Abstract
Delivering small interfering RNA (siRNA) to tumors using clinically viable formulations remains the primary technical hurdle that prevents the development of siRNA therapy for cancer treatment. Over the past several years, significant effort has been devoted to explore novel delivery strategies, whereas relatively little attention has been paid to understand the impact of physiological constrains such as tumor vasculature on the efficiency of siRNA delivery. Using the previously described positive-readout tumor models where successful siRNA delivery leads to an upregulation of β-galactosidase within tumor sections, we analyzed the spatial distribution of localized target knockdown within tumor sections relative to tumor hypoxia and found that stable nucleic acid lipid particle (SNALP), a lipid nanoparticle-based delivery system, predominantly delivers siRNA to areas adjacent to functional tumor blood vessels. Increasing tumor vascularity by ectopic expression of VEGF resulted in more efficient siRNA delivery to tumors using SNALP. SNALP-mediated delivery of a siRNA-targeting Ran GTPase led to target knockdown and significant antitumor efficacy in the highly vascularized HepG2-derived liver tumors, but not in the poorly vascularized HCT-116-derived liver tumors. These results highlight the significant impact of tumor vasculature on siRNA delivery and call for a more focused effort on addressing tumor penetration after extravasation, an area of only limited attention currently.
Keywords: siRNA, RNAi, delivery, cancer
Introduction
Small interfering RNA (siRNA) represents a promising, novel therapeutic modality for the treatment of complex diseases such as cancer.1, 2 However, delivering siRNA to tumors using clinically viable formulations remains the major technical hurdle. Over the past several years, significant effort has been devoted to explore novel delivery strategies,3, 4, 5, 6, 7, 8, 9 whereas relatively little attention has been paid to understand the impact of physiological constrains such as tumor vasculature on the efficiency of siRNA delivery. We have previously reported the successful development of positive-readout models for evaluating siRNA delivery to tumors.10 These models employ a siRNA designed to target the tet repressor (TetR) as a surrogate for siRNA delivery. Functional delivery of the TetR siRNA into tumors results in a robust upregulation of reporter signal that can be detected by immunohistochemistry (IHC; β-galactosidase model created using the TetR-ODC-LacZ cells) or bioluminescence imaging (luciferase model created using the TetR-ODC-Luc cells).10 Using these positive-readout tumor models, we screened various literature-described delivery systems and identified stable nucleic acid lipid particle (SNALP) as a promising siRNA delivery platform. Although SNALP mediates more efficient siRNA delivery than any other delivery systems that we examined, it only delivers siRNA to 10–15% of tumor cells in our in vivo models.10 In this study, we investigated the underlying causes for this limited tumor cell coverage and identified tumor vasculature as a key determinant for the efficiency of SNALP-mediated siRNA delivery.
