Abstract
Reactive oxygen species (ROS) arise from both endogenous and exogenous sources. These reactive molecules possess the ability to damage both the DNA nucleobases and the sugar phosphate backbone, leading to a wide spectrum of lesions, including non-bulky (8-oxoguanine and formamidopyrimidine) and bulky (cyclopurine and etheno adducts) base modifications, abasic sites, non-conventional single-strand breaks, protein-DNA adducts, and intra/interstrand DNA crosslinks. Unrepaired oxidative DNA damage can result in bypass mutagenesis during genome copying or gene expression, or blockage of the essential cellular processes of DNA replication or transcription. Such outcomes underlie numerous pathologies, including, but not limited to, carcinogenesis and neurodegeneration, as well as the aging process. Cells have adapted and evolved defense systems against the deleterious effects of ROS, and specifically devote a number of cellular DNA repair and tolerance pathways to combat oxidative DNA damage. Defects in these protective pathways trigger hereditary human diseases that exhibit increased cancer incidence, developmental defects, neurological abnormalities, and/or premature aging. We review herein classic and atypical oxidative DNA lesions, outcomes of encountering these damages during DNA replication and transcription, and the consequences of losing the ability to repair the different forms of oxidative DNA damage. We particularly focus on the hereditary human diseases Xeroderma Pigmentosum, Cockayne Syndrome and Fanconi Anemia, which may involve defects in the efficient repair of oxidative modifications to chromosomal DNA.
Keywords: Oxidative DNA Damage, Excision Repair, Interstrand Crosslink, Hereditary Disorder
Introduction
Reactive oxygen species (ROS) are thought to be a major driving force in human aging, disease, and carcinogenesis, and arise from both endogenous and exogenous sources. Normal cellular metabolism via oxidative phosphorylation by mitochondria [1], breakdown of long chain fatty acids by peroxisomes [2], and defense mechanisms employed by inflammatory cells (e.g., neutrophils, eosinophils, and macrophages) [3] are major sources of endogenous ROS. Ionizing radiation (IR), ultraviolet (UV) light, chemicals agents in food and the environment, and chemotherapeutics are exogenous agents that can induce ROS formation either through direct or indirect mechanisms. ROS are deleterious as they are highly reactive and can damage carbohydrates, proteins, lipids, and nucleic acids (DNA and RNA). Damage to nucleic acid, namely genomic DNA, is particularly detrimental, with modification occurring to both the nucleobases and the sugar phosphate backbone. Cells devote extensive resources to prevent (the so-called anti-oxidant defenses) and correct DNA damage caused by ROS. Base excision repair (BER), nucleotide excision repair (NER), strand break (single- and double-stranded) repair, homologous recombination (HR), and interstrand crosslink (ICL) repair pathways all act to remedy ROS-induced DNA damage, maintain genetic information, and genomic stability (Figure 1).
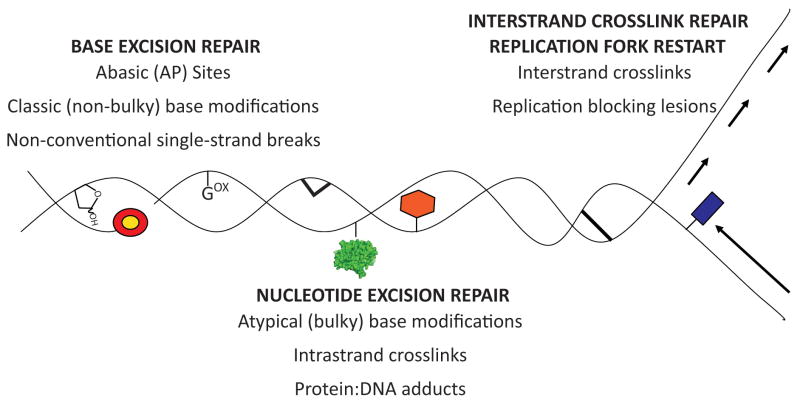
Figure 1.
Classic and atypical forms of oxidative DNA damage and associated DNA repair pathway(s). Gox = 8-oxoG; red/yellow circle = 5′ or 3′ terminal blocking group; green protein = protein-DNA adduct; red hexagon = bulky base modification; thick lines = intra- or inter-strand crosslink; blue rectangle = replication blocking lesion.
Classic oxidative DNA damage (non-bulky lesions)
Oxidative damage to DNA is thought mainly to involve modification of the nucleobases, generating primarily non-helix distorting (or non-bulky) lesions. All four nucleobases are susceptible to reaction with ROS, in particular the hydroxyl radical. Guanine is the most easily oxidized base [4] and produces principally 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxoG) [5] and 2,6-diamino-4-hydroxy-5-formamidopyrimidine (fapyG) [6]. Oxidation of adenine produces similar lesion types: 8-Oxo-7,8-dihydro-2′-deoxyadenosine (8-oxoA) and 4,6-diamino-5-formamidopyrimidine (fapyA) [7]. Free radical attack of cytosine produces cytosine glycol, which readily deaminates to form uracil glycol or dehydrates to form 5-hydroxycytosine [8]. Further deamination of 5-hydroxycytosine yields the 5-hydroxyuracil (5- OHU) base lesion [9]. Thymine oxidation yields thymine glycol (Tg), 5,6- dihydroxythymine [10], 5-hydroxymethyluracil, and 5-formyluracil [11; 12]. One important intermediate formed by either the action of a DNA repair glycosylase, which can excise a non-bulky base modification as part of BER (see later), or spontaneous hydrolysis of a normal or damaged nucleobase is an apurinic/apyrimidinic (AP) site [13]. In addition, direct free radical attack of the sugar ring of the DNA backbone can lead to the formation of oxidized AP sites, namely 2-deoxyribonolactone [14] and 2-deoxypentos-4-ulose [15], or single-strand breaks (SSBs) harboring non-conventional termini, such as a 3′-phosphate or 3′-phosphoglycolate [16].
“Atypical” oxidative DNA damage (“bulky” lesions)
Modification of nucleobases to yield helix distorting (or bulky) modified bases can occur via reactions with the hydroxyl radical or breakdown products of lipid peroxides. For example, tandem base lesions can arise through free radical attack of the C5 or C6 atoms of pyrimidine nucleobases, generating pyrimidine radicals that can attack a neighboring purine nucleobase to yield intrastrand nucleobase-nucleobase crosslinks [17]. Another group of atypical lesions caused by hydroxyl radical reactions are the 8,5′-cyclopurine-2′-deoxynucleosides [18]. Hydroxyl radical attack of the C5′ of 2-deoxy-adenosine or -guanosine, followed by attack of the C5′ radical at the C8 position results in C5′-C8 cyclization and formation of either 8,5′-cyclo-2-deoxyadenosine (cycloA) or 8-5′-cyclo-2-deoxyguanosine (cycloG), respectively. The covalent C5′-C8 bond causes sugar pucker [19], which changes the phosphodiester bond torsion angles, weakens the Watson-Crick hydrogen bonding, and disrupts the DNA helix near the lesion.
Endogenous aldehydes are formed by oxidative stress and lipid peroxidation, and can generate a wide range of DNA adducts. Such aldehydes can induce damage through direct interaction with the nucleobases, producing additional reactive intermediates. For example, 4-hydroxynonenal (4-HNE), an α,β unsaturated aldehyde, interacts with nucleobases to form etheno adducts, including N3-(3-oxypropyl)-guanine, as well as the cyclic adducts 1,N2- ethenoguanine (1,N2- ethenoG), 1,N6 ethanoadenine (1,N6 ethanoA), and 3,N4 ethenocytidine [20]. One of the 4-HNE 1,N2-guanosine adducts (6S,8R,11S) is capable of forming a DNA ICL through ring opening [21]. In addition, 4-Oxo-2-nonenal, another aldehyde generated by oxidation of polyunsaturated fatty acids, acts on purine nucleobases to create heptanone etheno-deoxypurine adducts [22]. Malondialdehyde (MDA), yet another endogenous lipid-derived reactive species, interacts with deoxyguanosine to form the cyclic 1,N2-G pyrimidopurinone, which converts to a ring open form (N2-(3-oxo-1-propentyl)-2′-guanine) when opposite cytidine. MDA can also react with adenine and cytidine to produce non-cyclic oxypropenyl adducts [23]. Acrolein, an α,β unsaturated aldehyde that is generated endogenously and is an environmental contaminant, forms cyclic 1,N2-3-hydroxypropano-guanine (1,N2-3-hydroxypropanoG) lesions, which have the ability to form DNA intrastrand crosslinks, ICLs in CpG sequence contexts, as well as protein-DNA crosslinks [24]. Finally, crotonaldehyde reacts with DNA to form α-methyl-γ-hydroxyl-1, N2-propano-guanine adducts that adopt an open ring configuration in double-stranded DNA that can react to generate ICLs [25].
Formaldehyde (FA), found in many exogenous sources and produced as a byproduct during metabolism, leads primarily to the formation of protein-DNA crosslink adducts through the establishment of a methylene bridge between deoxy-adenine, -guanine, or -cytidine of DNA and a lysine, histidine, cysteine or tryptophan residue within the protein. FA also interacts with DNA to form N6-methylene-adenine adducts and deoxyadenosine-methylene-deoxyadenosine DNA intrastrand crosslinks, as well as reacts with guanine to produce N2-hydroxymethyl -guanine and deoxyguanosine-deoxyguanosine DNA intrastrand crosslinks.
DNA and RNA polymerase action at oxidatively generated lesions
Oxidative DNA damage, regardless of its chemical complexity, has the potential, if left unrepaired, to adversely affect the essential biological processes of DNA replication and RNA transcription, leading to genome destabilization and gene expression defects that can promote cellular transformation or programmed cell death. For damage encountered by DNA polymerases during genome duplication, several scenarios can occur: (i) replicative polymerases are not blocked by the lesion and insert the correct nucleotide opposite (faithful replication), (ii) replicative polymerases are not blocked by the lesion, but insert the incorrect nucleotide opposite, leading to permanent genetic mutations that can drive oncogenesis, (iii) replicative polymerases are blocked by the lesion, yet specialized translesion DNA polymerases are recruited to facilitate bypass synthesis of the damage, and (iv) replicative polymerases are blocked by the lesion, resulting in a collapsed replication fork, which needs to be resolved by HR to prevent genetic instability.
Three polymerases, all of which are members of the B-family, are thought to act as the main mammalian replicative DNA polymerases for genome duplication during cell division (Polα, Polδ, and Polε). The remainder of the eukaryotic DNA polymerases, which are members of the A, B, Y, and X families, are thought to be principally involved in translesion synthesis and/or DNA repair. In some instances, translesion synthesis (scenario “iii” above) can occur faithfully, though in many situations, it is a mutagenic event. In this section, we review the most likely outcome for various DNA polymerases should a specific lesion be encountered on the template strand during DNA replication. A majority of the data reported below derives from in vitro reconstituted systems using human DNA polymerases (summarized in Table 1).
Table 1.
Action of DNA polymerases at various DNA lesions.
| DNA DAMAGE | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DNA Polymerase | 8-oxoG | Tg | 5-OHU | AP site | N2-isopropylG | N6-isopropylA | ||||||||
| Name | Gene | Family | I | E | I | E | I | E | I | E | I | E | I | E |
| α | POLA1 | B | A> C | Yes | A | No | A≫T,C,G | No | No | No | T≫A,C,G | Yes | ||
| δ | POLD1 | B | C,A | Yes | No | No | A≫T,G>C | Yes | ||||||
| ε | POLE | B | A ≫ C | Yes | No | No | ||||||||
| β | POLB | X | A,C | Yes | A,G,C,T | No | A | No | ||||||
| λ | POLL | X | C>A, G | Yes | A,G | No | Yes | Yes | ||||||
| μ | POLM | X | A | Yes | T>A | G>T,C | No | |||||||
| ζ | REV3L | B | A or C | Yes | A≫G>C,T | Yes | ||||||||
| REV1 | REV1 | Y | C | No | C>T,A,G | No | ||||||||
| η | POLH | Y | C>A,G | Yes | A>G>T,C | Yes | A,T>G,G | Yes | C≫T>A>G | Yes | T>C,A>G | Yes | ||
| ι | POLI | Y | A, G, >T | Yes | T>G>A≫C | Yes | T,G,A>C | No | C>T≫G>A | Yes | T≫A,C,G | Yes | ||
| κ | POLK | Y | A, C, or G | Yes | A>G,C,T | Yes | C,A≫G,T | Yes | ||||||
| θ | POLQ | A | Yes | Yes | A>G>T | Yes | ||||||||
| ν | POLN | A | A>G | Yes | No | No | ||||||||
| DNA DAMAGE | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DNA Polymerase | CycloA | 1, N6- ethanoA | 1,N2- ethenoG | 1, N2-hydroxypropanoG | Intrastrand (G-T) crosslink | N2-N2-Guanine ICL remnant | ||||||||
| Name | Gene | Family | I | E | I | E | I | E | I | E | I | E | I | E |
| α | POLA1 | B | A>T | No | ||||||||||
| δ | POLD1 | B | No | No | ||||||||||
| ε | POLE | B | A>T | Yes | ||||||||||
| β | POLB | X | No | No | ||||||||||
| λ | POLL | X | ||||||||||||
| μ | POLM | X | C≫A | No | ||||||||||
| ζ | REV3L | B | No | No | ||||||||||
| REV1 | REV1 | Y | C | No | C | No | ||||||||
| η | POLH | Y | A>T | Yes | A,T>G>C | Yes | G>A>C | Yes | A,G,T>C | Yes | A≫T,C,G/C≫T>A,G | Yes | ||
| ι | POLI | Y | C>T | No | C,T | No | ||||||||
| κ | POLK | Y | C,T | No | C | Yes | ||||||||
| θ | POLQ | A | ||||||||||||
| ν | POLN | A | ||||||||||||
DNA polymerase name, gene and family designation are indicated. Damages are defined in the text. I=Nucleotide insertion preference opposite the lesion; E=Ability to extend past lesion after nucleotide insertion; A=adenine; G=guanine; C=cytosine; T=thymine; U=uracil; AP=apurinic/apyrimidinic.
