Abstract
Despite persistent questions about the safety of black cohosh (Actaea racemosa L., syn. Cimicifuga racemosa L.), black cohosh products continue to be one of the most popular botanical supplements in the United States market. Black cohosh products have been associated with cases of liver toxicity, but subsequent evaluation found some products to be adulterated with other related plants from the same genus. US FDA regulations require that black cohosh products be unadulterated, and correct identification of different species of Actaea is a key first step for their good manufacturing practice. We have developed a phytochemical method to distinguish four different groups of Actaea, including: species other than A. racemosa; Asian species; A. racemosa; and North American species other than A. racemosa. Using HPLC-TOF-ESI-MS technique and principal component analysis, we identified 15 chemical markers (1–3, 5–6, 8–10, 12, 16–21). Three marker compounds were unambiguously identified using authentic standards, and twelve marker compounds were tentatively identified by comparison of fragmentation patterns with previously reported data. The presence of these marker compounds is critical for discrimination among the four groups of closely related plants. The use of metabolic profiling to distinguish black cohosh from related species of Actaea has broader implications in the identification of markers to help authenticate other important medicinal plants.
1. Introduction
Actaea (syn. Cimicifuga) is a small genus belonging to the Ranunculaceae, a family of plants native to temperate regions of the Northern Hemisphere. In North America, A. racemosa (black cohosh) is widely used as a dietary herbal supplement for the treatment of menopausal symptoms [1,2]. In 2008, black cohosh herbal products were ranked tenth in U.S. botanical sales [3]. In addition to black cohosh, there are eight other North American species of Actaea, including A. podocarpa, A pachypoda, A rubra, A laciniata, A americana, A elata, A arizonica, and A cordifolia, but none is used commonly for menopausal symptoms. Asian species of Actaea were also widely used as traditional medicines for hundreds of years. A. heracleifolia, A. dahurica, and A. cimicifuga (Syn. Cimicifuga foetida) are listed in the Chinese Pharmacopoeia and used for their anti-inflammatory, antipyretic and analgesic effects [4]. Modern research has revealed several bioactive properties of Asian species of Actaea, but no species is widely used for menopausal symptoms [5–10].
Black cohosh has different traditional usages with other North American and Asian species of Actaea. However, our studies have found that black cohosh products in the U.S. market are adulterated widely with other species of Actaea, which are lower-priced and exert unknown effect on menopausal symptoms [11]. It is critical that the black cohosh products be of high quality and be unadulterated. Health Canada has found that several cases of liver toxicity were associated with people who took black cohosh products adulterated with related herbal species [12]. The most commonly used species of Actaea for black cohosh adulteration include A. americana, A. cimicifuga, A. heracleifolia and A. dahurica [13]. Therefore, correct identification of the plant and the discovery of the suitable marker compounds to distinguish different species of Actaea becomes a key first step for good manufacturing practices of black cohosh products. Previously, these Actaea species have been distinguished, with various success, using DNA, HPLC-UV (DAD), HPLC-ELSD, HPLC-MS, GC-MS, high performance TLC (HPTLC)-densitometry, fourier transform near-infrared (FT-NIR) spectroscopy, and high-field NMR fingerprints [11,14–17]. Two marker compounds, cimiracemoside C and cimifugin, have been identified to distinguish black cohosh and other Asian species of Actaea [11,14]. These studies have resulted in either the different types of fingerprints to discriminate among different Actaea species without additional marker compounds, or marker compounds found based on available standards.
Recently, the use of HPLC coupled with time of flight mass spectrometry (TOF-MS) has been employed for the analysis of herbal medicines and metabolites, and proved to be powerful for the structural characterizations of constituents [18–21]. The TOF spectrometer can produce exact mass measurements. Based on these data, elemental compositions of marker molecular ions and their fragmental ions can be confirmed, which are very useful for elucidating the pathway of the mass fragmental cleavage.
In the present research, a multivariate statistical analysis method, principal component analysis (PCA), was introduced to analyze the HPLC-TOF-MS total ion current (TIC) chromatograms of Actaea species. We tried to find useful marker compounds like cimifugin derivatives, triterpene glycosides, and alkaloids, to distinguish different species of Actaea. Additionally, based on the molecular and fragmental ions analysis using a group of authentic standards which belong to different classes of aglycone structures, we aimed to first develop the proposed characteristic MS/MS fragmentation rules originated from the cleavage of the C(20)-C(27) side chain of these structures. This side chain shows the most structural variation among all the classes of triterpene glycoside aglycone skeleton. Applying these MS/MS fragmentation rules to compounds without standards, together with the data previously reported, most of structures of triterpene glycosides and alkaloid marker compounds would be tentatively elucidated.
2. Experimental
2.1. Chemicals
HPLC-MS grade acetonitrile, methanol, water (J. T. Baker, Phillipsburg, NJ) and formic acid (Sigma-Aldrich, Louis, MO) were used for HPLC-TOF-MS analysis. Guaranteed reagent grade methanol (EMD, Gibbstown, NJ) and deionized water were used for the extraction of Actaea plant material. Prim-O-glucosylcimifugin (1) was purchased from Shanghai Nature Standard Biotech Co. Ltd (Shanghai, China). Cimifugin (2), cimiracemoside A (13), actein (14) and 26-deoxyactein (15) were purchased from ChromaDex (Irvine, CA). Cimicifugoside H-1 (4), 23-O-acetylshengmanol 3-O-β-D-xylopyranoside (7), cimiracemoside C (10), and 25-acetylcimigenol 3-xyloside (11) were kindly provided by Dr. Stefan Gafner (Tom’s of Maine, ME).
2.2 Sample materials
Twenty-five samples of sixteen species of Actaea were collected in North America and Asia from 1999 to 2007. The collection locations, time, codes, and injection No. of TOF mass analysis for all the samples were shown in Table 1. Among them, A. americana and A. podocarpa were purchased from Botanical Liaison LLC (Boulder, DO). A. cordifolia was supplied by Dirk De Meyere at the National Botanic Garden of Belgium. A. laciniata, A. pachypoda, A. rubra, A. elata, A. elata var. alpestris, A. arizonica, and nine samples of A. racemosa were collected in North America. While the remaining seven samples, A. heracleifolia, A. mairei, A. dahurica, A. brachycarpa, A. simples, A. cimicifuga, and A. yunnanensis, were collected from Asia. Voucher specimens of Actaea samples used in this study were deposited at the City University of New York. Rhizomes of all Actaea plants were used in this study.
Table 1.
The general information of 25 samples (16 species) used in this researcha
| Name | Collection Place | Parts | Collection Date | Sample Codes | Injection ordersb |
|---|---|---|---|---|---|
| A. racemosa | Fox Lane (Lake), NY | Rhizome | 5/25/2000 | a1–a3 | 1st–3rd |
| A. racemosa | Ashevellie, NC | Rhizome | 1999 | b1–b3 | 4th–6th |
| A. racemosa | Fanfield, PA | Rhizome | 01/03/2006 | c1–c3 | 7th–9th |
| A. racemosa | Hunterdam Co. NJ | Rhizome | 09/10/2000 | d1–d3 | 10th–12th |
| A. racemosa | MD | Rhizome | Early 2000 | e1–e3 | 13th–15th |
| A. racemosa | Rippowam, NY | Rhizome | 5/25/2000 | f1–f3 | 16th–18th |
| A. racemosa | Pound Ridge, NY | Rhizome | 4/26/2007 | g1–g3 | 19th–21st |
| A. racemosa | Bedford, NY | Rhizome | 07/02/2006 | h1–h3 | 22nd–24th |
| A. racemosa | Buena Vista, VA | Rhizome | 10/03/2006 | i1–i3 | 25th–27th |
| A. americana | Botanical Liaison LLC | Rhizome | 2004 | j1–j3 | 28th–30th |
| A. laciniata | OR, WA | Rhizome | 08/2007 | k1–k3 | 31st–33rd |
| A. podocarpa | Botanical Liaison LLC, AZ | Rhizome | 2004 | l1–l3 | 34th–36th |
| A. pachypoda | Sugar Cove, NC | Rhizome | Before 2004 | m1–m3 | 37th–39th |
| A. rubra | Pineville, OR | Rhizome | Before 2004 | n1–n3 | 40th–42nd |
| A. elata | Crone Miller Lake, OSU Forest, OR | Rhizome | 08/2007 | o1–o3 | 43rd–45th |
| A. elata var. alpestris | OR | Rhizome | 10/2007 | p1–p3 | 46th–48th |
| A. arizonica | Flagstaff Arboretum, AZ | Rhizome | 2007 | q1–q3 | 49th–51st |
| A. cordifolia | National Botanic Garden of Belgium | Rhizome | 2007 | r1–r3 | 52nd–54th |
| A. heracleifolia | Liaoning, China | Rhizome | 08/2006 | s1–s3 | 55th–57th |
| A. mairei | Guizhou, China | Rhizome | 08/09/2006 | t1–t3 | 58th–60th |
| A. dahurica | Liaoning, China | Rhizome | 08/2006 | u1–u3 | 61st–63rd |
| A. brachycarpa | Yunnan, China | Rhizome | 08/2006 | v1–v3 | 64th–66th |
| A. simplex | Yunnan, China | Rhizome | 08/09/2006 | w1–w3 | 67th–69th |
| A. cimicifuga | Dali, Yuannan, China | Rhizome | 08/05/2006 | x1–x3 | 70th–72nd |
| A. yunnanensis | Yunnan, China | Rhizome | 07/2005 | y1–y3 | 73rd–75th |
Samples (codes from a–r) were collected from North America; Samples (codes from s–y) were collected from Asia;
These injection orders in this table are the same as those in Fig. 4.
2.3 Sample preparation
Air-dried rhizomes of each sample of Actaea plants (ca. 1 g) were extracted with 80% MeOH (10 mL) at room temperature. Extraction was facilitated by sonication for 60 min in a water bath, and the sample temperature not exceeding 40° C, and the resulting suspention was centrifuged at 3000 rpm for 15 min. The solvent was then removed, and the residue was re-extracted three times using the same method. The resulting extracts were combined and the solvent was evaporated under nitrogen. All remaining water was removed by lyophilization. The resulting extract was stored at −20°C until analysis. Prior to HPLC-TOF-MS analysis, each extract was dissolved in 70% MeOH (30 mL). In-source MS/MS fragmentation was conducted (with an aperture voltage of 60 V) on these extracts, following which extracts were diluted by ten-fold for standard MS analysis (with an aperture voltage of 0V). All the prepared samples were filtered through a 0.45 μm nylon membrane filter prior to analysis.
2.4 Liquid chromatography
Separation was achieved by HPLC using a Waters 2695 separations module (Milford, MA), equipped with a 2998 photodiode array detector (PDA). The separations were carried out on a 100 × 2.0 mm i.d., 2.5 μm Synergi C18 column (Phenomenex, Torrance, CA). All analyses were performed at 25 °C with a flow rate of 0.2 mL/min. The sample volume injected was 10 μL. Analysis was based on triple injections. Mobile phase was composed of 0.1% aqueous formic acid (A), acetonitrile containing 0.1% formic acid (B), and methanol containing 0.1% formic acid (C) using a stepwise gradient elution of 92%A/8%B/0%C–75%A/25%B/0%C at 0–16 min, 75%A/25%B/0%C–59%A/27%B/14%C at 16–26 min, 59%A/27%B/14%C–46%A/18%B/36%C at 26–27 min, 46%A/18%B/36%C–40.6%A/20.1%B/39.3%C at 27–44 min, 40.6%A/20.1%B/39.3%C–33%A/23%B/44%C at 44–62 min, 33%A/23%B/44%C–56%A/44%B/0%C at 62–62.5 min, 56%A/44%B/0%C–35%A/65%B/0%C at 62.5–75 min, 35%A/65%B/0%C–18%A/82%B/0%C at 75–90 min, 18%A/82%B/0%C–0%A/100%B/0%C at 90–105 min, and kept this proportion of solvent for 12 min. The UV/vis spectra were recorded from 190 to 500 nm.