Results and discussion
SNALP delivers siRNA to normoxic regions within tumor
In our earlier studies, we tested the ability of SNALP to deliver the TetR siRNA into tumors using the TetR-ODC-LacZ model. In this model, successful delivery of the TetR siRNA causes an increase of β-galactosidase expression that can be easily monitored via IHC detection of β-galactosidase in tumor sections. We found from these studies that, unlike doxycycline treatment, which induced β-galactosidase expression in all tumor cells, SNALP-mediated delivery of the TetR siRNA only induced β-galactosidase expression in a small fraction (∼10–15%) of cells in a tumor section,10 suggesting that SNALP delivers siRNA to some but not all areas within the TetR-ODC-LacZ tumor. We speculated that the differential responses to SNALP-mediated siRNA delivery in various areas within tumor may reflect the different accessibility of these regions to functional tumor vasculature. Our initial attempt on testing this hypothesis by correlating where functional siRNA delivery occurs (regions with positive β-galactosidase staining) with local microvessel density failed to establish a clear relationship between tumor vasculature and SNALP-mediated siRNA delivery (data not shown). We suspect that a direct examination of blood vessels within a particular region may not accurately reflect the accessibility of this region to functional circulation for both physiological and technical reasons. For example, tumors often contain abnormal and non-functional blood vessels, and vessels adjacent to a tumor area may not appear in the same tumor section depending on the orientation of the vessel (an adjacent vessel runs in parallel to the section could be missing from the section). To circumvent these issues, we used tumor hypoxia as a marker to assess the accessibility of a particular tumor area to functional circulation, with the assumption that hypoxic regions are farther away from functional tumor vessels relative to normoxic regions. We administered SNALP that contains the TetR siRNA to mice bearing the TetR-ODC-LacZ tumors. Before tumor harvesting, we dosed the mice with the hypoxia probe, pimonidazole HCl. IHC analysis of adjacent serial tumor sections was carried out with one section stained for β-galactosidase expression and the adjacent section stained for the hypoxia probe. Interestingly, β-galactosidase expression was almost exclusively detected in areas free of hypoxia probe staining (Figure 1). The apparent mutual exclusivity between the β-galactosidase staining (an indicator of successful siRNA delivery) and the hypoxia probe staining suggests that SNALP primarily delivers siRNA to the non-hypoxic regions within tumor, which are likely areas that are adjacent to functional tumor blood vessels.
Figure 1.
SNALP delivers siRNA to normoxic areas within tumor sections. Mice bearing the TetR-ODC-LacZ-derived liver tumors were administrated with SNALP liposomes containing the TetR siRNA on day 1 and day 4 (intravenous (i.v.), 2.5 mg siRNA per kg). In all, 0.2 ml pimonidazole HCl solution (6 mg per ml in saline) was administrated via i.v. injection 1 h before sample harvesting on day 7. Adjacent sequential serial sections of the same tumor were analyzed for β-galactosidase expression (right panel) and tumor hypoxia (left panel). Tumors from five mice were analyzed, and two representative fields were shown here. Equivalent regions in adjacent sections are highlighted using dotted lines. (a) Field 1 at × 5 magnification. (b) Field 2 at × 10 magnification.
Increasing tumor vascularity facilitates SNALP-mediated siRNA delivery
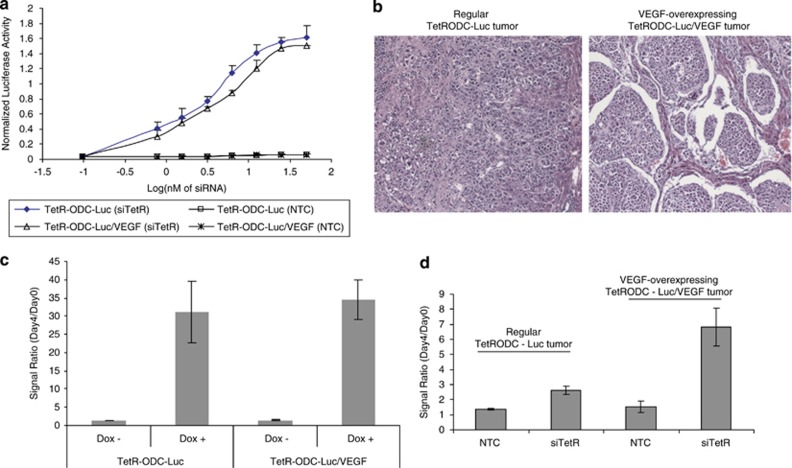
