Abstract
Nutrient and energy metabolism in mammals exhibits strong diurnal rhythm that aligns with the body clock. Circadian regulation of metabolism is mediated through reciprocal signaling between the clock and metabolic regulatory networks. Recent work has demonstrated that autophagy is rhythmically activated in a clock-dependent manner. As autophagy is a conserved biological process that contributes to nutrient and cellular homeostasis, its cyclic induction may provide a novel link between clock and metabolism. This review discusses the mechanisms underlying circadian autophagy regulation, the role of rhythmic autophagy in nutrient and energy metabolism, and its implications in physiology and metabolic disease.
Nutrient and energy metabolism is coupled to timing cues
Nutrient and energy metabolism is temporally organized in mammalian tissues to synchronize the storage and utilization of energy with light/dark cycles [1-3]. Circulating metabolites and hormones ebb and flow according to distinct diurnal patterns. In addition, rhythmic metabolic gene expression is prevalent in major metabolic tissues, such as the liver, adipose tissue, and skeletal muscle [4, 5]. As a consequence, the activities of many metabolic pathways are restricted not only to specific tissues in the body, but also to unique periods during the day. For example, hepatic gluconeogenesis, de novo lipogenesis, VLDL secretion, cholesterol biosynthesis, and xenobiotic detoxification are precisely timed and reach their respective peaks at different time [6-10]. These observations form the basis for the emerging concept that nutrient and energy metabolism is tightly coupled to the timing cues in mammalian tissues. The temporal restriction of metabolic functions may provide advantages for organisms as they anticipate and synchronize their feeding and activity cycles to the environment.
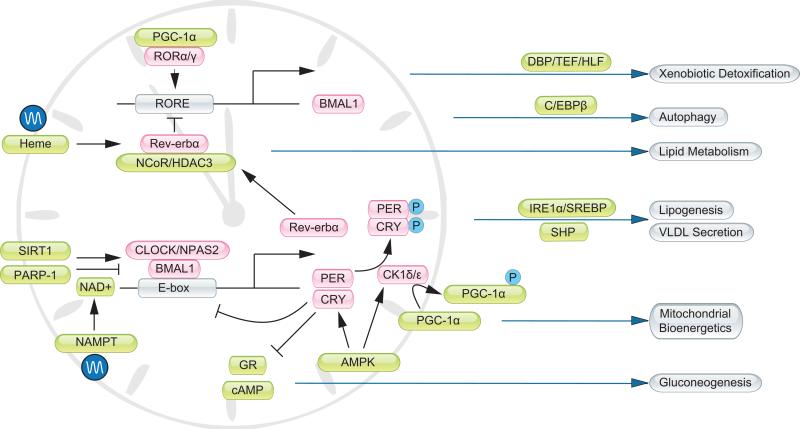
The integration of clock and metabolism is mediated through reciprocal crosstalk between these two regulatory networks (Figure 1). In mammals, the central clock in suprachiasmatic nucleus (SCN) responds to light and drives diverse behavioral and physiological cycles in the body [11]. This master clock effectively sets the phase of peripheral tissue clocks. At the molecular level, biological clock comprises factors that act in concert to drive rhythmic gene expression in the SCN and peripheral tissues (Box 1). Transcriptional profiling revealed that a large number of genes involved in glucose and lipid metabolism are temporally controlled [12-16]. Recent chromatin-immunoprecipitation sequencing studies support the notion that many of these rhythmically expressed genes are direct transcriptional targets of clock genes, such as Bmal1 and Rev-erbα [17, 18]. Clock exerts its physiological effects in part through these direct transcriptional targets. For example, diurnal regulation of xenobiotic detoxification is mediated through the circadian PAR-domain basic leucine zipper transcription factors DBP/TEF/HLF, all of which are clock targets [6]. Hepatic lipogenesis and lipoprotein secretion are rhythmically controlled by several factors, including the transcription factor SREBP, small heterodimer partner (SHP), and the histone deacetylase HDAC3 [10, 18, 19].
Figure 1. Integration of clock and metabolism.
Core components of the clock oscillator (pink) are gated by factors that relay nutrient and hormonal signals (green). In parallel, the timing cues are integrated with the metabolic regulatory network to drive rhythmic metabolic gene expression and output. GR, glucocorticoid receptor; RORE, Rev-erb/ROR responsive element.
Nuclear hormone receptors (NHR) are a family of transcriptional regulators that respond to diverse classes of metabolites and play important roles in metabolic regulation. The expression of many NHRs exhibits circadian regulation [20], some of which also directly interact with clock proteins [21, 22], potentially synchronizing the expression of clock and metabolic genes. For example, Rev-erbα, a NHR and also a core clock components, regulates circadian SREBP signaling and bile acid homeostasis [23]. NHRs control the expression of target genes through recruiting coactivator and corepressor proteins that participate in chromatin remodeling and epigenetic regulation [24, 25]. Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α) is a transcriptional coactivator initially found to stimulate mitochondrial biogenesis, fatty acid β-oxidation, brown fat thermogenesis, and hepatic gluconeogenesis [26, 27]. Recent work demonstrated that PGC-1α also directly regulates the expression of core clock genes, such as Bmal1 and Rev-erbα, and is indispensible for circadian pacemaker function [28] (Box 1). The expression of PGC-1α is diurnally regulated and is modulated by casein kinase 1δ [29], an important regulator of the clock oscillator. Similarly, HDAC3 is recruited to the NHR Rev-erbα and regulates a program of metabolic and clock gene expression in the liver [18, 30, 31]. As such, the regulatory networks that govern clock and metabolism are highly intertwined and integrated.