2.5 Mass spectrometry
High resolution electrospray ionization mass spectrometry (HR-ESI-MS) was performed using a LCT premier XE TOF mass spectrometer (Waters, Manifold, MA) equipped with an ESI interface and controlled by MassLynx V4.1 software. Mass spectra were acquired in both the positive and negative mode over the range m/z 100–1000. The capillary voltages were set at 3000 V (positive mode) and 2800 V (negative mode), respectively, and the cone voltage was 20 V. Nitrogen gas was used for both the nebulizer and in desolvation. The desolvation and cone gas flow rates were 600 and 20 L/h, respectively. The desolvation temperature was 400 °C, and the source temperature was 120 °C. For the dynamic range enhancement (DRE) lockmass, a solution of leucine enkephalin (Sigma-Aldrich, Steinheim, Germany) was infused by a secondary reference probe at 200 pg/mL in acetonitrile/water (1:1) containing 0.1% formic acid with the help of a second LC pump (Waters 515 HPLC pump). The reference mass was scanned once every five scans for each positive and negative data collection. Both positive and negative ESI data were collected using a scan time of 0.2 s, with an interscan time of 0.01 s, and a polarity switch time of 0.3 s. The full chromatograms were recorded at two different aperture voltages. The most intense fragmental ions and molecular ions could be obtained, when the aperture voltage were set at 60V and 0V, respectively. W-optics mode was used for increased resolution.
2.6 Chemometric data analysis
The HPLC-TOF-MS data of 25 samples from 16 species of Actaea was analyzed by PCA analysis to identify potential discriminate variables. Peak detection and alignment, and the filtering of raw data were carried out using the Markerlynx v4.1. The parameters used included a retention time range of 8–125 min, a mass range of 100–1000 Da, and a mass tolerance of 50 mDa. Isotopic peaks were excluded for analysis; noise elimination level was set at 1.00, the intensity threshold (counts) of collection parameters was set at 500; retention time tolerance was set at 0.4 min. The retention time and m/z data pair for each peak was determined by the software.
3. Results and discussion
3.1 Metabolic fingerprinting and PCA analysis
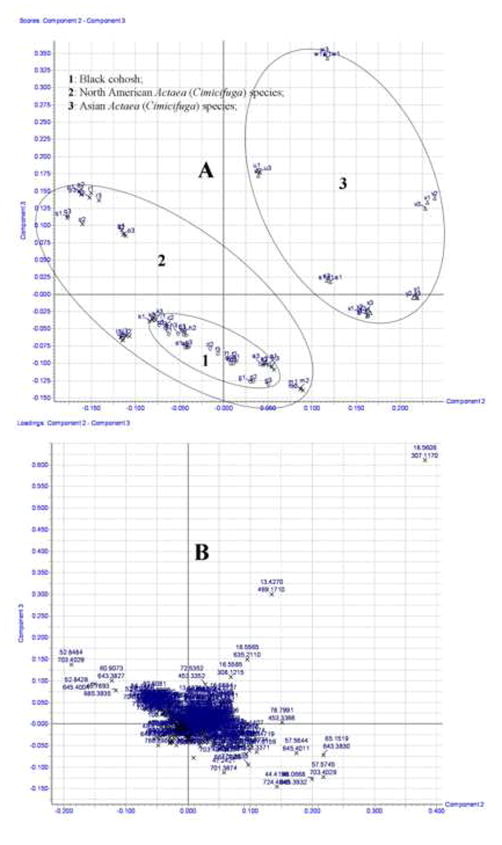
The TIC chromatograms for the reversed-phase LC-ESI-TOF-MS analysis of 25 samples extracted from different species of Actaea are shown in supplemental Fig. S1. Although some differences could be visually noted among these species of Actaea chromatograms, more subtle changes could be detected using a pattern recognition approach. The retention times, m/z value of mass fragmental ions, and their intensities were used to compare the phenotypic differences of the 16 species of Actaea using PCA, an unsupervised and therefore unbiased technique for multivariate analysis. Processed data displayed a clear differentiation of three clusters of the 16 species of Actaea which divided along geographic and botanical origin into Asian, North Amerian (other than A. racemosa), and true black cohosh (i.e. A. racemosa) groups (Fig. 1A). Each sample was injected three times, e.g. a1, a2, and a3. The samples are labeled alphabetically, corresponding to different Actaea, and numerically, corresponding to different LC injections (Table 1). All black cohosh species formed a centralized cluster within the larger North American group, because all black cohosh samples evolutionarily originated from the same species, and only differred in collection locations and harvest time. North American and Asian species also separated into distinct clusters (Fig. 1A). Using the corresponding loading plot, markers (ion-retention time pairs) most responsible for cluster formation, were identified as useful indicators of the botanical origin of each sample. (Fig. 1B). Fifteen molecular marker ions were found to be useful in distinguising species of Actaea and were selected for further identification by MS/MS fragmental and UV analysis.
Fig. 1.
Scores and loading plots of 25 Actaea samples by PCA analysis
3.2 Mass fragmental analysis of standards and marker compounds
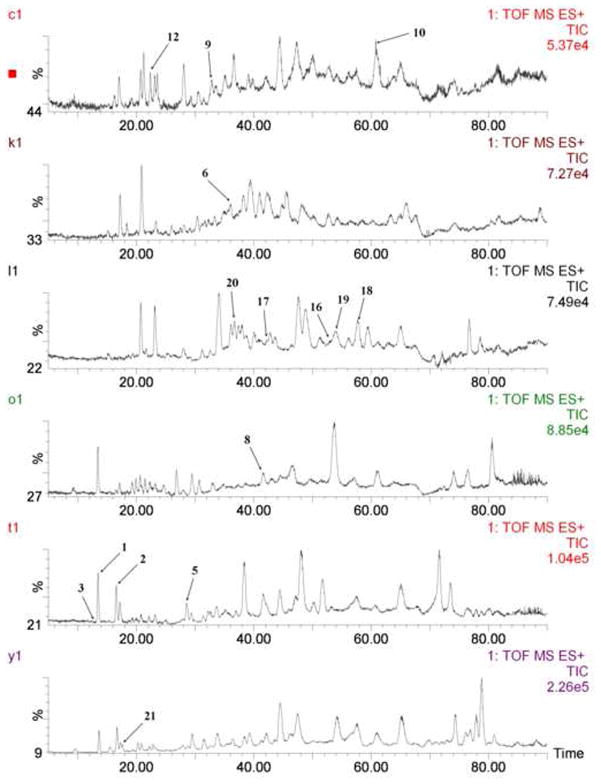
For each standard and Actaea species extract sample, TOF ESI-MS/MS spectra were recorded in both positive and negative ion modes; however, the more useful ion information was obtained in positive mode. Therefore, the positive ion mode was selected for analysis. TIC chromatograms of all identified marker compounds are shown in Fig. 2.
Fig. 2.
Fifteen marker compounds in the TIC chromatograms.
3.2.1 Analysis of cimifugin and its derivatives
Cimifugin derivatives are a group of linear dihydrofurochromone-class compounds (Fig. 3). Most of fragmentation occurrs on the dihydrofuran ring. The typical cleavages of this class of structure include fragmental losses of 18, 48 and 72 Da. These characteristic losses are useful in the identification of compounds with this skeleton [22].
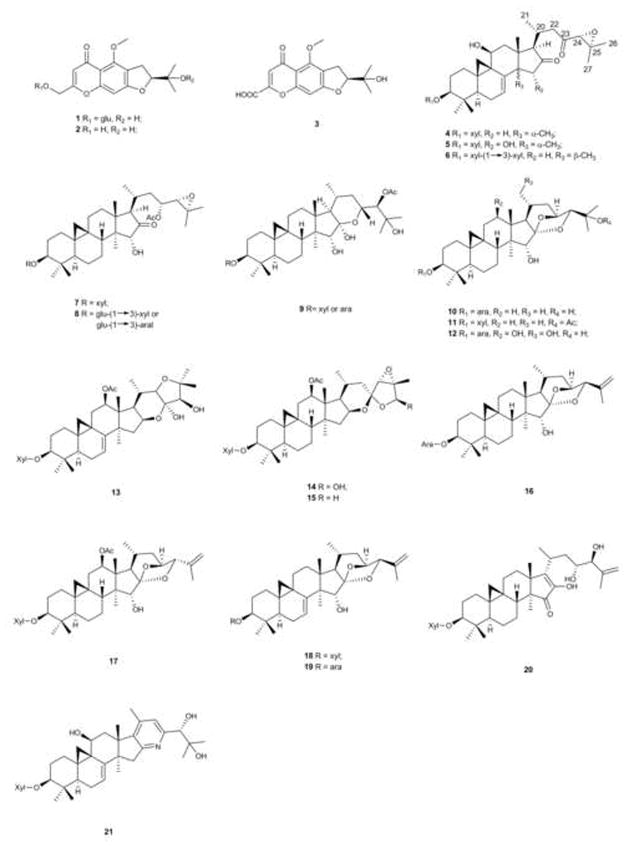
Fig. 3.
Chemical structures of standards and identified marker compounds for Actaea species.
Prim-O-glycosylcimifugin (1) and cimifugin (2) were identified using known standards. The UV spectra of these two compounds showed similar absorption maxima at 300.0, 254.2 (sh), 215.0 nm for 1 and 298.0, 254.3 (sh), 217.0 nm for 2 (supplemental Fig. S2A and S2B). When the aperture voltage was set at 60V, three cleavage fragmental ions at m/z 289.1068 [M + H – H2O]+, 259.0573 [M + H – H2O – 2 × ·CH3]+, and 235.0569 [M + H – 2,2-dimethylepoxyethane]+ with the losses of 18, 48 and 72 Da from the protonated molecular ion at m/z 307.1160 [M + H]+ were generated for 2 (Table 2). These similar losses of 18, 48 and 72 Da were also found in the fragmentation patterns of 1, indicated by fragmental ions at m/z 451.1602, 421.1099, 397.1128 from the molecular ion at m/z 469.1703, and m/z 289.1068, 259.0580, 235.0572 from the aglycone ion at m/z 307.1161 which was generated from the ion at m/z 469.1703 by the elimination of a glucosyl moiety (Table 2).
Table 2.