The replicative DNA polymerases are able to bypass classic non-bulky lesions like 8-oxoG, inserting an A or C opposite and then extending past the lesion [26; 27; 28]. Repair polymerase Polβ correctly inserts C opposite the 8-oxoG lesion and can also extend past [29]. REV1 can insert a C opposite 8-oxoG [30]. Translesion polymerases Polη, Polλ, Polκ, and Polι insert A, C, or G opposite 8- oxoG and proceed past [31], Polμ inserts A opposite and extends past [32], while Polζ inefficiently inserts opposite 8-oxoG, but efficiently extends past the lesion if insertion has occurred opposite [33]. At a Tg lesion, which can alter the DNA helical structure, Polα inserts A opposite, but does not extend past the damage [34]. Polβ inefficiently inserts any of the four nucleotides opposite Tg [35], with no extension past. Polη [36], Polκ [37; 38], Polλ [35], Polθ [39], and Polν [40] efficiently insert opposite Tg (typically A) and extend past the base modification, and Polζ can extend past Tg after insertion of a nucleotide by a different DNA polymerase [38]. Polι efficiently inserts T, G, or C opposite 5-OHU and extends past the base lesion [41]. AP sites pose potent blocks for the replicative DNA polymerases α and δ, though they can insert A opposite the lesion with low efficiency. AP sites impede extension by Polι, Polκ, and REV1 after insertion of a nucleotide opposite (T, G, or A; A; C, respectively) [42], while Polη and Polμ [32] are able to bypass an AP site.
In general, bulky base modifications pose blocks for the replicative DNA polymerases, and thus, most experimental efforts have focused on the action of translesion/repair polymerases at these lesion types (Table 1). Polη can effectively bypass an intrastrand crosslink and incorporate the correct nucleotides opposite each crosslinked base [43]. CycloA poses a particular problem, as only the R diastereomer has been found to be efficiently bypassed, specifically by Polη, which incorporates primarily A opposite, but requires a seldom incorporated T opposite for further extension to occur. The S diastereomer of cycloA can have A or T inserted opposite the lesion by Polη, yet no further extension takes place [44]. N2-isopropyl-purines, though blocks to Polα activity, do not impede progression by either Polη or Polι and are not mutagenic, as both polymerases insert the correct nucleotide opposite the lesion and can subsequently extend past [45].
The cyclic etheno adducts, 1,N2-ethanoA, and 1,N6 ethanoG block replicative DNA polymerases, but have differential effects on translesion/repair polymerases (Table 1). For example, 1,N6-ethanoA blocks Polβ and Polι, while Polμ and Polη are capable of lesion bypass, with Polη able to incorporate all four nucleotides opposite [46]. The 1,N2-ethanoG modification is permissible to insertion of T or C opposite by Polκ and Polι, while Polη efficiently bypasses the damage by inserting G, A, or C opposite [47]. The base lesion 1,N2-3-hydroxypropanoG is bypassed by Polη, which inserts C opposite [48]. During the process of ICL repair (see below), Polκ is able to accurately bypass an N2-N2 deoxyguanosine ICL remnant [49] and continue polymerization past. Polκ is also able to bypass an acrolein derived deoxyguanosine-protein DNA crosslink [50].
Mammalian RNA polymerase II (RNAPII) has been found in in vitro reconstituted transcription assays to bypass base lesions to varying extents, including Tg (~20 to 60%), 8-oxoG (~50–80%), and 5-OHU (~40–60%) [51]. RNAPII generally inserts A opposite 8-oxoG, yet can insert C opposite 8-oxoG to a lesser extent [52]. This co-called “transcriptional mutagenesis” caused by 8-oxoG has been shown to be biologically significant in a system specifically designed to look at the consequences of repair vs. transcriptional mutagenesis of codon 61 of the Ras oncogene. If a site-specifically introduced 8-oxoG lesion was repaired or if transcription was faithful, then wild-type Ras protein would be produced and downstream signaling events, such as ERK phosphorylation, would be normal. If, however, the 8-oxoG lesion was left unrepaired and RNAPII mis-inserted an A opposite the modified base during mRNA production, a constitutively active dominant mutant Ras protein would be generated that would drive increased constitutive ERK phosphorylation. Constitutive activation of Ras, found in nearly one third of all human cancers, and subsequent phosphorylation of ERK promotes cell proliferation, which then leads to an aberrant expansion of the cell population (tumorigenesis). Indeed, transcriptional mutagenesis by RNAPII in this system lead to the production of mutant Ras protein and constitutive phosphorylation of ERK, suggesting that transcriptional mutagenesis caused by oxidative DNA damage has the potential to activate oncogenic pathways and promote carcinogenesis [53].
RNAPII can bypass an atypical base lesion, i.e., cycloA [54]. RNAPII correctly inserts U opposite cycloA, then inserts A opposite the next nucleotide or, intriguingly, deletes 7, 13 or 21 nucleotides (nt) after uridine incorporation opposite the lesion in this transcriptional paradigm. RNAPII is generally blocked by more bulky base lesions, e.g., N2-ethyldeoxyguanosine, although the RNA polymerase can insert a C opposite such modifications in some instances [55]. Interference of RNA polymerase progression during transcription has been shown to activate certain cell death responses, and may be particularly relevant to non-dividing cell populations, such as neurons, and thus in diseases of the brain [56].
Oxidative DNA damage in human disease
Repair of non-bulky DNA damage
BER is the primary pathway for coping with non-bulky base modifications (e.g. 8-oxoG), abasic sites and a variety of chemical termini within a SSB, such as 3′-phosphoglycolates (Figure 1). Conventional BER is comprised of five major enzymatic steps: (i) substrate base removal by a DNA glycosylase, (ii) incision at the resulting abasic site by an endonuclease or lyase, (iii) clean-up of the 5′ and/or 3′ terminal strand break ends by a lyase or phosphodiesterase, respectively, (iv) gap-filling by a DNA polymerase, and (v) nick sealing by a DNA ligase (Table 2 and Figure2A). Studies of BER pathway defects have been hampered by the facts that: (i) many BER proteins are essential for embryonic development or (ii) gene deletion yields no pronounced phenotype. Homozygous mutations in the primary BER DNA polymerase Polβ [57], the AP endonuclease APE1 [58], and the SSB repair (SSBR) scaffold protein XRCC1 [59] all lead to early embryonic lethality in mice, as does deletion of both alleles of either DNA ligase I [60] or ligase III [61]. Conversely, knockouts of the DNA glycosylases that remove oxidative DNA base damage, e.g. the 8-oxoG glycosylase-OGG1 [62], the MutY homolog- MYH [63; 64], and the EndoIII homolog- NTH1 [65; 66], display no obvious, gross phenotypic defect, likely due to functional redundancy. One instance where knockout of a single oxidative DNA damage glycosylase in mice leads to an observable phenotype is for NEIL1 (endonuclease VIII-like). Intriguingly, this knockout does not result in susceptibility to cancer formation, but in metabolic syndrome, with mice displaying severe obesity, dyslipidemia, fatty liver disease, and a tendency to develop hyperinsulinemia [67].
Table 2.
The major steps and protein participants in BER.
| BER STEP | PROTEIN | BIOCHEMICAL ACTIVITY | DNA SUBSTRATE |
|---|---|---|---|
| GLYCOSYLASE | UNG | Glycosylase | U |
| SMUG1 | Glycosylase | U, hydroxyU, hydroxymethylU | |
| MBD4 | Glycosylase | U or T opposite G at CpG sites | |
| TDG | Glycosylase | U, T or ethenoC opposite G | |
| OGG1 | Glycosylase, AP β-lyase | 8-oxoG opposite C, FAPY | |
| MYH/MUTYH | Glycosylase | A opposite 8-oxoG | |
| NTH/NTHL1 | Glycosylase, AP β-lyase | Ring saturated or fragmented Py | |
| MPG | Glycosylase | 3-MeA, ethenoA, hypoxanthine | |
| NEIL1 | Glycosylase, AP β,δ-lyase | Tg, FAPY, hydroxyU | |
| NEIL2 | Glycosylase, AP β,δ-lyase | Oxidized Py | |
| NEIL3 | Glycosylase | Oxidized Py | |
| AP ENDONUCLEASE | APE1 | AP endonuclease | AP site |
| STRAND BREAK PROCESSING | POLβ | 5′-dRP lyase | 5′-dRP |
| APE1 | 3′-Phosphodiesterase | 3′-α,β-Unsaturated aldehyde | |
| PNKP | 3′-Phosphatase/5′-Kinase | 3′-Phosphate, 5′-hydroxyl | |
| TDP1 | 3′-Phosphodiesterase | 3′-Protein (TopoI) | |
| APRATAXIN | 5′-Hydrolase | 5′-AMP | |
| POLYMERIZATION | POLβ | Gap-filling polymerase | DNA Gap (1–6 nt) |
| POLδ (POLε) | Strand-displacement polymerase | DNA strand break | |
| FEN1 | 5′-Flap endonuclease | 5′-Flap | |
| LIGATION | XRCC1/LIG3α | Ligase complex | DNA nick |
| LIG1 | Ligase | DNA nick |
AP=apurinic/apyrimidinic; U=uracil; T=thymine; C=cytosine; G=guanine; FAPY=formamidopyrimidine; Py = pyrimidine; dRP = deoxyribose phosphate.
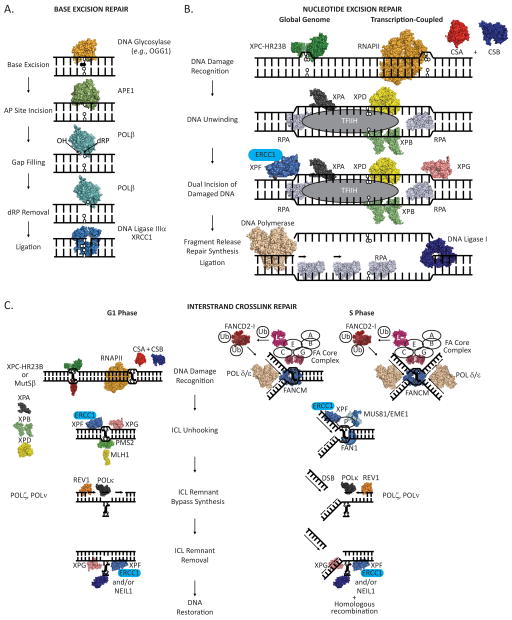
Figure 2.