Nutrient signaling exerts profound effects on the clock network (reviewed by Peek et al., in this issue). A notable example of nutrient sensing in the cell is via NAD+ and NAD+-dependent regulatory proteins. Sirtuin 1 (SIRT1) is an NAD+-dependent histone deacetylase that deacetylates several clock proteins [32, 33]. Poly (ADP-ribose) polymerase 1 (PARP-1), an NAD+-dependent ADP-ribosyltransferase, poly(ADP-ribosyl)ates Clock and alters the affinity of the Bmal1/Clock transcriptional complex (Box 1) to its target DNA [34]. PARP-1 also regulates SIRT1 activity indirectly through its modulation of NAD+ levels in the cell [35]. In parallel, the AMP-activated protein kinase (AMPK), a sensor for cellular AMP/ATP ratio, phosphorylates clock proteins such as Cry and CK1ε [36, 37]. Because intracellular NAD+ levels and energy charge are regulated by nutrient status, these studies highlight a direct role for metabolic signaling in fine-tuning pacemaker function. The reciprocal crosstalk between the clock and metabolic regulatory networks potentially provides a real-time mechanism for synchronizing cellular metabolism with other biological processes.
Perturbations of clock function have been associated with elevated risk for certain diseases in humans, including sleep disorder, metabolic syndrome, cardiovascular disease, rheumatoid arthritis, and cancer [2, 38]. Acute disruption of sleep rhythm in healthy individuals results in decreased insulin sensitivity while chronic circadian misalignment increases the risk for metabolic disorders in shift workers [39-41]. Various clock-deficient animal models have been generated and characterized in recent years. Clock mutant mice develop symptoms reminiscent of metabolic syndrome, whereas pancreatic islets lacking clock have impaired glucose-stimulated insulin secretion [42, 43]. Disruption of liver clock perturbs hepatic gluconeogenesis, lipid metabolism, and bile acid homeostasis [23, 44]. Exposure of mice to inverted circadian environment has also been shown to cause excess weight gain [45]. These studies underscore a potentially important role for circadian misalignment in the pathogenesis of metabolic disorders in humans.
Circadian regulation of autophagy
Autophagy literally means ‘self-eating’ and refers to the process that initiates with the formation of isolation membranes, which then elongate, engulf cytosolic components, and form enclosed vesicles called autophagosome [46]. The autophagosome subsequently fuses with lysosome to form autolysosome, where degradation occurs (Figure 2). The identification of factors that carry out autophagy has provided a molecular framework for autophagic degradation and its physiological significance [47] (Box 2). These studies have led to the conclusion that autophagy is critical for cellular homeostasis and nutrient metabolism in the starvation state. Autophagy is induced in neonatal tissues and in adult tissues in response to starvation [48, 49]. Defects in autophagy induction result in lower plasma glucose and amino acid levels and compromise survival during the early postnatal period. In parallel, autophagy is required for removing protein aggregates, damaged organelles, and certain pathogens [50]. Autophagy deficiency has been implicated in the pathogenesis of various disease conditions, such as cancer, diabetes, hepatic steatosis, skeletal myopathy and neurodegeneration [51-56].
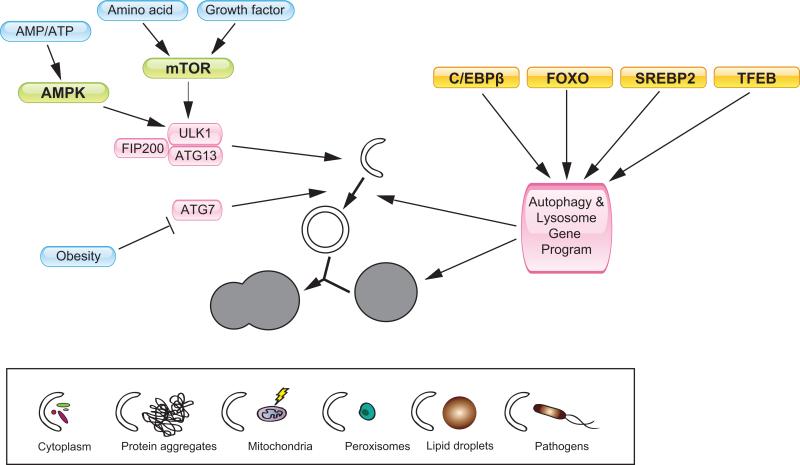
Figure 2. Transcriptional and post-translational regulation of autophagy.
Autophagy is involved in the degradation of certain cellular components, such as cytoplasmic material, protein aggregates, damaged mitochondria, peroxisomes, lipid droplets, and certain pathogens (lower box). Nutrient regulation of autophagy is mediated by mTOR and AMPK, which phosphorylate components of the ULK1-FIP200-ATG13 complex. Reduced Atg7 expression contributes to impaired autophagy in obesity state [89]. The autophagy and lysosome gene program is controlled by several transcription factors, including C/EBPβ, FOXO, SREBP2, and TFEB.