The summary results of the standards and marker compounds analyzed by HPLC-ESI-TOF-MS technique
| No. | RT (min) | Marker ion Exact Mass (ppm) | Selected fragmental ions Exact mass [Adduct molecular ions – neutral molecules or redicals]+ (molecular formula, ppm) | UV (λmax nm) | Identification | Type of Marker compoundsa |
|---|---|---|---|---|---|---|
| 1 | 13.44 | 469.1703 (−1.5) [M + H]+ (C22H29O11) | 451.1602 [M + H – H2O]+ (C22H27O10, −0.4); 421.1099 [M + H – H2O – 2CH3·]+ (C20H21O10, −8.5); 397.1128 [M + H – 2,2-dimethylepoxyethane]+ (C18H21O10, −1.8); 307.1161 [M + H – glucosyl group]+ (C16H19O6, −6.8); 290.1126 [M + H – glucosyl group – HO·]+ (C16H18O5, −9.6); 289.1060 [M + H – glucosyl group – H2O]+ (C16H17O5, −5.5); 261.1102 [M + H – glucosyl group – H2O – CO]+ (C14H11O5, −9.6); 259.0580 [M + H – glucosyl group – H2O – 2CH3·]+ (C14H11O5, −10.0); 235.0572 [M + H – glucosyl group – 2,2-dimethylepoxyethane]+ (C12H11O5, −14.5). | 300.0, 254.2sh, 215.0 | prim-O-glucosylcimifugin | A |
| 2 | 16.57 | 307.1160 (−7.2) [M + H]+ (C16H19O6) | 289.1068 [M + H – H2O]+ (C16H17O5, −2.8); 259.0573 [M + H – H2O – 2CH3·]+ (C14H11O5, −12.7); 235.0569 [M + H - 2,2-dimethylepoxyethane]+ (C12H11O5, −15.7); 217.0465 [M + H – 2,2-dimethylepoxyethane – H2O]+ (C12H9O4, −16.7); 207.0612 [M + H – 2,2-dimethylepoxyethane – CO]+ (C11H11O4, −21.7) | 298.0, 254.3sh, 217.0 | cimifugin | A |
| 3 | 12.41 | 321.0953 (−6.5) [M + H]+ (C16H17O7) | 303.0852 [M + H – H2O]+ (C16H15O6, −5.6); 273.0367 [M + H – H2O – 2CH3·]+ (C14H9O6, −11.7); 249.0362 [M + H - 2,2-dimethylepoxyethane]+ (C12H9O6, −14.9); 231.0222 [M + H – 2,2-dimethylepoxyethane – H2O]+ (C12H7O5, −17.7); 221.0414 [M + H – 2,2-dimethylepoxyethane – CO]+ (C11H9O5, −16.3) | 310.0, 255.0sh, 217.0 | divaricatacid | A |
| 4 | 38.39 | 639.3508 (−0.2) [M + Na]+ (C35H52O9Na) | 617.3694 [M + H]+ (C35H53O9, 0.6); 599.3591 [M + H – H2O]+ (C35H51O8, 1.2); 577.3390 [M + H – propa-1,2-diene]+ (C32H49O9, 2.3); 467.3165 [M + H – xylose]+ (C30H43O4, 0.9); 449.3050 [M + H – xylose – H2O]+ (C30H41O3, −1.3); 427.2861 [M + H – xylose – propa-1,2-diene]+ (C27H39O4, 3.0); 395.2580 [M + H – xylose – 2,2-dimethylepoxyethane]+ (C26H35O3, − 1.5); 377.2475 [M + H – xylose – H2O – 2,2-dimethylepoxyethane]+ (C26H33O2, −1.6); | cimicifugoside H-1 | Not marker compound | |
| 5 | 28.56 | 655.3448 (−1.5) [M + Na]+ (C35H52O10Na) | 633.3636 [M + H]+ (C35H53O10, −0.5); 615.3523 [M + H – H2O]+ (C35H51O9, −1.6); 575.3218 [M + H – H2O – propa-1,2-diene]+ (C32H47O9, −0.3); 561.3074 [M + H – 2,2-dimethylepoxyethane]+ (C31H45O9, 1.8); 483.3104 [M + H – xylose]+ (C30H43O5, −1.2); 465.3004 [M + H – xylose – H2O]+ (C30H41O4, −0.2); 447.2879 [M + H – xylose – 2H2O]+ (C30H39O3, −4.5) 443.2796 [M + H – xylose – propa-1,2-diene]+ (C27H39O5, −0.2); 425.2690 [M + H – xylose – H2O – propa-1,2-diene]+ (C27H37O4, −0.5); 411.2523 [M + H – xylose – 2,2-dimethylepoxyethane]+ (C26H35O4, −2.9); 393.2419 [M + H – xylose – H2O – 2,2-dimethylepoxyethane]+ (C26H33O3, −2.8); 375.2308 [M + H – xylose – 2H2O – 2,2-dimethylepoxyethane]+ (C26H31O2, − 4.3); | cimicifugoside H-5 | A | |
| 6 | 35.90 | 771.3942 (1.3) [M + Na]+ (C40H60O13Na) | 749.4130 [M + H]+ (C40H61O13, 2.4); 731.3984 [M + H – H2O]+ (C40H59O12, −3.1); 599.3588 [M + H – xylose]+ (C35H51O8, 0.7); 559.3263 [M + H – xylose – propa-1,2-diene]+ (C32H47O8, −1.4); 467.3158 [M + H – 2xylose]+ (C30H43O4, −0.6); 449.3054 [M + H – 2xylose – H2O]+ (C30H41O3, −0.4); 427.2846 [M + H – 2xylose – propa-1,2-diene]+ (C27H39O4, −0.5); 409.2753 [M + H – 2xylose – H2O – propa-1,2-diene]+ (C27H37O3, 2.4); 395.2620 [M + H – 2xylose – 2,2-dimethylepoxyethane]+ (C26H35O3, 8.6); 377.2483 [M + H – 2xylose – H2O – 2,2-dimethylepoxyethane]+ (C26H33O2, 0.5). | shengmacichun dixyloside | D | |
| 7 | 48.12 | 685.3895 (−4.8) [M + Na]+ (C37H58O10Na) | 663.4145 [M + H]+ (C37H59O10, 5.6); 645.3975 [M + H – H2O]+ (C37H57O9, −4.3); 603.3932 [M + H – acetic acid]+ (C35H55O8, 5.8); 585.3770 [M + H – acetic acid – H2O]+ (C35H53O7, −3.6); 545.3511 [M + H – acetic acid – H2O – propa-1,2-diene]+ (C32H49O7, 6.1); 531.3333 [M + H – acetic acid – 2,2-dimethylepoxyethane]+ (C31H47O7, 2.1); 513.3542 [M + H – xylose]+ (C32H49O5, −7.4); 495.3451 [M + H – xylose – H2O]+ (C32H47O4, 4.6); 453.3370 [M + H – xylose – acetic acid]+ (C30H45O3, 0.2); 435.3266 [M + H – xylose – acetic acid – H2O]+ (C30H43O2, 0.7); 413.3070 [M + H – xylose – acetic acid – propa-1,2-diene]+ (C27H41O3, 3.4); 395.2958 [M + H – xylose – acetic acid – H2O – propa-1,2-diene]+ (C27H39O2, 2.0); 381.2799 [M + H – xylose – acetic acid – 2,2-dimethylepoxyethane]+ (C26H37O2, 1.3); 363.2690 [M + H – xylose – acetic acid – H2O – 2,2-dimethylepoxyethane]+ (C26H35O, 0.6). | 23-O-acetylshengmanol 3-O-β-D-xylopyranoside | Not marker compound | |
| 8 | 41.59 | 847.4446 (−1.2) [M + Na]+ (C43H68O15Na) | 825.4619 [M + H]+ (C43H69O15, −2.1); 807.4501 [M + H – H2O]+ (C43H67O14, −3.7); 663.4095 [M + H – glucosyl group]+ (C37H59O10, −2.0); 645.4001 [M + H – glucosyl group – H2O]+ (C37H57O9, −0.3); 513.3581 [M + H – glucosyl group – xylose or arabinose]+ (C32H49O5, 0.2); 495.3475 [M + H – glucosyl group – xylose or arabinose – H2O]+ (C32H47O4, 0.2); 471.3484 [M + H – glucosyl group – xylose or arabinose – Ac]+ (C30H47O4, 2.1); 453.3388 [M + H – glucosyl group – xylose or arabinose – actetic acid]+ (C30H45O3, 4.2); 435.3284 [M + H – glucosyl group – xylose or arabinose – actetic acid – H2O]+ (C30H43O2, 4.8); 431.3179 [M + H – glucosyl group – xylose or arabinose – Ac – propa-1,2-diene]+ (C27H43O4, 4.2); 413.3058 [M + H – glucosyl group – xylose or arabinose – actetic acid – propa-1,2-diene]+ (C27H41O3, 0.5); 381.2804 [M + H – glucosyl group – xylose or arabinose – actetic acid – 2,2-dimethylepoxyethane]+ (C26H37O2, 2.6); 363.2682 [M + H – glucosyl group – xylose or arabinose – actetic acid – H2O – 2,2-dimethylepoxyethane]+ (C26H35O, −1.7) | 23-O-acetylshengmanol-3-O-β-D-glucopyranoside-(1–3)-β-D-xylopyranoside or arabinoside | D | |
| 9 | 32.92 | 703.4042 (1.3) [M + Na]+ (C37H60O11Na) | 663.4106 [M + H – H2O]+ (C37H59O10, −0.5); 645.4000 (C37H57O9, −0.8); 621.4014 [M + H – acetic acid]+ (C35H57O9, 2.9); 603.3905 [M + H – acetic acid – H2O]+ (C35H55O8, 2.2); 585.3795 [M + H – acetic acid – 2H2O]+ (C35H53O7, 1.2); 563.3603 [M + H – acetic acid – H2O – propa-1,2-diene]+ (C32H51O8, 6.0); 531.3677 [M + H – xylose or arabinose]+ (C32H51O6, −3.2); 513.3572 [M + H – xylose or arabinose – H2O]+ (C32H49O5, −3.1); 471.3472 [M + H – xylose or arabinose – acetic acid]+ (C30H47O4, −0.8); 453.3365 [M + H – xylose or arabinose – H2O – acetic acid]+ (C30H45O3, −2.0); 435.3264 [M + H – xylose or arabinose – 2H2O – acetic acid]+ (C30H43O2, 0.4); 399.2909 [M + H – xylose or arabinose – acetic acid – 2,2-dimethylepoxyethane]+ (C26H39O3, 6.2); 395.2963 [M + H – xylose or arabinose – 2H2O – acetic acid – propa-1,2-diene]+ (C27H39O2, 8.3); 381.2785 [M + H – xylose or arabinose – H2O – acetic acid – 2,2-dimethylepoxyethane]+ (C26H37O2, −6.3); 363.2677 [M + H – xylose or arabinose – 2H2O – acetic acid – 2,2-dimethylepoxyethane]+ (C26H35O, −8.3) | 24-O-acetylhydroshengmano l-3-O-β-D-xylopyranoside or arabinoside | C | |
| 10 | 60.93 | 643.3813 (1.4) [M + Na]+ (C35H56O9Na) | 621.4022 [M + H]+ (C35H57O9, 3.1); 603.3898 [M + H – H2O]+ (C35H55O8, −0.2); 585.3795 [M + H – 2H2O]+ (C35H53O7, 0.7); 531.3356 [M + H – H2O – 2,2-dimethylepoxyethane]+ (C31H47O7, 6.4); 471.3481 [M + H – arabinose]+ (C30H47O4, 1.5); 453.3388 [M + H – arabinose – H2O]+ (C30H45O3, 4.2); 435.3286 [M + H – arabinose – 2H2O]+ (C30H43O2, 5.3); 417.3160 [M + H – arabinose – 3H2O]+ (C30H41O, −1.7); 381.2786 [M + H – arabinose – H2O – 2,2-dimethylepoxyethane]+ (C26H37O2, −2.1); 363.2683 [M + H – arabinose – 2H2O – 2,2-dimethylepoxyethane]+ (C26H35O, −1.4) | cimiracemoside C | C | |