If SNALP-mediated siRNA delivery indeed occurs adjacent to functional tumor blood vessels, one would expect that better-vascularized tumors will respond better to SNALP-mediated siRNA delivery. To test this hypothesis, we took advantage of the previously described positive-readout luciferase model (tumor models generated using the TetR-ODC-Luc cells), which allows for rapid quantification of siRNA delivery to tumors in vivo, to compare the efficiency of SNALP-mediated siRNA delivery to tumors with different degrees of vascularization. We established TetR-ODC-Luc-derived cell lines that express high levels of VEGF (TetR-ODC-Luc/VEGF cells). Under in vitro tissue culture conditions, SNALP containing the non-targeted control siRNA (NTC) did not induce luciferase expression in either TetR-ODC-Luc cells or TetR-ODC-Luc/VEGF cells (Figure 2a, TetR-ODC-Luc (NTC) and TetR-ODC-Luc/VEGF (NTC)). Meanwhile, SNALP containing the TetR siRNA triggered a similar increase of luciferase expression in both cell lines (Figure 2a, TetR-ODC-Luc (siTetR) and TetR-ODC-Luc/VEGF (siTetR)). These results suggest that VEGF expression does not change the response of cultured cells to SNALP-mediated siRNA delivery. When inoculated in the liver, the VEGF-expressing TetR-ODC-Luc/VEGF cells formed more-vascularized tumors compared with tumors derived from the parental TetR-ODC-Luc cells (Figure 2b). Feeding mice with doxycycline, a small molecule inducer that is expected to trigger reporter expression in all tumor cells regardless of tumor vascularization pattern, caused a similar increase of luminescent signals in both the VEGF-expressing and non-expressing tumors, suggesting that VEGF-expression does not alter the intrinsic response of the reporter system in vivo (Figure 2c). By contrast, intravenous administration of SNALP-encapsulated TetR-siRNA led to a more dramatic increase of bioluminescent signal in the TetR-ODC-Luc/VEGF cell-derived tumors compared with tumors derived from the parental TetR-ODC-Luc cells (Figure 2d), suggesting that better siRNA delivery was achieved in the more-vascularized, VEGF-expressing tumors. Taken together, these results provided additional evidence that SNALP-mediated siRNA delivery appears dependent on the nature of the tumor vasculature, with better delivery occurring in highly vascularized tumors.
Figure 2.
Increasing tumor vascularization facilitates SNALP-mediated siRNA delivery. (a) The ability of SNALP liposome to deliver the TetR siRNA (siTetR) or the control siRNA (NTC) into TetR-ODC-Luc or TetR-ODC-Luc/VEGF cells in vitro was determined. Different amounts of SNALP liposomes containing siTetR or NTC were incubated with the TetR-ODC-Luc or TetR-ODC-Luc/VEGF cells for 72 h, and luciferase activity in cells was determined afterwards. The increase of luciferase activity indicates successful siRNA delivery. The x axis represents the dose of SNALP liposome in Log(nM of final siRNA concentration). SNALP-mediated delivery of TetR siRNA induced a similar dose–response curve in the TetR-ODC-Luc and the TetR-ODC-Luc/VEGF cells, indicating that high level of VEGF expression does not change the intrinsic response of the positive-readout system to siRNA delivery. (b) Representative tumor sections (hematoxylin and eosin staining) of the regular TetR-ODC-Luc tumors and the VEGF-expressing TetR-ODC-Luc/VEGF tumors. (c) The TetR-ODC-Luc or TetR-ODC-Luc/VEGF cells were used to create liver tumors via intrahepatic inoculation in scid mice. Mice were imaged on day 0, fed with regular water (Dox−) or doxycycline-containing water (Dox+) for 4 days, and reimaged on day 4. (d) Mice that bear TetR-ODC-Luc- or TetR-ODC-Luc/VEGF-derived liver tumors were administrated with SNALP liposomes containing the control siRNA (NTC) or the TetR siRNA (siTetR) via intravenous injection (2.5 mg siRNA kg−1) on day 1 and day 2. Mice were imaged 1 day before the start of dosing (day 0) and 2 days after the last dose (day 4). For both (c, d) N=5 for each treatment group. Each bar in the graph represents the average signal ratio±s.e.m. for each group.