In the 1970s, a series of electron microscopy studies demonstrated that the number of autophagic vacuoles varies throughout the day in several tissues, including the inner segment of retina rod cells, cardiomyocytes, hepatocytes, pancreatic acinar cells, and proximal tubules of kidney in rats [57, 58]. Using more specific molecular markers for autophagy, recent studies indicated that autophagy activity exhibits robust diurnal rhythm in several mouse tissues, including the liver, heart, and skeletal muscle [59]. When autophagy is activated, the rate of autophagosome formation, its conversion to autolysosome, and subsequent degradation in lysosome are increased. Autophagy flux can be estimated by examining the degradation rate of autophagic protein marker microtubule-associated protein1 light chain 3 (LC3). This measurement revealed that autophagy flux reached a peak in the afternoon and decreased to lower levels in the dark phase. In addition, the cyclic activation of autophagy flux in the liver is associated with changes in autophagosome abundance and rhythmic expression of autophagy genes. The expression of several genes in the autophagy and lysosomal pathways was also found to oscillate in the yeast during continuous growth under nutrient-limited conditions [60]. In this case, autophagy appears to be restricted to a specific temporal phase that is associated with reductive metabolic activities. While the timing cues that drive rhythmic autophagy activation in mammalian tissues and in yeast cells are likely different, the autophagy cycles may reflect a conserved property of cellular metabolism and homeostasis.
Transcriptional regulation of autophagy rhythm
The nature of timing cues that drive circadian autophagy appears to involve both clock and nutritional signals [59]. Liver-specific Bmal1 null mice have dampened rhythm of autophagy gene expression and autophagy flux, suggesting that clock exerts its effects on circadian autophagy, at least in part, through cell-autonomous mechanisms [59]. Nutritional status provides a strong entrainment signal for peripheral tissues. Restriction feeding resets the phase of peripheral clocks in rodents without affecting the central clock (Box 1) [61]. Notably, the phase of autophagy gene expression is also reversed following the feeding switch [59, 62]. While meal timing dominantly resets the phase of autophagy rhythm, it remains unknown whether this is secondary to the realignment of clock with feeding status.
Transcriptional regulation of autophagy genes is emerging as an important mechanism in the control of cellular autophagy activity. To date, several transcription factors have been identified that regulate various aspects of the autophagy gene program (Figure 2). In the context of circadian autophagy, C/EBPβ appears to play a crucial role in coordinating rhythmic expression of autophagy genes [59]. C/EBPβ is a basic leucine zipper transcription factor that regulates diverse biological processes, including immune response, cell differentiation, and metabolism [63-67]. The expression of C/EBPβ is highly rhythmic and is regulated by the liver clock in a tissue-autonomous manner. Adenoviral-mediated expression of C/EBPβ stimulates the program of autophagy gene expression and induces autophagic protein degradation in cultured hepatocytes. C/EBPβ directly binds to the promoters of autophagy genes and activates their transcription [59]. Similar to C/EBPβ, Forkhead transcription factor O3 (FoxO3) induces the expression of several autophagy genes in skeletal myocytes, including LC3B, Gabarapl1, Bnip3, and Bnip3l [68, 69]. This regulation of autophagic protein degradation by FoxO3 contributes to muscle atrophy induced by starvation. In addition to FoxO3, FoxO1 also regulates autophagy in cardiomyocytes [70]. A recent chromatin-immunoprecipitation sequencing study revealed an unexpected role for sterol regulatory element binding protein 2 (SREBP2) in the regulation of autophagy gene expression in the liver [71]. In this case, SREBP2 may provide a link between autophagy and cellular sterol homeostasis. Recent studies also demonstrate that the transcription factor TFEB controls a large number of genes involved in autophagy and lysosome dynamics in HeLa cells, and is sufficient to promote lysosome biogenesis, autophagy, and lysosomal exocytosis [72, 73]. Interestingly, TFEB is localized in cytosolic compartment under normal growth conditions, and translocates into the nucleus in response to lysosomal stress or nutrient limitation. Whether the FoxO transcription factors, SREBP2, and TFEB also contribute to circadian regulation of autophagy remains currently unknown.
In addition to the transcriptional mechanisms, it has been recognized that mTOR plays an important role in mediating nutrient regulation of autophagy. When nutrients are abundant, active mTOR physically interacts with the Ulk1-FIP200-Atg13 complex and phosphorylates Ulk1 and Atg13, thereby inhibiting Ulk1 kinase activity and autophagy induction [74-76].
Besides the mTOR complex, AMPK also regulates autophagy directly by phosphorylating Ulk1 and this particular phosphorylation increases the kinase activity of Ulk1 and promotes autophagy [77-79]. While the mTOR and AMPK pathways appear to undergo circadian regulation, their role in driving rhythmic autophagy activation in tissues remains to be explored [37, 80].
Rhythmic autophagy: a link between clock and metabolism?