| 11 | 78.81 | 685.3916 (−1.8) [M + Na]+ (C37H58O10Na) | 663.4117 [M + H]+ (C37H59O10, 1.4); 645.4024 [M + H – H2O]+ (C37H57O9, 3.3); 603.3891 [M + H – acetic acid]+ (C35H55O8, − 1.0); 585.3789 [M + H – acetic acid – H2O]+ (C35H53O7, −0.3); 513.3586 [M + H – xylose]+ (C32H49O5, 1.2); 495.3496 [M + H – xylose – H2O]+ (C32H47O4, 4.4); 453.3366 [M + H – xylose – acetic acid]+ (C30H45O3, −0.7); 435.3258 [M + H – xylose – acetic acid – H2O]+ (C30H43O2, −1.1); 381.2778 [M + H – xylose – acetic acid – 2,2-dimethylepoxyethane]+ (C26H37O2, −4.2); 363.2677 [M + H – xylose – acetic acid – H2O – 2,2-dimethylepoxyethane]+ (C26H35O, −3.0). | 25-O-acetylcimigenol 3-O-β-D-xylopyranoside | Not marker compound | |
| 12 | 22.41 | 675.3727 (1.0) [M + Na]+ (C35H56O11Na) | 635.3799 [M + H – H2O]+ (C35H55O10, 0.6); 617.3694 [M + H – 2H2O]+ (C35H53O9, 0.6); 599.3583 [M + H – 3H2O]+ (C35H51O8, −0.2); 581.3478 [M + H – 4H2O]+ (C35H49O7, 0.0); 545.3130 [M + H – 2H2O – 2,2-dimethylepoxyethane]+ (C31H45O8, 2.9); 503.3369 [M + H – arabinose]+ (C30H47O6, −0.8); 485.3271 [M + H – arabinose – H2O]+ (C30H45O5, 0.8); 467.3165 [M + H – arabinose – 2H2O]+ (C30H43O4, 0.9); 449.3062 [M + H – arabinose – 3H2O]+ (C30H41O3, 1.3); 413.2722 [M + H – arabinose – H2O – 2,2-dimethylepoxyethane]+ (C26H37O4, 7.3); 395.2588 [M + H – arabinose – 2H2O – 2,2-dimethylepoxyethane]+ (C26H35O3, 0.5); 377.2475 [M + H – arabinose – 3H2O – 2,2-dimethylepoxyethane]+ (C26H33O2, −1.6) | 12β,21-dihydroxycimigenol-3-O-L-arabinoside | C | |
| 13 | 36.65 | 677.3904 (0.4) [M + H]+ (C37H57O11) | 659.3799 [M + H – H2O]+ (C37H55O10, 0.6); 617.3688 [M + H – acetic acid]+ (C35H53O9, −0.4); 599.3578 [M + H – H2O – acetic acid]+ (C35H51O8, −1.0); 581.3455 [M + H – 2H2O – acetic acid]+ (C35H49O7, −4.0); 527.3363 [M + H – xylose]+ (C32H47O6, − 1.9); 467.3164 [M + H – xylose – acetic acid]+ (C30H43O4, 0.6); 449.3047 [M + H – xylose – acetic acid – H2O]+ (C30H41O3, − 2.0); 431.2951 [M + H – xylose – acetic acid – 2H2O]+ (C30H39O2, 0.2); 395.2586 [M + H – xylose – acetic acid – 2,2-dimethylepoxyethane]+ (C26H35O3, 0.0); 377.2465 [M + H – xylose – acetic acid – H2O – 2,2-dimethylepoxyethane]+ (C26H33O2, −4.2); 359.2328 [M + H – xylose – acetic acid – 2H2O – 2,2-dimethylepoxyethane]+ (C26H31O, −13.1) | cimiracemoside A | Not marker compound | |
| 14 | 47.29 | 677.3923 (3.2) [M + H]+ (C37H57O11) | 659.3782 [M + H – H2O]+ (C37H55O10, −2.0); 617.3701 [M + H – acetic acid]+ (C35H53O9, 1.8); 599.3586 [M + H – H2O – acetic acid]+ (C35H51O8, 0.3); 581.3487 [M + H – 2H2O – acetic acid]+ (C35H49O7, 1.5); 527.3391 [M + H – xylose]+ (C32H47O6, 3.4); 467.3171 [M + H – xylose – acetic acid]+ (C30H43O4, 2.1); 449.3063 [M + H – xylose – acetic acid – H2O]+ (C30H41O3, 1.6); 431.2971 [M + H – xylose – acetic acid – 2H2O]+ (C30H39O2, 4.9) | actein | Not marker compound | |
| 15 | 44.42 | 661.3952 (0.0) [M + H]+ (C37H57O10) | 643.3864 [M + H – H2O]+ (C37H55O9, 2.8); 601.3751 [M + H – acetic acid]+ (C35H53O8, 1.8); 583.3648 [M + H – H2O – acetic acid]+ (C35H51O7, 2.2); 565.3551 [M + H – 2H2O – acetic acid]+ (C35H49O6, 3.9); 511.3448 [M + H – xylose]+ (C32H47O5, 4.9); 451.3210 [M + H – xylose – acetic acid]+ (C30H43O3, −0.4); 433.3120 [M + H – xylose – acetic acid – H2O]+ (C30H41O2, 3.0); 415.3036 [M + H – xylose – acetic acid – 2H2O]+ (C30H39O, 8.4) | 26-deoxyactein | Not marker compound | |
| 16 | 52.84 | 625.3726 (1.6) [M + Na]+ (C35H54O8Na) | 603.3911 [M + H]+ (C35H55O8, 2.3); 585.3788 [M + H – H2O]+ (C35H53O7, −0.5); 561.3434 [M + H – H2O – propene]+ (C32H49O8, 1.2); 453.3376 [M + H – arabinose]+ (C30H45O3, 1.5); 435.3236 [M + H – arabinose – H2O]+ (C30H43O2, −6.2); 411.2885 [M + H – arabinose – propene]+ (C27H39O3, −3.4); 397.2731 [M + H – arabinose – 2-methylpropene]+ (C26H37O3, −3.0); 393.2791 [M + H – arabinose – H2O – propene]+ (C27H37O2, −0.8); 379.2625 [M + H – arabinose – H2O – 2-methylpropene]+ (C26H35O2, −3.2) | (23R,24S)-25-anhydrocimigenol-3-O-α-L-arabinose; | D | |
| 17 | 42.16 | 683.3776 (0.7) [M + Na]+ (C37H56O10Na) | 661.3954 [M + H]+ (C37H57O10, 0.3); 601.3738 [M + H – acetic acid]+ (C35H51O7, 2.5); 583.3615 [M + H – acetic acid – H2O]+ (C35H53O8, −0.3); 559.3281 [M + H – acetic acid – propene]+ (C32H47O8, 1.8); 511.3444 [M + H – xylose]+ (C32H47O5, 3.9); 469.3323 [M + H – xylose – Ac]+ (C30H45O4, 1.1); 451.3189 [M + H – xylose – acetic acid]+ (C30H43O3, −5.1); 433.3082 3189 [M + H – xylose – acetic acid – H2O]+ (C30H41O2, −5.8); 427.2848 [M + H – xylose – Ac – propene]+ (C27H39O4, 0.0); 409.2731 [M + H – xylose – acetic acid – propene]+ (C27H37O3, −2.9); 395.2586 [M + H – xylose – acetic acid – 2-methylpropene]+ (C26H35O3, 0.0); 377.2473 [M + H – xylose – acetic acid – H2O – 2-methylpropene]+ (C26H33O2, −2.1) | cimiracemoside K | D | |
| 18 | 57.91 | 623.3569 (1.4) [M + Na]+ (C35H52O8Na) | 601.3752 [M + H]+ (C35H53O8, 2.0); 583.3650 [M + H – H2O]+ (C35H51O7, 2.6); 559.3271 [M + H – propene]+ (C32H47O8, 0.0); 545.3136 [M + H – 2-methylpropene]+ (C31H45O8, 4.0); 527.2999 [M + H – H2O – 2-methylpropene]+ (C31H43O7, −1.9); 469.3320 [M + H – xylosyl group]+ (C30H45O4, 0.4); 451.3220 [M + H – xylose]+ (C30H43O3, 1.8); 433.3062 [M + H – xylose – H2O]+ (C30H41O2, −10.4); 427.2848 [M + H – xylosyl group – propene]+ (C27H39O4, 0.5); 409.2722 [M + H – xylose – propene]+ (C27H37O3, −5.1); 395.2585 [M + H – xylose – 2-methylpropene]+ (C26H35O3, −0.5); 377.2474 [M + H – xylose – H2O – 2-methylpropene]+ (C26H33O2, −1.9); | (23R,24R)-7-en-25-anhydrocimigenol-3-O-β-D-xylopyranoside | D | |
| 19 | 53.99 | 623.3569 (1.4) [M + Na]+ (C35H52O8Na) | 601.3745 [M + H]+ (C35H53O8, 0.8); 583.3651 [M + H – H2O]+ (C35H51O7, 2.7); 559.3276 [M + H – propene]+ (C32H47O8, 0.9); 527.3028 [M + H – H2O – 2-methylpropene]+ (C31H43O7, 3.6); 469.3314 [M + H – arabinosyl group]+ (C30H45O4, −0.9); 451.3222 [M + H – arabinose]+ (C30H43O3, 2.2); 433.3093 [M + H – arabinose – H2O]+ (C30H41O2, −3.2); 427.2842 [M + H – arabinosyl group – propene]+ (C27H39O4, −0.9); 409.2733 [M + H – arabinose – H2O – propene]+ (C27H37O3, −2.4); 395.2586 [M + H – arabinose – H2O – 2-methylpropene]+ (C26H35O3, −0.3); 377.2469 [M + H – arabinose – 2H2O – 2-methylpropene]+ (C26H33O2, −3.2); | (23R,24R)-7-en-25-anhydro cimigenol-3-O-α-L-arabinoside | D | |
| 20 | 36.75 | 643.3842 (3.1) [M + Na]+ (C35H56O9Na) | 621.4006 [M + H]+ (C35H57O9, 0.5); 603.3908 [M + H – H2O]+ (C35H55O8, 1.5); 585.3798 [M + H – 2H2O]+ (C35H53O7, 1.2); 579.3531 [M + H – propene]+ (C32H51O9, −0.3); 561.3434 [M + H – H2O – propene]+ (C32H49O8, 1.2); 471.3478 [M + H – xylose]+ (C30H47O4, 0.8); 453.3381 [M + H – xylose – H2O]+ (C30H45O3, 2.6); 435.3264 [M + H – xylose – 2H2O]+ (C30H43O2, 0.2); 429.2993 [M + H – xylose – propene]+ (C27H41O4, −0.7); 411.2888 [M + H – xylose – H2O – propene]+ (C27H39O3, −2.7); 397.2745 [M + H – xylose – H2O – 2-methylpropene]+ (C26H37O3, 0.5); 379.2635 [M + H – xylose – 2H2O – 2-methylpropene]+ (C26H35O2, 0.5) | Cimidahuside G | D | |
| 21 | 17.55 | 614.3679 (−2.3) [M + H]+ (C35H52NO8) | 596.3611 [M + H – H2O]+ (C35H50NO7, 4.0); 578.3476 [M + H – 2H2O]+ (C35H48NO6, −1.0); 560.3345 [M + H – 3H2O]+ (C35H46NO5, −5.5); 556.3260 [M + H – H2O – propa-1,2-diene]+ (C32H46NO7, −2.5); 464.3148 [M + H – xylose]+ (C30H42NO3, −3.7); 446.3062 [M + H – xylose – H2O]+ (C30H40NO2, 0.7); 428.2952 [M + H – xylose – 2H2O]+ (C30H38NO, −0.2) | Cimicifine A (syn. Cimicifugadine) | B |
A: Marker compounds for species other than Actaea racemosa; B: Marker compounds for Asian species; C: Marker compounds for Actaea racemosa; D: Marker compounds for North American species other than Actaea racemosa.
