Abstract
Mitochondria are responsible for generating ATP and metabolic intermediates for biosynthesis. These dual functions require the activity of the electron transport chain in the mitochondrial inner membrane. The performance of these electron carriers is imperfect, resulting in release of damaging reactive oxygen species. Thus, continued mitochondrial activity requires maintenance. There are numerous means by which this quality control is ensured. Autophagy and selective mitophagy are among them. However, the cell inevitably must compensate for declining quality control by activating a variety of adaptations that entail the signaling of the presence of mitochondrial dysfunction to the nucleus. The best known of these is the retrograde response. This signaling pathway is triggered by the loss of mitochondrial membrane potential, which engages a series of signal transduction proteins, and it culminates in the induction of a broad array of nuclear target genes. One of the hallmarks of the retrograde response is its capacity to extend the replicative life span of the cell. The retrograde signaling pathway interacts with several other signaling pathways, such as TOR and ceramide signaling. All of these pathways respond to stress, including metabolic stress. The retrograde response is also linked to both autophagy and mitophagy at the gene and protein activation levels. Another quality control mechanism involves age-asymmetry in the segregation of dysfunctional mitochondria. One of the processes that impinge on this age-asymmetry is related to biogenesis of the organelle. Altogether, it is apparent that mitochondrial quality control constitutes a complex network of processes, whose full understanding will require a systems approach.
Keywords: Yeast, Saccharomyces cerevisiae, Autophagy, Mitophagy, Age-asymmetry, Ceramide signaling, Replicative life span
1. Introduction
Maintenance of functional integrity operates at all levels of biological organization from the molecular to the organismic. Significant resources are dedicated to this activity to guarantee fitness in the face of natural selection. By the same token, the partitioning of resources to maintenance cannot be to the detriment of their allocation to reproduction. This dual prerogative, maintenance and reproduction, leads to the dwindling investment of resources in the former during the post-reproductive period and the emergence of biological aging, according to evolutionary theory [1]. These considerations serve as a backdrop when analyzing maintenance, because they point to the dynamic nature of maintenance processes which are subject to the ravages associated with age. Less obvious is the redundancy with which these processes are endowed. This redundancy constitutes a reservoir that is only depleted with time.
The term quality control has recently been applied to designate maintenance of functional integrity at the molecular and cellular levels. This is a useful way to encapsulate the notions of dynamism and fitness into one with maintenance itself. Quality control lends a relative flavor to maintenance, with emphasis on change over time. For this reason, this will be the term of choice in this review.
The focus of this article is the mitochondrion, and particularly the yeast Saccharomyces cerevisiae mitochondrion. Quality control in this organelle begins with its biogenesis, which is discussed in great detail in accompanying articles. Yeast and animal cells employ different strategies during mitochondrial biogenesis. In yeast, there is a coordination of transcription of mitochondrial DNA (mtDNA) and mitochondrial protein genes in the nuclear genome [2], while in animal cells deficient transcription of mtDNA results in the overproduction of nuclear-encoded mitochondrial proteins [3]. Mitochondrial quality control includes inner membrane protein and matrix protein surveillance, the purview of the m-AAA protease and the Lon protease, respectively, which are discussed elsewhere in this volume. The former activity is linked to the prohibitins, which form a ring structure in the inner membrane and impact mitochondrial dynamics through the dynamin-like GTPase Opa1 [4]. Damage and dysfunction accumulate nonetheless, and this triggers an adaptation called the retrograde response [5, 6], which will be discussed in some detail here. Although it is not strictly a quality control mechanism, it is a cellular maintenance mechanism that compensates for the loss of mitochondrial quality, which inevitably occurs with age. The retrograde response has regulatory links to autophagy, which constitutes an additional mechanism that can enhance mitochondrial quality. Asymmetric segregation of dysfunctional mitochondria is yet another quality control mechanism. One aspect of this asymmetry is surprisingly related to mitochondrial biogenesis, bringing the discussion full circle. Both autophagy and asymmetric segregation of mitochondria will be discussed briefly here.
2. The Retrograde Response
2.1. Ramifications of the accumulation of unusual RNA species in mtDNA-deficient yeast cells
In 1987, Parikh et al. [7] described the accumulation of a heterogeneous collection of nuclear DNA-encoded transcripts in various mitochondrial petite yeast strains, lacking intact mtDNA (rho− or rho0). This included polymerase II-derived polyadenylated RNA transcribed from rDNA [8]. The identification of the CIT2 gene transcript among these RNA species and the importance of its 5′-flanking sequences in this gene induction solidified the notion of retrograde signaling from the mitochondrion to the nucleus [9]. These studies also pointed to a physiologic cooperation or compensation for the loss of mtDNA in rho0 cells, by enhanced expression of the peroxisomal citrate synthase encoded by CIT2. Activation of this gene has become a diagnostic for retrograde signaling.
Subsequent studies [10, 11] on a genome-wide scale have demonstrated an extensive and varied set of transcriptional changes in rho0 compared to rho+ cells. They include the induction of a wide range of metabolic and stress response genes. Many of these gene regulatory events are also observed on treatment of cells with electron transport chain inhibitors and uncouplers. It is apparent that retrograde signaling responds to mitochondrial dysfunction by adapting cell metabolism to the loss of tricarboxylic acid (TCA) cycle activity. The term retrograde response was coined to denote this fact.
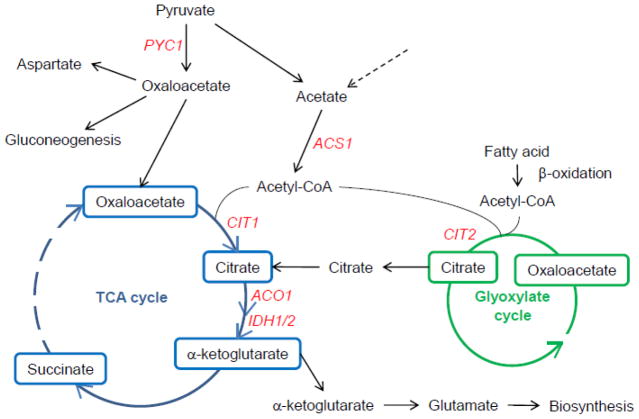
The key element in the retrograde response is the support of the activity of a truncated TCA cycle for production of biosynthetic intermediates (Fig. 1). Compromise of respiratory function precludes activity of succinate dehydrogenase in the TCA cycle. However, the first three enzymes of the cycle, citrate synthase, aconitase, and isocitrate dehydrogenase remain active to generate α-ketoglutarate, which is a source of glutamate. Glutamate is the ultimate donor of nitrogen in biosyntheses. This activity can only be sustained when there is a supply of citrate. This supply is maintained in cells lacking respiratory function through the activation of the glyoxylate cycle. CIT2 encodes the peroxisomal citrate synthase and together with aconitase and isocitrate lyase gives rise to glyoxylate that can be converted to malate by malate synthase through the addition of acetate. Malate dehydrogenase converts malate to oxaloacetate which accepts acetate to form citrate in the citrate synthase reaction, closing the glyoxylate cycle.
Fig. 1.
The retrograde response keys production of biosynthetic intermediates. Respiratory chain deficiencies render the reactions of the TCA cycle that convert succinate to oxaloacetate inoperable. However, the TCA cycle can be fueled by citrate generated in the glyoxylate cycle, which only requires a source of acetyl-CoA. This acetyl-CoA can be supplied by the β-oxidation of fatty acids or from other sources. This cooperation with the glyoxylate cycle allows the TCA cycle to remain as a net source of glutamate for biosynthesis. The TCA cycle can also be sustained by the anaplerotic conversion of pyruvate to oxaloacetate in reactions initiated by pyruvate carboxylase. The acetyl-CoA required to synthesize citrate from oxaloacetate in the TCA cycle is derived from pyruvate, as well. Oxaloacetate can also be a substrate for gluconeogenesis and for aspartate synthesis. The TCA cycle is located in the mitochondrial matrix, while the glyoxylate cycle and β-oxidation of fatty acids are found in peroxisomes. The remaining reactions occur in the cytoplasm. Retrograde response target genes are shown in red. ACS1, acetyl-coenzyme A synthetase; PYC1, pyruvate carboxylase; CIT1, mitochondrial citrate synthase; ACO1, aconitase; IDH1/2, isocitrate dehydrogenases 1 and 2; CIT2, peroxisomal citrate synthase.
Oxaloacetate is replenished in the anaplerotic conversion of pyruvate to oxaloacetate. This reaction is catalyzed by pyruvate carboxylase. The gene encoding this enzyme, PCY1, is also induced in the retrograde response. Phosphoenolpyruvate carboxykinase, in turn, converts oxaloacetate to phosphoenolpyruvate facilitating gluconeogenesis. The key to the anabolic processes of glutamate synthesis and gluconeogenesis is the glyoxylate cycle. It allows the utilization of simple, two-carbon compounds for the net synthesis of carbohydrate and protein. It does so by conserving the two carbons of acetate, rather than releasing them as carbon dioxide in the manner of the TCA cycle. Activation of the retrograde response is accompanied by a remarkable proliferation of peroxisomes [11]. This portends the stimulation of β-oxidation of fatty acids, which is confined to peroxisomes in yeast, and it is consistent with the generation of acetate to fuel the glyoxylate cycle.
The retrograde response has been characterized as a gene-regulatory process, and there have been no concerted metabolomics studies in support. However, metabolic changes consistent with the transcriptional events have been documented in yeast cells in which the retrograde response has been induced [9, 12]. Net utilization of the glyoxylate cycle occurs during severe nutrient limitation in yeast [13]. Thus, glyoxylate cycle activity can be equated with response to metabolic stress. This further supports the view that the retrograde response is an adaptation to metabolic stress in yeast.
2.2. The retrograde signaling pathway
A genetic screen identified three genes essential for transduction of the retrograde signal. These are the retrograde response genes RTG1, RTG2, and RTG3 [14–18]. RTG1 and RTG3 encode the subunits of a heterodimeric transcription factor belonging to the basic, helix-loop-helix, leucine zipper family. Rtg1–Rtg3 binds to the sequence GTCAC (R box) in the promoters of retrograde response target genes [14, 15].