SNALP triggers better target knockdown in highly vascularized tumors
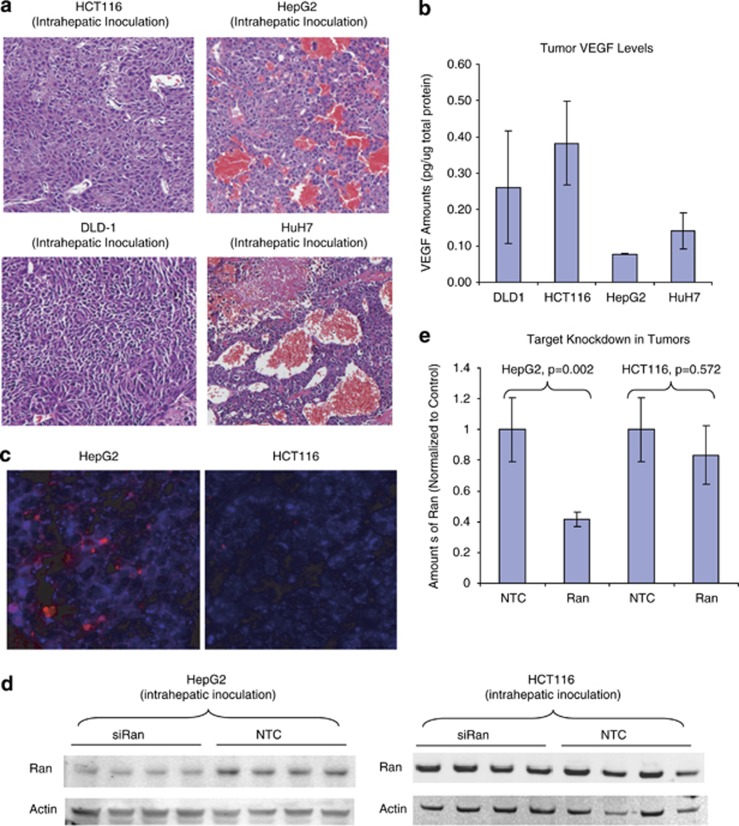
Both of the positive-readout cell lines, TetR-ODC-LacZ and TetR-ODC-Luc, are engineered cell lines originated from the MDA-MB435 parental cells. Although positive-readout models derived from these cell lines allowed us to take a close look at the spatial relationship of functional siRNA delivery and tumor vasculature, conclusions from these artificial systems need to be verified in more physiologically relevant models. Toward this end, we decided to examine the knockdown of an endogenous target in standard xenograft tumor models that have different degrees of vascularization. We generated liver tumors using standard cancer cell lines HCT116, DLD-1, HepG2, and HuH7. Hematoxylin and eosin staining of tumor sections revealed that these xenograft tumors appeared to have very different vasculature. The HCT116- and DLD-1-derived liver tumors have a compact structure and very few blood vessels, whereas HepG2 and HuH7-derived tumors contain a large number of vessels that are filled with red blood cells (Figure 3a). As VEGF is a key factor impacting tumor vascularity, we set out to determine whether the extensive vascularization in HepG2 and HuH7 tumors were driven by high levels of tumor VEGF. Surprisingly, lower levels of VEGF were present in HepG2 and HuH7 tumors compared with HCT116 and DLD-1 tumors, suggesting that the extensive vascularization in HuH7 and HepG2 tumors may result from a VEGF-independent mechanism (Figure 3b). Administrating SNALP containing DY647-labeled siRNA to the mice bearing HepG2 or HCT116 liver tumors resulted in clear DY647 fluorescent signal across the HepG2 tumor section. In contrast, only very weak DY647 signal was observed in HCT116 tumor section, suggesting that more SNALP liposome reaches the highly vascularized HepG2 tumor compared with the poorly vascularized HCT116 tumor (Figure 3c). Furthermore, administrating SNALP containing a potent siRNA (sense strand: 5′-GUGUGCCACCUCAUUAUUAdTdT-3′) that targets a previously described essential gene, Ran GTPase,11 led to a significant reduction of Ran protein in the highly vascularized, HepG2-derived liver tumors but not in the poorly vascularized, HCT116-derived liver tumors, indicating that better siRNA delivery was obtained in the more-vascularized tumors (Figures 3d and e). As controls, SNALP containing a NTC did not produce target knockdown in either of these models.