Divaricatacid (3) was tentatively identified by the similarity in fragmental loss patterns as those of 1 and 2. Additonally, 3 had a similar UV absorbance profile as 1 and 2 with maxima at 310.0, 255.0 (sh), and 217.0 nm (Table 2 and supplemental Fig. S2C). The typical fragmental ions by the losses of 18, 48, and 72 Da at m/z 303.0852 [M + H – H2O]+, 273.0367 [M + H – H2O – 2 × ·CH3]+, 249.0362 [M + H – 2,2-dimethylepoxyethane]+ were also found in the spectrum. An additional two fragmental ions at m/z 231.0222 [M + H – 2,2-dimethylepoxyethane - H2O]+ and 221.0414 [M + H – 2,2-dimethylepoxyethane - CO]+ were found in the spectrum (Table 2). All molecular and fragmental ions detected are in agreement with previous publication [22]. Although putative in identification, this is the first report for the presence of divaricatacid in Actaea species.
3.2.2 Analysis of triterpene glycosides
A number of different classes of triterpene glycosides, based on their aglycone structures, have been identified from Actaea species [23,24]. Some of these triterpene glycosides are known to have potent biological activity [25–28]. In our study, seven triterpene glycoside standards (4, 7, 10, 11, and 13–15) were used to aid in the characterization of marker compounds. These standards represent five classes of triterpene glycosides: 16,23-diketo-shengmanol, shengmanol, cimigenol, cimiracerol, and actetyl-acteol (Table 3) [24].
Table 3.
The characteristic molecular ions and fragmental cleavages in each standard and marker compounds in chromatogram.
| No. | Aglycone class | Abundance of molecular ions relative to base peak (%)a | Base peak (aperture voltage: 0 V) | Fragmental Base peak (aperture voltage: 60 V) | Aperture voltage: 60 Vd | Aperture voltage: 0 Vc | ||
|---|---|---|---|---|---|---|---|---|
| Characteristic fragmental ions [MW (MF, ppm) (AMBP)] produced by loss of C4H8Ob | Characteristic fragmental ions [MW (MF, ppm) (AMBP)] produced by loss of C4H8b | Characteristic fragmental ions [MW (MF, ppm) (AIFBP)] produced by loss of C3H4b | Characteristic fragmental ions [MW (MF, ppm) (AIFBP)] produced by loss of C3H6 b | |||||
| 4 | 16,23-diketo-shengmanol | [M + Na]+ 639.3508 (41) [M + H]+ 617.3694 (7) |
577.3390 (C32H49O9, [M + H – propa-1,2-diene]+) | 395.2580 (C26H35O3, [M + H – xylose – 2,2-dimethyl epoxyethane]+) | 467.3132 (C30H43O4, −6.2) (13) → 395.2580 (C26H35O3, −1.5) (100); 449.3050 (C30H41O3, −1.3) (19) → 377.2475 (C26H33O2, −1.6) (78); |
No | 617.3694 (C35H53O9, 0.6) (7) → 577.3390 (C32H49O9, 2.2) (100); 467.3165 (C30H43O4, 0.9) (6) → 427.2861 (C27H39O4, 3.0) (53); |
No |
| 5 | 16,23-diketo-shengmanol | [M + Na]+ 655.3448 (100) [M + H]+ 633.3636 (4) |
655.3448 (C35H52O10Na, [M + Na]+) | 393.2419 (C26H33O3, [M + H – xylose – H2O – 2,2-dimethyl epoxyethane]+) | 483.3077 (C30H43O5, −6.8) (12) → 411.2523 (C26H35O4, −2.9) (40); 465.2991 (C30H41O4, −3.0) (24) → 393.2419 (C26H33O3, −2.8) (100); 447.2879 (C30H39O3, −4.8) (11) → 375.2308 (C26H31O2, −4.3) (34); |
No | 615.3523 (C35H51O9, −1.6) (9) → 575.3218 (C32H47O9, −0.3) (5); 483.3104 (C30H43O5, −1.2) (17) → 443.2796 (C27H39O5, −0.2) (13); 465.3004 (C30H41O4, −0.2) (9) → 425.2690 (C27H37O4, −0.5) (7); |
No |
| 6 | 16,23-diketo-shengmanol | [M + Na]+ 771.3942 (100) [M + H]+ 749.4130 (4) |
771.3942 (C40H60O13Na, [M + Na]+) | 377.2483 (C26H33O2, [M + H – 2xylose – H2O – 2-methylprop-1-ene]+) | 467.3116 (C30H43O4, −9.6) (30) → 395.2620 (C26H35O3, 8.6) (77); 449.3036 (C30H41O3, −4.5) (52) → 377.2483 (C26H33O2, 0.5) (100); |
No | 599.3588 (C35H51O8, 0.7) (12) → 559.3263 (C32H47O8, −1.4) (2); 467.3158 (C30H43O4, −0.6) (13) → 427.2846 (C27H39O4, −0.5) (7); 449.3054 (C30H41O3, −0.4) (6) → 409.2753 (C27H37O3, 2.4) (5); |
No |
| 7 | shengmanol | [M + Na]+ 685.3895 (100) [M + H]+ 663.4145 (12) |
685.3895 (C37H58O10Na, [M + Na]+) | 435.3266 (C30H43O2, [M + H – xylose – acetic +) acid – H2O] | 453.3370 (C30H45O3, 0.2) (63) → 381.2799 (C26H37O2, 1.3) (76); 435.3266 (C30H43O2, 0.7) (100) → 363.2690 (C26H35O, 0.6) (67); |
No | 585.3770 (C35H53O7, −3.6) (20) → 545.3511 (C32H49O7, 6.1) (6); 453.3338 (C30H45O3, −6.8) (73) → 413.3070 (C27H41O3, 3.4) (9); 435.3239 (C30H43O2, −5.5) (37) → 395.2958 (C27H39O2, 2.0) (5); |
No |
| 8 | shengmanol | [M + Na]+ 847.4446 (100) [M + H]+ 825.4619 (8) |
847.4446 (C43H68O15Na, [M + Na]+) | 747.4294 (C41H63O12, [M + H – H2O - acetic acid]+) | 453.3388 (C30H45O3, 4.2) (51) → 381.2804 (C26H37O2, 2.6) (37); 435.3284 (C30H43O2, 4.8) (68) → 363.2682 (C26H35O, −1.7) (17); |
No | 471.3484 (C30H47O4, 2.1) (8) → 431.3179 (C27H43O4, 4.2) (2); 453.3375 (C30H45O3, 1.3) (27) → 413.3059 (C27H41O3, 0.7) (5); |
No |
| 9 | hydro-shengmanol | [M + Na]+ 703.4042 (100) [M + H]+ 681.4261 (2) |
703.4042 (C37H60O11Na, [M + Na]+) | 643.3813 (C35H56O9Na, [M + Na – acetic acid]+) | 471.3472 (C30H47O4, −0.4) (8) → 399.2929 (C26H39O3, 7.5) (16); 453.3365 (C30H45O3, −0.9) (64) → 381.2785 (C26H37O2, −2.4) (39); 435.3264 (C30H43O2, 0.2) (83) → 363.2677 (C26H35O, −3.0) (32); |
No | 603.3905 (C35H55O8, 1.3) (9) → 563.3603 (C32H51O8, 3.4) (2); 453.3380 (C30H45O3, 2.4) (24) → 413.3064 (C27H41O3, 1.9) (5); 435.3276 (C30H43O2, 3.0) (6) → 395.2963 (C27H39O2, 3.3) (1); |
No |
| 10 | cimigenol | [M + Na]+ 643.3813 (100) [M + H]+ 621.4022 (7) |
643.3813 (C35H56O9Na, [M + Na]+) | 453.3388 (C30H45O3, [M + H – arabinose – H2O]+) | 603.3914 (C35H55O8, 2.8) (32) → 531.3356 (C31H47O7, 6.4) (5); 453.3388 (C30H45O3, 4.2) (100) → 381.2786 (C26H37O2, −2.1) (25); 435.3286 (C30H43O2, 5.3) (83) → 363.2683 (C26H35O, −1.4) (22); |
No | No | No |
| 11 | cimigenol | [M + Na]+ 685.3931 (80) [M + H]+ 663.4052 (8) |
453.3376 (C30H45O3, [M + H – xylose – acetic acid]+) | 453.3376 (C30H45O3, [M + H – xylose – acetic acid]+) | 453.3366 (C30H45O3, −0.7) (78) → 381.2778 (C26H37O2, −4.2) (24); 435.3258 (C30H43O2, −1.1) (100) → 363.2677 (C26H35O, −3.0) (20); |
No | No | No |
| 12 | cimigenol | [M + Na]+ 675.3727 (100) [M + H]+ 653.3916 (3) |
675.3727 (C35H56O11Na, [M + Na]+) | 395.2588 (C26H35O3, [M + H – arabinose – 2H2O – 2,2-dimethyl epoxyethane]+) | 485.3271 (C30H45O5, 0.8) (17) → 413.2722 (C26H37O4, 7.3) (29); 467.3165 (C30H43O4, 0.9) (31) → 395.2588 (C26H35O3, 0.5) (100); 449.3062 (C30H41O3, 1.3) (53) → 377.2475 (C26H33O2, −1.6) (80); |
No | No | No |
| 13 | cimiracerol | [M + Na]+ 699.3719 (1) [M + H]+ 677.3901 (2) |
659.3799 (C37H55O10, [M + H – H2O]+) | 449.3047 (C37H55O10, [M + H – xylose – acetic acid – H2O]+) | 467.3164 (C30H43O4, 0.6) (33) → 395.2586 (C26H35O3, 0.0) (11); 449.3047 (C30H41O3, −2.0) (100) →377.2465 (C26H33O2, −4.2) (13); 431.2951 (C30H39O2, 0.2) (46) → 359.2328 (C26H31O, −13.1) (4); |
No | No | Not |
| 14 | actetyl-acteol | [M + Na]+ 699.3724 (4) [M + H]+ 677.3923 (6) |
659.3782 (C37H55O10, [M + H – H2O]+) | 449.3080 (C37H55O10, [M + H – xylose – acetic acid – H2O]+) | No | No | No | No |
| 15 | actetyl-acteol | [M + Na]+ 683.3781 (11) [M + H]+ 661.3952 (100) |
661.3952 (C37H57O10, [M + H]+) | 451.3204 (C30H43O3, [M + H – xylose – acetic acid]+) | No | No | No | No |
| 16 | anhydro-cimigenol | [M + Na]+ 625.3726 (100) [M + H]+ 603.3911 (27) |
625.3726 (C35H54O8Na, [M + Na]+) | 379.2625 (C26H35O2, [M + H – arabinose – H2O – 2 – methylprop-1-ene]+) | No | 453.3376 (C30H45O3, 1.5) (16) → 397.2731 (C26H37O3, −3.0) (59); 435.3236 (C30H43O2, −6.2) (12) → 379.2625 (C26H35O2, −3.2) (100); |
No | 603.3911 (C35H55O8, 2.3) (24) → 561.3434 (C32H49O8, 1.2) (57); 453.3376 (C30H45O3, 1.5) (7) → 411.2885 (C27H39O3, −3.4) (11); 435.3293 (C30H43O2, 6.9) (2) → 393.2791 (C27H37O2, −0.8) (5); |
| 17 | anhydro-cimigenol | [M + Na]+ 683.3776 (100) [M + H]+ 661.3954 (7) |
683.3776 (C37H56O10Na, [M + Na]+) | 377.2473 (C26H33O2, [M + H – xylose – acetic acid - H2O – 2 – methylprop-1-ene]+) | No | 451.3189 (C30H43O3, −5.1) (16) → 395.2588 (C26H35O3, 0.5) (96); 433.3082 (C30H41O2, −5.8) (12) → 377.2473 (C26H33O2, −2.1) (100); |
No | 601.3738 (C35H53O8, −0.3) (15) → 559.3281 (C32H47O8, 1.8) (10); 469.3323 (C30H45O4, 1.1) (15) → 427.2848 (C27H39O4, 0.0) (63); 451.3214 (C30H43O3, 0.4) (20) → 409.2731 (C27H37O3, −2.9) (11); |
| 18 | anhydro-cimigenol | [M + Na]+ 623.3569 (100) [M + H]+ 601.3752 (17) |
623.3569 (C35H52O8Na, [M + Na]+) | 377.2475 (C26H33O2, [M + H – xylose – H2O – 2 –methylprop-1-ene]+) | No | 451.3203 (C30H43O3, −2.0) (18) → 395.2585 (C26H35O3, −0.3) (56); 433.3062 (C30H41O2, −10.4) (11) → 377.2474 (C26H33O2, −1.9) (100); |
No | 601.3752 (C35H53O8, 2.0) (9) → 559.3271 (C32H47O8, 0.0) (10); 469.3320 (C30H45O4, 0.4) (7) → 427.2848 (C27H39O4, 0.0) (41); 451.3220 (C30H43O3, 1.8) (8) → 409.2722 (C27H37O3, −5.1) (9); |
| 19 | anhydro-cimigenol | [M + Na]+ 623.3569 (100) [M + H]+ 601.3746 (17) |
