Abstract
Mutators have been shown to hitchhike in asexual populations when the anticipated beneficial mutation supply rate of the mutator subpopulation, NUb (for subpopulation of size N and beneficial mutation rate Ub) exceeds that of the wild-type subpopulation. Here, we examine the effect of total population size on mutator dynamics in asexual experimental populations of Saccharomyces cerevisiae. Although mutators quickly hitchhike to fixation in smaller populations, mutator fixation requires more and more time as population size increases; this observed delay in mutator hitchhiking is consistent with the expected effect of clonal interference. Interestingly, despite their higher beneficial mutation supply rate, mutators are supplanted by the wild type in very large populations. We postulate that this striking reversal in mutator dynamics is caused by an interaction between clonal interference, the fitness cost of the mutator allele, and infrequent large-effect beneficial mutations in our experimental populations. Our work thus identifies a potential set of circumstances under which mutator hitchhiking can be inhibited in natural asexual populations, despite recent theoretical predictions that such populations should have a net tendency to evolve ever-higher genomic mutation rates.
Introduction
Alleles that increase the genomic mutation rate (mutators) experience indirect selection due to their associations with new fitness-affecting mutations arising at other loci (Kimura 1967; Leigh 1970; reviewed in Sniegowski et al. 2000; de Visser 2002). Indirect selection on mutators is particularly effective in asexual populations, because mutator alleles remain permanently linked to all new mutations in the genome. Given that the majority of fitness-affecting mutations are expected to be deleterious (Fisher 1930), mutators should be continually driven out of asexual populations because of the increased mutational influx they allow. Nevertheless, mutator alleles can occasionally increase in frequency by hitchhiking with linked beneficial mutations (Painter 1975; Taddei et al. 1997; Tenaillon et al. 1999; de Visser 2002). Evolution of higher mutation rates by mutator hitchhiking has been observed repeatedly in experimental microbial populations (e.g., Chao and Cox 1983; Giraud et al. 2001; Shaver et al. 2002; de Visser and Rozen 2006; Thompson et al. 2006; Gentile et al. 2011).
Several experimental studies have further shown that a mutator subpopulation will supplant a wild-type subpopulation if initially present above a certain threshold frequency (Chao and Cox 1983; de Visser and Rozen 2005; Thompson et al. 2006; Gentile et al. 2011), suggesting that mutator hitchhiking depends on the ratio of beneficial mutation supply rates of mutator and wild-type subpopulations (Tenaillon et al. 1999; Wylie et al. 2009; Desai and Fisher 2011). Here, we experimentally investigate the effect of total population size on mutator dynamics in asexual populations initiated at starting mutator-to-wild type ratios theoretically predicted to lead to mutator fixation. Our expectation was that in small populations with low beneficial mutation supply rates, mutators would always increase in frequency by hitchhiking with beneficial mutations. On the other hand, we hypothesized that clonal interference (Gerrish and Lenski 1998; Wilke 2004) could hinder mutator hitchhiking in large populations, in which beneficial mutations could be common in both the mutator and the wild-type subpopulations. Indeed, an early theoretical study (Painter 1975) predicted that clonal competition would provide temporary reprieves in mutator hitchhiking, while in more recent work André and Godelle (2006: page 620) speculated that such an effect alone might act to counter the evolutionary bias in favor of higher mutation rates in asexual populations.
To examine the effect of total population size on mutator dynamics, we competed strains carrying a deletion of the mismatch repair gene MSH2 against isogenic wild-type strains in asexual yeast populations of widely varying size. We have previously shown that the msh2Δ allele produces a strong mutator phenotype and has an immediate fitness cost (approximately 3% in a rich medium) potentially due to an increased influx of highly deleterious and lethal mutations (Raynes et al. 2011). As expected, msh2Δ rapidly hitchhiked to fixation in all small populations described in the present study. As population size increased, however, msh2Δ hitchhiking was prolonged while mutator trajectories became increasingly more variable. Notably, in very large populations, msh2Δ never increased in frequency and was instead quickly supplanted by the wild-type allele.