(A) Major steps in the short-patch base excision repair (BER) pathway. A damaged base is detected and excised from the DNA duplex by a DNA glycosylase, leaving an apurinic/apyrimidinic site that can be bound by APE1 endonuclease. APE1 then incises the DNA phosphodiester backbone 5′ to the abasic site leaving a 3′-hydroxyl (OH) and a 5′-deoxyribose phosphate (dRP). DNA Pol β removes the dRP moiety, via an intrinsic dRP lyase activity, and inserts a nucleotide. The remaining nick in the phosphodiester backbone is sealed through the action of a DNA ligase, leaving intact, undamaged duplex DNA. (B) Major steps in the nucleotide excision repair (NER) pathway. Helix distorting DNA damage (an oxidatively damaged DNA base in this instance) can be recognized by the XPC-HR23B protein complex (global genome-NER) or by an elongating RNA polymerase in concert with the CSA and CSB proteins (transcription-coupled-NER). XPA, RPA, TFIIH (including the XPB and XPD ATPases/helicases) are recruited to the site of the damage, and the DNA surrounding the damage is unwound. XPG and XPF/ERCC1 nucleases are recruited and cleave the DNA phosphodiester backbone on the 3′ and 5′ sides of the damage, respectively, releasing a damage containing DNA section. A DNA polymerase is then enlisted to fill the gap, leaving a nick in the phosphodiester backbone that is sealed by a DNA ligase, producing intact, undamaged duplex DNA. (C) Model of major steps in the repair of interstrand crosslinks (ICLs). In the G1 phase of the cell cycle, an ICL can be recognized by XPC-HR23B protein complex, MutSβ (MSH2-MSH3) protein complex, or by an elongating RNA polymerase in concert with the CSA and CSB proteins. Once the ICL has been recognized, it is unhooked from one strand by the action of the classic NER pathway, which involves the XPG and XPF/ERCC1 nucleases, or potentially through the endonuclease activity of the MLH1/PMS2 protein complex. The ICL remnant is then bypassed through the action of translesion DNA polymerases: REV1 then POLκ, and/or POLζ, POLν, and possibly additional DNA polymerases. At this point, a second round of DNA repair is initiated to remove the covalently linked ICL remnant from the second DNA strand. NER, involving the nuclease actions of XPG and XPF/ERCC1, likely acts to remove a DNA segment containing the physically linked ICL remnant, or the ICL remnant can be removed through the action of the NEIL1 DNA glycosylase. DNA can then be restored to the native duplex form through the activity of DNA polymerase(s), to fill in the gap, and a DNA ligase, to seal the nick in the phosphodiester backbone. During the S phase of a cell cycle, an ICL is detected by stalling of a single replication fork (left) or two converging replication forks (right). FANCM is initially recruited to the stalled replication fork complex and likely acts to regress and or stabilize the fork. The FA core complex, comprised of FANCA, B, C, E, F, G and L, recognizes the FANCM bound/stabilized replication fork and ubiquitinates the FANCD2/I protein complex, with FANCL acting as the E3 ubiquitin ligase. Ubiquitinated FANCD2/I then associates with chromatin. For simplicity, action at a single replication fork is shown further. FANCP coordinates endonucleolytic cleavage of the ICL stalled replication fork by XPF/ERCC1, MUS81/EME1, and FAN1. Cleavage by these nucleases unhooks the ICL from one DNA strand and generates a double strand break (DSB). DNA polymerization past the unhooked ICL remnant by translesion DNA polymerases, REV1 then POLκ, and/or POLζ, POLν, and possibly additional DNA polymerases, ensues. A second round of repair is initiated to remove the unhooked ICL remnant by NER and/or NEIL1. Homologous recombination then is used to reform the replication fork.
Studies combining multiple glycosylase knockouts have yielded more fruitful information on the roles of oxidative DNA glycosylases in tumorigenesis. OGG1−/− MYH−/− mice were found to be predisposed to cancers (e.g. lung, ovarian, and lymphomas) [63; 64], likely due to a global defect in the processing of the mutagenic lesion 8-oxoG in the genome. In addition, NTH−/−NEIL1−/− mice developed pulmonary and hepatocellular tumors [68], further highlighting the importance of repair of oxidized base lesions in DNA. Haploinsufficient mice have also been examined in instances where homozygous knockouts resulted in embryonic lethal. For instance, APE1+/− mice display increased sensitivity to oxidative stress, increased spontaneous mutagenesis, and develop lymphomas and adenocarcinomas [58; 69]. Unchallenged Polβ+/− mice display increased chromosomal aberrations, higher levels of SSBs compared to wild-type littermates, and, like the APE1+/− mice, develop spontaneous lymphomas and adenocarcinomas. Challenged Polβ+/ − mice as exhibit increased dimethyl sulfate induced mutagenesis and increased strand breaks caused by oxidative stress [70; 71]. XRCC1+/− haploinsufficient mice, while exhibiting normal lifespan and showing no signs of abnormal aging in the absence of environmental challenges, as assessed by several parameters, did display severe liver toxicity and an increased incidence of precancerous lesions in the colon following exposure to the alkylating agent azoxymethane [72].
To date, in humans, impaired MYH activity is the only case where a mutation in a DNA glycosylase has been found to be causative in a cancer syndrome [73]. MYH-associated colorectal polyposis (MAP) is an autosomal recessive disorder, typically with biallelic heterozygous mutations of MYH, and is characterized by the presence of multiple adenomatous polyps with an increased risk of colorectal cancer. Patients display elevated G:C to T:A transversions due to replication of unrepaired A:8-oxoG base pairs, which normally would be substrates for MYH glycosylase function and subsequent BER processing. Although not specific to repair of oxidative DNA damage, pathogenic mutations in the nuclear isoform of the uracil DNA glycosylase (UNG2) have been identified in patients suffering from Hyper-IgM syndrome, a disorder characterized by immunodeficiency that likely arises from the inability to generate high affinity antibodies via class switch recombination [74; 75]. Other DNA glycosylases, APE1, and Polβ gene mutations have been found to be disease-associated, though the relationship to disease pathology is largely unclear, except perhaps in a few situations involving Polβ tumor-associated variants, as has been reviewed elsewhere [76; 77].
Defects in SSBR proteins have been found to be causative in three autosomal recessive human disorders characterized by neurological abnormalities. The common theme to these diseases is that the mutated protein is directly involved in processing of non-conventional ends at DNA strand breaks. Mutation of TDP1, which encodes tyrosyl DNA phosphodiesterase 1, leads to spinocerebellar ataxia with axonal neuropathy (SCAN1) [78]. TDP1 functions to remove obstructions from 3′ ends of DNA breaks, including 3′-phosphoglycolates formed by free radical-mediated DNA cleavage and covalently trapped 3′-topoisomerase I moieties created at sites of DNA damage or by chemotherapeutics such as camptothecin [79; 80]. This action of TDP1 helps generate DNA termini that are ultimately suitable for phosphodiester bond re-formation (ligation). SCAN1 patients are rare and exhibit a late-childhood onset of symptoms of progressive cerebellar ataxia, areflexia, and peripheral neuropathy [81]. Another disease caused by mutation of a SSBR protein involved in DNA termini processing, i.e., polynucleotide kinase 3′-phosphatase (PNKP), is MCSZ (microcephaly and seizures) [82]. PNKP acts to phosphorylate 5′-hydroxyl ends and dephosphorylate 3′-phosphate termini, blocking lesions that arise mainly from free radical attack of DNA, to produce termini that are substrates for polymerization and ligation [83]. MCSZ patients are rare as well, exhibit microcephaly, early onset of seizures, and developmental delays. A lack of a functional aprataxin protein leads to autosomal recessive ataxia with oculomotor apraxia type 1 (AOA1) [84; 85]. Aprataxin acts to remove 5′-AMP groups from DNA caused by abortive ligation, which usually occurs in the context of oxidative SSBs, thereby cleaning up and generating a ligatable DNA end [86]. AOA1 patients have childhood onset of progressive cerebellar ataxia, followed by oculomotor apraxia, and primary motor peripheral axonal motor neuropathy [87]. Thus, the inability to efficiently process classic BER substrates, such as oxidative base modifications and non-conventional DNA strand breaks, can lead to biological outcomes such as mutagenesis and obstructed replication or transcription, cellular events that can drive cancer development and neurological disease.
Repair of bulky DNA lesions
Oxidative DNA damage has been proposed to play an important role in the etiology of human diseases that do not have an explicit defect in BER or SSBR. We review in this section the evidence that defects in the processing of oxidative DNA damage (particularly atypical lesions) contribute to the pathologies associated with the human genetic disorders Xeroderma Pigmentosum (XP), Cockayne Syndrome (CS), and Fanconi Anemia (FA). XP is a rare autosomal recessive disease caused by mutation of one of eight genes ( XP-A, B, C, D, E, F, G, or V) involved in NER (A–G), the major pathway for excising helix-distorting (bulky) base modifications, or DNA damage tolerance/lesion bypass (V) [88; 89]. CS is a rare autosomal recessive disease caused by mutation of either of two genes (CSA or CSB) critical for a subpathway of NER termed transcription coupled-NER (TC-NER), which preferentially deals with transcription-blocking (typically bulky) lesions within active regions of the genome. FA is a rare autosomal recessive or X-linked multisystem disease caused by a defect in one of fifteen genes [FANC-A, B, C, D1 (BRCA2), D2, E, F, G, J (BACH1/BRIP1), L, M, N (PALB2), P (SLX4), O (RAD51c)] critical for resolution of DNA ICLs in the S phase of the cell cycle [90].
Xeroderma pigmentosum
Strict XP patients primarily develop early sunlight-induced skin cancer, at an increased susceptibility of at least 1000-fold compared to the normal population. This increased skin cancer incidence is explained by the loss of the ability to remove bulky, helix-distorting UV-induced photoproducts in DNA by NER (Table 3 and Figure 2B). What is difficult to link to the inability to remove DNA photoproducts are the increased frequencies of internal cancers, including of the central nervous system and lung [91], and the neurological symptoms seen in some patients; neuronal degeneration, sensorineural hearing loss, ataxia, areflexia, and microcephaly [92]. In particular, these regions of the body are unlikely to receive significant UV irradiation from sunlight to produce photoproducts in DNA. It has therefore been hypothesized that the neurological disease associated with XP is the result of the failure to repair oxidative DNA damage, possibly certain “bulky” lesions via the NER pathway [93]. A defect in the repair of endogenous DNA damage could be the underlying cause of the internal cancers in XP patients as well.
Table 3.
The major steps and protein participants in global genome and transcription-coupled NER. Only the recognition step differs between these two sub-pathways of NER.
| NER STEP | PROTEIN | BIOCHEMICAL ACTIVITY | SUBSTRATE | |
|---|---|---|---|---|
| RECOGNITION | GLOBAL GENOME | XPC/RAD23B | DNA Binding/TFIIH recruitment | DNA helix distortion |
| DDB1/DDB2 (XPE) | DNA Binding | UV-damaged DNA | ||
| XPA | DNA Binding | XPC-TFIIH bound DNA | ||
| TRANSCRIPTION COUPLED | CSA | Unknown/Part of ubiquitin ligase complex | Stalled RNA Pol | |
| CSB | DNA-dependent ATPase | Stalled RNA Pol | ||
| DNA UNWINDING (TFIIH Multisubunit Complex*) | XPB (ERCC3) | 3′ to 5′ DNA helicase/ATPase | ||
| XPD (ERCC2) | 5′ to 3′ DNA helicase/ATPase | |||
| INCISION | XPG (ERCC5) | 3′-Incision | 5′-Flap, Bubble | |
| ERCC1/XPF (ERCC4) | 5′-Incision complex (catalyzed by XPF) | 3′-Flap, Bubble | ||
| POLYMERIZATION | POLδ (POLε) | PCNA-dependent replicative polymerase(s) | DNA gap (~25–30 nt) | |
| LIGATION | LIG1 | Ligase | DNA nick | |
| XRCC1/LIG3α | Ligase complex | DNA nick | ||
Of the multiple subunits that comprise the TFIIH transcription complex, only XPB and XPD are listed here. RPA (not listed) participates in NER by binding and protecting the single-stranded DNA regions generated upon duplex unwinding.
Consistent with the above hypothesis, XPB- and XPD-deficient lymphoblastoid cells are hypersensitive to H2O2 and display higher H2O2-induced DNA damage compared to wild type cells [94]. XPA-deficient cells also display defects in response to oxidative DNA damaging agents, including an increase in H2O2 genotoxicity, a lower threshold for H2O2-induced cell cycle arrest, and reduced repair of H2O2 generated DNA lesions [95]. Furthermore, XPC-deficient cells (keratinocytes and fibroblasts) are hypersensitive to agents that induce oxidative DNA lesions (e.g., IR and potassium bromate (KBrO3)). IR exposed XPC-deficient cells accumulate 8-oxoA, 8-oxoG, and the 8,5′-cyclopurine 2′-deoxynucleosides, (5′S)-cyclo dA, -cyclo dG and (5′R)-cyclo dG, implying a deficiency in removing a broad range of oxidative modifications [96]. In particular, compared to wild type cells, repair of the 8,5′-cyclopurine 2′-deoxynucleosides was not observed in XPC-deficient cells, leading to the proposal that these are the major endogenous lesions responsible for the non-UV driven pathologies in XP [97]. Additionally, it was found that repair of 8-oxoG was slower in XPC-deficient cells, that incision of an 8-oxoG containing oligonucleotide by XPC -deficient cell extracts was lower, and that XPC increased the affinity of OGG1 for damaged DNA, thereby stimulating 8-oxoG incision by the BER DNA glycosylase [96]. The NER 3′ endonuclease XPG and the glycosylase NTH1 have also been found to physically and functionally interact, with XPG stimulating Tg containing DNA binding by NTH1 and subsequent Tg incision by this glycosylase [98; 99]. These data indicate that the NER pathway acts both in the repair of endogenous helix distorting DNA lesions, namely the cyclopurines, and in supporting the repair of classic oxidative DNA damages.