Figure 3.
SNALP triggers better target knockdown in highly vascularized tumors. (a) Liver tumors were established via intrahepatic injection of HCT116 or HepG2 cells in scid mice. Representative hematoxylin and eosin sections of the HCT116- and HepG2-derived liver tumors were shown. (b) VEGF levels in tumor homogenates (N=5 for each tumor type) were determined using the QuantiGlo human VEGF kit and normalized to the total protein amount in the same sample. (c) SNALP liposome containing DY647-labeled TetR siRNA were administrated to tumor-bearing mice (intravenous (i.v.), 2.5 mg siRNA kg−1). Tumors were harvested 20 h later and cryosections of the tumor were stained with an antibody that recognizes a human specific mitochondria marker. Representative confocal image of resulted HepG2 or HCT116 sections were shown. Blue: human mitochondria staining that specifically labels xenograft tumor tissue. Red: DY647 signal that indicates the presence of SNALP(siRNA) delivery. (d) Mice that bear HCT116- and HepG2-derived liver tumors were administrated with SNALPs-containing NTC or siRAN at 2 mg siRNA kg−1, i.v., q.d. × 2. Tumors were collected 48 h after last dosing and analyzed by western blotting for the expression of Ran and Actin. N=4 for each treatment group. (e) Densitometry quantification of western results in (d). The y axis represents the average amount of Ran (normalized to the amount of actin in each sample).
Therapeutic siRNA delivered using SNALP exhibits better efficacy against highly vascularized tumors
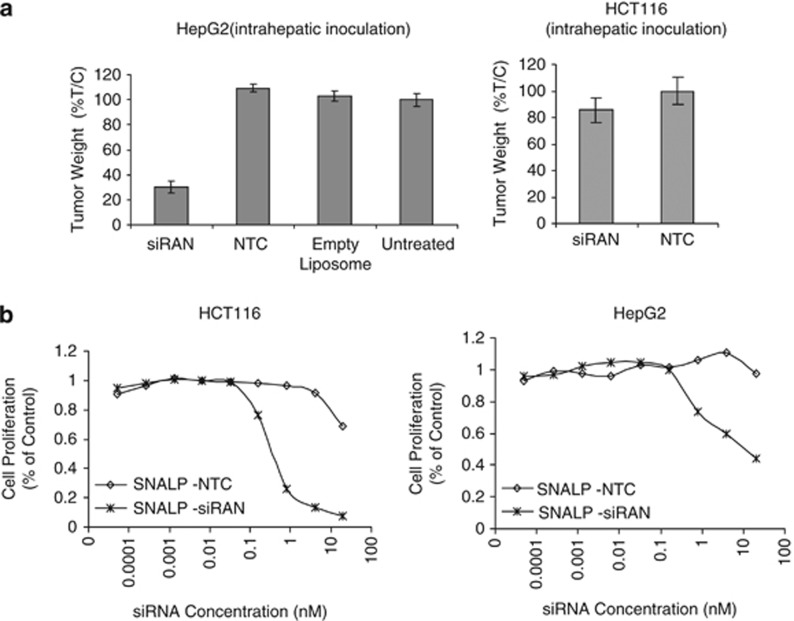
To further determine to what degree tumor vasculature will impact therapeutic efficacy, we compared the antitumor efficacy of SNALP-siRAN in the HepG2-derived liver tumor model and the HCT116-derived liver tumor model. Intravenous administration of SNALP-siRAN at 2 mg siRNA per kg, twice per week for three weeks caused significant tumor growth inhibition in the HepG2 model but not in the HCT116 model (Figure 4a). In contrast, under in vitro culture conditions, SNALP-siRAN induced significant cell death in both HCT116 (half maximal effective concentration (EC 50) ∼0.2 nM) and HepG2 cells (EC 50∼1 nM) with more robust cell killing observed in the HCT116 cells over HepG2 cells at similar SNALP-RAN concentrations (Figure 4b). These results suggest that HCT116 cells are more sensitive to SNALP-siRAN than HepG2 cells when exposed to similar level of SNALP-siRAN. Therefore, the lack of in vivo efficacy in HCT116 tumors likely reflects less efficient siRNA delivery to the poorly vascularized HCT116 tumors rather than a result of intrinsic resistance of the HCT116 tumor to SNALP-siRAN.