Results
Mutator dynamics
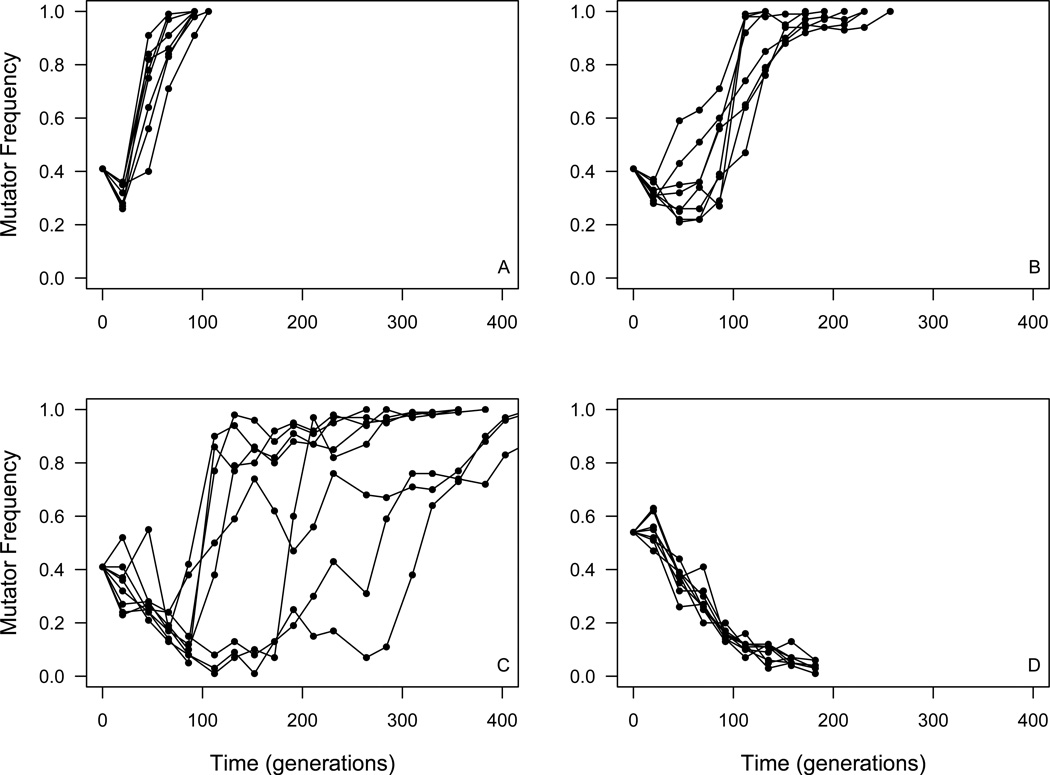
Figure 1 shows mutator dynamics at four effective population sizes (Ne). At Ne = 3.3×105 mutators uniformly increased in frequency after an initial lag period of about 20 generations and fixed at the expense of the wild type in all populations in approximately 100 generations (Figure 1A). Mutator fixation required more time once population size was increased to Ne = 4.4×106, but the mutator still fixed in all replicate populations after about 200 generations of propagation (Figure 1B). A further increase in the size of experimental populations to Ne =4.4×107 resulted in both a further delay in mutator fixation (to approximately 300–400 generations) and marked variability in mutator frequency dynamics (Figure 1C). Finally, at Ne = 2.2×108 the mutator strains uniformly declined in frequency in all replicate populations, approaching extinction in fewer than 200 generations (Figure 1D).
Figure 1. Mutator dynamics in experimental populations.
Asexual populations of yeast were propagated at four different sizes: (A) Ne = 3.3×105, (B) Ne = 4.4×106, (C) Ne =4.4 ×107, and (D) Ne = 2.2×108.