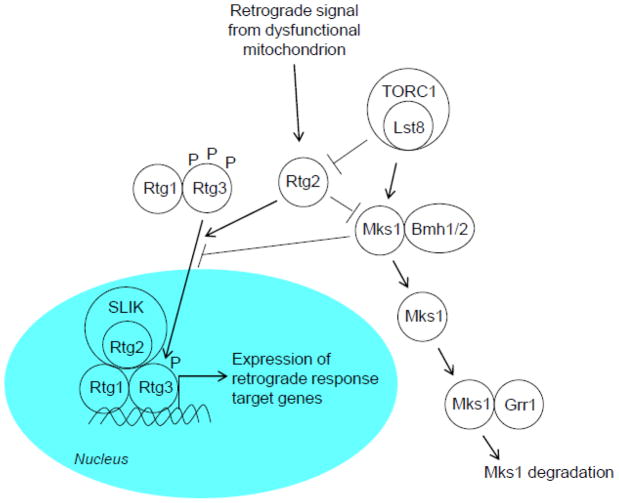
Rtg1–Rtg3 resides in the cytoplasm (Fig. 2). Activation of the retrograde response results in its translocation to the nucleus [16, 17, 19]. This translocation requires Rtg2, which alters the phosphorylation status of Rtg3 presumably due to phosphatase activity [17]. Rtg2 is thus the focal point of retrograde regulation. Fully phosphorylated Rtg3 cannot enter the nucleus; however, partial phosphorylation is necessary for its transcription-promoting activity.
Fig. 2.
The retrograde signaling pathway. A signal from dysfunctional mitochondria is transduced by Rtg2 to the heterodimeric transcription factor Rtg1–Rtg3. The phosphatase activity of Rtg2 may partially dephosphorylate Rtg3, facilitating the translocation of Rtg1–Rtg3 into the nucleus, where it induces the expression of a wide array of retrograde response target genes. The retrograde transcription factor is aided in this gene induction by the transcriptional co-activator complex SLIK (SAGA-like), which contains Rtg2 as an integral component. The translocation of Rtg1–Rtg3 into the nucleus is inhibited by Mks1, which in a complex with the 14-3-3 proteins Bmh1/2 promotes hyperphosphorylation of Rtg3. Rtg2 can prevent this by sequestering Mks1. Mks1 is degraded by a ubiquitin-dependent mechanism, involving the ubiquitin ligase Grr1. The nutrient-sensing TORC1, one subunit of which is the WD-protein Lst8 a know retrograde response regulator, inhibits retrograde signaling both upstream and downstream of Rtg2. It negatively regulates Rtg2 and positively regulates Mks1.
MKS1 is a negative regulator of the retrograde response [18, 20–23]. It reversibly binds to Rtg2, which prevents Mks1 from promoting hyperphosphorylation of Rtg3 and blocking Rtg1–Rtg3 translocation to the nucleus. This is another way in which Rtg2 has a positive effect on retrograde signaling. Mks1 is removed by ubiquitin-mediated degradation, facilitated by the action of Grr1 which is a component of ubiquitin ligase [24]. Binding of Mks1 by the 14-3-3 protein Bmh1/2 prevents this degradation [18, 24]. Thus, Grr1 and Bmh1/2 are positive and negative retrograde response regulators, respectively.
The retrograde response is activated in rho0 cells. A more severe mitochondrial defect results from deletion of CRD1 in rho0 cells. CRD1, the cardiolipin synthase gene, is not essential for yeast viability at optimal growth temperatures. However, in a rho0 background, deletion of the gene leads to a variety of phenotypes, one of which is cell division delay [25]. This delay is caused by the elevated expression of SWE1, which encodes a morphogenesis checkpoint protein. The induction of SWE1 in the rho0 crd1 mutant is dependent on RTG2 and RTG3 [25].
One clue to the enlarged mitochondrial dysfunction in rho0 cells carrying a deletion in CRD1 comes from the finding that phosphatidyl glycerol and cardiolipin are needed for the activation of inositolsphingolipid phospholipase C activity of Isc1 in late logarithmic growth phase [26]. Isc1 translocates from the endoplasmic reticulum to the mitochondrion in late logarithmic phase, and its activity increases dramatically due to enzyme activation for which the cardiolipin could be uniquely required [27]. Isc1 is similar to mammalian sphingomyelinases in generating ceramide, which is likely to possess downstream targets. The deletion of ISC1 results in a failure to up-regulate a wide array of genes associated with the late-logarithmic growth phase metabolic adaptation called the diauxic shift in yeast [28]. This gene-regulation defect is similar to that found in rho0 cells. However, the retrograde response is not induced in the isc1 mutant. Thus, ceramide signaling mediated by Isc1 may activate a pathway parallel to retrograde signaling.
It has recently been shown that Isc1 is an upstream regulator of Sit4, the catalytic subunit of the protein phosphatase 2A which is activated by ceramide [29]. Protein phosphatase 2A is also regulated by target of rapamycin complex 1 (TORC1). Thus, the pathway parallel to retrograde signaling in which ceramide functions may be mediated by TORC1. Ceramide has long been known to arrest cells in the G1 phase of the cell cycle by activating protein phosphatase 2A [30]. ISC1 interacts with cell cycle checkpoint genes and in particular with SWE1 [31]. This may be another way in which ceramide can arrest cell cycle progression. Thus, retrograde signaling may affect expression of SWE1, as indicated above, while ISC1-mediated ceramide signaling would affect the activity of the Swe1 protein.
Recently, two other pathways of signaling from the mitochondrion to the nucleus have been identified. One of them has been named mitochondrial back-signaling [32]. This pathway is activated upon deletion of the AFO1/MRPL25 gene, which encodes a protein found in mitochondrial ribosomes. Activation of mitochondrial back-signaling results in the extension of replicative life span, similar to what occurs in the retrograde response, as discussed below. This effect of back-signaling requires an active TORC1 and the transcription factor Sfp1, which regulates cytoplasmic ribosome synthesis. This signaling pathway is activated in rho0 cells grown on glucose in a strain in which retrograde response signaling is repressed by this carbon source. The retrograde response is activated in rho0 cells of this strain on the non-repressive carbon source raffinose. However, the interaction of retrograde signaling and back-signaling was not examined. It will be interesting to determine the relationship of these two pathways.
Another presumed pathway that appears to involve communication between the mitochondrion and the nucleus involves the set of mitochondrial proteins that activate translation of mtDNA-encoded proteins [33]. These protein activators are called the mitochondrial translation complex (MTC). The deletion of the nuclear genes encoding these specialized translation factors leads to replicative life span extension, which is dependent on the Sir2 histone deacetylase. This effect appears independent of retrograde signaling. However, any interaction between the two pathways has not been examined. It should be pointed out that downregulation of mitochondrial protein synthesis by erythromycin was shown quite some time ago to extend yeast replicative life span [34].
2.3. Crosstalk between retrograde and other signaling pathways
Deletion of RAS2 abrogates the retrograde response [35]. This G-protein can operate in a cAMP-dependent or independent pathway [36]. It is not clear which of these is important for retrograde signaling. MKS1 was first identified as a negative regulator of the Ras2-cAMP pathway [37]. This would be consistent with the involvement of the Ras2-cAMP pathway in retrograde signaling, but Mks1 could function independently in both Ras2 pathways. As discussed in more detail below, activation of the retrograde response extends yeast replicative life span [35]. RAS2 has a positive effect on yeast replicative life span [36], as well. However, this positive effect is exerted through the cAMP-independent pathway. This would suggest that it is the cAMP-independent pathway that impinges on retrograde signaling. As discussed above for Mks1, however, these pathways may not converge. Thus, the involvement of the Ras2-cAMP-dependent versus independent pathways in the retrograde response remains an open question. Indeed, it is possible that each plays a role.
TORC1 signaling inhibits the retrograde response. Inhibition of TORC1 by rapamycin activates retrograde response target genes in an Rtg2-dependent manner [38]. Other studies point to TORC1 inhibition of retrograde signaling at the level of Rtg1–Rtg3 [39, 40]. The WD-protein Lst8 is a component of the TORC1 complex [41–43]. It was identified as a negative regulator of retrograde signaling, acting both upstream and downstream of Rtg2 [44]. Thus, it is probable that TORC1 inhibits the retrograde response at multiple points.
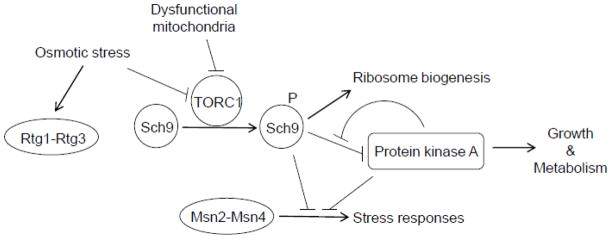
There appears to be negative feedback from dysfunctional mitochondria to TORC1 [45]. In rho0 cells or in rho+ cells treated with a mitochondrial uncoupler, the TORC1-mediated phosphorylation of Sch9, an AGC protein kinase family member, is down-regulated. Sch9 is normally activated by phosphorylation. Its activity antagonizes stress responses under the control of the Msn2–Msn4 transcription factor and promotes ribosome biogenesis [46]. Sch9 negatively regulates protein kinase A activity, exerting an effect on cell growth, metabolism, and stress resistance [47]. Protein kinase A negatively regulates the Msn2–Msn4 mediated stress responses. However, it also feedback inhibits its own activation, explaining the chain of events following the effect of dysfunctional mitochondria on TORC1. Interestingly, osmotic stress transiently reduces Sch9 phosphorylation by TORC1 [46]. The response to osmotic stress recruits the Rtg1–Rtg3 transcription factor [48]. In summary, the observations described here suggest a complex effect of mitochondrial dysfunction on the signaling pathways that regulate metabolism and stress responses (Fig. 3). They also indicate that retrograde signaling responds not only to metabolic stress but to other types of stress as well.
Fig. 3.
Effects of mitochondrial dysfunction on metabolism and stress responses. Mitochondrial dysfunction not only positively activates the retrograde response, but it also inhibits TORC1, which is a negative regulator of the retrograde response. One of the effects of TORC1 is phosphorylation of the protein kinase Sch9, which results in its activation. Active Sch9 promotes ribosome biogenesis and suppresses a variety of stress responses mediated by the transcription factor Msn2–Msn4. Sch9 also phosphorylates protein kinase A (PKA), resulting in its inhibition. Active PKA stimulates growth and metabolic activity by phosphorylating many different proteins and prevents the activation of stress response genes by Msn2–Msn4. Thus, Sch9 paradoxically would inhibit growth and metabolism in the presence of nutrients. However, this does not occur because PKA feedback inhibits its own activation. Like mitochondrial dysfunction, osmotic stress inhibits TORC1 activity, albeit transiently. Osmotic stress also activates retrograde response target genes by recruiting Rtg1–Rtg3.
