unique roles for p110β in cellular metabolism and oncogenesis.
In the liver, p110α appears to be the predominant PI3K isoform activated following insulin receptor stimulation (Foukas et al., 2006). Indeed, Jia et al. (2008) show that deletion of p110β does not affect the ability of insulin to activate Akt in the liver, which supports the results of prior pharmacologic studies showing that p110β does not play a significant role in insulin-stimulated Akt activation (Knight et al., 2006). However, Jia et al. (2008) found that the absence of p110β impairs insulin repression of gluconeogenic genes and leads to glucose intolerance. Many insulin-repressed genes in the liver contain binding sites for FOXO transcription factors, and expression of constitutively active FOXO leads to increased gluconeogenic gene expression (Zhang et al., 2006). Notably, mice heterozygous for both p110α and p110β exhibit glucose intolerance despite intact insulin-stimulated AKT activity, suggesting that factors in addition to Akt are required for insulin signaling in the liver (Brachmann et al., 2005). Jia et al. (2008) did not examine FOXO phosphorylation, and other kinases, in addition to Akt, can phosphory-late FOXO. However, FOXO is one of many transcription factors contributing to PEPCK expression; therefore, full repression in the presence of insulin may require activation of an as-yet-unidentified signaling complex by p110β and may occur in a PIP3-independent manner (see Figure 1A). Alternatively, p110β catalytic activity and PIP3 production might be required for repression of PEPCK by insulin. It is known that pharmacologic inhibition of p110β does not block acute Akt phosphorylation, but it does significantly inhibit PIP3 generation. This observation raises the possibility that while p110α activation alone produces more than sufficient PIP3 for acute AKT activation, additional PIP3-dependent responses that require higher levels of this lipid or that require PIP3 production at a unique location by p110β play additional roles in suppression of gluconeogenesis.
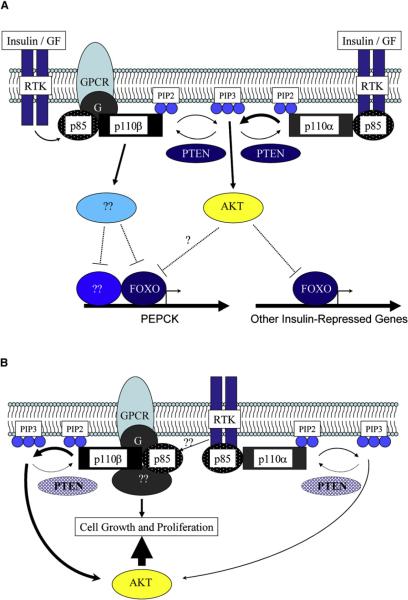
Figure 1. Potential Roles of p110β in Metabolism and Oncogenesis.
(A) Under conditions where PTEN is present, Akt is activated predominantly by the RTK-p110α pathway, as occurs in the liver after insulin stimulation. Insulin-induced activation of Akt results in the phosphorylation of FOXO transcription factors and ultimately, the inhibtion of target gene expression. However, p110β is required for full repression of glucoeneogenic gene expression (e.g., the PEPCK gene) by insulin, suggesting that this isoform activates downstream kinases in addition to Akt and/or regulates transcription factors in addition to FOXO. The specific activation of non-Akt kinases by p110β may require either the generation of spacially restricted pools of PIP3 within the cell or the generation of quantities of PIP3 exceeding those required for Akt activation.
(B) Under conditions where PTEN expression is reduced, as occurs in many human tumors, the basal level of PIP3 is increased due to unopposed p110β activity. These increased basal levels of PIP3 result in increased Akt activity and increased cell growth. In both scenarios (A and B) p110β may also function in a PIP3-independent manner such that its catalytic activity is not required for regulation of downstream signaling. In this model, p110β serves as a “molecular scaffold” for recruiting additional catalytically active signaling molecules. See text for further details.
In addition to their roles in metabolism, both p110α and p110β are also essential for normal mammalian growth and development: germline deletion of either isoform results in embryonic lethality (Bi et al., 2002). Jia et al. (2008) showed that deletion of p110β in mouse embryo fibroblasts (MEFs) retarded cell proliferation but had negligible effect on Akt phosphorylation in response to insulin and EGF, suggesting an Akt-independent role for p110β in mitogenesis. Interestingly, reconstitution of p110β-KO MEFs with a kinase-dead p110β restored normal cell proliferation, suggesting a kinase-independent scaffolding function for p110β in growth and proliferation. Previous work has shown that p110β can serve as a signaling conduit for G protein coupled receptor (GPCR)-linked PI3K signaling and that this isoform is less important than p110α in RTK signaling (Guillermet-Guibert et al., 2008). A model to explain this observation is that p110β has an additional kinase-independent “adaptor” function in signaling, coupling an as-yet-unidentified signaling molecule to GPCRs and RTKs (Figure 1B). Furthermore, p110β has been associated with Rab5 and clathrin-coated endocytic vesicles (Shin et al., 2005), and phosphoinositides regulate the endocytic pathway. Consistent with these observations, Jia et al. (2008) showed that transferrin uptake, a surrogate marker for receptor-mediated endocytosis, was defective in p110β KO cells. This endocytic defect was also rescued by the kinase-dead p110β, suggesting an analogous scaffolding function for p110β in regulation of receptor-mediated endocytosis. Taken together, these data suggest that p110β likely recruits additional protein(s) important for both GPCR-linked PI3K signaling and endocytosis, a function that is largely independent of its lipid kinase activity.