Parallel fitness gains
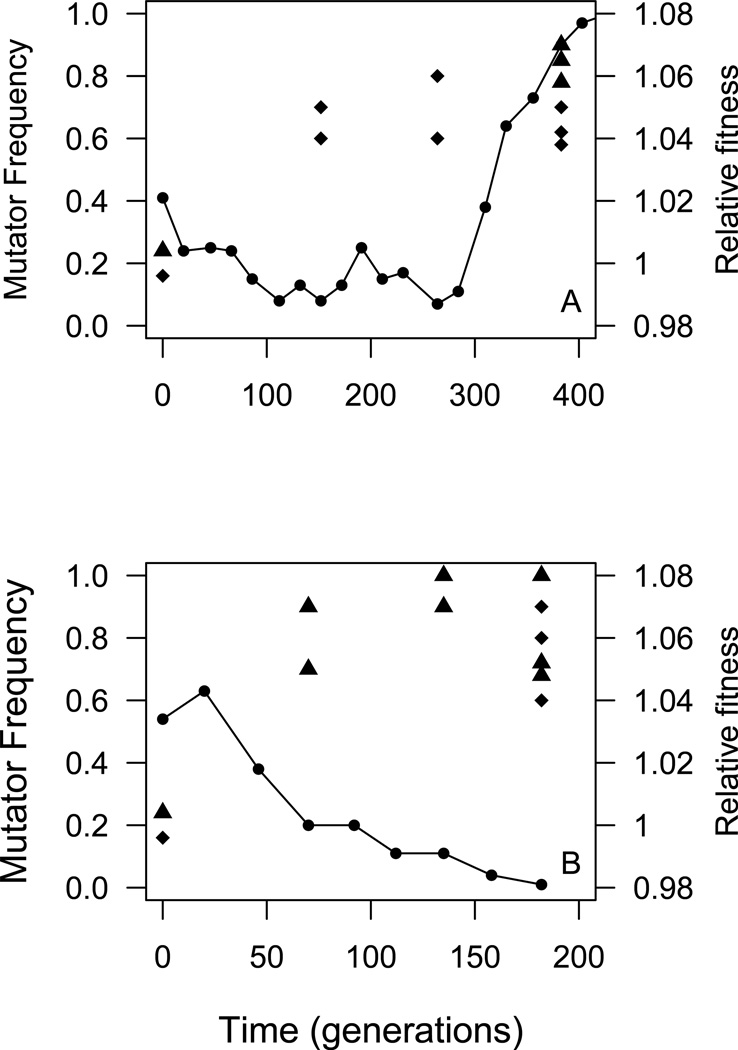
To test whether beneficial mutations were concurrently arising and spreading in both wild-type and mutator backgrounds during the experiment, we measured the competitive fitnesses of evolved wild-type and mutator clones, relative to ancestral wild-type and mutator strains respectively, isolated from two randomly chosen large populations. Figure 2A shows relative fitnesses of the evolved wild-type and mutator clones isolated from a population propagated at Ne = 4.4×107. Both mutator (F1,19 = 21.80, p<<0.01) and wild-type (F1,19 = 39.79, p<<0.01) clones isolated from approximately generation 400 had significantly higher relative fitnesses than their ancestral strains. Furthermore, mutator clones isolated from approximately generation 130 (F1,14 = 27.26, p <<0.01) and 250 (F1,14 = 44.53, p<<0.01) also had significantly higher fitnesses than the ancestral mutator strain. Figure 2B shows the relative fitness estimates of evolved mutator and wild-type clones isolated from a population propagated at Ne = 2.2×108. Wild-type clones isolated during the experiment were significantly more fit than the ancestral wild-type strain at both generation 70 (F1,13 = 18.60, p <<0.01) and 135 (F1,19 = 55.64, p <<0.01). At the end of propagation (generation 180), the observed increases in fitness are highly statistically significant in both mutator (F1,18 = 54.30, p<<0.01) and wild-type (F1,19 = 62.41, p<<0.01) backgrounds.
Figure 2. Relative fitness measurements.