2.4. External signals impinging upon retrograde signaling
The protein complexes TORC1 and TORC2 are distinct in that the former contains both Tor1 and Tor2 and is sensitive to rapamycin while the latter only possesses Tor2 and is resistant to this drug [49]. Nutrients have a positive effect on TORC1/2 signaling. However, it is not clear how they exert this effect. Similarly, availability of glucose activates the Ras-cAMP pathway in yeast, but the manner in which this occurs is not well understood. Thus, the involvement of TORC1 and Ras2 in the retrograde response may or may not be the result of nutrient sensing. There are also other ways in which nutrients are known to impinge on retrograde signaling.
The amino acid sensor SPS, composed of the amino acid sensor in the plasma membrane Ssy1 and the downstream effectors Ptr3 and Ssy5, responds to the presence of the amino acids glutamate and glutamine in the growth medium [50]. Certain LST8 mutants activate the retrograde response and depending on the particular mutation do this in an RTG2-dependent or independent manner [51]. The former are similar to mutants in the SPS sensor [52], suggesting a link between nutrient sensing and retrograde signaling. Glutamate and aspartate have a repressive effect on the retrograde response, consistent with its role in providing glutamate in the absence of an active TCA cycle [5], while RTG gene mutants are glutamate auxotrophs [14, 15]. It is also well known that retrograde signaling is repressed by glucose, at least in some yeast strains [9] though the mechanism is not known. It is possible that this is somehow tied into the Ras2 pathway.
2.5. Rtg2 has multiple roles
Rtg2 has the molecular features of a phosphatase, including an N-terminal ATP binding domain [53]. This ATP binding domain is essential for the function of Rtg2 [18], including Rtg3 dephosphorylation and translocation of the Rtg1–Rtg3 transcription factor to the nucleus. There is more to the function of Rtg2 in retrograde signaling, however.
Rtg2 has been found to be an integral component of the SLIK (SAGA-like) transcriptional activation complex, which contains the histone acetyltranferase Gcn5 [54]. In addition to the absence of Rtg2, SLIK differs from SAGA by the absence of Spt8 and the truncation of Spt7. SAGA is recruited to gene promoters by the AD1 domains of transcription factors [55], and Rtg3 possesses this motif suggesting that SLIK and Rtg3 interact. SLIK (with Rtg2) binds to the CIT2 promoter in vivo suggesting that chromatin modification plays a role in the induction of at least some retrograde response target genes. SLIK and SAGA activate only partially overlapping sets of genes; thus, SLIK may be the primary activator of retrograde response target genes. Certainly, chromatin modification is necessary for the retrograde response, as deletion of GCN5 prevents CIT2 induction and also curtails yeast replicative life span in rho0 cells [56].
A curious feature of activation of the retrograde response is the production and accumulation of episomal circles derived from rDNA [57], which are termed extrachromosomal ribosomal DNA circles (ERC). This is a form of genome instability. High ERC levels curtail the yeast replicative life span [58]. ERC accumulate progressively during the replicative life span whether the cells are rho+ or rho0, although the levels are always higher in the latter, and the rho+ cells do not become rho0 during replicative aging [59]. In parallel to this ERC accumulation, the retrograde response is progressively activated during the replicative life span [59]. This indicates that a source of mitochondrial dysfunction other than loss of mtDNA normally triggers the retrograde response and ERC accumulation. Indeed, activation of the retrograde response can occur in the presence of mtDNA, as noted above. One of the retrograde response target genes is ACO1. This gene not only performs a metabolic function, but it also promotes mtDNA stability, as Aco1 is a component of the mitochondrial nucleoid, in which it can replace Abf2 function [60].
Loss of heterozygosity, another form of genome instability, and petite formation accelerate in the daughters of diploid yeast cells as the mother cells become replicatively older [61]. Loss of heterozygosity occurs when one of the alleles of a gene normally present is missing. The relationship between ERC production and loss of heterozygosity in normally aging cells is not clear at present, and it may not be correct to equate events in diploid with those in haploid cells, which have been used in most yeast aging studies. As will be discussed below, age asymmetry and its progressive loss during the replicative life span are features of normal yeast aging, and ERC are segregated asymmetrically during yeast cell division [58, 62] although the mechanism is not entirely clear [63–65].
The relationship between ERC and retrograde signaling is mechanistic. Rtg2 not only is a signal transducer in the retrograde response, but it also is involved in suppression of genome instability. This was initially found as its ability to prevent the expansion of trinucleotide repeats that were artificially introduced into the yeast genome [66]. It was further shown that this protein suppresses ERC formation [59]. Rtg2 lies at a branch point in the retrograde pathway; it either extends the replicative life span by transducing the retrograde response signal, or it suppresses genome instability. It is not present in amounts sufficient to perform both functions effectively [59]. This explains the accumulation of ERC in rho0 cells. However, it is not known whether Rtg2 acts alone in preventing ERC accumulation. It does not appear to do so as a component of SLIK. This is because deletion of GCN5 suppresses ERC formation, while abrogating the induction of CIT2 and life span extension [56]. This implies that disruption of SLIK and release of Rtg2 on deletion of GCN5 makes Rtg2 available to suppress ERC formation.
2.6. Loss of mitochondrial membrane potential triggers the retrograde pathway
Activation of the retrograde response occurs during normal replicative aging in rho+ yeast cells [59]. Together with the fact that its constitutive activation in certain yeast strains extends their replicative life span, this suggests that yeast cells live as long as they do because they respond to mitochondrial dysfunction during aging by activating a compensatory adaptation, the retrograde response. What is the signal generated by dysfunctional mitochondria that triggers the retrograde signaling pathway? This has been a missing piece to the retrograde signaling puzzle until recently.
With increasing replicative age, yeast mitochondria lose membrane potential [67]. These mitochondrial changes are associated with an increase in reactive oxygen species (ROS) production [68] and protein oxidation [69]. Ultimately yeast cells die apoptotically at the end of their replicative life spans [68]. Changes in mitochondrial membrane potential (MMP) can result in ROS production [70], and interruption of the electron transport chain at virtually any point can result in ROS generation [71, 72]. Thus, either decreased MMP alone and/or ROS production are candidates for the mitochondria-proximal trigger of the retrograde response. This issue was resolved recently.
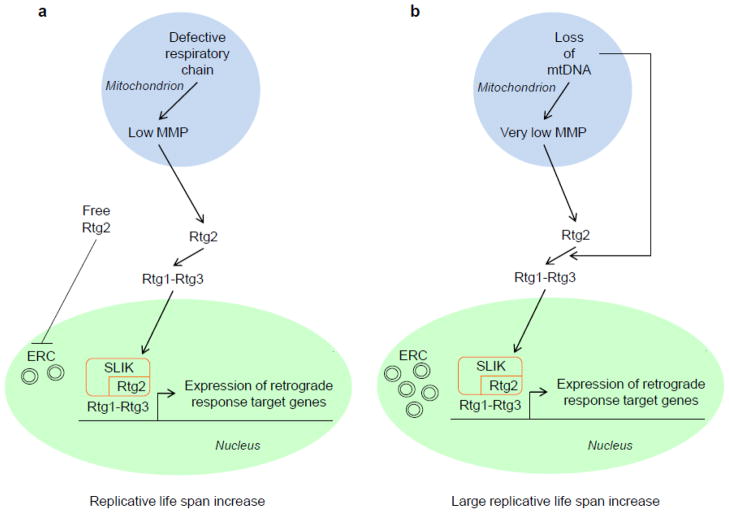
MMP was manipulated in yeast cells regardless of the presence or absence of mtDNA [73]. Raising the MMP in rho0 cells by expression of a hyperactive F1-ATPase prevented translocation of Rtg3 to the nucleus and induction of the four retrograde response target genes tested. It also suppressed replicative life span extension seen in the rho0 cells. Lowering the MMP in rho+ cells by deletion of COX4 stimulated Rtg3 translocation to the nucleus and CIT2 expression, while extending life span. Treatment of cells with the free radical scavenger phenyl tert butyl nitrone (PBN) in concentrations that eliminate ROS substantially had no effect on CIT2 induction or life span extension. These results indicate that falling MMP is the mitochondrial signal that elicits the retrograde response and that this effect is not mediated by elevated ROS. The question remains how this signal is “read” by Rtg2, the signal transducer proximal to the mitochondrion in the pathway.
The decrease in MMP in rho0 cells is associated with the accumulation of ERC, as indicated earlier. As expected, raising the MMP in these cells by expression of the hyperactive F1-ATPase prevents this accumulation [73]. Interestingly, lowering the MMP in rho+ cells by deletion of the COX4 gene did not result in an increase in ERC production, even though the retrograde response was induced. This induction is weaker than in rho0 cells, and it may not fully engage Rtg2 leaving some available to suppress ERC production. The situation is a bit more complicated, however. The results suggest that there are signals generated by mtDNA or mitochondrial nucleoid defects that contribute to retrograde signaling. These signals may reflect the action of so-called inter-genomic signaling, which appears to by-pass Rtg2 and to act directly upon Rtg3 to extend replicative life span in rho0 cells [74]. The model in Fig. 4 takes into account these nuances and provides a framework for further experiments.
Fig. 4.
Cellular response to retrograde signaling from the mitochondrion to the nucleus. (a) Mitochondrial dysfunction associated with respiratory chain defects in the presence of mtDNA (rho+). Localized deficiencies in respiratory chain complexes lead to reduction in mitochondrial membrane potential (MMP). This is the signal that is transduced to the retrograde signaling protein Rtg2, which in turn activates the retrograde transcription factor, a heterodimer of proteins Rtg1 and Rtg3, and results in its translocation to the nucleus where it activates retrograde response target genes. This activation is aided by the co-activator complex SLIK (SAGA-like complex), which contains Rtg2 as an integral component and possesses histone transacetylase activity Free Rtg2 remains available to perform its function to suppress extrachromosomal ribosomal DNA circle (ERC) accumulation. The net effect of the retrograde response is an increase in replicative life span. (b) Mitochondrial dysfunction associated with the loss of mtDNA (rho0). Loss of mtDNA leads to pervasive defects that result in a very low MMP. This engages the available pool of Rtg2 in retrograde signaling, leaving insufficient amounts of the protein to suppress ERC accumulation. Loss of mtDNA in addition activates Rtg1–Rtg3 in a poorly understood mechanism that by-passes Rtg2. The accumulation of ERC does not prevent the large increase in replicative life span, because the retrograde response includes functions that appear to compensate for their deleterious effects.