623.3569 (C35H52O8Na, [M + Na]+) | 377.2469 (C26H33O2, [M + H – arabinose – H2O – 2 – methylprop-1-ene]+) | No | 451.3207 (C30H43O3, −1.1) (23) → 395.2586 (C26H35O3, 0.0) (38); 433.3093 (C30H41O2, −3.2) (13) → 377.2469 (C26H33O2, −3.2) (100); |
No | 601.3745 (C35H53O8, 0.8) (17) → 559.3276 (C32H47O8, 0.9) (50); 469.3314 (C30H45O4, −0.9) (11) → 427.2842 (C27H39O4, −1.4) (16); 451.3222 (C30H43O3, 2.2) (9) → 409.2733 (C27H37O3, −2.4) (20); |
| 20 | andydro-hydroshengmanol | [M + Na]+ 643.3842 (100) [M + H]+ 621.4006 (12) |
643.3842 (C35H56O9Na, [M + Na]+) | 379.2625 (C26H35O2, [M + H – xylose – 2H2O – 2-methylprop-1-ene]+) | No | 453.3352 (C30H45O3, −3.8) (13) → 397.2745 (C26H37O3, 0.5) (52); 435.3264 (C30H43O2, 0.2) (7) → 379.2635 (C26H35O2, 0.5) (100); |
No | 621.4006 (C35H57O9, 0.5) (22) → 579.3531 (C32H51O9, −0.3) (9); 603.3908 (C35H55O8, 1.8) (20) → 561.3434 (C32H49O8, 1.2) (28); 471.3478 (C30H47O4, 0.8) (6) → 429.2993 (C27H41O4, −0.7) (12); 453.3381 (C30H45O3, 2.6) (6) → 411.2888 (C27H39O3, −2.7) (7); |
| 21 | alkaloid | [M + H]+ 614.3679 (100) | 614.3679 (C35H52NO8, [M + H]+) | 596.3611 (C35H50NO7, [M + H – H2O]+) | No | No | 596.3611 (C35H50NO7, 4.0) (100) → 556.3260 (C32H46NO7, −2.5) (5)e | No |
Aperture voltage: 0V; The abundances were calculated on base peak;
MW: molecular weight; AMBP: abundance of molecular ions relative to base peak; MF: molecular formula; AIFBP: abundance of the ions relative to fragmental base peak (%); C4H8O: 2,2-dimethylepoxyethane; C3H4: propa-1,2-diene; C4H8: 2-methylpropene; C3H4: propene;
The abundances were calculated on base peak;
The abundances were calculated on fragmental base peak;
The aperture voltage used for compound 21 was 60 V.
The highly oxygenated Actaea triterpene glycosides are readily decomposed during ionization by the loss of neutral molecules such as water, sugar, acetic acid, and occasionally an acetyl group. The loss of an acetyl group is a rare occurrence usually observed in esters and acetylamines [29–32]. When the aperture voltage was set to 0V, most of the cleavages, such as the loss of neutral water, pentose (xylose or arabinose), acetic acid, or ketene (acetyl group) loss, were found in the spectra (Table 2). Many triterpenoids exist as isomers in Actaea species, but many of these isomers belong to different structural classes. These isomers were subjected to these kinds of losses, and they can not be distinguished only by such cleavages analyses. Furthermore, standards of all these triterpene glycosides are not commercially available. To address this issue, we developed a method of MS/MS fragmental analysis from the available authentic standards to distinguish these triterpene glycosides without standards. Although an unambiguous identification of each compound cannot always be achieved, we were able to assign a structural class for most of the triterpene glycosides we analyzed. These triterpene glycosides are similar in structure and their diversity is attributed to variation in the structure of C(20)-C(27), it is in this region that we focused our fragmentation analysis (Fig. 3). In order to study this region of structural variation (C(20)-C(27)) we conducted in-source fragmentation by increasing the aperature voltage to 60 V, this provided additional information for structure elucidation. Different classes of triterpene glycosides showed different capacities to lose 2,2-dimethylepoxyethane, propa-1,2-diene, 2-methylpropene, and propene, when fragmentation occurred using different aperture voltages. Relative abundance of molecular ions and base peaks was also used to distinguish among classes of triterpene glycosides (Table 3).
16,23-diketoshengmanol class (4–6)
This class of triterpene glycosides contained a highly abundant sodium adduct peak in the spectra (Table 3). At an aperture voltage of 60V, these compounds lost 2,2-dimethylepoxyethane (C4H8O). For example, in the spectrum of cimicifugoside H-1 (4), the cleavage from the fragmental ion at m/z 467.3132 [M + H – xylose]+ (C30H43O4) to m/z 395.2580 [M + H – xylose – 2,2-dimethylepoxyethane]+ (C26H35O3), and from the fragment ion m/z 449.3050 [M + H – xylose – H2O]+ (C30H41O3) to m/z 377.2475 [M + H – xylose – H2O – 2,2-dimethylepoxyethane]+ (C26H33O2). The relative abundance of the product ions (m/z 395.2580 and 377.2475) were much greater than those of their respective parent ions (m/z 467.3132 and 449.3050) (Table 3). Compounds in this class also showed losses of propa-1,2-diene (C3H4). At an aperture voltage of 0V, it was easy to find the molecular ion at m/z 617.3694 [M + H]+ (C35H53O9) losing a C3H4 sturcture to m/z 577.3390 [M + H – propa-1,2-diene]+ (C32H49O9), and the aglycone fragment at m/z 467.3165 [M + H – xylose]+ (C30H43O4) losing a propa-1,2-diene structure to m/z 427.2861 [M + H – xylose – propa-1,2-diene]+ (C27H39O4) (Table 3). The proposed mechanisms of loss of 2,2-dimethylepoxyethane and propa-1,2-diene are explained in supplemental Fig. S3A. The loss of 2,2-dimethylepoxyethane was due to a clevage of the C(23)-C(24) bond and the rearrangement of the proton on C(22) to C(24). Simultaneously, a double bond between C(22) and C(23) formed. The loss of propa-1,2-diene was due to clevages of the C(24)-C(25) and C(25)-O bonds, and the rearrangements of the H(26) and H(27) protons to C(24) and the oxygen atom attached at C(24) and C(25).
Compounds 5 and 6 were tentatively identified as cimicifugoside H-5 and shengmacichun dixyloside (Fig. 3), based on the 2,2-dimethylepoxyethane and propa-1,2-diene losses and the abundances of the much higher product ions than those of the parent ions (Table 3), together with the presence in species of Actaea reported by previous publication [33–35].
Shengmanol class (7 and 8)
Both the 16,23-diketoshengmanol and shengmanol classes of triterpene glycosides contain a 2,2-dimethylepoxyethanyl group in their structures (Fig. 3). An important difference between them is the presence of a ketone group on C(23) in 16,23-diketoshengmanol where there is a acetate group (CH3COO) group in shengmanol class compounds. In the mass spectra, 7 and 8 also showed losses of 2,2-dimethylepoxyethane and propa-1,2-diene; however the abundance ratios of product ions to parent ions were much smaller compared to those in compounds 4–6 (Table 3). For example, in compound 7, the fragmental ions m/z 453.3370 [M + H – xylose – acetic acid]+ (C30H45O3) and 435.3266 [M + H – xylose – acetic acid – H2O]+ (C30H43O2) each lose a 2,2-dimethylepoxyethane producing fragments at m/z 381.2799 [M + H – xylose – acetic acid – 2,2-dimethylepoxyethane]+ (C26H37O2) and m/z 363.2690 [M + H – xylose – acetic acid – H2O – 2,2-dimethylepoxyethane]+ (C26H35O), repectively. The relative abundance of the product ions at m/z 381.2799 and 363.3690 compared with parent ions at m/z 453.3370 and 435.3266, were much less than those of 4–6. The ratio of the abundance of the product ion to its parent ion is a useful characteristic to distinguish shengmanol and 16,23-diketoshengmanol classes of triterpene glycosides. The MS/MS spectra of 23-O-acetylshengmanol-3-O-β-D-gluco-pyranoside-(1–3)-β-D-xylopyranoside or arabinoside (8) also displayed this charactercteristic fragmentation and was identified by combining this data with previously published results [36]. The proposed mechanisms of two losses of 2,2-dimethylepoxyethane and propa-1,2-diene for shangmanol class triterpene glycosides was outlined in supplemental Fig. S3B.
Hydroshengmanol class (9)
Kusano hypothesized that hydroshengmanol class of triterpene glycosides in Actaea species originated from shengmanol glycoside by biosynthesis [23]. The mass spectrum of 9 showed the presence of a pentose and an acetyl group in the structure (Table 2). Based on the previous publication, this compound was tentatively identified as 24β-O-acetylhydroshengmanol-3-O-β-D-xylopyranoside or arabinoside [37–39]. The characteristic fragmental loss of 2,2-dimethyl epoxyethane and propa-1,2-diene is very similar to those in the shengmanol class of triterpene glycosides (Table 3). A loss of 2,2-dimethylepoxyethane occurred in response to the formation of a bond between C(24) and the oxygen present at C(25), a clevage of the C(23)-C(24) bond, and a rearrangement of the proton on C(22) to the AcO group cleaved from C(24) (supplemental Fig. S3C). Propa-1,2-diene was lost following the cleavages of C(24)-C(25) and C(25)-O, and rearrangements of the two protons at C(26) and C(27) to acetate and hydroxyl groups to form acetic acid and water to leave, respectively (supplemental Fig. S3C).