Mutator and wild-type clones isolated from populations at (A) Ne =4.4 ×107 and (B) Ne = 2.2×108. Each point represents an independently isolated clone – mutators (diamonds) and wild types (triangles). Error bars were omitted for clarity. Notice, however, that fitnesses of evolved mutator and wild-type clones were not significantly different.
Effect of glucose concentration
In order to achieve the range of Ne values used in this study, experimental populations were propagated in media with different glucose concentrations. Smaller populations (Figure 1A and 1B) were propagated at 0.002% and 0.02% glucose respectively while larger populations (Figure 1C and 1D), in which the effect of clonal interference appeared most pronounced, were propagated at 2% glucose (see Methods). To test whether glucose concentration differences alone, rather than effective population size differences, could explain our results, we propagated 1 ml populations of size Ne = 4.4×106 (comparable to populations in Figure 1B) at 2% glucose. Mutator strains quickly hitchhiked to fixation in all six replicate populations in this experiment (see Supporting Figure). The difference between average mutator fixation times in populations of size Ne = 4.4×106 propagated at different glucose concentrations was not quite statistically significant (t12 = 1.9582, p > 0.08). Importantly, the average mutator fixation time at Ne = 4.4×106 (at 2% glucose) was still significantly longer than that at Ne = 4.4×105 (t12 = 12.7849, p << 0.01) and significantly shorter than that at Ne = 4.4×107 (t11 = 8.7535, p << 0.01).
Furthermore, we measured the competitive fitness of a single mutator clone, isolated from a population of size Ne = 4.4×107, relative to the ancestral wild-type strain in media containing 2% and 0.02% glucose. In both environments, the mutator clone was significantly more fit than the ancestral wild type, but, importantly, the difference between mutator relative fitnesses in the two environments was not statistically significant (t18 = 1.0470, p > 0.30).
Discussion
Here, we have investigated the effect of population size on the dynamics of a costly mutator allele in asexual populations of yeast. Despite the fact that the initial mutator/wild type NUb ratios were kept within a narrow range for all of the populations, mutator dynamics varied dramatically with population size. This observation cannot be explained merely as a consequence of changing glucose concentration: relative fitness of an evolved clone isolated from one of the populations was not significantly different between media with different glucose concentrations indicating that glucose concentration did not appreciably affect the distribution of available beneficial mutations, while small populations of size Ne = 4.4×106 were propagated in both high and low glucose concentrations and exhibited very similar mutator dynamics.
Instead, our results provide several lines of evidence that mutator dynamics were affected by clonal interference between beneficial mutations arising concurrently in mutator and wild-type subpopulations. As population size increased from Ne = 3.3×105 to 4.4×106 to 4.4×107 and beneficial mutations presumably became more common, mutator hitchhiking to fixation required significantly more time (F2,22 = 70.65, p < 0.0001), consistent with more intense competition from beneficial mutations in wild-type lineages (Gerrish and Lenski 1998; Miralles et al. 1999; de Visser and Rozen 2005). The dynamics of mutator frequency also became more variable as population size increased, consistent with both subpopulations acquiring new beneficial mutations at random times during propagation (Shaver et al. 2002; de Visser and Rozen 2006; Kao and Sherlock 2008). Increased variability in mutator dynamics was reflected in the increased standard deviation of fixation times among the populations with successful mutators: sd ≈ 5.34, 51.25, and 61.49 generations for Ne = 3.3×105, Ne = 4.4×106, and Ne = 4.4×107 respectively.