Alterations that lead to increased PI3K signaling confer a survival and growth advantage and are frequent in human tumors. Activating point mutations and gene amplification of the p110α isoform have been detected in a variety of cancers (Engelman et al., 2006). Additionally, PTEN, a tumor suppressor that antagonizes PI3K activity by dephosphorylating PIP3, is deleted or mutated in many cancers. Since activating mutations in p110β have not been observed in human cancers, one might predict that p110α would be the principal class IA PI3K involved in producing PIP3 in oncogene-induced tumors. Surprisingly, Jia et al. (2008) show that p110β appears to play a predominant role in oncogenesis. Loss of p110β abrogated transformation of immortalized MEFs by mutant Ras and mutant EGFR, while loss of p110α had a less pronounced effect. In a mouse model of prostate cancer induced by PTEN loss, concomitant ablation of p110β—but not p110α—led to decreased Akt phosphorylation in the prostate and prevented the development of high-grade prostatic intraepithelial neoplasia (PIN). These findings are consistent with the model put forth by Knight et al. (2006) that p110β generates a basal pool of PIP3 that defines a threshold for p110α activation necessary for signaling. The authors posit that inactivation of PTEN raises the basal PIP3 levels generated by p110β, thereby lowering the threshold for Akt activation and transformation.
Prior studies have shown that overexpression of p110β is sufficient to induce transformation of chicken embryo fibroblasts (Kang et al., 2006). Loss of p110α may fail to abrogate tumor formation due to loss of PTEN because the basal levels of PIP3 are sufficiently high in PTEN−/− cells for activation of Akt and for maintenance of the transformed phenotype. Notably, Jia et al. (2008) evaluated prostate tumorigenesis at 12 weeks of age. It would be interesting to assess these mice at later time points, when prostate-specific PTEN loss has been shown to result in more aggressive cancers (Wang et al., 2003). Perhaps over time the absence of p110β will be insufficient to overcome the elevated basal PIP3 levels, and the proliferative effects of unopposed p110α signaling will drive tumor development. In this setting, loss of both p110α and p110β might be necessary to prevent tumorigenesis. Alternatively, differential signaling from unidentified upstream GPCRs or RTKs, distinct interactions with signaling components, or phosphorylation of unique downstream effectors could explain the differences observed following deletion of p110α versus p110β (Figure 1B). To further elucidate the kinase-independent “scaffolding” function of p110β in vivo, it would be interesting to determine whether prostate-specific knockin of the kinase-dead mutant p110β fails to abrogate PTEN-induced tumorigenesis.
In summary, Jia et al. (2008) present intriguing evidence for p110β as a mediator of diverse cellular processes. They propose that, in contrast to p110α, p110β has a kinase-dependent role in oncogenic transformation that is distinct from its kinase-independent roles in endocytosis, cell proliferation, and potentially, glucose homeostasis, making this isoform of PI3K an attractive target for therapeutic intervention that could perhaps minimize potential side effects of global PI3K inhibition.
REFERENCES
- Bi L, Okabe I, Bernard DJ, Nussbaum RL. Mamm. Genome. 2002;13:169–172. doi: 10.1007/BF02684023. [DOI] [PubMed] [Google Scholar]
- Brachmann SM, Ueki K, Engelman JA, Kahn RC, Cantley LC. Mol. Cell. Biol. 2005;25:1596–1607. doi: 10.1128/MCB.25.5.1596-1607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA, Luo J, Cantley LC. Nat. Rev. Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- Foukas LC, Claret M, Pearce W, Okkenhaug K, Meek S, Peskett E, Sancho S, Smith AJ, Withers DJ, Vanhaesebroeck B. Nature. 2006;441:366–370. doi: 10.1038/nature04694. [DOI] [PubMed] [Google Scholar]
- Guillermet-Guibert J, Bjorklof K, Salpekar A, Gonella C, Ramadani F, Bilancio A, Meek S, Smith AJ, Okkenhaug K, Vanhaesebroeck B, et al. Proc. Natl. Acad. Sci. USA. 2008;105:8292–8297. doi: 10.1073/pnas.0707761105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazeki O, Okada T, Kurosu H, Takasuga S, Suzuki T, Katada T. Life Sci. 1998;62:1555–1559. doi: 10.1016/s0024-3205(98)00106-4. [DOI] [PubMed] [Google Scholar]
- Jia S, Liu Z, Zhang S, Liu P, Zhang L, Lee SH, Zhang J, Signoretti S, Loda M, Roberts TM, Zhao JJ. Nature. 2008;454:776–779. doi: 10.1038/nature07091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S, Denley A, Vanhaesebroeck B, Vogt PK. Proc. Natl. Acad. Sci. USA. 2006;103:1289–1294. doi: 10.1073/pnas.0510772103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight ZA, Gonzalez B, Feldman ME, Zunder ER, Goldenberg DD, Williams O, Loewith R, Stokoe D, Balla A, Toth B, et al. Cell. 2006;125:733–747. doi: 10.1016/j.cell.2006.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin HW, Hayami M, Christoforidis S, Lacas-Gervais S, Hoepfner S, Wenk MR, Modregger J, Uttenweiler-Joseph S, Wilm M, Nystuen A, et al. J. Cell Biol. 2005;170:607–618. doi: 10.1083/jcb.200505128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Gao J, Lei Q, Rozengurt N, Pritchard C, Jiao J, Thomas GV, Li G, Roy-Burman P, Nelson PS, et al. Cancer Cell. 2003;4:209–221. doi: 10.1016/s1535-6108(03)00215-0. [DOI] [PubMed] [Google Scholar]
- Zhang W, Patil S, Chauhan B, Guo S, Powell DR, Le J, Klotsas A, Matika R, Xiao X, Franks R, et al. J. Biol. Chem. 2006;281:10105–10117. doi: 10.1074/jbc.M600272200. [DOI] [PubMed] [Google Scholar]