2.7. A retrograde response in organisms other than yeast
It was proposed some time ago that a retrograde response exists in Caenorhabditis elegans and that it is involved in determining its life span [35]. This conclusion was based on the up-regulation of the glyoxylate cycle during aging in this worm, as well as in certain mutants that show an increased life span [75]. Later, life span extension was observed on knockdown of certain mitochondrial protein genes by RNAi [76, 77]. Finally, a systematic search for genes whose knockdown extends worm life span has led to the conclusion that a retrograde response indeed extends worm longevity [78].
Recent studies shed new light on the “retrograde response” in C. elegans. Knockdown of anyone of several respiratory chain genes including cco-1 extends life span and activates the hypoxia-inducible transcription factor HIF-1 [79]. This effect appears to be mediated by elevation of ROS during respiratory stress. In another study, it was shown that downregulation of COX4 (cco-1) by RNAi extends life span and concomitantly activates the mitochondrial unfolded protein response in the worm [80]. The induction of nuclear gene expression requires the transcription factors UBL-5 and DVE-1 in this case, and it is cell-non-autonomous implying the secretion of a “mitokine” by certain cells to which other cells respond. In a third study, an RNAi screen identified the predicted transcription factor CEH-23 as uniquely responsible for the longevity increase in worms with reduced mitochondrial electron transport chain function [81]. In each of the above studies, the respective transcription factors were shown to be necessary and sufficient for life span extension, begging the question of the nature of the mutual relationships of the responses observed.
Recent studies in Drosophila melanogaster also point to a retrograde response. The first characterized a variety of RNAi strains that displayed knockdown of several respiratory chain components. Many of these displayed an increase in life span [82]. A mutant in a gene called sbo that is involved in coenzyme Q biosynthesis was isolated and demonstrated to extend life span as well [83].
Studies in mice also point to a retrograde response, though it is not likely that it involves a glyoxylate cycle. Reduced activity of MCLK1, needed for coenzyme Q biosynthesis, results in a compromised electron transport chain and markedly increased life span with no apparent tradeoff in growth or fertility [84, 85]. Disruption of complex IV assembly in a SURF1 knockout also substantially increased longevity [86]. It is likely that these mouse retrograde responses utilize very different signal transduction proteins than the yeast RTG genes [6].
Mitochondrial respiratory defects have been known for quite some time to elicit expression of nuclear genes in mammalian cells [6]. Several potential signaling pathways have been implicated, including calcium-signaling and NFκB-signaling. NFκB is a master regulator that is conserved in phylogeny, and it responds to a wide range of stress signals including ROS [87]. It is also associated with mitochondrial biogenesis [87]. A mitochondria-specific stress response activated by aggregation of ornithine decarboxylase has also been described [88]. This response may be similar to the mitochondrial unfolded protein response in C elegans, described above; however, it specifically involves the CHOP transcription factor.
Loss of mtDNA in human cells results in many gene expression changes that differ from one study to the next [6]. A side-by-side comparison of gene expression differences in rho+/rho0 pairs of three different human cell types showed that these changes are quite heterogeneous between cell types and may reflect cellular pathology [89, 90]. However, there were a handful of gene expression changes that was consistent across cell types. They reflect a compensation for loss of respiratory function through stimulation of glycolysis, enhanced protection from ROS, and compensation for genome instability. In many ways, these gene expression changes mirror the physiologic events in yeast cells in which the retrograde response is activated. Among the genes activated across cell types was c-Myc, a basic, helix-loop-helix, leucine zipper transcription factor. A bioinformatics analysis has shown that the Myc-Max heterodimer is homologous to the heterodimer of the basic, helix-loop-helix, leucine zipper transcription factor Rtg1–Rtg3 [87]. Interestingly, NFκB has two binding sites for Myc, suggesting that the mammalian retrograde response may not choose between NFκB and Myc. However, homologues of Rtg2 have not yet been found in mammals, thus this critical element in yeast retrograde signaling is still missing.
The replicative senescence of normal human diploid fibroblasts is delayed by mild mitochondrial uncoupling using dinitrophenol to lower MMP [91]. This treatment lowers ROS production, reduces telomere shortening, prevents the appearance of DNA repair foci in the nucleus, and induces a variety of gene expression changes. It also extends cell replicative life span. Thus, mammalian cells not only display many of the molecular features of yeast retrograde signaling, but they also present the cellular outcome of extended life span that characterizes the retrograde response.
3. Autophagy and mitophagy in mitochondrial quality control
3.1. Role of the retrograde response and ceramide signaling in autophagy
Autophagy is a gene-regulated process by which diverse cytoplasmic components, including mitochondria, are delivered to the lysosome, in yeast represented by the vacuole, for degradation [92]. This process has at least three related roles: During periods of starvation, it provides building blocks for biosynthesis, in particular amino acids for protein synthesis. It facilitates remodeling of the cell in response to stress, including metabolic stress. Importantly for this discussion, it serves to remove damaged and dysfunctional molecules and organelles, such as mitochondria. General autophagy is a non-selective process.
Not surprisingly given its role in nutrient sensing, TORC1 regulates autophagy. Inhibition of TORC1 in fact triggers it, because the cell senses this as a starvation state. TORC1 blocks autophagy by phosphorylating Atg13, which is an essential component of the Atg1 kinase complex that is required for autophagy [93]. Nitrogen starvation induces autophagy in yeast. Interestingly, deletion of IPT1 and SKN1 augments this autophagy without inducing apoptosis [94]. These genes encode the enzymes involved in the synthesis of yeast sphingolipids, the mannosyldiinositolphorphylceramides. Thus, these complex ceramides may modulate autophagy, limiting its extent. The direct precursor of sphingolipids in mammalian cells is ceramide. Along with sphingosine-1-phosphate, ceramide stimulates autophagy in mammalian cells [95]. Sphingosine and ceramide are precursors of complex sphingolipids. This suggests that the balance in sphingolipid biosynthetic activity can tip the scale in autophagy from quality control to wholesale degradation and remodeling. However, the mechanisms underlying the effects of the various sphingolipid species on autophagy are poorly understood.
TORC1 is not the only TOR complex that can regulate autophagy. TORC2 also has this potential because it regulates ceramide synthase activity [96]. It does so through the Ypk2 protein kinase, an AGC protein kinase family member. TORC2 in the presence of nutrients stimulates ceramide synthesis. This would provide the substrate for mannosyldiinositolphosphorylceramide, which attenuates autophagy [94]. In contrast, stress inhibits ceramide synthase activity through calcineurin, a calcium/calmodulin-dependent protein phosphatase [96]. Thus, stress would potentiate autophagy. Taken together the findings described above suggest that TORC1 is a major regulator of autophagy, while TORC2 fine tunes the process.
The response to mitochondrial dysfunction may be linked to ceramide synthase activity, located in the endoplasmic reticulum membrane. Two homologous proteins possess ceramide synthase activity in yeast, Lag1 and Lac1 [97–99]. They are encoded by the longevity assurance gene LAG1 and its homolog LAC1 [97, 100]. Their orthologs in human, LASS1–6, encode the six human ceramide synthases, Lass1–6 [101–103]. The LAC1 gene is coordinately regulated with PDR5 and other members of the multi-drug resistance family of membrane transporters in yeast [104]. PDR5 is activated in rho0 cells by retrograde signaling and by mechanisms independent of the RTG genes in the presence of other mitochondrial defects [105]. This is also the case for the other transporters in this family [106]. It is possible that LAC1 also responds to certain mitochondrial defects. A connection between the mitochondrion and the endoplasmic reticulum involving a physical junction has been identified, and it has been proposed to regulate mitochondrial biology [107]. This regulation may involve ceramide synthase. Ceramide signaling plays an important role in a variety of cellular processes, including cell cycle control and stress resistance [108]. The LAG1 ceramide synthase gene in yeast determines yeast replicative life span [100], and this function is finely tuned [109].
3.1. The retrograde response and mitophagy
Mitophagy is the term given to selective removal of mitochondria by autophagy [110, 111]. Mitophagy utilizes the same machinery as autophagy; however, the process becomes selective through the participation of Atg32, which tags mitochondria for elimination [112–116]. The mitochondrial fission-fusion cycle appears to play a supporting role in mitophagy. Deletion of the DNM1 gene, which is required for fission, reduces mitophagy without eliminating it entirely [114]. It also extends yeast replicative life span [117], which would potentially retain the function of mitophagy in mitochondrial quality control. Thus, the operation of mitophagy, at least at a reduced level, is compatible with an increase in yeast fitness, while extensive mitophagy may not be beneficial.
Retrograde signaling appears to be important for mitophagy, at least in stationary phase [118]. Induction of mitophagy goes hand in hand with the activation of the retrograde response, and this process requires the AUP1 gene. Furthermore, deletion of AUP1 prevents retrograde signaling and expression of retrograde target genes, and deletion of RTG3 suppresses stationary phase mitophagy. The phosphorylation pattern on Rtg3 is dependent on Aup1. AUP1 encodes a protein phosphatase localized to the intramembrane space in the mitochondrion. The relationship of this protein and its potential interactions with Rtg2 remain to be examined. It is possible that it may by-pass Rtg2 to alter Rtg3 phosphorylation status.
4. Asymmetric segregation of mitochondria and quality control
Mitochondrial dynamics are highly complex, and they are responsive to growth state and stress [119]. These dynamics involve the fission-fusion cycle already mentioned earlier, and they engage many proteins operating in multiple pathways to facilitate mitochondrial inheritance [120]. During cell division, approximately equal amounts of mitochondria find their way into the mother and daughter cells [121]. Is this all that there is to mitochondrial segregation?
Mitochondria are constantly exposed to ROS, resulting in damage, which is mitigated through the quality control processes discussed earlier. This quality control may be sufficient from the perspective of the individual cell; however, it apparently does not suffice for the yeast clone or population. The retrograde response which is progressively activated to compensate for accumulating mitochondrial dysfunction during the yeast replicative life span attests to this. However, the survival of the clone requires that daughter cells have the capacity for a full replicative life span; they must be born young [122], a process that breaks down with advancing age of the mother cell [123, 124].