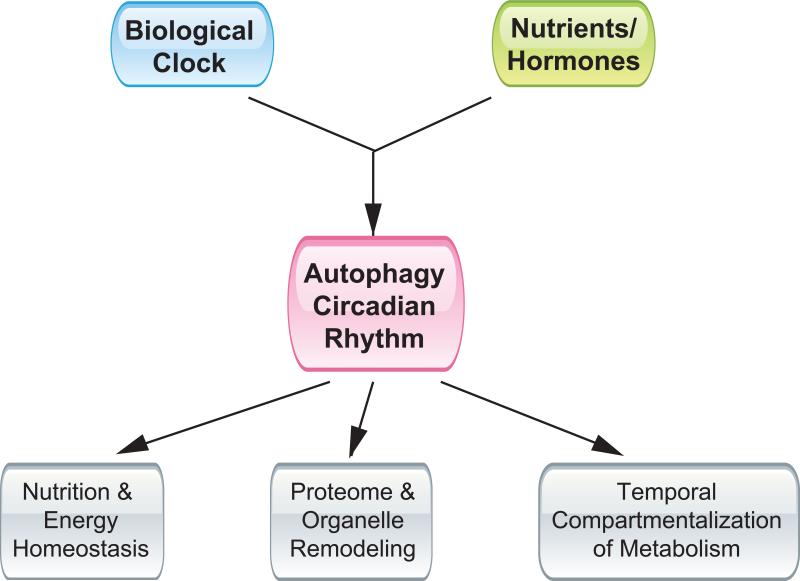
While it is evident that autophagy is rhythmically activated in the body, the significance of autophagy cycles in physiology and disease is far from clear. Conceptually, close coupling of autophagic degradation to the biological clock may provide distinct advantages for multicellular organisms to maintain nutrient and energy homeostasis, remodel proteomes and organelles, and achieve temporal compartmentalization of tissue metabolism.
Nutrient and energy homeostasis
A major function of autophagy is to degrade cellular components when nutrients become limited. The concentrations of plasma amino acids and metabolites exhibit robust circadian oscillation [81, 82]. Interestingly, diurnal regulation of branched-chain amino acid levels in plasma, the liver, and skeletal muscle in fasted state is diminished in liver-specific Atg7-deficient mice [82]. Disruption of circadian autophagy rhythm in liver-specific Bmal1 null mice is also associated with impaired hepatic gluconeogenesis and hypoglycemia during late light phase [44], which coincides with peak autophagy flux. Recent studies have implicated autophagy in the catabolism of triglycerides stored in lipid droplets [53]. Because the expression of genes involved in de novo lipogenesis, cholesterol biosynthesis, and fatty acid β-oxidation is highly rhythmic in the liver, it is likely that the circadian regulation of these metabolic cycles is coordinated with autophagy to optimize the supply of nutrients for storage or oxidation.
Proteome and organelle remodeling
Rhythmic autophagic induction may be important for temporal remodeling of proteomes and organelles. This dynamic regulation of cellular proteomes and organelles enables the cells to adjust their functions in response to specific physiological needs. A surprising finding with regard to the circadian regulation of the hepatic proteome and transcriptome came from comparative analysis of diurnal regulation of protein and mRNA expression [83]. While up to 20% of soluble proteins assayed in mouse liver exhibit circadian oscillation, nearly half of them lack corresponding mRNA cycles. Additionally, these oscillated proteins tend to peak in the dark phase when autophagy activity is lower [57, 59, 83], raising the possibility that circadian autophagy could play a role in selective proteome remodeling in the liver. Interestingly, the abundance of mitochondria, peroxisomes, and endoplasmic reticulum varies throughout the day, which is likely mediated through cyclic activation of autophagy-mediated turnover [84, 85]. The periodical removal of mitochondria and other organelles may facilitate the adjustment of the bioenergetic properties throughout different circadian phases.
Temporal compartmentalization of tissue metabolism
Autophagy-mediated proteome and organelle turnover may provide an important mechanism for the temporal compartmentalization of tissue metabolism. Biological rhythm is an intrinsic cellular property that is conserved from single-cell eukaryotes to different kingdoms of multicellular eukaryotes. Yeast grown under nutrient-limited condition exhibits robust cycles of oxygen consumption and redox changes in the cell [60]. In each phase, a subset of oscillating genes peaks, including those involved in ubiquitin/proteasome function and autophagy. It is possible that this rhythmic induction of degradation pathways is necessary for large-scale removal of cellular components that paves the way for metabolic phase transition. As such, the metabolic functions in higher organisms are not only restricted to specific tissues, but also compartmentalized along the temporal axis. The cyclic activation of autophagy may alter the composition and/or functional characteristics of cellular proteomes and organelles, thus defining distinct temporal compartments of nutrient and energy metabolism.
Implications of autophagy rhythm in metabolic disease
Autophagy is a fundamental cellular process that has been implicated in various disease conditions [50]. Deletion of Beclin 1, also known as autophagy-related gene 6 (Atg6), a protein required for the initiation of autophagosome formation, was found in patients with breast cancer [54]. Genetic deletion of Atg5 or Atg7 in the liver leads to the development of benign liver adenomas, likely as a result of mitochondrial dysfunction, oxidative stress, and impaired DNA damage response [86]. Because autophagy is critical for the removal of protein aggregates, defects in autophagy have also been linked to the pathogenesis of neurodegenerative disease, muscular dystrophy as well as liver damage caused by mutations in α1-antitrypsin [55, 56, 87]. Genetic and pharmacological activation of autophagy alleviates disease progression and severity in these animal models.