Cockayne syndrome
Another human disease originally thought to be caused by a defect in an NER-related pathway is CS. In particular, a hallmark feature of CS cells is the inability to recover RNA synthesis following UV irradiation, leading to the conclusion that the disorder stems from a deficiency in TC-NER of transcription-blocking lesions [100] (Table 3 and Figure 2B). However, like XP, many of the pathological phenotypes of CS cannot be easily explained by a defect in a response system relevant to external exposures, such as UV. CS patients, while interestingly exhibiting no increased cancer incidence, display segmental progeria, drastic lifespan reduction (~12.5 years on average), cachectic dwarfism, sensorineural hearing loss, dental caries, and neuroabnormalities; including microcephaly, brain calcification, gait ataxia, and de- or dys-myelination. Thus, it has been hypothesized that some form of endogenous DNA damage contributes to, and may be the driving force behind, CS.
Consistent with this model, CS cells from both complementation groups (A and B) display hypersensitivity to IR-induced oxidative DNA damage [101], with an accumulation of 8-oxoG and 8-oxoA found in IR-treated CSB-deficient cells [102; 103; 104]. CS cells are also hypersensitive to the oxidative DNA damaging effects of H2O2 [105] and KBrO3 [106]. After KBrO3-induced oxidative stress, CSA-deficient fibroblasts displayed inefficient repair of 8-oxoG and (5′S)-cyclo dA [107], while CSB fibroblasts displayed elevated levels of DNA breakage, suggesting inefficient processing of several forms of oxidative DNA damage. In addition, CSB-deficient cells exhibit decreased survival when treated with either paraquat or menadione [108], agents that lead to increased generation of ROS in cells. Tissues from CSB-deficient mice show an increase in the steady state levels of formamidopyrimidines [109] (brain and liver) and 8- oxoG (liver) [108] in nuclear DNA. Additionally, mitochondrial DNA from CSB-deficient mice display increased steady state levels of fapyA [109] and 8-oxoG [108], suggesting defects in both nuclear and mitochondrial oxidative DNA damage processing.
At a molecular level, the link between CS and oxidative DNA damage repair goes even further. A physical and functional interaction between the SSBR protein poly-ADP ribose-polymerase 1 (PARP1) and CSB has been identified. After oxidative stress, CSB localizes to sites of PARP1, and the two proteins physically interact through the N-terminus of CSB. PARP1 can poly(ADP-ribosyl)ate CSB to regulate its double-strand DNA-dependent ATPase activity. Additionally, CSB-deficient cells are hypersensitive to PARP inhibition [110], suggesting that CSB and PARP1 cooperate in the repair of endogenous cytotoxic DNA strand breaks. CSB has also been shown to interact with and stimulate formamidopyrimidine and 5-OHU incision activity of the NEIL1 DNA glycosylase [109], and the AP site incision activity of APE1 [111]. In all, there is substantial evidence that CS proteins regulate the cellular response to classic oxidative DNA damage, and thus, that such damage contributes significantly to the CS pathologies. Whether atypical oxidative lesions, in addition to the cyclopurines noted above, play a specific role in the etiology of CS, either due to defects in global genome or TC-repair, needs to be addressed in future studies. The fact that CS patients lack cancer predisposition, yet exhibit segmental premature aging phenotypes, has lead to the hypothesis that persistent DNA damage in CSA- or CSB-deficient cells promotes cell death or senescence, likely through a mechanism that is dependent on transcription interference, and not wide-spread mutagenesis, as presumably occurs in XP patients [112].
Fanconi anemia
FA is characterized by congenital malformations, short stature, progressive bone marrow failure, and cancer predisposition (hematological and squamous cell carcinomas) [113]. The predominant role of the FA pathway appears to be in the repair of DNA ICLs, which as described earlier, can be formed from reactions with a number of endogenous products. ICLs, which covalently link both strands of DNA, are particularly detrimental, as they create absolute blocks to the DNA replication and transcription machinery [114]. Such damage must undergo multiple rounds of enzyme action to repair the modification to each strand, with the DNA repair pathways called upon being influenced by the cell cycle phase (Table 4 and Figure 2C). Repair of ICLs is complex and would appear to involve proteins from the FA, NER, DSB repair, and HR pathways, as well as translesion DNA polymerases. Recognition can occur directly in duplex DNA, in some instances by blockage of the transcription machinery [115; 116; 117; 118], or by inhibition of the replisome [119; 120].
Table 4.
The major steps and protein participants in G1 phase and S phase ICL repair.
| ICL REPAIR STEP | PROTEIN | BIOCHEMICAL ACTIVITY | SUBSTRATE | |
|---|---|---|---|---|
| RECOGNITION | G1 Phase | XPC | (See Table 3, general genome NER) | Helix distorting DNA damage |
| MUTSβ (MSH2-MSH3) | DNA binding | DNA Mismatch/insertion-deletion | ||
| S Phase | FANCM/FAAP24 | ATPase/replication fork regression | Collapsed replication fork | |
| FA Core Complex (A,B,C,E,F,G, FAAP100) | DNA damage signaling | FANCM-Replication fork complex | ||
| FANCL + FA Core Complex | Ubiquitin ligase | FANCD2-I | ||
| ICL UNHOOKING | G1 Phase | MLH1-PMS2 | Incision | |
| ERCC1/XPF (ERCC4) | Incision (catalyzed by XPF) | 3′-Flap | ||
| S Phase | ERCC1/XPF (ERCC4) | Incision (catalyzed by XPF) | 3′-Flap | |
| MUS81/EME1 | Incision | 3′-Flap | ||
| FAN1 | Incision | |||
| ICL BYPASS POLYMERIZATION* | REV1 | Deoxycytosine transferase | ICL remnant-containing DNA gap | |
| POLζ | Translesion DNA polymerase | ICL remnant-containing DNA gap | ||
| POLκ | Translesion DNA polymerase | ICL remnant-containing DNA gap | ||
| POLν | Translesion DNA polymerase | ICL remnant-containing DNA gap | ||
| ICL REMNANT REMOVAL | NER | (See Table 3, general genome NER) | Three-stranded ICL remnant | |
| NEIL1 | (See Table 2, glycosylase step) | Three-stranded ICL remnant | ||
Several translesion DNA polymerases may participate in bypass synthesis. In addition, homologous recombination (HR) events and proteins (not listed) are critical in S phase resolution of ICLs and reestablishment of a functional replication fork.
In S phase, ICL detection occurs by stalling, blockage, or collapse of a replication fork [121] or two converging replication forks [122] at the site of the lesion. FANCM acts to regress the blocked/collapsed replication fork(s) [123; 124], and the remainder of the FA core complex (FANCA, B, C, E, F, G, L) functions as a ubiquitin ligase [125; 126; 127] for the FANCD2-I complex [128; 129]. Monoubiquitinated FANCD2-I complex then translocates to chromatin [130; 131], where it associates with additional FA proteins including, FANCD1 (BRCA2)[132], FANCN (PALB2)[133], FANCP (SLX4) [134; 135; 136], and FANCO (Rad51c) [137], and proteins from other DNA metabolic pathways, such as the replication factor PCNA [138], the HR protein Rad51 [139], and the translesion bypass polymerase REV1 [140]. FANCP [141; 142] then coordinates blocked/collapsed replication fork cleavage by the nuclease activities of the Mus81-Eme1 complex [143], the XPF-ERCC1 complex (which also functions in NER) [144], and FAN1 [145; 146; 147]. These incision events generate a DSB/HR intermediate that is stabilized by a collection of FA proteins and processed further to complete unhooking of the ICL from one DNA strand. The translesion activities of REV1 [148; 149], which inserts a C opposite the unhooked crosslink remnant [150; 151], and Polζ probably carry out the repair synthesis step [122]. At this stage, a second round of repair is required to remove the crosslink remnant from the second DNA strand. It is thought that classic NER is responsible for excision of the remaining crosslink remnant, as this intermediate is believed to be viewed by the cellular DNA repair machinery as a bulky monoadduct. This model is supported by previous work demonstrating that inclusion of recombinant XPA protein to cell extracts from XPA-deficient cells supported removal of the unhooked remnant and generated duplex DNA that could be cleaved by a restriction endonuclease [152]. Such data also indicate that NER components play an important role in the removal of additional atypical oxidative lesions, namely ICLs (see above).
Other work examining repair of the ICL remnant has implicated the BER pathway. Specifically, NEIL1 was shown to cleave the glycosidic bond between the psoralen-thymine adduct and deoxyribose sugar in a three-stranded ICL remnant containing substrate, yielding a free thymine base cross-linked to a oligonucleotide and duplex DNA containing a single nucleotide gap. Inclusion of APE1, to remove the 3′-phosphate and a DNA polymerase and ligase resulted in formation of a fully repaired duplex DNA [153]. Additionally, the NEIL1 DNA glycosylase has been shown to excise psoralen induced monoadducts in duplex DNA, with the APE1 endonuclease subsequently able to remove the 3′phosphate generated by NEIL1 AP site incision activity [154]. It has also been observed that the level of NEIL1 in cells is contingent on an intact FA pathway, as cells depleted for FANCA, C, or D2 or FA patient cells from complementation groups A, C, D2, and E have significantly lower NEIL1 protein levels that can be restored, at least for FA-C cells, by expression of the defective FA protein [155].
There is evidence that FA cells are sensitive to oxidative stress induced by non-crosslinking agents and that this sensitivity may contribute to FA pathologies, specifically hematopoeitic failure, suggesting roles for the FA proteins beyond ICL repair. In particular, cultured FA cells have been shown to have oxygen-induced chromosomal aberrations [156], and FA fibroblasts display better growth under hypoxic conditions [157]. FANCC−/− mouse embryonic fibroblasts and hematopoietic progenitor cells have also been found to be hypersensitive to H2O2-induced oxidative stress [158]. Furthermore, FANCC−/− hematopoietic progenitor cells display impaired repair of oxidative DNA damage, as measured by delayed clearance of DNA damage markers (p53Ser20 and γ-H2AX) and increased chromosomal aberrations [159], and senesce prematurely when exposed to repeated hypoxia-reoxygenation cycles [160]. Thus, there is evidence that FA pathologies may arise from defects in the repair of a spectrum of oxidative DNA lesions, including ICLs.
Conclusions
Oxidative damage to DNA can result in a large and diverse spectrum of lesions ranging from simple oxidized bases, cyclic bases, and bases irreversibly physically linked to one another (crosslinks) [161]. Some of these DNA lesions can be tolerated by cells, although mutagenesis, loss of genetic information, and genomic instability can be the molecular consequences. Faithful repair of oxidative DNA damage is thus preferential and requires the activity and interaction of different DNA repair pathways, including BER, NER, and ICL repair, depending on the type and structure of the lesion(s) present (Figure 1). When components of these repair and tolerance pathways are absent or impaired, the accumulation of un- or mis-repaired DNA damage can drive human disease, including carcinogenesis, neuroabnormalities/neurodegeneration, and aging (for additional information on oxidative DNA damage and disease please see [162]). Human hereditary diseases that result because of a loss of DNA repair proteins highlight the importance of the ability to repair DNA damage during development, as well as the penalties associated with the loss of this ability. We have described herein the cooperation between proteins of different repair pathways, via physical and functional interactions, to avoid the mutagenic and genotoxic potential of oxidative DNA damage. We hope to have brought to light two major points in the review: (i) that oxidative DNA damage includes a wide spectrum of lesions, beyond those most commonly described, i.e., classic damages, and (ii) that future research should consider both classic and atypical oxidative DNA damage in their assessment of pathogenic causes, particularly for certain human hereditary diseases, such as XP, CS and FA.
Finally, we note that oxidative DNA damage, such as base modifications, abasic sites, and/or SSBs, can arise in close proximity. In particular, a signature of IR exposure is the formation of clustered DNA damage (a.k.a., multiply damaged sites), which is defined as two or more lesions within 1–2 helical turns of DNA; such clustered damage can also be introduced through the action of certain chemical genotoxins [163]. If the lesions are positioned in close proximity on opposing strands, they can give rise to DNA double-strand breaks. Alternatively, if the lesions are on the same strand, are positioned in an arrangement that blocks repair processing and strand break formation, or are sufficiently apart, they will comprise the larger pool of non-double-strand break clustered lesions that are created. Multiple factors, including the exact location of the lesion(s), the ability of enzymes to carry out processing, and the stage of the cell cycle, will dictate the precise cellular response to the particular lesion constellation. Since the percentage of naturally-occurring endogenous DNA damage that exists in a clustered arrangement is presently unclear, we have not covered this topic herein, but direct you to a few recent reviews that discuss in greater detail the potential repair mechanisms for and contributions of clustered lesions in carcinogenesis [164; 165; 166].