Cimigenol class (10–12)
This class of compounds lost a 2,2-dimethylepoxyethane moiety when the aperture voltage was increased to 60V. The proposed mechanism for this cleavage is shown in supplemental Fig. S3D. For compound 11, prior to the loss of 2,2-dimethylepoxyethane, a ketene was eliminated from the acetyl group to form a hydroxyl group. The loss of propa-1,2-diene observed in the 16,23-diketoshengmanol, shengmanol, and hydroshengmanol classes was not found in the fragmentation of cimigenol compounds, and helped differentiate this class of triterpene glycosides from others (Table 3). Compounds 10 and 11 were unambiguously identified as cimiracemoside C and 25-O-acetylcimigenol 3-O-β-D-xylopyranoside by comparison of retention time, mass, and fragmentation patterns with those of authentic standards. Compound 12 was tentatively identified as 12β,21-dihydroxycimigenol-3-O-L-arabinoside based on the characteristic cleavages as observed in 10 and 11 and reported from a previous study [40].
Cimiracerol class (13)
The aglycone fragment in this class displayed the same characteristic cleavage as in the cimigenol class such as the loss of 2,2-dimethylepoxyethane, however the loss of a propa-1,2-diene was not observed. The proposed mechanism of the loss of 2,2-dimethylepoxyethane is shown in supplemental Fig. S3E. The differentiating factor between the cimiracerol and cimigenol classes is the sodium adduct abundance of the molecular ion. For the cimigenol class of compounds (10–12), when the aperture voltage was set to 0V, the abundance of sodium adduct ions was much greater than those of protonated ions, and most of the sodium adduct peaks were the base peak in the whole spectra. However, only a small abundance of the sodium adduct was found in the spectra of 13 (Table 3).
Acetyl-acteol class (14 and 15)
This class of triterpene glycosides is unique that the 2,2-dimethylepoxyethane and the propa-1,2-diene moiety losses are not present in the spectra, this differentiates this class of aglycones from all others in this study. Simmilar to the cimiracerol class, sodium adducts are found in low abundance (Table 3).
Anhydrocimigenol class (16–19)
This class of triterpene glycosides differs from the cimigenol class by the loss of water which occurred from the hydroxyl group on C(25) and the proton on C(26) (Fig. 3). In the mass spectra of this class, the unique fragmental losses of 2-methylpropene, and propene were observed, when the aperture voltage was set to 60V and 0V, respectively, whereas in the 16,23-diketoshengmanol, shengmanol, hydroshengmanol, cimigenol, and cimiracerol classes, 2,2-dimethylepoxyethane and propa-1,2-diene fragmentation occurred (Table 3). This can be explained by the absence of oxygen on C(25). The proposed mechanisms of anhydrocimigenol triterpene glycosides losses of 2-methylpropene and propene are outlined in supplemental Fig. S3F. The cleavage of the C(24)-O and C(16)-O(23) bonds leads to the formation of ketones on C(24) and C(16). The cleavage of the C(23)-C(24) bond and the rearrangement of the two protons from C(22) and C(23) to C(24) induced the loss of 2-methylpropene. The cleavage of the C(24)-C(25) bond and the rearrangement of the C(23) proton to C(25) led the loss of propene, which produced a double bond between C(23) and C(24). Compounds 16–19 showed the same mass fragmental cleavage profiles. The aglycones of 16 and 17 were tentatively identified as 25-anhydrocimigenol and 12-O-acetyl-25-anhydrocimigenol, respectively. Compounds 18 and 19 both contain the (23R,24R)-7-en-25-anhydrocimigenol aglycone. The sugar residues of 16–18 are likely to be either xylose or arabinose due to the natural abundance of these sugars in Actaea triterpene glycosides; however, in a previous study, the Actaea triterpene arabinosides had a shorter retention time than triterpene xylosides with the same aglycone using a solvent system of either acetonitrile-water with 0.1% acetic acid or acetonitrile-methanol-water [24,41]. Therefore, using relative retention times we tentatively identified 16–18 as 25-anhydrocimigenol-3-O-α-L-arabinose [42], 12-O-acetyl-25-anhydrocimigenol-3-O-β-D-xylopyranoside (cimiracemoside K) [33], and (23R,24R)-7-en-25-anhydro cimigenol-3-O-β-D-xylopyranoside [43], respectively. Compound 19 possessed the same molecular formula and similar mass spectral pattern as 18; however it eluted at a shorter retention time, we therefore infer the presence of arabinose, and tentatively identified its structure as (23R,24R)-7-en-25-anhydrocimigenol-3-O-α-L-arabinoside (Table 3).
Anhydroshengmanol class (20)
Similar to anhydroshengmanol class compounds, compound 20 contains a 2-methyl-3-propenyl group on carbon-24. Due to this 2-methyl-3-propenyl group, compound 20 displays the similar losses of 2-methylpropene and propene typically observed in the anhydroshengmanol class compounds (Fig. 3). Based on this data and comparison with previous studies, we tentatively identified 20 as Cimidahuside G (Table 3) [7]. The proposed mechanism of the losses of 2-methylpropene and propene are outlined in supplemental Fig. S3G.
Alkaloid class (21)
A single alkaloid, cimicifugadine (syn. cimicifine A), was identified by MS/MS which matched spectral values well with the previous reports (Table 2) [44,45]. When the aperture voltage was increased to 60V, no 2,2-dimethylepoxyethane moiety loss was observed in the mass spectrum. However, the propa-1,2-diene moiety loss is present in the spectrum, when the aperture voltage was set to 0V (Table 3). The proposed mechanism of the losses of propa-1,2-diene is outlined in supplemental Fig. S3H.
3.3 Different types of marker compounds
Marker compounds refer to chemical constituents within a medicinal material that can be used to verify its potency or identity. For some medicinal plants, marker compounds can be described as active ingredients. In other instances, marker compounds of interest are chemicals that confirm the correct botanical identity of original plant material [46]. In this study, we have identified 15 marker compounds which are useful for the correct identification of Actaea plant material. These compounds were informative in the division of samples into four distinct categories: Actaea species other than A. racemosa (OAR); Asian species (AS); A. racemosa (AR); and North American species other than A. racemosa (NAOAR).
The classification of the four types of marker compounds was mainly based on the trend view plot produced by PCA analysis. Using the ESI probe, two types of molecular ions, the protonated ion and sodium adduct, were formed for every marker compound. The trend views were based on data from either the protonated molecular ion or sodium adduct molecular ion. Comparing these two types of trend view plots for each marker compound, the trend view that showed the presence of a marker among more samples was selected for marker evaluation.
3.3.1 Marker compounds for OAR
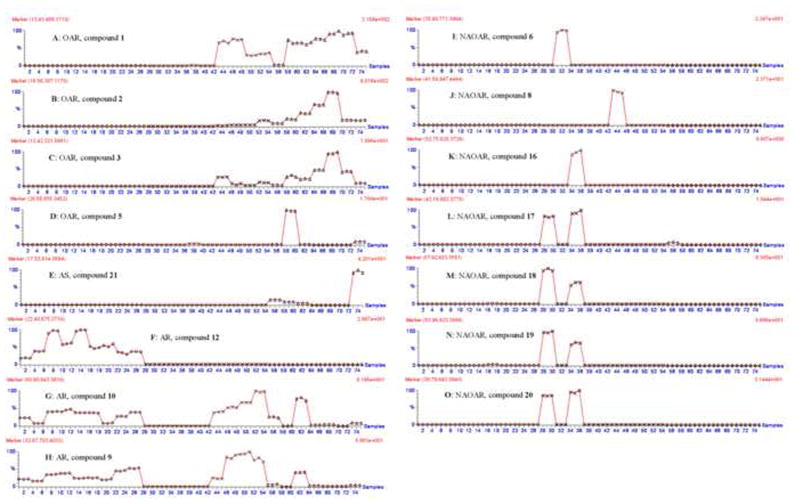
Marker compounds for OAR could not be detected in A. racemosa. This type of marker compound is useful to determine the presence of Actaea species other than black cohosh, which are potential adulterants in black cohosh dietary supplements. Three cimifugin derivatives (1–3), and one 16,23-diketoshengmanol class triterpene glycoside (5), were found to be useful OAR markers. For example, prim-O-cimifugin (1) was found in all Asian and North American species with the exception of A. americana, A. laciniata, A. podocarpa, and A. racemosa (Fig. 4A). The detection of compound 1 in black cohosh products would indicate the presence of other Actaea species, and therefore the adulteration of the black cohosh product.
Fig. 4.
Selected trend plots of marker compounds for Actaea species. Triple injections for each sample, from first to twenty-seventh injections (labeled “○”) are for black cohosh samples from different locations; From twenty-eighth to fifty-fourth injections (labeled “×”) are for North American species of Actaea other than black cohosh; From fifty-fifth to seventy-eighth injections (labeled “△”) are for Asian species of Actaea.
This is the first report of the detection of OAR marker compounds 1 and 2 in A. heracleifolia (Figs. 4A and 4B). A previous study was unable to detect the presence of these two compounds in A. heracleifolia [24]. We associate this finding with our use of highly sensitive methods in the TOF-MS analysis we conducted.
3.3.2 Marker compounds for AS
Compound 21, the alkaloid, was found to be a useful marker only present in Asian species of Actaea (AS) (Fig. 4E). This marker compound is useful in identifying the presence of Asian Actaea species. Compound 21 was detected in A. heracleifolia, A. mairei, A. dahurica, and A. yunnanensis. Among them, A. heracleifolia and A. dahuric are most commonly used in Chinese traditional medicine. This marker compound may be very useful in identification of Asian species of Actaea in black cohosh products as these species are often mistaken with black cohosh.
3.3.3 Marker compounds for AR
In order to confirm the presence of Actaea racemosa in black cohosh products or other Actaea samples, a marker compound that correlates to the presence of black cohosh and no other species would be very useful. Compound 12 was found in high concentrations in all the black cohosh samples collected from different locations, but was not be detected in any other Actaea species. Compound 10 has been previously reported to be a useful marker in the identification of black cohosh species [14]. In this work we detected the presence of compound 10 in black cohosh as well as other North American Actaea species including A. elata, A. elata var. alpestris, A. arizonica, A. cordifolia, and some Asian Actaea species including A. heracleifolia, A. dahurica, and A. yunnanensis (Fig. 4G). On the basis of our findings we determined that compound 10 alone is not a suitable marker compound for black cohosh identification. Compound 10 was useful as an AR marker when combined with additional marker compounds in a two-marker-compound method for identification. This method is based on combining two types of marker compounds to aid in the identification of black cohosh species within a black cohosh product. Type I marker compounds are found in black cohosh species, but not in A. americana, A. laciniata, A. podocarpa, A. pachypoda, and A. rubra species, such as compounds 9 and 10 (Figs. 4G and 4H). Type II marker compounds are cimifugin derivatives (1–3), which are not found in A. americana, A. laciniata, A. podocarpa, and black cohosh species (Figs. 4A, 4B, and 4C). If no cimifugin derivatives (1–3) are detected, A. americana, A. laciniata, A. podocarpa, and black cohosh may be present within a product. If a type I marker compound is present in a sample, but no type II marker can be found, we can determine that black cohosh is the only species present in a black cohosh product.