Potential involvement of autophagy in the pathogenesis of metabolic diseases is drawing increasing attention. Autophagy activity appears to be reduced in the liver in diet-induced and genetically obese mice [88, 89]. Importantly, rescue of autophagy function in the liver restores hepatic insulin signaling and glucose homeostasis. Autophagy also plays a direct role in the hydrolysis of triglycerides stored in lipid droplets [53]. In this case, lysosomal hydrolysis of triglycerides provides a previously unappreciated mechanism for lipid hydrolysis and fatty acid β-oxidation. As hepatic steatosis is a common feature in insulin resistant state, it is possible that defects in autophagy may contribute to excess triglyceride accumulation in the liver. The extent to which reduced autophagy contributes to hepatic steatosis and potentially non-alcoholic steatohepatitis remains to be established. A second pathway that links autophagy to hepatic lipid metabolism is through autophagy-mediated degradation of Apolipoprotein B (ApoB). Under physiological conditions, a significant proportion of nascent ApoB-containing VLDL particles is diverted from the secretory pathway and towards autophagic degradation [90]. It is possible that defective clearance of these lipid-containing particles may further aggravate hepatic steatosis. Hepatic VLDL secretion is itself diurnally regulated, and as such, autophagy-mediated ApoB degradation may be coupled to the circadian regulation of VLDL secretion [91]. Finally, autophagy is also required for adipogenesis, insulin secretion by β-cells as well as muscle metabolism and function [51, 52, 55, 92, 93]. The coupling of autophagy and metabolism, and importantly, their alignment to the body clock are emerging as a novel factor underlying metabolic homeostasis and disease.
Concluding remarks
Nutrient and energy metabolism in mammalian tissues exhibits strong diurnal rhythms. The orchestration of metabolic rhythm requires time-dependent transitions between metabolic states and their alignment to the biological clock. In this context, cyclic activation of autophagy throughout light/dark cycles could in principle fulfill several critical roles. Autophagic degradation provides energy and biosynthetic intermediates to help maintain cellular and systemic homeostasis between feeding cycles. The periodic activation of autophagy may contribute to proteome turnover and organelle homeostasis, which collectively define different metabolic phases and the transition among these temporal compartments in the tissue. Because autophagy is emerging as an important process for metabolic homeostasis, future work is needed to assess the significance of autophagy cycles in physiology and the extent to which its disruption contributes to the pathogenesis of metabolic disease.
Box 1. The core components of the mammalian clock.
To adapt to the daily light/dark cycle, some prokaryotes and most eukaryotes have evolved mechanisms to synchronize their behavior and physiology to the environment. Although the exact molecular players are not the same, all of these biological timing systems consist of positive and negative regulators assembled into auto-regulatory feedback loops [94-96]. In mammals, the Bmal1/Clock and Bmal1/Npas2 transcriptional complexes are key activators that constitute the positive arm of the molecular clock. The expression of Bmal1 itself is under the control of nuclear receptor RORα/γ and transcriptional coactivator PGC-1α. Major target genes of Bmal1 include Period 1 (Per1), Per2, Per3, Cryptochrome 1 (Cry1), and Cry2, which form repressor complexes that inhibit the transcriptional activity of Bmal1. In addition, Bmal1 also induces the expression of Rev-erbα, which forms repressor complexes with NCoR and HDAC3 and inhibits Bmal1 expression [30, 97]. These negative feedback loops serve as the molecular basis for the generation of transcriptional rhythm. Casein kinase 1δ (CK1δ) and 1ε phosphorylate Per and Cry proteins, leading to their proteasomal degradation. In addition to phosphorylation, the components of the core clock are regulated by other post-translational mechanisms including acetylation, deacetylation, and ubiquitination, which modulate the stability and/or activities of clock proteins [98].
The molecular clock exists in diverse cell types in the body [95, 96]. The central clock refers to the circadian pacemaker residing in the SCN, which is essential for light entrainment of behavioral and physiological cycles. The clocks in peripheral tissues, such as the liver, lung and kidney, are highly responsive to nutrient and hormonal signals derived from feeding. Under restriction feeding conditions, the peripheral clocks can be entrained by meal timing independently and become uncoupled from the central clock [61]. In humans, disruption of circadian rhythm has been associated with increased risk for obesity and cardiovascular disease [99]. Genetic disruption of core clock components results in metabolic perturbations reminiscent of metabolic syndrome, including obesity, α-cell dysfunction and hyperlipidemia [43, 44, 100].
Box 2. Molecular machinery of autophagy.
Autophagy is a highly conserved cellular process that defends the cell against acute nutrient deprivation and certain stresses. Under conditions of limited nutrient access, such as starvation and the interruption of placental nutrient supply in neonates, autophagy can be rapidly and robustly induced in multiple tissues [48, 49]. Restriction of various types of nutrients, such as amino acids, growth factors, oxygen, and energy, can also induce autophagy in the mammalian cells [50]. This induction of autophagy is in part mediated by the mTOR and AMPK signaling pathways [101]. Certain stress conditions, such as hypoxia, mitochondria damage, ER stress, and immune signals can also induce autophagy through distinct downstream mediators [102, 103]. In addition to the post-translational regulation, recent studies suggest transcriptional regulation is also important underlying mechanisms of autophagy activation [59, 68, 69, 72].
Autophagosomes are generated in close proximity to the endoplasmic reticulum (ER), with membrane supply from Golgi complex, mitochondria, and plasma membrane [47, 101]. A number of factors have been identified to play an essential role in autophagosome formation. They constitute five macromolecular complexes: Ulk1-FIP200-Atg13-Atg101 (Ulk1 complex), Beclin-PI3-kinase-Atg14, Atg9-vacuole membrane protein 1 (VMP1), Atg5-Atg12-Atg16L1, and the LC3-phosphatidylethanolamine (PE) conjugation complex [101]. The Ulk1 complex is regulated by mTORC1 and AMPK through phosphorylation and serves as a potential gatekeeper for the autophagy machinery [79, 104-108]. Upon autophagy induction, the Ulk1 complex translocates to discrete domains on ER, where it regulates the Beclin-PI3-kinase-Atg14 complex. Formation of phosphatidylinositol 3-phosphate (PI3P) by Beclin-PI3-kinase-Atg14 complex recruits double FYVE-containing protein 1 and WIPI proteins. These proteins promote the formation and maturation of isolation membrane, the precursor of autophagosome [50], while the Atg5-Atg12-Atg16L1 complex and LC3-PE conjugation complex promotes the elongation and closure of the isolation membrane. A number of adaptor proteins, such as p62 and Bnip3l (NIX), mediate the degradation of selective cargoes through autophagy, such as ubiquitinated protein aggregates, damaged mitochondria, and pathogens [50] (Figure 2).