Figure 4.
Therapeutic siRNA delivered using SNALP exhibits better efficacy against highly vascularized tumors. (a) HepG2 and HCT116 cells were used to create liver tumors in scid mice via intrahepatic inoculation. At 72 h after inoculation, mice were either untreated or administrated with empty SNALP liposome, SNALP liposomes containing NTC or siRNA-targeting Ran (siRAN; intravenous, 2.5 mg siRNA kg−1, twice per week) for 3 weeks. Tumor weight in each mouse was determined 72 h after the last dosing. %T/C (the ratio between the mean tumor weight of treated mice (T) and that of control mice (C)) was calculated as the average tumor weight in treated mice versus the average tumor weight in untreated group. N=15 for each treatment group, and the average %T/C ±s.e.m. was plotted. (b) HCT116 and HepG2 cells were incubated with different amounts of SNALP containing a NTC or siRAN, and the number of viable cells was determined 72 h later using the CellTiter Glo kit.
In summary, in addition to serving as a frontline screening tool to identify promising delivery platforms for further optimization, the positive-readout tumor model makes it possible to distinguish between areas within tumors where effective delivery occurs versus areas that are less accessible to the delivery. By studying the spatial distribution of localized target knockdown within tumor sections relative to tumor hypoxia, we found that SNALP appears to predominantly deliver siRNA to areas adjacent to functional tumor blood vessels. This result is largely in line with the notion that large particles can only travel a short distance after extravasation from tumor blood vessels. Because of this limitation, tumor vasculature becomes a key determinant for the efficiency of SNALP-mediated siRNA delivery. The significant impact of tumor vasculature on nanoparticle-mediated siRNA delivery described here could have important ramifications in choosing the optimal tumor types and siRNA targets for a therapeutic siRNA program. Because many nanoparticle-based delivery vehicles are expected to be restricted to cells adjacent to tumor vasculature like SNALP, these delivery systems may be better suited to deliver an antiangiogenic siRNA that targets the tumor vasculature versus delivering a cytotoxic siRNA that targets the tumor cells. Alternatively, therapeutic formulations containing a cytotoxic siRNA may be more efficacious in patients with highly vascularized tumors where the delivery formulation can reach a sufficient number of tumor cells to produce significant antitumor efficacy. It is noteworthy that the HCT116- and HepG2-derived liver tumors exhibited dramatically different responses to SNALP-mediated delivery of a therapeutic siRNA despite that both tumors were grown in the liver. This result suggests that despite the prevailing enthusiasm of targeting liver tumors in the therapeutic siRNA field, some liver tumors may not be viable candidates for nanoparticle-based siRNA therapy. The differential response of HCT116- and HepG2-derived liver tumors suggest that better therapeutic response to SNALP or other nanoparticle-based therapeutic siRNA formulations will be more likely obtained in typically highly vascularized primary hepatocellular carcinomas than in less-vascularized tumors such as liver metastases of colorectal carcinomas. Although, in a general sense, the impact of tumor vasculature on nanoparticle-mediated delivery is not surprising, it is enlightening that the delivery efficiency of SNALP, the perceived current state of art in siRNA delivery, is still severely limited by tumor vascularity. Understanding this limitation not only has important implications on patient selection and choice of therapeutic targets as discussed above, it also calls for a more focused effort on addressing tumor penetration after extravasation, which is an area of limited attention currently.