This is a good point for a brief digression. Mention has been made of the yeast replicative life span. This requires some definition. An individual yeast cell is not immortal. It divides a limited number of times and dies [122]. The daughters that are produced at each cell division have in principle the capacity for a full replicative life span, while the mothers have an ever decreasing remaining replicative life span. I have called this the cell spiral to denote the progression of consecutive cell cycles [125, 126]. This leads to the concept of age asymmetry [67, 127], which as indicated above breaks down with advancing age of the mother cell [123, 124]. The distinction between mother and daughter is easily made in yeast, because the daughter cell is smaller at the time of birth than its mother. This makes possible the measurement of yeast replicative life spans which are determined for individual cells by micromanipulation. Asymmetry, whether structural or functional, forms the basis for a theory of aging [127]. The asymmetry may be like that found between mother and daughter yeast cell or between the soma and the germ line.
Old yeast cells show high levels of ROS production [68]. This is a progressive feature of yeast aging [128]. During the replicative life span, there is a loss of MMP [67] and a concomitant increase in carbonylated aconitase [69]. This is accompanied by fragmentation of mitochondria [67, 128]. These manifestations of mitochondrial dysfunction are not inherited by the daughters [67, 69, 128], at least until later in the life span of the mother cells. The accumulation of aberrant mitochondria and their segregation to daughter cells is accelerated in so-called yeast age-asymmetry mutants, one of which was characterized in detail [67]. This conditional mutant in the ATP2 gene produced daughter cells that possessed the same replicative age as the mother cell at birth; thus, the daughters were born old. Asymmetric segregation of bulk mitochondria between mother and daughter cells may not occur [121]. However, a mutation in MMR1, which encodes a protein that tethers mitochondria to the bud tip, disrupts the segregation of dysfunctional mitochondria to the mother and produces heterogeneity in replicative life span [129]. The studies cited here suggest that asymmetric segregation of damaged and dysfunctional mitochondria constitutes another quality control mechanism.
The ATP2 age-asymmetry mutant mentioned here does not affect protein function of the βsubunit of ATP synthase which it encodes [67]. Instead, it has a subtle effect on the kinetics of import of the protein into mitochondria [130]. It takes several generations for this defect to manifest itself phenotypically. The ATP2 mutant can be suppressed by multiple copies of the PEX6 gene, which encodes a peroxin. Pex6 performs the function of a chaperone in peroxisomal biogenesis. The ortholog of Pex6 in human is mutated in Zellweger syndrome. The suppression of the atp2 mutant, both in vivo and in vitro, indicates that Pex6 also can perform a chaperone function in mitochondrial biogenesis, which requires its ATP binding and hydrolysis activity [130]. Thus, this aspect of mitochondrial quality control through mechanisms involving age-asymmetry in segregation of damaged and dysfunctional mitochondria is related to the biogenesis of this organelle.
5. Conclusions
Mitochondria are not generated de novo by cells. Some mitochondrial material must be present to provide a template of sorts. Thus, mitochondrial quality control is of great importance in maintaining cell function. Autophagy and the selective process of mitophagy play significant roles in this quality control. This emerging insight expands the scope of mitophagy and autophagy, which have been considered mainly as a response to starvation or as an adaptation to changing metabolic needs. Studies in yeast have been particularly illuminating in this regard. There appears to be a fine balance to the extent to which autophagy and mitophagy are induced. This balance is struck through the action of retrograde and ceramide signaling, both of which determine yeast replicative life span. The implication is that some but not excessive recycling of mitochondria has a salutary effect on longevity.
The retrograde response compensates for accumulating mitochondrial dysfunction as yeasts progress through their replicative life span. This raises the notion that mitochondrial quality control is not sufficient to maintain this organelle over a cell’s lifetime. It is consistent with the decline in the force of natural selection with replicative age and the necessity to devote resources to the production of daughter cells and not only to maintenance. Curiously, there is a link between the retrograde response and mitochondrial quality control in the form of autophagy and mitophagy, suggesting coordination in the cell’s responses to mitochondrial damage.
The retrograde response signaling pathway is subject to crosstalk with several other signaling pathways in the cell. Thus, it is embedded in a complex network of cellular responses. These responses appear related to various types of stress, including metabolic stress. The retrograde response is progressively induced as mitochondrial dysfunction increases. There is a threshold of dysfunction that appears to augment retrograde signaling with another mitochondrial response involving ceramide signaling by Isc1 that can directly impact the cell division cycle in yeast. In addition to the typical retrograde response, there are at least two other types of response to mitochondrial dysfunction that are independent of the retrograde signal transducers. Their relationship to retrograde signaling is not clear at present.
Age-asymmetry in the segregation of dysfunctional mitochondria is yet another form of mitochondrial quality control. This age-asymmetry mechanism is responsible for the fact that daughters are born young by receiving only functional mitochondria. It represents the cost of reproduction in yeast, because the mothers retain the dysfunctional mitochondria. Even this mechanism, however, breaks down with replicative age. Interestingly, maintenance of age-asymmetry is at least in part related to mitochondrial biogenesis. This brings the processes of mitochondrial quality control full circle to the accurate biogenesis of this organelle in the first place.
Our understanding of mitochondrial quality control has benefited greatly from genetic dissection in yeast of the several pathways and processes it entails. It is clear that these pathways and processes are coordinated. However, our ability to make sense of this coordination in a way that allows us to predict outcomes is limited due to the complexity of this network. A systems approach, which includes mathematical modeling and computer simulation, will be useful in predicting the behavior of this nonlinear system. This approach will facilitate the formulation of new hypotheses for experimental testing.
Highlights.
The retrograde response compensates for mitochondrial dysfunction accumulating over a cell’s lifetime.
Autophagy and mitophagy provide mitochondrial quality control.
Age-asymmetry in segregation of dysfunctional mitochondria is another quality control mechanism.
Mitochondrial quality control constitutes a complex network of processes that requires a systems approach to understand fully.
Acknowledgments
The research in the author’s laboratory was supported in part by grants from the National Institute on Aging of the National Institutes of Health (U.S.P.H.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kirkwood TB, Rose MR. Evolution of senescence: late survival sacrificed for reproduction. Philos Trans R Soc Lond B Biol Sci. 1991;332:15–24. doi: 10.1098/rstb.1991.0028. [DOI] [PubMed] [Google Scholar]
- 2.Poyton RO, McEwen JE. Crosstalk between nuclear and mitochondrial genomes. Annu Rev Biochem. 1996;65:563–607. doi: 10.1146/annurev.bi.65.070196.003023. [DOI] [PubMed] [Google Scholar]
- 3.Heddi A, Lestienne P, Wallace DC, Stepien G. Mitochondrial DNA expression in mitochondrial myopathies and coordinated expression of nuclear genes involved in ATP production. J Biol Chem. 1993;268:12156–12163. [PubMed] [Google Scholar]
- 4.Merkwirth C, Langer T. Prohibitin function within mitochondria: essential roles for cell proliferation and cristae morphogenesis. Biochim Biophys Acta. 2009;1793:27–32. doi: 10.1016/j.bbamcr.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 5.Liu Z, Butow RA. Mitochondrial retrograde signaling. Annu Rev Genet. 2006;40:159–185. doi: 10.1146/annurev.genet.40.110405.090613. [DOI] [PubMed] [Google Scholar]
- 6.Butow RA, Avadhani NG. Mitochondrial signaling: the retrograde response. Mol Cell. 2004;14:1–15. doi: 10.1016/s1097-2765(04)00179-0. [DOI] [PubMed] [Google Scholar]
- 7.Parikh VS, Morgan MM, Scott R, Clements LS, Butow RA. The mitochondrial genotype can influence nuclear gene expression in yeast. Science. 1987;235:576–580. doi: 10.1126/science.3027892. [DOI] [PubMed] [Google Scholar]
- 8.Parikh VS, Conrad-Webb H, Docherty R, Butow RA. Interaction between the yeast mitochondrial and nuclear genomes influences the abundance of novel transcripts derived from the spacer region of the nuclear ribosomal DNA repeat. Mol Cell Biol. 1989;9:1897–1907. doi: 10.1128/mcb.9.5.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liao XS, Small WC, Srere PA, Butow RA. Intramitochondrial functions regulate nonmitochondrial citrate synthase (CIT2) expression in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:38–46. doi: 10.1128/mcb.11.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Traven A, Wong JM, Xu D, Sopta M, Ingles CJ. Interorganellar communication. Altered nuclear gene expression profiles in a yeast mitochondrial DNA mutant. J Biol Chem. 2001;276:4020–4027. doi: 10.1074/jbc.M006807200. [DOI] [PubMed] [Google Scholar]