Figure 3. Autophagy rhythm and diurnal metabolic homeostasis.
Rhythmic activation of autophagy is controlled by the biological clock as well as nutritional signals, and may contribute to nutritional and energy homeostasis through light/dark cycles, proteome and organelle remodeling, and the temporal compartmentalization of tissue metabolism.
Acknowledgments
We would like to thank Drs. Daniel Klionsky and Jun-Lin Guan for discussions. This work was supported by the National Institutes of Health (DK077086, HL097738), American Diabetes Association, and American Heart Association. We apologize to colleagues whose relevant work was not cited here due to space limitation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Asher G, Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 2011;13:125–137. doi: 10.1016/j.cmet.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 2.Green CB, et al. The meter of metabolism. Cell. 2008;134:728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rutter J, et al. Metabolism and the control of circadian rhythms. Annu Rev Biochem. 2002;71:307–331. doi: 10.1146/annurev.biochem.71.090501.142857. [DOI] [PubMed] [Google Scholar]
- 4.Lowrey PL, Takahashi JS. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baggs JE, Hogenesch JB. Genomics and systems approaches in the mammalian circadian clock. Curr Opin Genet Dev. 2010;20:581–587. doi: 10.1016/j.gde.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gachon F, et al. The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metab. 2006;4:25–36. doi: 10.1016/j.cmet.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 7.Hems DA, et al. Fatty acid synthesis in liver and adipose tissue of normal and genetically obese (ob/ob) mice during the 24-hour cycle. Biochem J. 1975;150:167–173. doi: 10.1042/bj1500167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phillips LJ, Berry LJ. Circadian rhythm of mouse liver phosphoenolpyruvate carboxykinase. Am J Physiol. 1970;218:1440–1444. doi: 10.1152/ajplegacy.1970.218.5.1440. [DOI] [PubMed] [Google Scholar]
- 9.Edwards PA, et al. In vivo demonstration of the circadian thythm of cholesterol biosynthesis in the liver and intestine of the rat. J Lipid Res. 1972;13:396–401. [PubMed] [Google Scholar]
- 10.Pan X, et al. Diurnal regulation of MTP and plasma triglyceride by CLOCK is mediated by SHP. Cell Metab. 2010;12:174–186. doi: 10.1016/j.cmet.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reppert SM, Weaver DR. Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol. 2001;63:647–676. doi: 10.1146/annurev.physiol.63.1.647. [DOI] [PubMed] [Google Scholar]
- 12.McCarthy JJ, et al. Identification of the circadian transcriptome in adult mouse skeletal muscle. Physiol Genomics. 2007;31:86–95. doi: 10.1152/physiolgenomics.00066.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Panda S, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 14.Storch KF, et al. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 15.Ueda HR, et al. A transcription factor response element for gene expression during circadian night. Nature. 2002;418:534–539. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- 16.Zvonic S, et al. Characterization of peripheral circadian clocks in adipose tissues. Diabetes. 2006;55:962–970. doi: 10.2337/diabetes.55.04.06.db05-0873. [DOI] [PubMed] [Google Scholar]
- 17.Rey G, et al. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol. 2011;9:e1000595. doi: 10.1371/journal.pbio.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng D, et al. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331:1315–1319. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cretenet G, et al. Circadian clock-coordinated 12 Hr period rhythmic activation of the IRE1alpha pathway controls lipid metabolism in mouse liver. Cell Metab. 2010;11:47–57. doi: 10.1016/j.cmet.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 20.Yang X, et al. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 21.Schmutz I, et al. The mammalian clock component PERIOD2 coordinates circadian output by interaction with nuclear receptors. Genes Dev. 2011;24:345–357. doi: 10.1101/gad.564110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lamia KA, et al. Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature. 2011 doi: 10.1038/nature10700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Martelot G, et al. REV-ERBalpha participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biol. 2009;7:e1000181. doi: 10.1371/journal.pbio.1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosenfeld MG, et al. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes & development. 2006;20:1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- 25.Spiegelman BM, Heinrich R. Biological control through regulated transcriptional coactivators. Cell. 2004;119:157–167. doi: 10.1016/j.cell.2004.09.037. [DOI] [PubMed] [Google Scholar]
- 26.Finck BN, Kelly DP. PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. J Clin Invest. 2006;116:615–622. doi: 10.1172/JCI27794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin J, et al. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 28.Liu C, et al. Transcriptional coactivator PGC-1alpha integrates the mammalian clock and energy metabolism. Nature. 2007;447:477–481. doi: 10.1038/nature05767. [DOI] [PubMed] [Google Scholar]
- 29.Li S, et al. Circadian Metabolic Regulation through Crosstalk between Casein Kinase 1delta and Transcriptional Coactivator PGC-1alpha. Mol Endocrinol. 2011;25:2084–2093. doi: 10.1210/me.2011-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yin L, Lazar MA. The orphan nuclear receptor Rev-erbalpha recruits the N-CoR/histone deacetylase 3 corepressor to regulate the circadian Bmal1 gene. Mol Endocrinol. 2005;19:1452–1459. doi: 10.1210/me.2005-0057. [DOI] [PubMed] [Google Scholar]