Materials and methods
Cell culture and reagents
The HCT116, DLD-1, HepG2 and HuH7 cells were obtained from ATCC (Manassas, VA, USA) and cultured in vendor-recommended media. The TetR-ODC-Luc and TetR-ODC-LacZ cells were MDA-MB435-derived positive-readout cell lines that were described earlier.10 To generate the TetR-ODC-Luc/VEGF cells, TetR-ODC-Luc cells were transfected with pcDNA6 plasmid that encoding the VEGF gene, and stable clones were selected and tested for VEGF expression. Cell proliferation was determined use the CellTiter Glo kit (Promega, Madison, WI, USA). All siRNAs were purchased from Dharmacon (Lafayette, CO, USA).
Tumor models
The animal studies were carried out in accordance with the internal Institutional Animal Care and Use Committee guidelines at Abbott Laboratories. Cancer cells were directly injected into the liver of scid female mice to generate liver tumor models. All animals were obtained from Charles River Laboratory (Wilmington, MA, USA). All cell lines used for creating xenograft tumors were subjected to the IMPACT profile I test (18 agents) at the University of Missouri Research Animal Diagnostic and Investigative Laboratory, and all cell lines were found negative for the 18 infectious agents tested.
Animal dosing and sample harvesting
For in vivo knockdown evaluation in positive-readout models, treatments were started 3–4 weeks after tumor inoculation for liver tumors. For doxycycline treatment in the TetR-ODC-Luc model, doxycycline (Sigma, St Louis, WA, USA) was supplied in drinking water at a concentration of 1 mg ml−1 for 4 days before imaging. For siRNA treatment in the positive-readout models, formulated siRNAs were administrated via tail vein injection at the indicated dose and schedule. For antitumor efficacy evaluation, at the end of the efficacy trial, mice were humanly euthanized, liver tumor nodules were surgically excised and weighted. For mice without visible tumors, the tumor weight was recorded as 0.
IHC analysis
IHC was carried out as previously described.12 Briefly, tumors were excised, cut into pieces of <3 mm in thickness and immediately fixed in buffered formalin solution with neutral pH (Sigma, St Louis, MO, USA). The formalin-fixed and paraffin-embedded tumor sections were then used for staining. The mouse anti-β-galactosidase mAb (Promega) was used to detect β-galactosidase in tumor sections. IHC analysis of tumor hypoxia was carried out using the Hypoxyprobe-1 kit (Natural Pharmacia International Inc., Burlington, MA, USA) according to the manufacturer's suggested protocol. Briefly, 0.2 ml pimonidazole HCl solution (6 mg ml−1 in saline) was administrated via intravenous injection 1 h before tumor harvesting. The hypoxic areas within tumor sections were detected using mouse antibody recognizing the protein adduct formed by hypoxyprobe. For all IHC studies,3,3′-diaminobenzidine was used as the chromogen, and IHC images were acquired using the Nikon TE2000 inverted microscope.
Bioluminescence imaging and analysis
In vivo bioluminescence imaging and analysis were conducted on the IVIS 200 system using the Living Image acquisition and analysis software (Caliper Life Science, Hopkinton, MA, USA) as previously described.10 Briefly, after intraperitoneal injection of luciferin (Promege, Madison, WI, USA) at 150 mg kg−1, mice were anesthetized with isofluorane. At 4 min after the injection of luciferin, a series of time-lapse images were acquired at 2 min intervals for a total of 10 min. Regions of interest were drawn around the tumors and signal intensity was quantified as the sum of photon counts per second within the Regions of interest after the subtraction of background luminescence. The peak reading during the 10-min imaging period was used for calculating the signal ratio before and after siRNA delivery.