Acknowledgments
This research was supported in part by the Intramural Research Program of the NIH, National Institute on Aging.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Imlay JA, Linn S. DNA damage and oxygen radical toxicity. Science. 1988;240:1302–1309. doi: 10.1126/science.3287616. [DOI] [PubMed] [Google Scholar]
- 2.Mannaerts GP, Van Veldhoven PP. Metabolic pathways in mammalian peroxisomes. Biochimie. 1993;75:147–158. doi: 10.1016/0300-9084(93)90072-z. [DOI] [PubMed] [Google Scholar]
- 3.Shen Z, Wu W, Hazen SL. Activated leukocytes oxidatively damage DNA, RNA, and the nucleotide pool through halide-dependent formation of hydroxyl radical. Biochemistry. 2000;39:5474–5482. doi: 10.1021/bi992809y. [DOI] [PubMed] [Google Scholar]
- 4.Kovacic P, Wakelin LP. Review: DNA molecular electrostatic potential: novel perspectives for the mechanism of action of anticancer drugs involving electron transfer and oxidative stress. Anticancer Drug Des. 2001;16:175–184. [PubMed] [Google Scholar]
- 5.Wood ML, Dizdaroglu M, Gajewski E, Essigmann JM. Mechanistic studies of ionizing radiation and oxidative mutagenesis: genetic effects of a single 8-hydroxyguanine (7-hydro-8-oxoguanine) residue inserted at a unique site in a viral genome. Biochemistry. 1990;29:7024–7032. doi: 10.1021/bi00482a011. [DOI] [PubMed] [Google Scholar]
- 6.Gajewski E, Rao G, Nackerdien Z, Dizdaroglu M. Modification of DNA bases in mammalian chromatin by radiation-generated free radicals. Biochemistry. 1990;29:7876–7882. doi: 10.1021/bi00486a014. [DOI] [PubMed] [Google Scholar]
- 7.Raoul S, Bardet M, Cadet J. Gamma irradiation of 2′-deoxyadenosine in oxygen-free aqueous solutions: identification and conformational features of formamidopyrimidine nucleoside derivatives. Chem Res Toxicol. 1995;8:924–933. doi: 10.1021/tx00049a005. [DOI] [PubMed] [Google Scholar]
- 8.Tremblay S, Douki T, Cadet J, Wagner JR. 2′-Deoxycytidine glycols, a missing link in the free radical-mediated oxidation of DNA. J Biol Chem. 1999;274:20833–20838. doi: 10.1074/jbc.274.30.20833. [DOI] [PubMed] [Google Scholar]
- 9.Dizdaroglu M, Holwitt E, Hagan MP, Blakely WF. Formation of cytosine glycol and 5,6- dihydroxycytosine in deoxyribonucleic acid on treatment with osmium tetroxide. Biochem J. 1986;235:531–536. doi: 10.1042/bj2350531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teoule R, Bert C, Bonicel A. Thymine fragment damage retained in the DNA polynucleotide chain after gamma irradiation in aerated solutions. II. Radiat Res. 1977;72:190–200. [PubMed] [Google Scholar]
- 11.Teebor GW, Frenkel K, Goldstein MS. Ionizing radiation and tritium transmutation both cause formation of 5-hydroxymethyl-2′-deoxyuridine in cellular DNA. Proc Natl Acad Sci U S A. 1984;81:318–321. doi: 10.1073/pnas.81.2.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frenkel K, Cummings A, Solomon J, Cadet J, Steinberg JJ, Teebor GW. Quantitative determination of the 5-(hydroxymethyl)uracil moiety in the DNA of gamma-irradiated cells. Biochemistry. 1985;24:4527–4533. doi: 10.1021/bi00338a007. [DOI] [PubMed] [Google Scholar]
- 13.Lindahl T, Ljungquist S. Apurinic and apyrimidinic sites in DNA. Basic Life Sci. 1975;5A:31–38. doi: 10.1007/978-1-4684-2895-7_5. [DOI] [PubMed] [Google Scholar]
- 14.Decarroz C, Wagner JR, Cadet J. Specific deprotonation reactions of the pyrimidine radical cation resulting from the menadione mediated photosensitization of 2′-deoxycytidine. Free Radic Res Commun. 1987;2:295–301. doi: 10.3109/10715768709065295. [DOI] [PubMed] [Google Scholar]
- 15.Isildar M, Schuchmann MN, Schulte-Frohlinde D, von Sonntag C. gamma-Radiolysis of DNA in oxygenated aqueous solutions: alterations at the sugar moiety. Int J Radiat Biol Relat Stud Phys Chem Med. 1981;40:347–354. doi: 10.1080/09553008114551301. [DOI] [PubMed] [Google Scholar]
- 16.Henner WD, Rodriguez LO, Hecht SM, Haseltine WA. gamma Ray induced deoxyribonucleic acid strand breaks. 3′ Glycolate termini. J Biol Chem. 1983;258:711–713. [PubMed] [Google Scholar]
- 17.Bellon S, Ravanat JL, Gasparutto D, Cadet J. Cross-linked thymine-purine base tandem lesions: synthesis, characterization, and measurement in gamma-irradiated isolated DNA. Chem Res Toxicol. 2002;15:598–606. doi: 10.1021/tx015594d. [DOI] [PubMed] [Google Scholar]
- 18.Raleigh JA, Kremers W, Whitehouse R. Radiation chemistry of nucleotides: 8,5′-cyclonucleotide formation and phosphate release initiated by hydroxyl radical attack on adenosine monophosphates. Radiat Res. 1976;65:414–422. [PubMed] [Google Scholar]
- 19.Miaskiewicz K, Miller JH, Fuciarelli AF. Theoretical analysis of DNA intrastrand cross linking by formation of 8,5′-cyclodeoxyadenosine. Nucleic Acids Res. 1995;23:515–521. doi: 10.1093/nar/23.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minko IG, Kozekov ID, Harris TM, Rizzo CJ, Lloyd RS, Stone MP. Chemistry and biology of DNA containing 1,N(2)-deoxyguanosine adducts of the alpha,beta-unsaturated aldehydes acrolein, crotonaldehyde, and 4-hydroxynonenal. Chem Res Toxicol. 2009;22:759–778. doi: 10.1021/tx9000489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang H, Kozekov ID, Harris TM, Rizzo CJ. Site-specific synthesis and reactivity of oligonucleotides containing stereochemically defined 1,N2-deoxyguanosine adducts of the lipid peroxidation product trans-4-hydroxynonenal. J Am Chem Soc. 2003;125:5687–5700. doi: 10.1021/ja0288800. [DOI] [PubMed] [Google Scholar]
- 22.Blair IA. Lipid hydroperoxide-mediated DNA damage. Exp Gerontol. 2001;36:1473–1481. doi: 10.1016/s0531-5565(01)00133-4. [DOI] [PubMed] [Google Scholar]
- 23.Marnett LJ. Lipid peroxidation-DNA damage by malondialdehyde. Mutat Res. 1999;424:83–95. doi: 10.1016/s0027-5107(99)00010-x. [DOI] [PubMed] [Google Scholar]
- 24.Minetti CA, Remeta DP, Johnson F, Iden CR, Breslauer KJ. Impact of alpha-hydroxypropanodeoxyguanine adducts on DNA duplex energetics: opposite base modulation and implications for mutagenicity and genotoxicity. Biopolymers. 2010;93:370–382. doi: 10.1002/bip.21355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho YJ, Wang H, Kozekov ID, Kurtz AJ, Jacob J, Voehler M, Smith J, Harris TM, Lloyd RS, Rizzo CJ, Stone MP. Stereospecific formation of interstrand carbinolamine DNA cross-links by crotonaldehyde- and acetaldehyde-derived alpha-CH3-gamma-OH-1,N2-propano-2′-deoxyguanosine adducts in the 5′-CpG-3′ sequence. Chem Res Toxicol. 2006;19:195–208. doi: 10.1021/tx050239z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Einolf HJ, Guengerich FP. Fidelity of nucleotide insertion at 8-oxo-7,8-dihydroguanine by mammalian DNA polymerase delta. Steady-state and pre-steady-state kinetic analysis. J Biol Chem. 2001;276:3764–3771. doi: 10.1074/jbc.M006696200. [DOI] [PubMed] [Google Scholar]
- 27.Duarte V, Muller JG, Burrows CJ. Insertion of dGMP and dAMP during in vitro DNA synthesis opposite an oxidized form of 7,8-dihydro-8-oxoguanine. Nucleic Acids Res. 1999;27:496–502. doi: 10.1093/nar/27.2.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sabouri N, Viberg J, Goyal DK, Johansson E, Chabes A. Evidence for lesion bypass by yeast replicative DNA polymerases during DNA damage. Nucleic Acids Res. 2008;36:5660–5667. doi: 10.1093/nar/gkn555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shibutani S, Takeshita M, Grollman AP. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y, Wu X, Rechkoblit O, Geacintov NE, Taylor JS, Wang Z. Response of human REV1 to different DNA damage: preferential dCMP insertion opposite the lesion. Nucleic Acids Res. 2002;30:1630–1638. doi: 10.1093/nar/30.7.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maga G, Villani G, Crespan E, Wimmer U, Ferrari E, Bertocci B, Hubscher U. 8-oxo-guanine bypass by human DNA polymerases in the presence of auxiliary proteins. Nature. 2007;447:606–608. doi: 10.1038/nature05843. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, Wu X, Guo D, Rechkoblit O, Taylor JS, Geacintov NE, Wang Z. Lesion bypass activities of human DNA polymerase mu. J Biol Chem. 2002;277:44582–44587. doi: 10.1074/jbc.M207297200. [DOI] [PubMed] [Google Scholar]
- 33.Kamiya H, Satou K, Hori M, Yamaguchi A, Harashima H. Roles of specialized DNA polymerases in mutagenesis by oxidized guanine. Nucleic Acids Symp Ser (Oxf) 2009:223–224. doi: 10.1093/nass/nrp112. [DOI] [PubMed] [Google Scholar]
- 34.Clark JM, Beardsley GP. Template length, sequence context, and 3′-5′ exonuclease activity modulate replicative bypass of thymine glycol lesions in vitro. Biochemistry. 1989;28:775–779. doi: 10.1021/bi00428a054. [DOI] [PubMed] [Google Scholar]
- 35.Belousova EA, Maga G, Fan Y, Kubareva EA, Romanova EA, Lebedeva NA, Oretskaya TS, Lavrik OI. DNA polymerases beta and lambda bypass thymine glycol in gapped DNA structures. Biochemistry. 2010;49:4695–4704. doi: 10.1021/bi901792c. [DOI] [PubMed] [Google Scholar]
- 36.Kusumoto R, Masutani C, Iwai S, Hanaoka F. Translesion synthesis by human DNA polymerase eta across thymine glycol lesions. Biochemistry. 2002;41:6090–6099. doi: 10.1021/bi025549k. [DOI] [PubMed] [Google Scholar]
- 37.Fischhaber PL, Gerlach VL, Feaver WJ, Hatahet Z, Wallace SS, Friedberg EC. Human DNA polymerase kappa bypasses and extends beyond thymine glycols during translesion synthesis in vitro, preferentially incorporating correct nucleotides. J Biol Chem. 2002;277:37604–37611. doi: 10.1074/jbc.M206027200. [DOI] [PubMed] [Google Scholar]
- 38.Yoon JH, Bhatia G, Prakash S, Prakash L. Error-free replicative bypass of thymine glycol by the combined action of DNA polymerases kappa and zeta in human cells. Proc Natl Acad Sci U S A. 2010;107:14116–14121. doi: 10.1073/pnas.1007795107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seki M, Masutani C, Yang LW, Schuffert A, Iwai S, Bahar I, Wood RD. High-efficiency bypass of DNA damage by human DNA polymerase Q. EMBO J. 2004;23:4484–4494. doi: 10.1038/sj.emboj.7600424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takata K, Shimizu T, Iwai S, Wood RD. Human DNA polymerase N (POLN) is a low fidelity enzyme capable of error-free bypass of 5S-thymine glycol. J Biol Chem. 2006;281:23445–23455. doi: 10.1074/jbc.M604317200. [DOI] [PubMed] [Google Scholar]
- 41.Vaisman A, Woodgate R. Unique misinsertion specificity of poliota may decrease the mutagenic potential of deaminated cytosines. EMBO J. 2001;20:6520–6529. doi: 10.1093/emboj/20.22.6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choi JY, Lim S, Kim EJ, Jo A, Guengerich FP. Translesion synthesis across abasic lesions by human B-family and Y-family DNA polymerases alpha, delta, eta, iota, kappa, and REV1. J Mol Biol. 2010;404:34–44. doi: 10.1016/j.jmb.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Colis LC, Raychaudhury P, Basu AK. Mutational specificity of gamma-radiation-induced guanine-thymine and thymine-guanine intrastrand cross-links in mammalian cells and translesion synthesis past the guanine-thymine lesion by human DNA polymerase eta. Biochemistry. 2008;47:8070–8079. doi: 10.1021/bi800529f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuraoka I, Robins P, Masutani C, Hanaoka F, Gasparutto D, Cadet J, Wood RD, Lindahl T. Oxygen free radical damage to DNA. Translesion synthesis by human DNA polymerase eta and resistance to exonuclease action at cyclopurine deoxynucleoside residues. J Biol Chem. 2001;276:49283–49288. doi: 10.1074/jbc.M107779200. [DOI] [PubMed] [Google Scholar]