Finally, in a clear confirmation that beneficial mutations appeared in both subpopulations during propagation, both mutator and wild-type clones isolated at the end of experimental propagation from two of the larger populations (Ne = 4.4×107 and 2.2×108) showed significant increases in fitness of 4–8% (Figure 2). Notably, as Figure 2A shows, mutator frequency did not start rising for almost 300 generations at Ne = 4.4×107 despite the fact that mutator clones isolated at generation 130 had already increased in fitness relative to their ancestors, again consistent with a delay in mutator hitchhiking due to clonal interference. It is important to note that the three sets of populations propagated at Ne = 3.3×105, Ne = 4.4×106, and Ne = 4.4×107 were all started from the same mixed culture (see Methods) and, therefore, may have all been inoculated with the same beneficial mutations that arose prior to the beginning of co-propagation. If beneficial mutations were already present in the original inoculum, we may expect mutator subpopulations at all three sizes to carry beneficial mutations at the same initial frequency and therefore require the same time to fix these beneficial mutations before starting to supplant the competing wild-type subpopulations. However, we observed only a short lag period before mutators started rising in frequency in the smallest populations and a much longer lag period coupled with an initial decline in mutator frequency in populations of Ne = 4.4×106 and 4.4×107, suggesting that beneficial mutations were more likely acquired after the beginning of propagation.
Clonal interference alone, nevertheless, seems insufficient to explain the reversal in mutator dynamics reflected in our largest populations propagated at Ne = 2.2×108 (Figure 1D). Even if mutators experienced intense competition from the wild types in these populations, it seems unlikely that such an effect could have done more than merely delay mutator hitchhiking. Suppose, however, that large-effect beneficial mutations capable of compensating for the mutator’s inherent fitness cost are rare. In such a situation, a costly mutator like msh2Δ (Raynes et al. 2011) could be faced with a period of net selective disadvantage once the more accessible of such large-effect beneficial mutations had arisen on the wild-type background, consistent with the high-fitness wild-type clones isolated early in the propagation (Figure 2B), and before less accessible mutations could appear on the mutator background. Because the supply of easily accessible large-effect beneficial mutations should be exhausted more quickly in large than in small populations, the scenario described above provides a plausible explanation for the reversal in mutator dynamics that occurred as population size increased in our experiments.
In general, it seems likely that mutator alleles will present a spectrum of fitness costs, both direct and indirect. Clonal interference has previously been shown to delay the fixation of low-cost mutator alleles (Shaver et al. 2002; de Visser and Rozen 2006; Gentile et al. 2011). The results we have presented here show that when a mutator has a considerable fitness cost, clonal interference may conspire with other factors to prevent mutator hitchhiking altogether. Recent theoretical studies indicate that asexual populations may be biased towards accumulation of mutator alleles through repeated hitchhiking events (Andre and Godelle 2006; Gerrish et al. 2007). Furthermore, there is growing evidence in support of high beneficial mutation rates, especially in experimental populations (Joseph and Hall 2004; Desai et al. 2007; Perfeito et al. 2007; Hall et al. 2008); thus, it seems rather likely that mutator hitchhiking in large microbial populations is commonly affected by clonal interference. Our results suggest that the interaction between clonal interference and mutator cost can provide a mechanism that stabilizes mutation rates by preventing mutator hitchhiking in some asexual populations.
Methods
Strains and media
The wild-type and mutator S. cerevisiae strains used in these experiments have been described previously (Raynes et al. 2011). The wild-type strain, YPS3343 (ho::nat, MSH2, MAT a), was isolated from Mettler's Woods, NJ (Kuehne et al. 2007). The isogenic mutator strain YPS3460 was constructed by replacing the coding sequence of the mismatch repair locus MSH2 in YPS3343 with an msh2::kan allele (generously provided by Clifford Zeyl). The kanamycin cassette confers dominant resistance to the drug geneticin and acted as the mutator marker in the evolution experiments. Populations were propagated in a standard SD minimal medium (Rose et al. 1990) prepared with different glucose concentrations. Populations at Ne = 3.3×105 were propagated at 0.002% glucose, Ne = 4.4×106 at 0.02% glucose and Ne = 4.4×107 and Ne = 2.2×108 at 2% glucose. Populations of size Ne = 4.4×106 were also propagated in 2% glucose by daily 1:100 dilutions to 1ml (instead of 10ml) of fresh medium.