- 11.Epstein CB, Waddle JA, Hale Wt, Dave V, Thornton J, Macatee TL, Garner HR, Butow RA. Genome-wide responses to mitochondrial dysfunction. Mol Biol Cell. 2001;12:297–308. doi: 10.1091/mbc.12.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Small WC, Brodeur RD, Sandor A, Fedorova N, Li G, Butow RA, Srere PA. Enzymatic and metabolic studies on retrograde regulation mutants of yeast. Biochemistry. 1995;34:5569–5576. doi: 10.1021/bi00016a031. [DOI] [PubMed] [Google Scholar]
- 13.Wang J, Jiang JC, Jazwinski SM. Gene regulatory changes in yeast during life extension by nutrient limitation. Exp Gerontol. 2010;45:621–631. doi: 10.1016/j.exger.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liao X, Butow RA. RTG1 and RTG2: two yeast genes required for a novel path of communication from mitochondria to the nucleus. Cell. 1993;72:61–71. doi: 10.1016/0092-8674(93)90050-z. [DOI] [PubMed] [Google Scholar]
- 15.Jia Y, Rothermel B, Thornton J, Butow RA. A basic helix-loop-helix-leucine zipper transcription complex in yeast functions in a signaling pathway from mitochondria to the nucleus. Mol Cell Biol. 1997;17:1110–1117. doi: 10.1128/mcb.17.3.1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rothermel BA, Thornton JL, Butow RA. Rtg3p, a basic helix-loop-helix/leucine zipper protein that functions in mitochondrial-induced changes in gene expression, contains independent activation domains. J Biol Chem. 1997;272:19801–19807. doi: 10.1074/jbc.272.32.19801. [DOI] [PubMed] [Google Scholar]
- 17.Sekito T, Thornton J, Butow RA. Mitochondria-to-nuclear signaling is regulated by the subcellular localization of the transcription factors Rtg1p and Rtg3p. Mol Biol Cell. 2000;11:2103–2115. doi: 10.1091/mbc.11.6.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Z, Sekito T, Spirek M, Thornton J, Butow RA. Retrograde signaling is regulated by the dynamic interaction between Rtg2p and Mks1p. Mol Cell. 2003;12:401–411. doi: 10.1016/s1097-2765(03)00285-5. [DOI] [PubMed] [Google Scholar]
- 19.Rothermel BA, Shyjan AW, Etheredge JL, Butow RA. Transactivation by Rtg1p, a basic helix-loop-helix protein that functions in communication between mitochondria and the nucleus in yeast. J Biol Chem. 1995;270:29476–29482. doi: 10.1074/jbc.270.49.29476. [DOI] [PubMed] [Google Scholar]
- 20.Dilova I, Chen CY, Powers T. Mks1 in concert with TOR signaling negatively regulates RTG target gene expression in S. cerevisiae. Curr Biol. 2002;12:389–395. doi: 10.1016/s0960-9822(02)00677-2. [DOI] [PubMed] [Google Scholar]
- 21.Dilova I, Aronova S, Chen JC, Powers T. Tor signaling and nutrient-based signals converge on Mks1p phosphorylation to regulate expression of Rtg1–Rtg3p-dependent target genes. J Biol Chem. 2004;279:46527–46535. doi: 10.1074/jbc.M409012200. [DOI] [PubMed] [Google Scholar]
- 22.Sekito T, Liu Z, Thornton J, Butow RA. RTG-dependent mitochondria-to-nucleus signaling is regulated by MKS1 and is linked to formation of yeast prion [URE3] Mol Biol Cell. 2002;13:795–804. doi: 10.1091/mbc.01-09-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tate JJ, Cox KH, Rai R, Cooper TG. Mks1p is required for negative regulation of retrograde gene expression in Saccharomyces cerevisiae but does not affect nitrogen catabolite repression-sensitive gene expression. J Biol Chem. 2002;277:20477–20482. doi: 10.1074/jbc.M200962200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Z, Spirek M, Thornton J, Butow RA. A novel degron-mediated degradation of the RTG pathway regulator, Mks1p, by SCFGrr1. Mol Biol Cell. 2005;16:4893–4904. doi: 10.1091/mbc.E05-06-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen S, Liu D, Finley RL, Jr, Greenberg ML. Loss of mitochondrial DNA in the yeast cardiolipin synthase crd1 mutant leads to up-regulation of the protein kinase Swe1p that regulates the G2/M transition. J Biol Chem. 2010;285:10397–10407. doi: 10.1074/jbc.M110.100784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vaena de Avalos S, Su X, Zhang M, Okamoto Y, Dowhan W, Hannun YA. The phosphatidylglycerol/cardiolipin biosynthetic pathway is required for the activation of inositol phosphosphingolipid phospholipase C, Isc1p, during growth of Saccharomyces cerevisiae. J Biol Chem. 2005;280:7170–7177. doi: 10.1074/jbc.M411058200. [DOI] [PubMed] [Google Scholar]
- 27.Vaena de Avalos S, Okamoto Y, Hannun YA. Activation and localization of inositol phosphosphingolipid phospholipase C, Isc1p, to the mitochondria during growth of Saccharomyces cerevisiae. J Biol Chem. 2004;279:11537–11545. doi: 10.1074/jbc.M309586200. [DOI] [PubMed] [Google Scholar]
- 28.Kitagaki H, Cowart LA, Matmati N, Montefusco D, Gandy J, de Avalos SV, Novgorodov SA, Zheng J, Obeid LM, Hannun YA. ISC1-dependent metabolic adaptation reveals an indispensable role for mitochondria in induction of nuclear genes during the diauxic shift in Saccharomyces cerevisiae. J Biol Chem. 2009;284:10818–10830. doi: 10.1074/jbc.M805029200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barbosa AD, Osorio H, Sims KJ, Almeida T, Alves M, Bielawski J, Amorim MA, Moradas-Ferreira P, Hannun YA, Costa V. Role for Sit4p-dependent mitochondrial dysfunction in mediating the shortened chronological lifespan and oxidative stress sensitivity of Isc1p-deficient cells. Mol Microbiol. 2011;81:515–527. doi: 10.1111/j.1365-2958.2011.07714.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nickels JT, Broach JR. A ceramide-activated protein phosphatase mediates ceramide-induced G1 arrest of Saccharomyces cerevisiae. Genes Dev. 1996;10:382–394. doi: 10.1101/gad.10.4.382. [DOI] [PubMed] [Google Scholar]
- 31.Tripathi K, Matmati N, Zheng WJ, Hannun YA, Mohanty BK. Cellular morphogenesis under stress is influenced by the sphingolipid pathway gene ISC1 and DNA integrity checkpoint genes in Saccharomyces cerevisiae. Genetics. 2011;189:533–547. doi: 10.1534/genetics.111.132092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heeren G, Rinnerthaler M, Laun P, von Seyerl P, Kossler S, Klinger H, Hager M, Bogengruber E, Jarolim S, Simon-Nobbe B, Schuller C, Carmona-Gutierrez D, Breitenbach-Koller L, Muck C, Jansen-Durr P, Criollo A, Kroemer G, Madeo F, Breitenbach M. The mitochondrial ribosomal protein of the large subunit, Afo1p, determines cellular longevity through mitochondrial back-signaling via TOR1. Aging (Albany NY) 2009;1:622–636. doi: 10.18632/aging.100065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caballero A, Ugidos A, Liu B, Oling D, Kvint K, Hao X, Mignat C, Nachin L, Molin M, Nystrom T. Absence of mitochondrial translation control proteins extends life span by activating sirtuin-dependent silencing. Mol Cell. 2011;42:390–400. doi: 10.1016/j.molcel.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 34.Holbrook MA, Menninger JR. Erythromycin slows aging of Saccharomyces cerevisiae. J Gerontol Series A Biol Sci Med Sci. 2002;57:B29–36. doi: 10.1093/gerona/57.1.b29. [DOI] [PubMed] [Google Scholar]
- 35.Kirchman PA, Kim S, Lai CY, Jazwinski SM. Interorganelle signaling is a determinant of longevity in Saccharomyces cerevisiae. Genetics. 1999;152:179–190. doi: 10.1093/genetics/152.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun J, Kale SP, Childress AM, Pinswasdi C, Jazwinski SM. Divergent roles of RAS1 and RAS2 in yeast longevity. J Biol Chem. 1994;269:18638–18645. [PubMed] [Google Scholar]
- 37.Matsuura A, Anraku Y. Characterization of the MKS1 gene, a new negative regulator of the Ras-cyclic AMP pathway in Saccharomyces cerevisiae. Mol Gen Genet. 1993;238:6–16. doi: 10.1007/BF00279524. [DOI] [PubMed] [Google Scholar]
- 38.Komeili A, Wedaman KP, O’Shea EK, Powers T. Mechanism of metabolic control. Target of rapamycin signaling links nitrogen quality to the activity of the Rtg1 and Rtg3 transcription factors. J Cell Biol. 2000;151:863–878. doi: 10.1083/jcb.151.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giannattasio S, Liu Z, Thornton J, Butow RA. Retrograde response to mitochondrial dysfunction is separable from TOR1/2 regulation of retrograde gene expression. J Biol Chem. 2005;280:42528–42535. doi: 10.1074/jbc.M509187200. [DOI] [PubMed] [Google Scholar]
- 40.Breitkreutz A, Choi H, Sharom JR, Boucher L, Neduva V, Larsen B, Lin ZY, Breitkreutz BJ, Stark C, Liu G, Ahn J, Dewar-Darch D, Reguly T, Tang X, Almeida R, Qin ZS, Pawson T, Gingras AC, Nesvizhskii AI, Tyers M. A global protein kinase and phosphatase interaction network in yeast. Science. 2010;328:1043–1046. doi: 10.1126/science.1176495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, Oppliger W, Jenoe P, Hall MN. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell. 2002;10:457–468. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- 42.Wedaman KP, Reinke A, Anderson S, Yates J, 3rd, McCaffery JM, Powers T. Tor kinases are in distinct membrane-associated protein complexes in Saccharomyces cerevisiae. Mol Biol Cell. 2003;14:1204–1220. doi: 10.1091/mbc.E02-09-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reinke A, Anderson S, McCaffery JM, Yates J, 3rd, Aronova S, Chu S, Fairclough S, Iverson C, Wedaman KP, Powers T. TOR complex 1 includes a novel component, Tco89p (YPL180w), and cooperates with Ssd1p to maintain cellular integrity in Saccharomyces cerevisiae. J Biol Chem. 2004;279:14752–14762. doi: 10.1074/jbc.M313062200. [DOI] [PubMed] [Google Scholar]
- 44.Liu Z, Sekito T, Epstein CB, Butow RA. RTG-dependent mitochondria to nucleus signaling is negatively regulated by the seven WD-repeat protein Lst8p. EMBO J. 2001;20:7209–7219. doi: 10.1093/emboj/20.24.7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kawai S, Urban J, Piccolis M, Panchaud N, De Virgilio C, Loewith R. Mitochondrial genomic dysfunction causes dephosphorylation of Sch9 in the yeast Saccharomyces cerevisiae. Eukaryot Cell. 2011;10:1367–1369. doi: 10.1128/EC.05157-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Urban J, Soulard A, Huber A, Lippman S, Mukhopadhyay D, Deloche O, Wanke V, Anrather D, Ammerer G, Riezman H, Broach JR, De Virgilio C, Hall MN, Loewith R. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol Cell. 2007;26:663–674. doi: 10.1016/j.molcel.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 47.Zhang A, Shen Y, Gao W, Dong J. Role of Sch9 in regulating Ras-cAMP signal pathway in Saccharomyces cerevisiae. FEBS Lett. 2011;585:3026–3032. doi: 10.1016/j.febslet.2011.08.023. [DOI] [PubMed] [Google Scholar]