- 45.Perrino FW, Harvey S, Blans P, Gelhaus S, Lacourse WR, Fishbein JC. Polymerization past the N2-isopropylguanine and the N6-isopropyladenine DNA lesions with the translesion synthesis DNA polymerases eta and iota and the replicative DNA polymerase alpha. Chem Res Toxicol. 2005;18:1451–1461. doi: 10.1021/tx050114u. [DOI] [PubMed] [Google Scholar]
- 46.Hang B, Chenna A, Guliaev AB, Singer B. Miscoding properties of 1,N6-ethanoadenine, a DNA adduct derived from reaction with the antitumor agent 1,3-bis(2-chloroethyl)-1-nitrosourea. Mutat Res. 2003;531:191–203. doi: 10.1016/j.mrfmmm.2003.07.006. [DOI] [PubMed] [Google Scholar]
- 47.Choi JY, Zang H, Angel KC, Kozekov ID, Goodenough AK, Rizzo CJ, Guengerich FP. Translesion synthesis across 1,N2-ethenoguanine by human DNA polymerases. Chem Res Toxicol. 2006;19:879–886. doi: 10.1021/tx060051v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Minko IG, Washington MT, Kanuri M, Prakash L, Prakash S, Lloyd RS. Translesion synthesis past acrolein-derived DNA adduct, gamma -hydroxypropanodeoxyguanosine, by yeast and human DNA polymerase eta. J Biol Chem. 2003;278:784–790. doi: 10.1074/jbc.M207774200. [DOI] [PubMed] [Google Scholar]
- 49.Minko IG, Harbut MB, Kozekov ID, Kozekova A, Jakobs PM, Olson SB, Moses RE, Harris TM, Rizzo CJ, Lloyd RS. Role for DNA polymerase kappa in the processing of N2-N2-guanine interstrand cross-links. J Biol Chem. 2008;283:17075–17082. doi: 10.1074/jbc.M801238200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Minko IG, Kozekov ID, Kozekova A, Harris TM, Rizzo CJ, Lloyd RS. Mutagenic potential of DNA-peptide crosslinks mediated by acrolein-derived DNA adducts. Mutat Res. 2008;637:161–172. doi: 10.1016/j.mrfmmm.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Charlet-Berguerand N, Feuerhahn S, Kong SE, Ziserman H, Conaway JW, Conaway R, Egly JM. RNA polymerase II bypass of oxidative DNA damage is regulated by transcription elongation factors. EMBO J. 2006;25:5481–5491. doi: 10.1038/sj.emboj.7601403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bregeon D, Peignon PA, Sarasin A. Transcriptional mutagenesis induced by 8-oxoguanine in mammalian cells. PLoS Genet. 2009;5:e1000577. doi: 10.1371/journal.pgen.1000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saxowsky TT, Meadows KL, Klungland A, Doetsch PW. 8-Oxoguanine-mediated transcriptional mutagenesis causes Ras activation in mammalian cells. Proc Natl Acad Sci U S A. 2008;105:18877–18882. doi: 10.1073/pnas.0806464105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marietta C, Brooks PJ. Transcriptional bypass of bulky DNA lesions causes new mutant RNA transcripts in human cells. EMBO Rep. 2007;8:388–393. doi: 10.1038/sj.embor.7400932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cheng TF, Hu X, Gnatt A, Brooks PJ. Differential blocking effects of the acetaldehyde-derived DNA lesion N2-ethyl-2′-deoxyguanosine on transcription by multisubunit and single subunit RNA polymerases. J Biol Chem. 2008;283:27820–27828. doi: 10.1074/jbc.M804086200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brooks PJ, Cheng TF, Cooper L. Do all of the neurologic diseases in patients with DNA repair gene mutations result from the accumulation of DNA damage? DNA Repair (Amst) 2008;7:834–848. doi: 10.1016/j.dnarep.2008.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sobol RW, Horton JK, Kuhn R, Gu H, Singhal RK, Prasad R, Rajewsky K, Wilson SH. Requirement of mammalian DNA polymerase-beta in base-excision repair. Nature. 1996;379:183–186. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- 58.Meira LB, Devaraj S, Kisby GE, Burns DK, Daniel RL, Hammer RE, Grundy S, Jialal I, Friedberg EC. Heterozygosity for the mouse Apex gene results in phenotypes associated with oxidative stress. Cancer Res. 2001;61:5552–5557. [PubMed] [Google Scholar]
- 59.Tebbs RS, Flannery ML, Meneses JJ, Hartmann A, Tucker JD, Thompson LH, Cleaver JE, Pedersen RA. Requirement for the Xrcc1 DNA base excision repair gene during early mouse development. Dev Biol. 1999;208:513–529. doi: 10.1006/dbio.1999.9232. [DOI] [PubMed] [Google Scholar]
- 60.Bentley D, Selfridge J, Millar JK, Samuel K, Hole N, Ansell JD, Melton DW. DNA ligase I is required for fetal liver erythropoiesis but is not essential for mammalian cell viability. Nat Genet. 1996;13:489–491. doi: 10.1038/ng0896-489. [DOI] [PubMed] [Google Scholar]
- 61.Puebla-Osorio N, Lacey DB, Alt FW, Zhu C. Early embryonic lethality due to targeted inactivation of DNA ligase III. Mol Cell Biol. 2006;26:3935–3941. doi: 10.1128/MCB.26.10.3935-3941.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Klungland A, Rosewell I, Hollenbach S, Larsen E, Daly G, Epe B, Seeberg E, Lindahl T, Barnes DE. Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc Natl Acad Sci U S A. 1999;96:13300–13305. doi: 10.1073/pnas.96.23.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xie Y, Yang H, Cunanan C, Okamoto K, Shibata D, Pan J, Barnes DE, Lindahl T, McIlhatton M, Fishel R, Miller JH. Deficiencies in mouse Myh and Ogg1 result in tumor predisposition and G to T mutations in codon 12 of the K-ras oncogene in lung tumors. Cancer Res. 2004;64:3096–3102. doi: 10.1158/0008-5472.can-03-3834. [DOI] [PubMed] [Google Scholar]
- 64.Russo MT, De Luca G, Degan P, Parlanti E, Dogliotti E, Barnes DE, Lindahl T, Yang H, Miller JH, Bignami M. Accumulation of the oxidative base lesion 8-hydroxyguanine in DNA of tumor-prone mice defective in both the Myh and Ogg1 DNA glycosylases. Cancer Res. 2004;64:4411–4414. doi: 10.1158/0008-5472.CAN-04-0355. [DOI] [PubMed] [Google Scholar]
- 65.Takao M, Kanno S, Shiromoto T, Hasegawa R, Ide H, Ikeda S, Sarker AH, Seki S, Xing JZ, Le XC, Weinfeld M, Kobayashi K, Miyazaki J, Muijtjens M, Hoeijmakers JH, van der Horst G, Yasui A. Novel nuclear and mitochondrial glycosylases revealed by disruption of the mouse Nth1 gene encoding an endonuclease III homolog for repair of thymine glycols. EMBO J. 2002;21:3486–3493. doi: 10.1093/emboj/cdf350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ocampo MT, Chaung W, Marenstein DR, Chan MK, Altamirano A, Basu AK, Boorstein RJ, Cunningham RP, Teebor GW. Targeted deletion of mNth1 reveals a novel DNA repair enzyme activity. Mol Cell Biol. 2002;22:6111–6121. doi: 10.1128/MCB.22.17.6111-6121.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vartanian V, Lowell B, Minko IG, Wood TG, Ceci JD, George S, Ballinger SW, Corless CL, McCullough AK, Lloyd RS. The metabolic syndrome resulting from a knockout of the NEIL1 DNA glycosylase. Proc Natl Acad Sci U S A. 2006;103:1864–1869. doi: 10.1073/pnas.0507444103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chan MK, Ocampo-Hafalla MT, Vartanian V, Jaruga P, Kirkali G, Koenig KL, Brown S, Lloyd RS, Dizdaroglu M, Teebor GW. Targeted deletion of the genes encoding NTH1 and NEIL1 DNA N-glycosylases reveals the existence of novel carcinogenic oxidative damage to DNA. DNA Repair (Amst) 2009;8:786–794. doi: 10.1016/j.dnarep.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huamani J, McMahan CA, Herbert DC, Reddick R, McCarrey JR, MacInnes MI, Chen DJ, Walter CA. Spontaneous mutagenesis is enhanced in Apex heterozygous mice. Mol Cell Biol. 2004;24:8145–8153. doi: 10.1128/MCB.24.18.8145-8153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cabelof DC, Guo Z, Raffoul JJ, Sobol RW, Wilson SH, Richardson A, Heydari AR. Base excision repair deficiency caused by polymerase beta haploinsufficiency: accelerated DNA damage and increased mutational response to carcinogens. Cancer Res. 2003;63:5799–5807. [PubMed] [Google Scholar]
- 71.Cabelof DC, Ikeno Y, Nyska A, Busuttil RA, Anyangwe N, Vijg J, Matherly LH, Tucker JD, Wilson SH, Richardson A, Heydari AR. Haploinsufficiency in DNA polymerase beta increases cancer risk with age and alters mortality rate. Cancer Res. 2006;66:7460–7465. doi: 10.1158/0008-5472.CAN-06-1177. [DOI] [PubMed] [Google Scholar]
- 72.McNeill DR, Lin PC, Miller MG, Pistell PJ, de Souza-Pinto NC, Fishbein KW, Spencer RG, Liu Y, Pettan-Brewer C, Ladiges WC, Wilson DM., 3rd XRCC1 haploinsufficiency in mice has little effect on aging, but adversely modifies exposure-dependent susceptibility. Nucleic Acids Res. 2011;39:7992–8004. doi: 10.1093/nar/gkr280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Baglioni S, Melean G, Gensini F, Santucci M, Scatizzi M, Papi L, Genuardi M. A kindred with MYH-associated polyposis and pilomatricomas. Am J Med Genet A. 2005;134A:212–214. doi: 10.1002/ajmg.a.30585. [DOI] [PubMed] [Google Scholar]
- 74.Imai K, Slupphaug G, Lee WI, Revy P, Nonoyama S, Catalan N, Yel L, Forveille M, Kavli B, Krokan HE, Ochs HD, Fischer A, Durandy A. Human uracil-DNA glycosylase deficiency associated with profoundly impaired immunoglobulin class-switch recombination. Nat Immunol. 2003;4:1023–1028. doi: 10.1038/ni974. [DOI] [PubMed] [Google Scholar]
- 75.Kavli B, Andersen S, Otterlei M, Liabakk NB, Imai K, Fischer A, Durandy A, Krokan HE, Slupphaug G. B cells from hyper-IgM patients carrying UNG mutations lack ability to remove uracil from ssDNA and have elevated genomic uracil. J Exp Med. 2005;201:2011–2021. doi: 10.1084/jem.20050042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wilson DM, 3rd, Kim D, Berquist BR, Sigurdson AJ. Variation in base excision repair capacity. Mutat Res. 2011;711:100–112. doi: 10.1016/j.mrfmmm.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nemec AA, Wallace SS, Sweasy JB. Variant base excision repair proteins: contributors to genomic instability. Semin Cancer Biol. 2010;20:320–328. doi: 10.1016/j.semcancer.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Takashima H, Boerkoel CF, John J, Saifi GM, Salih MA, Armstrong D, Mao Y, Quiocho FA, Roa BB, Nakagawa M, Stockton DW, Lupski JR. Mutation of TDP1, encoding a topoisomerase I-dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nat Genet. 2002;32:267–272. doi: 10.1038/ng987. [DOI] [PubMed] [Google Scholar]
- 79.Interthal H, Pouliot JJ, Champoux JJ. The tyrosyl-DNA phosphodiesterase Tdp1 is a member of the phospholipase D superfamily. Proc Natl Acad Sci U S A. 2001;98:12009–12014. doi: 10.1073/pnas.211429198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Inamdar KV, Pouliot JJ, Zhou T, Lees-Miller SP, Rasouli-Nia A, Povirk LF. Conversion of phosphoglycolate to phosphate termini on 3′ overhangs of DNA double strand breaks by the human tyrosyl-DNA phosphodiesterase hTdp1. J Biol Chem. 2002;277:27162–27168. doi: 10.1074/jbc.M204688200. [DOI] [PubMed] [Google Scholar]
- 81.Walton C, Interthal H, Hirano R, Salih MA, Takashima H, Boerkoel CF. Spinocerebellar ataxia with axonal neuropathy. Adv Exp Med Biol. 2010;685:75–83. doi: 10.1007/978-1-4419-6448-9_7. [DOI] [PubMed] [Google Scholar]