Experimental propagation
Both YPS 3343 and YPS 3460 were started from frozen stocks and grown overnight. After 24 hours of growth, mutator and wild-type strains were combined at roughly equal frequencies and the mixture was used to establish replicate experimental populations in 10ml of the appropriate medium. The three sets of populations at lower sizes were started from the same mutator/wild type mixture (resulting in the same initial mutator frequency) and propagated by daily 1:100 dilutions to 9.9ml of fresh medium resulting in about 6.6 generations of growth per day. To achieve the large population size of Ne = 2.2×108, experimental populations were only diluted 1:10 which resulted in only about 3.3 generations a day. Populations were propagated at 30°C with shaking at 200 rpm.
Relative fitness measurements
Individual evolved mutator and wild-type clones and ancestral strains (YPS3343 and YPS 3460) were inoculated into liquid SD (2% glucose), grown overnight, and introduced into common cultures after a 200-fold dilution. Each competition was replicated 6 times (in a few cases one of the replicates was lost and these competitions ended up being replicated 5 times). After 24 hours of growth, all competitions were transferred into fresh SD after 1:100 dilution for another cycle of propagation. All cultures were plated on YPD plates at the beginning and at the end of the 2nd day of propagation. Mutator frequencies were estimated by replica plating about 200–500 colonies per competition onto YPD+geneticin plates. Relative fitnesses were calculated from the change in mutator frequencies during competitions using standard population genetics theory (Crow and Kimura 1970).
Supplementary Material
Acknowledgements
We thank P.J. Gerrish, K. Sprouffske, C. Gentile, M. Eghbal, T. Singh, A. Halstead, D. Katzianer and K. Kerpen for valuable discussion and comments on the manuscript and C.W. Zeyl for sharing yeast strains. We also thank two anonymous reviewers for their constructive comments. This work was supported by National Institutes of Health Grant GM079483-01A2.
Literature Cited
- Andre JB, Godelle B. The evolution of mutation rate in finite asexual populations. Genetics. 2006;172:611–626. doi: 10.1534/genetics.105.046680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao L, Cox EC. Competition between high and low mutating strains of Escherichia coli. Evolution. 1983;37:125–134. doi: 10.1111/j.1558-5646.1983.tb05521.x. [DOI] [PubMed] [Google Scholar]
- Crow JF, Kimura M. An Introduction to Population Genetics Theory. New York: Harper and Row; 1970. [Google Scholar]
- de Visser JAGM. The fate of microbial mutators. Microbiology. 2002;148:1247–1252. doi: 10.1099/00221287-148-5-1247. [DOI] [PubMed] [Google Scholar]
- de Visser JAGM, Rozen DE. Limits to adaptation in asexual populations. Journal of Evolutionary Biology. 2005;18:779–788. doi: 10.1111/j.1420-9101.2005.00879.x. [DOI] [PubMed] [Google Scholar]
- de Visser JAGM, Rozen DE. Clonal interference and the periodic selection of new beneficial mutations in Escherichia coli. Genetics. 2006;172:2093–2100. doi: 10.1534/genetics.105.052373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai MM, Fisher DS. The balance between mutators and nonmutators in asexual populations. Genetics. 2011 doi: 10.1534/genetics.111.128116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai MM, Fisher DS, Murray AW. The speed of evolution and maintenance of variation in asexual populations. Current Biology. 2007;17:385–394. doi: 10.1016/j.cub.2007.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA. The Genetical Theory of Natural Selection. Oxford: Dover Publications; 1930. [Google Scholar]
- Gentile CF, Yu SC, Serrano SA, Gerrish PJ, Sniegowski PD. Competition between high- and higher-mutating strains of Escherichia coli. Biology Letters. 2011;7:422–424. doi: 10.1098/rsbl.2010.1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrish PJ, Colato A, Perelson AS, Sniegowski PD. Complete genetic linkage can subvert natural selection. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:6266–6271. doi: 10.1073/pnas.0607280104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrish PJ, Lenski RE. The fate of competing beneficial mutations in an asexual population. Genetica. 1998;103:127–144. [PubMed] [Google Scholar]