3.3.4 Marker compounds for NAOAR
Compounds 6, 8, and 16 could only be detected in A. laciniata, A. elata and A. podocarpa, respectively (Figs. 4I, 4J and 4K), and are therefore useful for identification of each of these three North American species. Compound 20 was found in A. americana and A. podocarpa and no other species we analyzed; it is therefore a useful marker compound for these species (Fig. 4O). Marker compounds 17, 18, and 19 were detected in large concentrations in A. americana and A. podocarpa species. Compound 17 was also detected in small amounts in A. heracleifolia. Compounds 18 and 19 were also detected in small amounts in one sample of A. racemosa (sample code: f) (Figs. 4L, 4M and 4N). While compounds 6, 8, 16, and 20 can each be used individually as marker compounds for NAOAR species, compounds 17, 18, and 19 must be combined with additional markers, compounds 9 or 10, for complete identification. If compounds 17, 18, or 19 are present in a sample, but neither compounds 9 or 10 cannot be found, we can determine that NAOAR species are present in a black cohosh product. (Figs. 4G, 4H, 4L, 4M and 4N).
4. Conclusion
In this study, we used HPLC-ESI-TOF-MS/MS and chemometric analysis to identify marker compounds useful in the assessment of 16 species of Actaea. A total of 15 marker compounds including three cimifugin derivatives, eleven triterpene glycosides and one alkaloid, were identified using a rapid extraction process in which no sample pre-treatment was required. Discovery and identification of these marker compounds were achieved using time of flight mass spectrometry and multivariate principle component analysis. By varying the MS aperture voltage, we were able to achieve efficient fragmentation. Using the accurate mass measurements recorded for each of these fragments we were able to produce strong evidence for structural identification.
Principle component analysis was used to direct our search for marker compounds from 25 samples representing 16 species of Actaea. The utility of these marker compounds for species and group identification has been determined under the experimental conditions described. It is therefore important to adhere to these conditions in order to make full use of the marker compounds described here for accurate identifications.
Using characteristic fragmental cleavage we developed original rules for the identification of triterpene glycosides in species of Actaea. These fragmental rules focus on the differences observed in the C(20)-C(27) region of triterpenes, where previous publications only report simple losses of water, acetic acid, and sugars. These cleavage rules add useful ways to analyze or identify the minor triterpene glycosides in Actaea species.
The methods we developed have implications beyond the identification of Actaea species and can be useful for a number of applications. HPLC-ESI-TOF and chemometric analysis are powerful tools for the identification of chemical markers which can be very useful in the identification of components of botanical products. The adulteration of botanical supplements is a major problem that efficacy of the original product can be decreased and even toxicity can become a concern [12]. Therefore, identification of markers compounds may provide useful tools in quality control of botanical supplements.
Supplementary Material
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Borrelli F, Izzo AA, Ernst E. Life Sci. 2003;73:1215. doi: 10.1016/s0024-3205(03)00378-3. [DOI] [PubMed] [Google Scholar]
- 2.Kronenberg F, Fugh-Berman A. Ann Intern Med. 2002;137:805. doi: 10.7326/0003-4819-137-10-200211190-00009. [DOI] [PubMed] [Google Scholar]
- 3.Cavaliere PRC, Lynch ME, Blumenthal M. HerbalGram. 2009;82:58. [Google Scholar]
- 4.New Medicinal College of Jiangsu. Dictionary of Chinese Materia Medica. Shanghai Scientific and Technological Press; Shanghai: 1977. [Google Scholar]
- 5.Shin TY, Seo HM, Chae BS. Yakhak Hoechi. 1998;42:403. [Google Scholar]
- 6.Shibata M, Sakurai N, Onoda M. Yakugaku Zasshi. 1977;97:911. doi: 10.1248/yakushi1947.97.8_911. [DOI] [PubMed] [Google Scholar]
- 7.Tian Z, Si J, Chang Q, Zhou L, Chen S, Xiao P, Wu E. BMC Cancer. 2007;7:237. doi: 10.1186/1471-2407-7-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lal B, Kansal VK, Singh R, Sankar C, Kulkarni AS, Gund VG. Indian J Chem, Sect B: Org Chem Incl Med Chem. 1998;37B:881. [Google Scholar]
- 9.Qiu M, Kim JH, Lee HK, Min BS. Phytother Res. 2006;20:945. doi: 10.1002/ptr.1982. [DOI] [PubMed] [Google Scholar]
- 10.Tian Z, Pan R, Chang Q, Si J, Xiao P, Wu E. J Ethnopharmacol. 2007;114:227. doi: 10.1016/j.jep.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 11.Jiang B, Kronenberg F, Nuntanakorn P, Qiu MH, Kennelly EJ. J Agric Food Chem. 2006;54:3242. doi: 10.1021/jf0606149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Painter D, Perwaiz S, Murty M. Canadian Adverse Reaction Newsletter. 2010;20:1. [Google Scholar]
- 13.Ankli A, Reich E, Steiner M. J AOAC Int. 2008;91:1257. [PubMed] [Google Scholar]
- 14.He K, Zheng B, Kim CH, Rogers L, Zheng Q. Planta Med. 2000;66:635. doi: 10.1055/s-2000-8619. [DOI] [PubMed] [Google Scholar]
- 15.Gafner S, Sudberg S, Sudberg EM, Villinski JR, Gauthier R, Bergeron C. Acta Hortic. 2006;720:83. [Google Scholar]
- 16.Zerega NJC, Mori S, Lindqvist C, Zheng Q, Motley TJ. Econ Bot. 2002;56:154. [Google Scholar]
- 17.Compton JA, Culham A, Gibbings JG, Jury SL. Biochem Syst Ecol. 1998;26:185. [Google Scholar]
- 18.Nagele E, Fandino AS. Journal of Chromatography A. 2007;1156:196. doi: 10.1016/j.chroma.2007.01.024. [DOI] [PubMed] [Google Scholar]
- 19.Zheng X, Shi P, Cheng Y, Qu H. J Chromatogr A. 2008;1206:140. doi: 10.1016/j.chroma.2008.08.038. [DOI] [PubMed] [Google Scholar]
- 20.Qi LW, Wen XD, Cao J, Li CY, Li P, Yi L, Wang YX, Cheng XL, Ge XX. Rapid Commun Mass Spectrom. 2008;22:2493. doi: 10.1002/rcm.3638. [DOI] [PubMed] [Google Scholar]
- 21.Inagaki S, Noda T, Min JZ, Toyo’oka T. Journal of Chromatography A. 2007;1176:94. doi: 10.1016/j.chroma.2007.10.065. [DOI] [PubMed] [Google Scholar]
- 22.Kang J, Sun JH, Zhou L, Ye M, Han J, Wang BR, Guo DA. Rapid Commun Mass Spectrom. 2008;22:1899. doi: 10.1002/rcm.3559. [DOI] [PubMed] [Google Scholar]
- 23.Kusano G. Yakugaku Zasshi. 2001;121:497. doi: 10.1248/yakushi.121.497. [DOI] [PubMed] [Google Scholar]
- 24.He K, Pauli GF, Zheng B, Wang H, Bai N, Peng T, Roller M, Zheng Q. J Chromatogr A. 2006;1112:241. doi: 10.1016/j.chroma.2006.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao JC, Zhang JC, Lu ZJ, Zhu GY, Yang MS, Xiao PG. Biochemical Systematics and Ecology. 2006;34:710. [Google Scholar]
- 26.Einbond LS, Wen-Cai Y, He K, Wu HA, Cruz E, Roller M, Kronenberg F. Phytomedicine. 2008;15:504. doi: 10.1016/j.phymed.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tian Z, Yang M, Huang F, Li K, Si J, Shi L, Chen S, Xiao P. Cancer Lett. 2005;226:65. doi: 10.1016/j.canlet.2004.11.019. [DOI] [PubMed] [Google Scholar]
- 28.Sakurai N, Wu JH, Sashida Y, Mimaki Y, Nikaido T, Koike K, Itokawa H, Lee KH. Bioorg Med Chem Lett. 2004;14:1329. doi: 10.1016/j.bmcl.2003.12.035. [DOI] [PubMed] [Google Scholar]
- 29.Jiang H, Timmermann BN, Gang DR. Rapid Commun Mass Spectrom. 2007;21:509. doi: 10.1002/rcm.2858. [DOI] [PubMed] [Google Scholar]
- 30.Adeuya A, Price NP. Rapid Commun Mass Spectrom. 2009;23:1173. doi: 10.1002/rcm.3980. [DOI] [PubMed] [Google Scholar]
- 31.de Oca Porto RM, Perez AR, Correa Vidal MT. Rapid Commun Mass Spectrom. 2007;21:1871. doi: 10.1002/rcm.3028. [DOI] [PubMed] [Google Scholar]
- 32.Fenaille F, Tabet JC, Guy PA. Rapid Commun Mass Spectrom. 2004;18:67. doi: 10.1002/rcm.1283. [DOI] [PubMed] [Google Scholar]
- 33.Nian Y, Chen JC, Lu L, Zhang XM, Zhou L, Qiu MH. Helv Chim Acta. 2009;92:112. [Google Scholar]
- 34.Koeda M, Aoki H, Sakurai N, Nagai M. Chem Pharm Bull. 1995;43:771. doi: 10.1248/cpb.43.771. [DOI] [PubMed] [Google Scholar]
- 35.Chen D, Si J, Zhao X, Shen L. CN1281698. Patent.
- 36.Pan RL, Chen DH, Si JY, Zhao XH, Shen LG. J Asian Nat Prod Res. 2004;6:63. doi: 10.1080/1028602031000135530. [DOI] [PubMed] [Google Scholar]
- 37.Ali Z, Khan SI, Pawar RS, Ferreira D, Khan IA. J Nat Prod. 2007;70:107. doi: 10.1021/np060152t. [DOI] [PubMed] [Google Scholar]
- 38.Ali Z, Khan SI, Khan IA. Planta Med. 2006;72:1350. doi: 10.1055/s-2006-951696. [DOI] [PubMed] [Google Scholar]
- 39.Findeis M. 2008136863. Patent WO.
- 40.Watanabe K, Mimaki Y, Sakagami H, Sashida Y. Chem Pharm Bull. 2002;50:121. doi: 10.1248/cpb.50.121. [DOI] [PubMed] [Google Scholar]
- 41.Cicek SS, Aberham A, Ganzera M, Stuppner H. Anal Bioanal Chem. doi: 10.1007/s00216-010-4068-y. Published online http://www.springerlink.com/content/5w18608h36930115/fulltext.pdf. [DOI] [PubMed]
- 42.Chen SN, Fabricant DS, Lu ZZ, Fong HHS, Farnsworth NR. J Nat Prod. 2002;65:1391. doi: 10.1021/np0200818. [DOI] [PubMed] [Google Scholar]
- 43.Nanba T, Kadota S, Myahara T. 09030977 Patent JP.
- 44.Dan C, Zhou Y, Ye D, Peng S, Ding L, Gross ML, Qiu SX. Org Lett. 2007;9:1813. doi: 10.1021/ol070542m. [DOI] [PubMed] [Google Scholar]
- 45.Sun LR, Yan J, Lu L, Pei SJ, Li ZR, Zhou L, Zhang XM, Qiu MH. Helv Chim Acta. 2007;90:1313. [Google Scholar]
- 46.Brand E. What are marker compounds? [last accessed 10th Aug. 2010).];Blue Poppy webpage, Blue Poppy Enterprises, Inc. (Available via a link found at http://www.bluepoppy.com/blog/blogs/blog1.php/2010/03/08/what-are-marker-compounds.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.