- 31.Duez H, Staels B. Rev-erb-alpha: an integrator of circadian rhythms and metabolism. J Appl Physiol. 2009;107:1972–1980. doi: 10.1152/japplphysiol.00570.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Asher G, et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 33.Nakahata Y, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Asher G, et al. Poly(ADP-ribose) polymerase 1 participates in the phase entrainment of circadian clocks to feeding. Cell. 2010;142:943–953. doi: 10.1016/j.cell.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 35.Bai P, et al. PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell Metab. 2011;13:461–468. doi: 10.1016/j.cmet.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Um JH, et al. Activation of 5'-AMP-activated kinase with diabetes drug metformin induces casein kinase Iepsilon (CKIepsilon)-dependent degradation of clock protein mPer2. J Biol Chem. 2007;282:20794–20798. doi: 10.1074/jbc.C700070200. [DOI] [PubMed] [Google Scholar]
- 37.Lamia KA, et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326:437–440. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Copinschi G, et al. Pathophysiology of human circadian rhythms. Novartis Found Symp. 2000;227:143–157. doi: 10.1002/0470870796.ch9. discussion 157-162. [DOI] [PubMed] [Google Scholar]
- 39.Scheer FA, et al. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc Natl Acad Sci U S A. 2009;106:4453–4458. doi: 10.1073/pnas.0808180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spiegel K, et al. Impact of sleep debt on metabolic and endocrine function. Lancet. 1999;354:1435–1439. doi: 10.1016/S0140-6736(99)01376-8. [DOI] [PubMed] [Google Scholar]
- 41.Antunes LC, et al. Obesity and shift work: chronobiological aspects. Nutr Res Rev. 2010;23:155–168. doi: 10.1017/S0954422410000016. [DOI] [PubMed] [Google Scholar]
- 42.Marcheva B, et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466:627–631. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turek FW, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lamia KA, et al. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci U S A. 2008;105:15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fonken LK, et al. Light at night increases body mass by shifting the time of food intake. Proc Natl Acad Sci U S A. 2010;107:18664–18669. doi: 10.1073/pnas.1008734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuma A, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 49.Mizushima N, et al. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 51.Ebato C, et al. Autophagy is important in islet homeostasis and compensatory increase of beta cell mass in response to high-fat diet. Cell Metab. 2008;8:325–332. doi: 10.1016/j.cmet.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 52.Jung HS, et al. Loss of autophagy diminishes pancreatic beta cell mass and function with resultant hyperglycemia. Cell Metab. 2008;8:318–324. doi: 10.1016/j.cmet.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 53.Singh R, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liang XH, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 55.Grumati P, et al. Autophagy is defective in collagen VI muscular dystrophies, and its reactivation rescues myofiber degeneration. Nat Med. 2010;16:1313–1320. doi: 10.1038/nm.2247. [DOI] [PubMed] [Google Scholar]
- 56.Komatsu M, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 57.Pfeifer U, Strauss P. Autophagic vacuoles in heart muscle and liver. A comparative morphometric study including circadian variations in meal-fed rats. J Mol Cell Cardiol. 1981;13:37–49. doi: 10.1016/0022-2828(81)90227-3. [DOI] [PubMed] [Google Scholar]
- 58.Pfeifer U, Scheller H. A morphometric study of cellular autophagy including diurnal variations in kidney tubules of normal rats. J Cell Biol. 1975;64:608–621. doi: 10.1083/jcb.64.3.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ma D, et al. Temporal orchestration of circadian autophagy rhythm by C/EBPbeta. EMBO J. 2011;30:4642–4651. doi: 10.1038/emboj.2011.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tu BP, et al. Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes. Science. 2005;310:1152–1158. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- 61.Damiola F, et al. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pfeifer U. Inverted diurnal rhythm of cellular autophagy in liver cells of rats fed a single daily meal. Virchows Arch B Cell Pathol. 1972;10:1–3. doi: 10.1007/BF02899710. [DOI] [PubMed] [Google Scholar]
- 63.Tanaka T, et al. Targeted disruption of the NF-IL6 gene discloses its essential role in bacteria killing and tumor cytotoxicity by macrophages. Cell. 1995;80:353–361. doi: 10.1016/0092-8674(95)90418-2. [DOI] [PubMed] [Google Scholar]
- 64.Croniger CM, et al. Mice with a deletion in the gene for CCAAT/enhancer-binding protein beta have an attenuated response to cAMP and impaired carbohydrate metabolism. J Biol Chem. 2001;276:629–638. doi: 10.1074/jbc.M007576200. [DOI] [PubMed] [Google Scholar]
- 65.Cao Z, et al. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev. 1991;5:1538–1552. doi: 10.1101/gad.5.9.1538. [DOI] [PubMed] [Google Scholar]
- 66.Akira S, et al. A nuclear factor for IL-6 expression (NF-IL6) is a member of a C/EBP family. EMBO J. 1990;9:1897–1906. doi: 10.1002/j.1460-2075.1990.tb08316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang L, et al. Increased insulin receptor substrate-1 and enhanced skeletal muscle insulin sensitivity in mice lacking CCAAT/enhancer-binding protein beta. J Biol Chem. 2000;275:14173–14181. doi: 10.1074/jbc.m000764200. [DOI] [PubMed] [Google Scholar]