- Giraud A, Matic I, Tenaillon O, Clara A, Radman M, Fons M, Taddei F. Costs and benefits of high mutation rates: Adaptive evolution of bacteria in the mouse gut. Science. 2001;291:2606–2608. doi: 10.1126/science.1056421. [DOI] [PubMed] [Google Scholar]
- Hall DW, Mahmoudizad R, Hurd AW, Joseph SB. Spontaneous mutations in diploid Saccharomyces cerevisiae: another thousand cell generations. Genetical Research. 2008;90:229–241. doi: 10.1017/S0016672308009324. [DOI] [PubMed] [Google Scholar]
- Joseph SB, Hall DW. Spontaneous mutations in diploid Saccharomyces cerevisiae: More beneficial than expected. Genetics. 2004;168:1817–1825. doi: 10.1534/genetics.104.033761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao KC, Sherlock G. Molecular characterization of clonal interference during adaptive evolution in asexual populations of Saccharomyces cerevisiae. Nat Genet. 2008;40:1499–1504. doi: 10.1038/ng.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. On the evolutionary adjustment of spontaneous mutation rates. Genetical Research. 1967;9:23–34. [Google Scholar]
- Kuehne HA, Murphy HA, Francis CA, Sniegowski PD. Allopatric divergence, secondary contact and genetic isolation in wild yeast populations. Current Biology. 2007;17:407–411. doi: 10.1016/j.cub.2006.12.047. [DOI] [PubMed] [Google Scholar]
- Leigh EG. Natural selection and mutability. The American Naturalist. 1970;104:301–305. [Google Scholar]
- Miralles R, Gerrish PJ, Moya A, Elena SF. Clonal interference and the evolution of RNA viruses. Science. 1999;285:1745–1747. doi: 10.1126/science.285.5434.1745. [DOI] [PubMed] [Google Scholar]
- Painter PR. Mutator genes and selection for mutation rate in bacteria. Genetics. 1975;79:649–660. doi: 10.1093/genetics/79.4.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfeito L, Fernandes L, Mota C, Gordo I. Adaptive mutations in bacteria: high rate and small effects. Science. 2007;317:813–815. doi: 10.1126/science.1142284. [DOI] [PubMed] [Google Scholar]
- Raynes Y, Gazzara M, Sniegowski P. Mutator dynamics in sexual and asexual experimental populations of yeast. BMC Evolutionary Biology. 2011;11:158. doi: 10.1186/1471-2148-11-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Shaver AC, Dombrowski PG, Sweeney JY, Treis T, Zappala RM, Sniegowski PD. Fitness evolution and the rise of mutator alleles in experimental Escherichia coli populations. Genetics. 2002;162:557–566. doi: 10.1093/genetics/162.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sniegowski PD, Gerrish PJ, Johnson T, Shaver A. The evolution of mutation rates: separating causes from consequences. BioEssays. 2000;22:1057–1066. doi: 10.1002/1521-1878(200012)22:12<1057::AID-BIES3>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Taddei F, Radman M, Maynard-Smith J, Toupance B, Gouyon PH, Godelle B. Role of mutator alleles in adaptive evolution. Nature. 1997;387:700–702. doi: 10.1038/42696. [DOI] [PubMed] [Google Scholar]
- Tenaillon O, Toupance B, Le Nagard H, Taddei F, Godelle B. Mutators, population size, adaptive landscape and the adaptation of asexual populations of bacteria. Genetics. 1999;152:485–493. doi: 10.1093/genetics/152.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DA, Desai MM, Murray AW. Ploidy controls the success of mutators and nature of mutations during budding yeast evolution. Current Biology. 2006;16:1581–1590. doi: 10.1016/j.cub.2006.06.070. [DOI] [PubMed] [Google Scholar]
- Wilke CO. The speed of adaptation in large asexual populations. Genetics. 2004;167:2045–2053. doi: 10.1534/genetics.104.027136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie CS, Ghim CM, Kessler D, Levine H. The fixation probability of rare mutators in finite asexual populations. Genetics. 2009;181:1595–1612. doi: 10.1534/genetics.108.094532. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.