- 82.Shen J, Gilmore EC, Marshall CA, Haddadin M, Reynolds JJ, Eyaid W, Bodell A, Barry B, Gleason D, Allen K, Ganesh VS, Chang BS, Grix A, Hill RS, Topcu M, Caldecott KW, Barkovich AJ, Walsh CA. Mutations in PNKP cause microcephaly, seizures and defects in DNA repair. Nat Genet. 2010;42:245–249. doi: 10.1038/ng.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jilani A, Ramotar D, Slack C, Ong C, Yang XM, Scherer SW, Lasko DD. Molecular cloning of the human gene, PNKP, encoding a polynucleotide kinase 3′-phosphatase and evidence for its role in repair of DNA strand breaks caused by oxidative damage. J Biol Chem. 1999;274:24176–24186. doi: 10.1074/jbc.274.34.24176. [DOI] [PubMed] [Google Scholar]
- 84.Moreira MC, Barbot C, Tachi N, Kozuka N, Uchida E, Gibson T, Mendonca P, Costa M, Barros J, Yanagisawa T, Watanabe M, Ikeda Y, Aoki M, Nagata T, Coutinho P, Sequeiros J, Koenig M. The gene mutated in ataxia-ocular apraxia 1 encodes the new HIT/Zn-finger protein aprataxin. Nat Genet. 2001;29:189–193. doi: 10.1038/ng1001-189. [DOI] [PubMed] [Google Scholar]
- 85.Date H, Onodera O, Tanaka H, Iwabuchi K, Uekawa K, Igarashi S, Koike R, Hiroi T, Yuasa T, Awaya Y, Sakai T, Takahashi T, Nagatomo H, Sekijima Y, Kawachi I, Takiyama Y, Nishizawa M, Fukuhara N, Saito K, Sugano S, Tsuji S. Early-onset ataxia with ocular motor apraxia and hypoalbuminemia is caused by mutations in a new HIT superfamily gene. Nat Genet. 2001;29:184–188. doi: 10.1038/ng1001-184. [DOI] [PubMed] [Google Scholar]
- 86.Ahel I, Rass U, El-Khamisy SF, Katyal S, Clements PM, McKinnon PJ, Caldecott KW, West SC. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature. 2006;443:713–716. doi: 10.1038/nature05164. [DOI] [PubMed] [Google Scholar]
- 87.Coutinho P, Barbot C. In: Ataxia with Oculomotor Apraxia Type 1. Pagon RA, Bird TD, Dolan CR, Stephens K, editors. GeneReviews; Seattle (WA): 1993. [Google Scholar]
- 88.Lehmann AR, McGibbon D, Stefanini M. Xeroderma pigmentosum. Orphanet J Rare Dis. 2011;6:70. doi: 10.1186/1750-1172-6-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kraemer KH, DiGiovanna JJ. In: Xeroderma Pigmentosum. Pagon RA, Bird TD, Dolan CR, Stephens K, editors. GeneReviews; Seattle (WA): 1993. [Google Scholar]
- 90.Su X, Huang J. The Fanconi anemia pathway and DNA interstrand cross-link repair. Protein Cell. 2011;2:704–711. doi: 10.1007/s13238-011-1098-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kamenisch Y, Berneburg M. Progeroid syndromes and UV-induced oxidative DNA damage. J Investig Dermatol Symp Proc. 2009;14:8–14. doi: 10.1038/jidsymp.2009.6. [DOI] [PubMed] [Google Scholar]
- 92.Bradford PT, Goldstein AM, Tamura D, Khan SG, Ueda T, Boyle J, Oh KS, Imoto K, Inui H, Moriwaki S, Emmert S, Pike KM, Raziuddin A, Plona TM, DiGiovanna JJ, Tucker MA, Kraemer KH. Cancer and neurologic degeneration in xeroderma pigmentosum: long term follow-up characterises the role of DNA repair. J Med Genet. 2011;48:168–176. doi: 10.1136/jmg.2010.083022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Brooks PJ. The 8,5′-cyclopurine-2′-deoxynucleosides: candidate neurodegenerative DNA lesions in xeroderma pigmentosum, and unique probes of transcription and nucleotide excision repair. DNA Repair (Amst) 2008;7:1168–1179. doi: 10.1016/j.dnarep.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gopalakrishnan K, Low GK, Ting AP, Srikanth P, Slijepcevic P, Hande MP. Hydrogen peroxide induced genomic instability in nucleotide excision repair-deficient lymphoblastoid cells. Genome Integr. 2010;1:16. doi: 10.1186/2041-9414-1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Low GK, Fok ED, Ting AP, Hande MP. Oxidative damage induced genotoxic effects in human fibroblasts from Xeroderma Pigmentosum group A patients. Int J Biochem Cell Biol. 2008;40:2583–2595. doi: 10.1016/j.biocel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 96.D′Errico M, Parlanti E, Teson M, de Jesus BM, Degan P, Calcagnile A, Jaruga P, Bjoras M, Crescenzi M, Pedrini AM, Egly JM, Zambruno G, Stefanini M, Dizdaroglu M, Dogliotti E. New functions of XPC in the protection of human skin cells from oxidative damage. EMBO J. 2006;25:4305–4315. doi: 10.1038/sj.emboj.7601277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Brooks PJ. The case for 8,5′-cyclopurine-2′-deoxynucleosides as endogenous DNA lesions that cause neurodegeneration in xeroderma pigmentosum. Neuroscience. 2007;145:1407–1417. doi: 10.1016/j.neuroscience.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bessho T. Nucleotide excision repair 3′ endonuclease XPG stimulates the activity of base excision repairenzyme thymine glycol DNA glycosylase. Nucleic Acids Res. 1999;27:979–983. doi: 10.1093/nar/27.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Klungland A, Hoss M, Gunz D, Constantinou A, Clarkson SG, Doetsch PW, Bolton PH, Wood RD, Lindahl T. Base excision repair of oxidative DNA damage activated by XPG protein. Mol Cell. 1999;3:33–42. doi: 10.1016/s1097-2765(00)80172-0. [DOI] [PubMed] [Google Scholar]
- 100.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 101.Cramers P, Verhoeven EE, Filon AR, Rockx DA, Santos SJ, van der Leer AA, Kleinjans JC, van Zeeland AA, Mullenders LH. Impaired repair of ionizing radiation-induced DNA damage in Cockayne syndrome cells. Radiat Res. 2011;175:432–443. doi: 10.1667/RR1972.1. [DOI] [PubMed] [Google Scholar]
- 102.Tuo J, Jaruga P, Rodriguez H, Bohr VA, Dizdaroglu M. Primary fibroblasts of Cockayne syndrome patients are defective in cellular repair of 8-hydroxyguanine and 8-hydroxyadenine resulting from oxidative stress. FASEB J. 2003;17:668–674. doi: 10.1096/fj.02-0851com. [DOI] [PubMed] [Google Scholar]
- 103.Tuo J, Jaruga P, Rodriguez H, Dizdaroglu M, Bohr VA. The cockayne syndrome group B gene product is involved in cellular repair of 8-hydroxyadenine in DNA. J Biol Chem. 2002;277:30832–30837. doi: 10.1074/jbc.M204814200. [DOI] [PubMed] [Google Scholar]
- 104.Tuo J, Muftuoglu M, Chen C, Jaruga P, Selzer RR, Brosh RM, Jr, Rodriguez H, Dizdaroglu M, Bohr VA. The Cockayne Syndrome group B gene product is involved in general genome base excision repair of 8-hydroxyguanine in DNA. J Biol Chem. 2001;276:45772–45779. doi: 10.1074/jbc.M107888200. [DOI] [PubMed] [Google Scholar]
- 105.Spivak G, Hanawalt PC. Host cell reactivation of plasmids containing oxidative DNA lesions is defective in Cockayne syndrome but normal in UV-sensitive syndrome fibroblasts. DNA Repair (Amst) 2006;5:13–22. doi: 10.1016/j.dnarep.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 106.Ropolo M, Degan P, Foresta M, D’Errico M, Lasiglie D, Dogliotti E, Casartelli G, Zupo S, Poggi A, Frosina G. Complementation of the oxidatively damaged DNA repair defect in Cockayne syndrome A and B cells by Escherichia coli formamidopyrimidine DNA glycosylase. Free Radic Biol Med. 2007;42:1807–1817. doi: 10.1016/j.freeradbiomed.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 107.D’Errico M, Parlanti E, Teson M, Degan P, Lemma T, Calcagnile A, Iavarone I, Jaruga P, Ropolo M, Pedrini AM, Orioli D, Frosina G, Zambruno G, Dizdaroglu M, Stefanini M, Dogliotti E. The role of CSA in the response to oxidative DNA damage in human cells. Oncogene. 2007;26:4336–4343. doi: 10.1038/sj.onc.1210232. [DOI] [PubMed] [Google Scholar]
- 108.Osenbroch PO, Auk-Emblem P, Halsne R, Strand J, Forstrom RJ, van der Pluijm I, Eide L. Accumulation of mitochondrial DNA damage and bioenergetic dysfunction in CSB defective cells. FEBS J. 2009;276:2811–2821. doi: 10.1111/j.1742-4658.2009.07004.x. [DOI] [PubMed] [Google Scholar]
- 109.Muftuoglu M, de Souza-Pinto NC, Dogan A, Aamann M, Stevnsner T, Rybanska I, Kirkali G, Dizdaroglu M, Bohr VA. Cockayne syndrome group B protein stimulates repair of formamidopyrimidines by NEIL1 DNA glycosylase. J Biol Chem. 2009;284:9270–9279. doi: 10.1074/jbc.M807006200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Thorslund T, von Kobbe C, Harrigan JA, Indig FE, Christiansen M, Stevnsner T, Bohr VA. Cooperation of the Cockayne syndrome group B protein and poly(ADP-ribose) polymerase 1 in the response to oxidative stress. Mol Cell Biol. 2005;25:7625–7636. doi: 10.1128/MCB.25.17.7625-7636.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wong HK, Muftuoglu M, Beck G, Imam SZ, Bohr VA, Wilson DM., 3rd Cockayne syndrome B protein stimulates apurinic endonuclease 1 activity and protects against agents that introduce base excision repair intermediates. Nucleic Acids Res. 2007;35:4103–4113. doi: 10.1093/nar/gkm404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hoeijmakers JH. DNA damage, aging, and cancer. N Engl J Med. 2009;361:1475–1485. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- 113.D’Andrea AD. Susceptibility pathways in Fanconi’s anemia and breast cancer. N Engl J Med. 2010;362:1909–1919. doi: 10.1056/NEJMra0809889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Deans AJ, West SC. DNA interstrand crosslink repair and cancer. Nat Rev Cancer. 2011;11:467–480. doi: 10.1038/nrc3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jones JC, Zhen WP, Reed E, Parker RJ, Sancar A, Bohr VA. Gene-specific formation and repair of cisplatin intrastrand adducts and interstrand cross-links in Chinese hamster ovary cells. J Biol Chem. 1991;266:7101–7107. [PubMed] [Google Scholar]
- 116.May A, Nairn RS, Okumoto DS, Wassermann K, Stevnsner T, Jones JC, Bohr VA. Repair of individual DNA strands in the hamster dihydrofolate reductase gene after treatment with ultraviolet light, alkylating agents, and cisplatin. J Biol Chem. 1993;268:1650–1657. [PubMed] [Google Scholar]
- 117.Islas AL, Baker FJ, Hanawalt PC. Transcription-coupled repair of psoralen cross-links but not monoadducts in Chinese hamster ovary cells. Biochemistry. 1994;33:10794–10799. doi: 10.1021/bi00201a029. [DOI] [PubMed] [Google Scholar]
- 118.Vos JM, Hanawalt PC. Processing of psoralen adducts in an active human gene: repair and replication of DNA containing monoadducts and interstrand cross-links. Cell. 1987;50:789–799. 27. doi: 10.1016/0092-8674(87)90337-0. [DOI] [PubMed] [Google Scholar]
- 119.Sala-Trepat M, Rouillard D, Escarceller M, Laquerbe A, Moustacchi E, Papadopoulo D. Arrest of S-phase progression is impaired in Fanconi anemia cells. Exp Cell Res. 2000;260:208–215. doi: 10.1006/excr.2000.4994. [DOI] [PubMed] [Google Scholar]
- 120.Rothfuss A, Grompe M. Repair kinetics of genomic interstrand DNA cross-links: evidence for DNA double-strand break-dependent activation of the Fanconi anemia/BRCA pathway. Mol Cell Biol. 2004;24:123–134. doi: 10.1128/MCB.24.1.123-134.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.McHugh PJ, Sones WR, Hartley JA. Repair of intermediate structures produced at DNA interstrand cross-links in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:3425–3433. doi: 10.1128/mcb.20.10.3425-3433.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Raschle M, Knipscheer P, Enoiu M, Angelov T, Sun J, Griffith JD, Ellenberger TE, Scharer OD, Walter JC. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell. 2008;134:969–980. doi: 10.1016/j.cell.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gari K, Decaillet C, Delannoy M, Wu L, Constantinou A. Remodeling of DNA replication structures by the branch point translocase FANCM. Proc Natl Acad Sci U S A. 2008;105:16107–16112. doi: 10.1073/pnas.0804777105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gari K, Decaillet C, Stasiak AZ, Stasiak A, Constantinou A. The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks. Mol Cell. 2008;29:141–148. doi: 10.1016/j.molcel.2007.11.032. [DOI] [PubMed] [Google Scholar]