- 68.Mammucari C, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 69.Zhao J, et al. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 70.Sengupta A, et al. FoxO transcription factors promote autophagy in cardiomyocytes. J Biol Chem. 2009;284:28319–28331. doi: 10.1074/jbc.M109.024406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Seo YK, et al. Genome-wide localization of SREBP-2 in hepatic chromatin predicts a role in autophagy. Cell metabolism. 2011;13:367–375. doi: 10.1016/j.cmet.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Settembre C, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sardiello M, et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 74.Ganley IG, et al. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284:12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jung CH, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hosokawa N, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shang L, et al. Nutrient starvation elicits an acute autophagic response mediated by Ulk1 dephosphorylation and its subsequent dissociation from AMPK. Proc Natl Acad Sci U S A. 2011;108:4788–4793. doi: 10.1073/pnas.1100844108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim J, et al. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Egan DF, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Loboda A, et al. Diurnal variation of the human adipose transcriptome and the link to metabolic disease. BMC Med Genomics. 2009;2:7. doi: 10.1186/1755-8794-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Minami Y, et al. Measurement of internal body time by blood metabolomics. Proc Natl Acad Sci U S A. 2009;106:9890–9895. doi: 10.1073/pnas.0900617106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ezaki J, et al. Liver autophagy contributes to the maintenance of blood glucose and amino acid levels. Autophagy. 2011;7:727–736. doi: 10.4161/auto.7.7.15371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Reddy AB, et al. Circadian orchestration of the hepatic proteome. Curr Biol. 2006;16:1107–1115. doi: 10.1016/j.cub.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 84.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Uchiyama Y. Rhythms in morphology and function of hepatocytes. J Gastroenterol Hepatol. 1990;5:321–333. doi: 10.1111/j.1440-1746.1990.tb01634.x. [DOI] [PubMed] [Google Scholar]
- 86.Takamura A, et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hidvegi T, et al. An autophagy-enhancing drug promotes degradation of mutant alpha1-antitrypsin Z and reduces hepatic fibrosis. Science. 2010;329:229–232. doi: 10.1126/science.1190354. [DOI] [PubMed] [Google Scholar]
- 88.Liu HY, et al. Hepatic autophagy is suppressed in the presence of insulin resistance and hyperinsulinemia: inhibition of FoxO1-dependent expression of key autophagy genes by insulin. J Biol Chem. 2009;284:31484–31492. doi: 10.1074/jbc.M109.033936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang L, et al. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 2010;11:467–478. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ohsaki Y, et al. Cytoplasmic lipid droplets are sites of convergence of proteasomal and autophagic degradation of apolipoprotein B. Mol Biol Cell. 2006;17:2674–2683. doi: 10.1091/mbc.E05-07-0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pan X, Hussain MM. Diurnal regulation of microsomal triglyceride transfer protein and plasma lipid levels. J Biol Chem. 2007;282:24707–24719. doi: 10.1074/jbc.M701305200. [DOI] [PubMed] [Google Scholar]
- 92.Zhang Y, et al. Adipose-specific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. Proc Natl Acad Sci U S A. 2009;106:19860–19865. doi: 10.1073/pnas.0906048106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Singh R, et al. Autophagy regulates adipose mass and differentiation in mice. J Clin Invest. 2009;119:3329–3339. doi: 10.1172/JCI39228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ukai H, Ueda HR. Systems biology of mammalian circadian clocks. Annual review of physiology. 2010;72:579–603. doi: 10.1146/annurev-physiol-073109-130051. [DOI] [PubMed] [Google Scholar]
- 95.Dibner C, et al. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annual review of physiology. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- 96.Welsh DK, et al. Suprachiasmatic nucleus: cell autonomy and network properties. Annual review of physiology. 2010;72:551–577. doi: 10.1146/annurev-physiol-021909-135919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yin L, et al. Nuclear receptor Rev-erbalpha is a critical lithium-sensitive component of the circadian clock. Science. 2006;311:1002–1005. doi: 10.1126/science.1121613. [DOI] [PubMed] [Google Scholar]
- 98.Mehra A, et al. Post-translational modifications in circadian rhythms. Trends Biochem Sci. 2009;34:483–490. doi: 10.1016/j.tibs.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Leproult R, Van Cauter E. Role of sleep and sleep loss in hormonal release and metabolism. Endocr Dev. 2010;17:11–21. doi: 10.1159/000262524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rudic RD, et al. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2:e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mizushima N, et al. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 102.Kroemer G, et al. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mehrpour M, et al. Overview of macroautophagy regulation in mammalian cells. Cell Res. 2010;20:748–762. doi: 10.1038/cr.2010.82. [DOI] [PubMed] [Google Scholar]
- 104.Hosokawa N, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Molecular biology of the cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jung CH, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Molecular biology of the cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ganley IG, et al. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. The Journal of biological chemistry. 2009;284:12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kim J, et al. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shang L, et al. Nutrient starvation elicits an acute autophagic response mediated by Ulk1 dephosphorylation and its subsequent dissociation from AMPK. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:4788–4793. doi: 10.1073/pnas.1100844108. [DOI] [PMC free article] [PubMed] [Google Scholar]