- 48.Pastor MM, Proft M, Pascual-Ahuir A. Mitochondrial function is an inducible determinant of osmotic stress adaptation in yeast. J Biol Chem. 2009;284:30307–30317. doi: 10.1074/jbc.M109.050682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 50.Forsberg H, Ljungdahl PO. Sensors of extracellular nutrients in Saccharomyces cerevisiae. Curr Genet. 2001;40:91–109. doi: 10.1007/s002940100244. [DOI] [PubMed] [Google Scholar]
- 51.Chen EJ, Kaiser CA. LST8 negatively regulates amino acid biosynthesis as a component of the TOR pathway. J Cell Biol. 2003;161:333–347. doi: 10.1083/jcb.200210141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roberg KJ, Bickel S, Rowley N, Kaiser CA. Control of amino acid permease sorting in the late secretory pathway of Saccharomyces cerevisiae by SEC13, LST4,LST7 and LST8. Genetics. 1997;147:1569–1584. doi: 10.1093/genetics/147.4.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koonin EV. Yeast protein controlling inter-organelle communication is related to bacterial phosphatases containing the Hsp 70-type ATP-binding domain. Trends Biochem Sci. 1994;19:156–157. doi: 10.1016/0968-0004(94)90275-5. [DOI] [PubMed] [Google Scholar]
- 54.Pray-Grant MG, Schieltz D, McMahon SJ, Wood JM, Kennedy EL, Cook RG, Workman JL, Yates JR, 3rd, Grant PA. The novel SLIK histone acetyltransferase complex functions in the yeast retrograde response pathway. Mol Cell Biol. 2002;22:8774–8786. doi: 10.1128/MCB.22.24.8774-8786.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Massari ME, Grant PA, Pray-Grant MG, Berger SL, Workman JL, Murre C. A conserved motif present in a class of helix-loop-helix proteins activates transcription by direct recruitment of the SAGA complex. Mol Cell. 1999;4:63–73. doi: 10.1016/s1097-2765(00)80188-4. [DOI] [PubMed] [Google Scholar]
- 56.Kim S, Ohkuni K, Couplan E, Jazwinski SM. The histone acetyltransferase GCN5 modulates the retrograde response and genome stability determining yeast longevity. Biogerontology. 2004;5:305–316. doi: 10.1007/s10522-004-2568-x. [DOI] [PubMed] [Google Scholar]
- 57.Conrad-Webb H, Butow RA. A polymerase switch in the synthesis of rRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:2420–2428. doi: 10.1128/mcb.15.5.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sinclair DA, Guarente L. Extrachromosomal rDNA circles--a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 59.Borghouts C, Benguria A, Wawryn J, Jazwinski SM. Rtg2 protein links metabolism and genome stability in yeast longevity. Genetics. 2004;166:765–777. doi: 10.1093/genetics/166.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen XJ, Wang X, Kaufman BA, Butow RA. Aconitase couples metabolic regulation to mitochondrial DNA maintenance. Science. 2005;307:714–717. doi: 10.1126/science.1106391. [DOI] [PubMed] [Google Scholar]
- 61.Veatch JR, McMurray MA, Nelson ZW, Gottschling DE. Mitochondrial dysfunction leads to nuclear genome instability via an iron-sulfur cluster defect. Cell. 2009;137:1247–1258. doi: 10.1016/j.cell.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shcheprova Z, Baldi S, Frei SB, Gonnet G, Barral Y. A mechanism for asymmetric segregation of age during yeast budding. Nature. 2008;454:728–734. doi: 10.1038/nature07212. [DOI] [PubMed] [Google Scholar]
- 63.Khmelinskii A, Keller PJ, Lorenz H, Schiebel E, Knop M. Segregation of yeast nuclear pores. Nature. 2010;466:E1. doi: 10.1038/nature09255. [DOI] [PubMed] [Google Scholar]
- 64.Khmelinskii A, Meurer M, Knop M, Schiebel E. Artificial tethering to nuclear pores promotes partitioning of extrachromosomal DNA during yeast asymmetric cell division. Curr Biol. 2011;21:R17–18. doi: 10.1016/j.cub.2010.11.034. [DOI] [PubMed] [Google Scholar]
- 65.Gehlen LR, Nagai S, Shimada K, Meister P, Taddei A, Gasser SM. Nuclear geometry and rapid mitosis ensure asymmetric episome segregation in yeast. Curr Biol. 2011;21:25–33. doi: 10.1016/j.cub.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 66.Bhattacharyya S, Rolfsmeier ML, Dixon MJ, Wagoner K, Lahue RS. Identification of RTG2 as a modifier gene for CTG*CAG repeat instability in Saccharomyces cerevisiae. Genetics. 2002;162:579–589. doi: 10.1093/genetics/162.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lai CY, Jaruga E, Borghouts C, Jazwinski SM. A mutation in the ATP2 gene abrogates the age asymmetry between mother and daughter cells of the yeast Saccharomyces cerevisiae. Genetics. 2002;162:73–87. doi: 10.1093/genetics/162.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Laun P, Pichova A, Madeo F, Fuchs J, Ellinger A, Kohlwein S, Dawes I, Frohlich KU, Breitenbach M. Aged mother cells of Saccharomyces cerevisiae show markers of oxidative stress and apoptosis. Mol Microbiol. 2001;39:1166–1173. [PubMed] [Google Scholar]
- 69.Klinger H, Rinnerthaler M, Lam YT, Laun P, Heeren G, Klocker A, Simon-Nobbe B, Dickinson JR, Dawes IW, Breitenbach M. Quantitation of (a)symmetric inheritance of functional and of oxidatively damaged mitochondrial aconitase in the cell division of old yeast mother cells. Exp Gerontol. 2010;45:533–542. doi: 10.1016/j.exger.2010.03.016. [DOI] [PubMed] [Google Scholar]
- 70.Gourlay CW, Carpp LN, Timpson P, Winder SJ, Ayscough KR. A role for the actin cytoskeleton in cell death and aging in yeast. J Cell Biol. 2004;164:803–809. doi: 10.1083/jcb.200310148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Barros MH, Bandy B, Tahara EB, Kowaltowski AJ. Higher respiratory activity decreases mitochondrial reactive oxygen release and increases life span in Saccharomyces cerevisiae. J Biol Chem. 2004;279:49883–49888. doi: 10.1074/jbc.M408918200. [DOI] [PubMed] [Google Scholar]
- 72.Davidson JF, Schiestl RH. Mitochondrial respiratory electron carriers are involved in oxidative stress during heat stress in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:8483–8489. doi: 10.1128/MCB.21.24.8483-8489.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Miceli MV, Jiang JC, Tiwari A, Rodriguez-Quiñones JF, Jazwinski SM. Loss of mitochondrial membrane potential triggers the retrograde response extending yeast replicative lifespan. Front Genet. 2011;2:102. doi: 10.3389/fgene.2011.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Woo DK, Phang TL, Trawick JD, Poyton RO. Multiple pathways of mitochondrial-nuclear communication in yeast: intergenomic signaling involves ABF1 and affects a different set of genes than retrograde regulation. Biochim Biophys Acta. 2009;1789:135–145. doi: 10.1016/j.bbagrm.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 75.Vanfleteren JR, De Vreese A. The gerontogenes age-1 and daf-2 determine metabolic rate potential in aging Caenorhabditis elegans. FASEB J. 1995;9:1355–1361. doi: 10.1096/fasebj.9.13.7557026. [DOI] [PubMed] [Google Scholar]
- 76.Dillin A, Hsu AL, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298:2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- 77.Lee SS, Lee RY, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat Genet. 2003;33:40–48. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- 78.Cristina D, Cary M, Lunceford A, Clarke C, Kenyon C. A regulated response to impaired respiration slows behavioral rates and increases lifespan in Caenorhabditis elegans. PLoS Genet. 2009;5:e1000450. doi: 10.1371/journal.pgen.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee SJ, Hwang AB, Kenyon C. Inhibition of respiration extends C. elegans life span via reactive oxygen species that increase HIF-1 activity. Curr Biol. 2010;20:2131–2136. doi: 10.1016/j.cub.2010.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144:79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Walter L, Baruah A, Chang HW, Pace HM, Lee SS. The homeobox protein CEH-23 mediates prolonged longevity in response to impaired mitochondrial electron transport chain in C. elegans. PLoS Biol. 2011;9:e1001084. doi: 10.1371/journal.pbio.1001084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Copeland JM, Cho J, Lo T, Jr, Hur JH, Bahadorani S, Arabyan T, Rabie J, Soh J, Walker DW. Extension of Drosophila life span by RNAi of the mitochondrial respiratory chain. Curr Biol. 2009;19:1591–1598. doi: 10.1016/j.cub.2009.08.016. [DOI] [PubMed] [Google Scholar]
- 83.Liu J, Wu Q, He D, Ma T, Du L, Dui W, Guo X, Jiao R. Drosophila sbo regulates lifespan through its function in the synthesis of coenzyme Q in vivo. J Genet Genomics. 2011;38:225–234. doi: 10.1016/j.jgg.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 84.Lapointe J, Hekimi S. Early mitochondrial dysfunction in long-lived Mclk1+/− mice. J Biol Chem. 2008;283:26217–26227. doi: 10.1074/jbc.M803287200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu X, Jiang N, Hughes B, Bigras E, Shoubridge E, Hekimi S. Evolutionary conservation of the clk-1-dependent mechanism of longevity: loss of Mclk1 increases cellular fitness and lifespan in mice. Genes Dev. 2005;19:2424–2434. doi: 10.1101/gad.1352905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dell’agnello C, Leo S, Agostino A, Szabadkai G, Tiveron C, Zulian A, Prelle A, Roubertoux P, Rizzuto R, Zeviani M. Increased longevity and refractoriness to Ca(2+)-dependent neurodegeneration in Surf1 knockout mice. Hum Mol Genet. 2007;16:431–444. doi: 10.1093/hmg/ddl477. [DOI] [PubMed] [Google Scholar]