Liposome formulation
The SNALP liposomes were prepared according to the literature-described procedure.6 D-Lin-DMA and PEG-C-DMA were synthesized in-house and formulated with distearoylphosphatidylcholine (DSPC), Cholesterol and siRNA using a 25:1 lipid/siRNA ratio and a 48/40/10/2 molar ratio of Cholesterol/D-Lin-DMA/DSPC/PEG-C-DMA. The resulted SNALP liposomes have similar characteristics to what was described in the literature (80–100 nm in size, PDI<0.1, and a close to neutral zeta potential).
Quantitation of human VEGF within xenograft tumors
Tumors were homogenized in CelLytic-M (Sigma) with protease inhibitors. After centrifugation, the concentration of human VEGF in supernatant were measured using the QuantiGlo human VEGF kit (R&D systems, Minneapolis, MN, USA). The total protein concentration of lysate was used to normalize the human VEGF level in each tumor.
The authors declare no conflict of interest.
References
- de Fougerolles A, Vornlocher HP, Maraganore J, Lieberman J. Interfering with disease: a progress report on siRNA-based therapeutics. Nat Rev Drug Discov. 2007;6:443–453. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y. Advances in the development of siRNA-based therapeutics for cancer. IDrugs. 2008;11:572–578. [PubMed] [Google Scholar]
- Akinc A, Zumbuehl A, Goldberg M, Leshchiner ES, Busini V, Hossain N, et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat Biotechnol. 2008;26:561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleku M, Schulz P, Keil O, Santel A, Schaeper U, Dieckhoff B, et al. Atu027, a liposomal small interfering RNA formulation targeting protein kinase N3, inhibits cancer progression. Cancer Res. 2008;68:9788–9798. doi: 10.1158/0008-5472.CAN-08-2428. [DOI] [PubMed] [Google Scholar]
- Hu-Lieskovan S, Heidel JD, Bartlett DW, Davis ME, Triche TJ. Sequence-specific knockdown of EWS-FLI1 by targeted, nonviral delivery of small interfering RNA inhibits tumor growth in a murine model of metastatic Ewing's sarcoma. Cancer Res. 2005;65:8984–8992. doi: 10.1158/0008-5472.CAN-05-0565. [DOI] [PubMed] [Google Scholar]
- Zimmermann TS, Lee AC, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN, et al. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- Rozema DB, Lewis DL, Wakefield DH, Wong SC, Klein JJ, Roesch PL, et al. Dynamic PolyConjugates for targeted in vivo delivery of siRNA to hepatocytes. Proc Natl Acad Sci USA. 2007;104:12982–12987. doi: 10.1073/pnas.0703778104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judge AD, Robbins M, Tavakoli I, Levi J, Hu L, Fronda A, et al. Confirming the RNAi-mediated mechanism of action of siRNA-based cancer therapeutics in mice. J Clin Invest. 2009;119:661–673. doi: 10.1172/JCI37515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple SC, Akinc A, Chen J, Sandhu AP, Mui BL, Cho CK, et al. Rational design of cationic lipids for siRNA delivery. Nat Biotechnol. 2010;28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- Lin X, Li L, Wang R, Wilcox D, Zhao X, Song J, et al. A robust in vivo positive-readout system for monitoring siRNA delivery to xenograft tumors. RNA. 2011;17:603–612. doi: 10.1261/rna.2546011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan-Lappe SE, Tucker LA, Huang X, Zhang Q, Sarthy AV, Zakula D, et al. Identification of Ras-related nuclear protein, targeting protein for xenopus kinesin-like protein 2, and stearoyl-CoA desaturase 1 as promising cancer targets from an RNAi-based screen. Cancer Res. 2007;67:4390–4398. doi: 10.1158/0008-5472.CAN-06-4132. [DOI] [PubMed] [Google Scholar]
- Li L, Lin X, Staver M, Shoemaker A, Semizarov D, Fesik SW, et al. Evaluating hypoxia-inducible factor-1alpha as a cancer therapeutic target via inducible RNA interference in vivo. Cancer Res. 2005;65:7249–7258. doi: 10.1158/0008-5472.CAN-04-4426. [DOI] [PubMed] [Google Scholar]