- 125.Meetei AR, de Winter JP, Medhurst AL, Wallisch M, Waisfisz Q, van de Vrugt HJ, Oostra AB, Yan Z, Ling C, Bishop CE, Hoatlin ME, Joenje H, Wang W. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat Genet. 2003;35:165–170. doi: 10.1038/ng1241. [DOI] [PubMed] [Google Scholar]
- 126.Meetei AR, Yan Z, Wang W. FANCL replaces BRCA1 as the likely ubiquitin ligase responsible for FANCD2 monoubiquitination. Cell Cycle. 2004;3:179–181. [PubMed] [Google Scholar]
- 127.Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, Grompe M, D’Andrea AD. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 128.Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER, 3rd, Hurov KE, Luo J, Ballif BA, Gygi SP, Hofmann K, D’Andrea AD, Elledge SJ. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sims AE, Spiteri E, Sims RJ, 3rd, Arita AG, Lach FP, Landers T, Wurm M, Freund M, Neveling K, Hanenberg H, Auerbach AD, Huang TT. FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nat Struct Mol Biol. 2007;14:564–567. doi: 10.1038/nsmb1252. [DOI] [PubMed] [Google Scholar]
- 130.Yuan F, El Hokayem J, Zhou W, Zhang Y. FANCI protein binds to DNA and interacts with FANCD2 to recognize branched structures. J Biol Chem. 2009;284:24443–24452. doi: 10.1074/jbc.M109.016006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Longerich S, San Filippo J, Liu D, Sung P. FANCI binds branched DNA and is monoubiquitinated by UBE2T-FANCL. J Biol Chem. 2009;284:23182–23186. doi: 10.1074/jbc.C109.038075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Howlett NG, Taniguchi T, Olson S, Cox B, Waisfisz Q, De Die-Smulders C, Persky N, Grompe M, Joenje H, Pals G, Ikeda H, Fox EA, D’Andrea AD. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297:606–609. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- 133.Reid S, Schindler D, Hanenberg H, Barker K, Hanks S, Kalb R, Neveling K, Kelly P, Seal S, Freund M, Wurm M, Batish SD, Lach FP, Yetgin S, Neitzel H, Ariffin H, Tischkowitz M, Mathew CG, Auerbach AD, Rahman N. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet. 2007;39:162–164. doi: 10.1038/ng1947. [DOI] [PubMed] [Google Scholar]
- 134.Kim Y, Lach FP, Desetty R, Hanenberg H, Auerbach AD, Smogorzewska A. Mutations of the SLX4 gene in Fanconi anemia. Nat Genet. 2011;43:142–146. doi: 10.1038/ng.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Crossan GP, van der Weyden L, Rosado IV, Langevin F, Gaillard PH, McIntyre RE, Gallagher F, Kettunen MI, Lewis DY, Brindle K, Arends MJ, Adams DJ, Patel KJ. Disruption of mouse Slx4, a regulator of structure-specific nucleases, phenocopies Fanconi anemia. Nat Genet. 2011;43:147–152. doi: 10.1038/ng.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Stoepker C, Hain K, Schuster B, Hilhorst-Hofstee Y, Rooimans MA, Steltenpool J, Oostra AB, Eirich K, Korthof ET, Nieuwint AW, Jaspers NG, Bettecken T, Joenje H, Schindler D, Rouse J, de Winter JP. SLX4, a coordinator of structure-specific endonucleases, is mutated in a new Fanconi anemia subtype. Nat Genet. 2011;43:138–141. doi: 10.1038/ng.751. [DOI] [PubMed] [Google Scholar]
- 137.Vaz F, Hanenberg H, Schuster B, Barker K, Wiek C, Erven V, Neveling K, Endt D, Kesterton I, Autore F, Fraternali F, Freund M, Hartmann L, Grimwade D, Roberts RG, Schaal H, Mohammed S, Rahman N, Schindler D, Mathew CG. Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat Genet. 2010;42:406–409. doi: 10.1038/ng.570. [DOI] [PubMed] [Google Scholar]
- 138.Hussain S, Wilson JB, Medhurst AL, Hejna J, Witt E, Ananth S, Davies A, Masson JY, Moses R, West SC, de Winter JP, Ashworth A, Jones NJ, Mathew CG. Direct interaction of FANCD2 with BRCA2 in DNA damage response pathways. Hum Mol Genet. 2004;13:1241–1248. doi: 10.1093/hmg/ddh135. [DOI] [PubMed] [Google Scholar]
- 139.Digweed M, Rothe S, Demuth I, Scholz R, Schindler D, Stumm M, Grompe M, Jordan A, Sperling K. Attenuation of the formation of DNA-repair foci containing RAD51 in Fanconi anaemia. Carcinogenesis. 2002;23:1121–1126. doi: 10.1093/carcin/23.7.1121. [DOI] [PubMed] [Google Scholar]
- 140.Howlett NG, Taniguchi T, Durkin SG, D’Andrea AD, Glover TW. The Fanconi anemia pathway is required for the DNA replication stress response and for the regulation of common fragile site stability. Hum Mol Genet. 2005;14:693–701. doi: 10.1093/hmg/ddi065. [DOI] [PubMed] [Google Scholar]
- 141.Yamamoto KN, Kobayashi S, Tsuda M, Kurumizaka H, Takata M, Kono K, Jiricny J, Takeda S, Hirota K. Involvement of SLX4 in interstrand cross-link repair is regulated by the Fanconi anemia pathway. Proc Natl Acad Sci U S A. 2011;108:6492–6496. doi: 10.1073/pnas.1018487108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Munoz IM, Hain K, Declais AC, Gardiner M, Toh GW, Sanchez-Pulido L, Heuckmann JM, Toth R, Macartney T, Eppink B, Kanaar R, Ponting CP, Lilley DM, Rouse J. Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Mol Cell. 2009;35:116–127. doi: 10.1016/j.molcel.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 143.Hanada K, Budzowska M, Modesti M, Maas A, Wyman C, Essers J, Kanaar R. The structure-specific endonuclease Mus81-Eme1 promotes conversion of interstrand DNA crosslinks into double-strands breaks. EMBO J. 2006;25:4921–4932. doi: 10.1038/sj.emboj.7601344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bhagwat N, Olsen AL, Wang AT, Hanada K, Stuckert P, Kanaar R, D’Andrea A, Niedernhofer LJ, McHugh PJ. XPF-ERCC1 participates in the Fanconi anemia pathway of cross-link repair. Mol Cell Biol. 2009;29:6427–6437. doi: 10.1128/MCB.00086-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.MacKay C, Declais AC, Lundin C, Agostinho A, Deans AJ, MacArtney TJ, Hofmann K, Gartner A, West SC, Helleday T, Lilley DM, Rouse J. Identification of KIAA1018/FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2. Cell. 2010;142:65–76. doi: 10.1016/j.cell.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kratz K, Schopf B, Kaden S, Sendoel A, Eberhard R, Lademann C, Cannavo E, Sartori AA, Hengartner MO, Jiricny J. Deficiency of FANCD2-associated nuclease KIAA1018/FAN1 sensitizes cells to interstrand crosslinking agents. Cell. 2010;142:77–88. doi: 10.1016/j.cell.2010.06.022. [DOI] [PubMed] [Google Scholar]
- 147.Smogorzewska A, Desetty R, Saito TT, Schlabach M, Lach FP, Sowa ME, Clark AB, Kunkel TA, Harper JW, Colaiacovo MP, Elledge SJ. A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol Cell. 2010;39:36–47. doi: 10.1016/j.molcel.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Mirchandani KD, McCaffrey RM, D’Andrea AD. The Fanconi anemia core complex is required for efficient point mutagenesis and Rev1 foci assembly. DNA Repair (Amst) 2008;7:902–911. doi: 10.1016/j.dnarep.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hicks JK, Chute CL, Paulsen MT, Ragland RL, Howlett NG, Gueranger Q, Glover TW, Canman CE. Differential roles for DNA polymerases eta, zeta, and REV1 in lesion bypass of intrastrand versus interstrand DNA cross-links. Mol Cell Biol. 2010;30:1217–1230. doi: 10.1128/MCB.00993-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Masuda Y, Ohmae M, Masuda K, Kamiya K. Structure and enzymatic properties of a stable complex of the human REV1 and REV7 proteins. J Biol Chem. 2003;278:12356–12360. doi: 10.1074/jbc.M211765200. [DOI] [PubMed] [Google Scholar]
- 151.Hlavin EM, Smeaton MB, Noronha AM, Wilds CJ, Miller PS. Cross-link structure affects replication-independent DNA interstrand cross-link repair in mammalian cells. Biochemistry. 2010;49:3977–3988. doi: 10.1021/bi902169q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Cipak L, Watanabe N, Bessho T. The role of BRCA2 in replication-coupled DNA interstrand cross-link repair in vitro. Nat Struct Mol Biol. 2006;13:729–733. doi: 10.1038/nsmb1120. [DOI] [PubMed] [Google Scholar]
- 153.Couve S, Mace-Aime G, Rosselli F, Saparbaev MK. The human oxidative DNA glycosylase NEIL1 excises psoralen-induced interstrand DNA cross-links in a three-stranded DNA structure. J Biol Chem. 2009;284:11963–11970. doi: 10.1074/jbc.M900746200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Couve-Privat S, Mace G, Rosselli F, Saparbaev MK. Psoralen-induced DNA adducts are substrates for the base excision repair pathway in human cells. Nucleic Acids Res. 2007;35:5672–5682. doi: 10.1093/nar/gkm592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Mace-Aime G, Couve S, Khassenov B, Rosselli F, Saparbaev MK. The Fanconi anemia pathway promotes DNA glycosylase-dependent excision of interstrand DNA crosslinks. Environ Mol Mutagen. 2010;51:508–519. doi: 10.1002/em.20548. [DOI] [PubMed] [Google Scholar]
- 156.Joenje H, Arwert F, Eriksson AW, de Koning H, Oostra AB. Oxygen-dependence of chromosomal aberrations in Fanconi’s anaemia. Nature. 1981;290:142–143. doi: 10.1038/290142a0. [DOI] [PubMed] [Google Scholar]
- 157.Schindler D, Hoehn H. Fanconi anemia mutation causes cellular susceptibility to ambient oxygen. Am J Hum Genet. 1988;43:429–435. [PMC free article] [PubMed] [Google Scholar]
- 158.Saadatzadeh MR, Bijangi-Vishehsaraei K, Hong P, Bergmann H, Haneline LS. Oxidant hypersensitivity of Fanconi anemia type C-deficient cells is dependent on a redox-regulated apoptotic pathway. J Biol Chem. 2004;279:16805–16812. doi: 10.1074/jbc.M313721200. [DOI] [PubMed] [Google Scholar]
- 159.Zhang X, Sejas DP, Qiu Y, Williams DA, Pang Q. Inflammatory ROS promote and cooperate with the Fanconi anemia mutation for hematopoietic senescence. J Cell Sci. 2007;120:1572–1583. doi: 10.1242/jcs.003152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zhang X, Li J, Sejas DP, Pang Q. Hypoxia-reoxygenation induces premature senescence in FA bone marrow hematopoietic cells. Blood. 2005;106:75–85. doi: 10.1182/blood-2004-08-3033. [DOI] [PubMed] [Google Scholar]
- 161.Cadet J, Douki T, Ravanat JL. Oxidatively generated base damage to cellular DNA. Free Radic Biol Med. 2010;49:9–21. doi: 10.1016/j.freeradbiomed.2010.03.025. [DOI] [PubMed] [Google Scholar]
- 162.Evans MD, Dizdaroglu M, Cooke MS. Oxidative DNA damage and disease: induction, repair and significance. Mutat Res. 2004;567:1–61. doi: 10.1016/j.mrrev.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 163.Ward JF. The complexity of DNA damage: relevance to biological consequences. Int J Radiat Biol. 1994;66:427–432. doi: 10.1080/09553009414551401. [DOI] [PubMed] [Google Scholar]
- 164.Kryston TB, Georgiev AB, Pissis P, Georgakilas AG. Role of oxidative stress and DNA damage in human carcinogenesis. Mutat Res. 2011;711:193–201. doi: 10.1016/j.mrfmmm.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 165.Sage E, Harrison L. Clustered DNA lesion repair in eukaryotes: relevance to mutagenesis and cell survival. Mutat Res. 2011;711:123–133. doi: 10.1016/j.mrfmmm.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Eccles LJ, O’Neill P, Lomax ME. Delayed repair of radiation induced clustered DNA damage: friend or foe? Mutat Res. 2011;711:134–141. doi: 10.1016/j.mrfmmm.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]