- 87.Srinivasan V, Kriete A, Sacan A, Jazwinski SM. Comparing the yeast retrograde response and NFκB stress responses: implications for aging. Aging Cell. 2010;9:933–941. doi: 10.1111/j.1474-9726.2010.00622.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhao Q, Wang J, Levichkin IV, Stasinopoulos S, Ryan MT, Hoogenraad NJ. A mitochondrial specific stress response in mammalian cells. EMBO J. 2002;21:4411–4419. doi: 10.1093/emboj/cdf445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Miceli MV, Jazwinski SM. Nuclear gene expression changes due to mitochondrial dysfunction in ARPE-19 cells: implications for age-related macular degeneration. Invest Ophthalmol Vis Sci. 2005;46:1765–1773. doi: 10.1167/iovs.04-1327. [DOI] [PubMed] [Google Scholar]
- 90.Miceli MV, Jazwinski SM. Common and cell type-specific responses of human cells to mitochondrial dysfunction. Exp Cell Res. 2005;302:270–280. doi: 10.1016/j.yexcr.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 91.Passos JF, Saretzki G, Ahmed S, Nelson G, Richter T, Peters H, Wappler I, Birket MJ, Harold G, Schaeuble K, Birch-Machin MA, Kirkwood TB, von Zglinicki T. Mitochondrial dysfunction accounts for the stochastic heterogeneity in telomere-dependent senescence. PLoS Biol. 2007;5:e110. doi: 10.1371/journal.pbio.0050110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 93.Kamada Y, Yoshino K, Kondo C, Kawamata T, Oshiro N, Yonezawa K, Ohsumi Y. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol Cell Biol. 2010;30:1049–1058. doi: 10.1128/MCB.01344-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Thevissen K, Yen WL, Carmona-Gutierrez D, Idkowiak-Baldys J, Aerts AM, Francois IE, Madeo F, Klionsky DJ, Hannun YA, Cammue BP. Skn1 and Ipt1 negatively regulate autophagy in Saccharomyces cerevisiae. FEMS Microbiol Lett. 2010;303:163–168. doi: 10.1111/j.1574-6968.2009.01869.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lavieu G, Scarlatti F, Sala G, Carpentier S, Levade T, Ghidoni R, Botti J, Codogno P. Sphingolipids in macroautophagy. Methods Mol Biol. 2008;445:159–173. doi: 10.1007/978-1-59745-157-4_11. [DOI] [PubMed] [Google Scholar]
- 96.Aronova S, Wedaman K, Aronov PA, Fontes K, Ramos K, Hammock BD, Powers T. Regulation of ceramide biosynthesis by TOR complex 2. Cell Metab. 2008;7:148–158. doi: 10.1016/j.cmet.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jiang JC, Kirchman PA, Zagulski M, Hunt J, Jazwinski SM. Homologs of the yeast longevity gene LAG1 in Caenorhabditis elegans and human. Genome Res. 1998;8:1259–1272. doi: 10.1101/gr.8.12.1259. [DOI] [PubMed] [Google Scholar]
- 98.Guillas I, Kirchman PA, Chuard R, Pfefferli M, Jiang JC, Jazwinski SM, Conzelmann A. C26-CoA-dependent ceramide synthesis of Saccharomyces cerevisiae is operated by Lag1p and Lac1p. EMBO J. 2001;20:2655–2665. doi: 10.1093/emboj/20.11.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schorling S, Vallee B, Barz WP, Riezman H, Oesterhelt D. Lag1p and Lac1p Are Essential for the Acyl-CoA-dependent Ceramide Synthase Reaction in Saccharomyces cerevisae. Mol Biol Cell. 2001;12:3417–3427. doi: 10.1091/mbc.12.11.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.D’Mello PN, Childress AM, Franklin DS, Kale SP, Pinswasdi C, Jazwinski SM. Cloning and characterization of LAG1, a longevity-assurance gene in yeast. J Biol Chem. 1994;269:15451–15459. [PubMed] [Google Scholar]
- 101.Guillas I, Jiang JC, Vionnet C, Roubaty C, Uldry D, Chuard R, Wang J, Jazwinski SM, Conzelmann A. Human homologues of LAG1 reconstitute Acyl-CoA-dependent ceramide synthesis in yeast. J Biol Chem. 2003;278:37083–37091. doi: 10.1074/jbc.M307554200. [DOI] [PubMed] [Google Scholar]
- 102.Venkataraman K, Riebeling C, Bodennec J, Riezman H, Allegood JC, Sullards MC, Merrill AH, Jr, Futerman AH. Upstream of growth and differentiation factor 1 (uog1), a mammalian homolog of the yeast longevity assurance gene 1 (LAG1),regulates N-stearoyl-sphinganine (C18-(dihydro)ceramide) synthesis in a fumonisin B1-independent manner in mammalian cells. J Biol Chem. 2002;277:35642–35649. doi: 10.1074/jbc.M205211200. [DOI] [PubMed] [Google Scholar]
- 103.Teufel A, Maass T, Galle PR, Malik N. The longevity assurance homologue of yeast LAG1 (Lass) gene family. Int J Mol Med. 2009;23:135–140. [PubMed] [Google Scholar]
- 104.Kolaczkowski M, Kolaczkowska A, Gaigg B, Schneiter R, Moye-Rowley WS. Differential regulation of ceramide synthase components LAC1 and LAG1 in Saccharomyces cerevisiae. Eukaryot Cell. 2004;3:880–892. doi: 10.1128/EC.3.4.880-892.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hallstrom TC, Moye-Rowley WS. Multiple signals from dysfunctional mitochondria activate the pleiotropic drug resistance pathway in Saccharomyces cerevisiae. J Biol Chem. 2000;275:37347–37356. doi: 10.1074/jbc.M007338200. [DOI] [PubMed] [Google Scholar]
- 106.Moye-Rowley WS. Retrograde regulation of multidrug resistance in Saccharomyces cerevisiae. Gene. 2005;354:15–21. doi: 10.1016/j.gene.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 107.Kornmann B, Walter P. ERMES-mediated ER-mitochondria contacts: molecular hubs for the regulation of mitochondrial biology. J Cell Sci. 2010;123:1389–1393. doi: 10.1242/jcs.058636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dickson RC. Roles for sphingolipids in Saccharomyces cerevisiae. Adv Exp Med Biol. 2010;688:217–231. doi: 10.1007/978-1-4419-6741-1_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jiang JC, Kirchman PA, Allen M, Jazwinski SM. Suppressor analysis points to the subtle role of the LAG1 ceramide synthase gene in determining yeast longevity. Exp Gerontol. 2004;39:999–1009. doi: 10.1016/j.exger.2004.03.026. [DOI] [PubMed] [Google Scholar]
- 110.Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kissova I, Salin B, Schaeffer J, Bhatia S, Manon S, Camougrand N. Selective and non-selective autophagic degradation of mitochondria in yeast. Autophagy. 2007;3:329–336. doi: 10.4161/auto.4034. [DOI] [PubMed] [Google Scholar]
- 112.Okamoto K, Kondo-Okamoto N, Ohsumi Y. A landmark protein essential for mitophagy: Atg32 recruits the autophagic machinery to mitochondria. Autophagy. 2009;5:1203–1205. doi: 10.4161/auto.5.8.9830. [DOI] [PubMed] [Google Scholar]
- 113.Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. De Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 114.Kanki T, Wang K, Baba M, Bartholomew CR, Lynch-Day MA, Du Z, Geng J, Mao K, Yang Z, Yen WL, Klionsky DJ. A genomic screen for yeast mutants defective in selective mitochondria autophagy. Mol Biol Cell. 2009;20:4730–4738. doi: 10.1091/mbc.E09-03-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kanki T, Klionsky DJ. Atg32 is a tag for mitochondria degradation in yeast. Autophagy. 2009;5:1201–1202. doi: 10.4161/auto.5.8.9747. [DOI] [PubMed] [Google Scholar]
- 116.Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Scheckhuber CQ, Erjavec N, Tinazli A, Hamann A, Nystrom T, Osiewacz HD. Reducing mitochondrial fission results in increased life span and fitness of two fungal ageing models. Nat Cell Biol. 2007;9:99–105. doi: 10.1038/ncb1524. [DOI] [PubMed] [Google Scholar]
- 118.Journo D, Mor A, Abeliovich H. Aup1-mediated regulation of Rtg3 during mitophagy. J Biol Chem. 2009;284:35885–35895. doi: 10.1074/jbc.M109.048140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Okamoto K, Shaw JM. Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annu Rev Genet. 2005;39:503–536. doi: 10.1146/annurev.genet.38.072902.093019. [DOI] [PubMed] [Google Scholar]
- 120.Frederick RL, Okamoto K, Shaw JM. Multiple pathways influence mitochondrial inheritance in budding yeast. Genetics. 2008;178:825–837. doi: 10.1534/genetics.107.083055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Simon VR, Karmon SL, Pon LA. Mitochondrial inheritance: cell cycle and actin cable dependence of polarized mitochondrial movements in Saccharomyces cerevisiae. Cell Motil Cytoskeleton. 1997;37:199–210. doi: 10.1002/(SICI)1097-0169(1997)37:3<199::AID-CM2>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 122.Mortimer RK, Johnston JR. Life span of individual yeast cells. Nature. 1959;183:1751–1752. doi: 10.1038/1831751a0. [DOI] [PubMed] [Google Scholar]
- 123.Egilmez NK, Jazwinski SM. Evidence for the involvement of a cytoplasmic factor in the aging of the yeast Saccharomyces cerevisiae. J Bacteriol. 1989;171:37–42. doi: 10.1128/jb.171.1.37-42.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kennedy BK, Austriaco NR, Jr, Guarente L. Daughter cells of Saccharomyces cerevisiae from old mothers display a reduced life span. J Cell Biol. 1994;127:1985–1993. doi: 10.1083/jcb.127.6.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jazwinski SM, Egilmez NK, Chen JB. Replication control and cellular life span. Exp Gerontol. 1989;24:423–436. doi: 10.1016/0531-5565(89)90049-1. [DOI] [PubMed] [Google Scholar]
- 126.Jazwinski SM. Longevity, genes, and aging. Science. 1996;273:54–59. doi: 10.1126/science.273.5271.54. [DOI] [PubMed] [Google Scholar]
- 127.Jazwinski SM. The genetics of aging in the yeast Saccharomyces cerevisiae. Genetica. 1993;91:35–51. doi: 10.1007/BF01435986. [DOI] [PubMed] [Google Scholar]
- 128.Lam YT, Aung-Htut MT, Lim YL, Yang H, Dawes IW. Changes in reactive oxygen species begin early during replicative aging of Saccharomyces cerevisiae cells. Free Radic Biol Med. 2011;50:963–970. doi: 10.1016/j.freeradbiomed.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 129.McFaline-Figueroa JR, Vevea J, Swayne TC, Zhou C, Liu C, Leung G, Boldogh IR, Pon LA. Mitochondrial quality control during inheritance is associated with lifespan and mother-daughter age asymmetry in budding yeast. Aging Cell. 2011;10:885–895. doi: 10.1111/j.1474-9726.2011.00731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Seo JG, Lai CY, Miceli MV, Jazwinski SM. A novel role of peroxin PEX6: suppression of aging defects in mitochondria. Aging Cell. 2007;6:405–413. doi: 10.1111/j.1474-9726.2007.00291.x. [DOI] [PubMed] [Google Scholar]