Abstract
Summary: Sequencing by hybridization to oligonucleotides has evolved into an inexpensive, reliable and fast technology for targeted sequencing. Hundreds of human genes can now be sequenced within a day using a single hybridization to a resequencing microarray. However, several issues inherent to these arrays (e.g. cross-hybridization, variable probe/target affinity) cause sequencing errors and have prevented more widespread applications. We developed an R package for resequencing microarray data analysis that integrates a novel statistical algorithm, sequence robust multi-array analysis (SRMA), for rare variant detection with high sensitivity (false negative rate, FNR 5%) and accuracy (false positive rate, FPR 1×10−5). The SRMA package consists of five modules for quality control, data normalization, single array analysis, multi-array analysis and output analysis. The entire workflow is efficient and identifies rare DNA single nucleotide variations and structural changes such as gene deletions with high accuracy and sensitivity.
Availability: http://cran.r-project.org/, http://odin.mdacc.tmc.edu/~wwang7/SRMAIndex.html
Contact: wwang7@mdanderson.org
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
To understand the specific DNA variants that contribute to the inheritance of human diseases is a major goal of human genetics (Bodmer and Bonilla, 2008). Medical resequencing has greatly accelerated the identification of disease-related DNA variants. Resequencing array technology utilizes differential hybridization of target DNA to oligonucleotide probes to decode individual DNA sequences. It has been successful in identifying novel and very rare variants in disease candidate genes (Shen et al., 2011; Wang et al., 2011; Wilkins et al., 2012). However, low-variant frequency (1/1000 bp), variable data quality as well as technical and experimental limitations have the potential to create sequencing errors. There is a need for improved statistical methods for array-based resequencing. We have recently developed sequence robust multi-array analysis (SRMA) for resequencing array data analysis (Wang et al., 2011). By improving preprocessing procedures, borrowing strength across samples and targeting unique features of rare variations, SRMA achieved a false discovery rate of 2% (FPR 1.2×10−5, FNR 5%), which is comparable to that of next-generation sequencing technologies. Here, we have established an R package (R Development Core Team, 2010) called ‘SRMA’ that fully implements these methods and provides an automated analysis pipeline for medical resequencing array data with high accuracy of calling rare variants. (System requirements, file structure, package installation, and a description of the analysis of resequencing array data are provided in the Supplementary Material.)
2 AVAILABLE FUNCTIONALITY
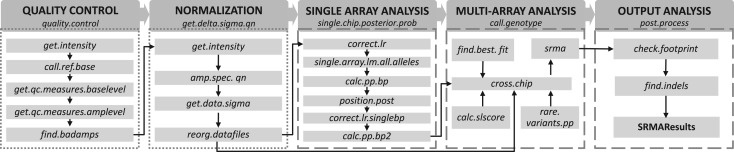
We describe five modules and their principal functions for the resequencing microarray data analysis in Figure 1. To take full advantage of the SRMA algorithm for rare DNA variant discovery, we recommend using a larger sample size (>20).
Fig. 1.
Workflow of SRMA. We provide wrapper functions for the five modules as indicated in the dark gray boxes. Under each module, we list the main functions that are called by the wrapper function. The functions in dotted boxes require aroma.affymetrix file structure and those in dashed boxes do not. The arrows suggest the order in which each function is used in the analysis pipeline. The main inputs of the first function ‘quality.control()’ are array related information and the data frame mapping amplicons to exons. The outputs are a Boolean matrix indicating the failed amplicons in samples and the chosen quality control metrics. The function ‘get.delta.sigma.qn()’ performs normalization. The main outputs of the function are arrays of normalized data organized by bases, RM/AM basepairs, samples and strands. The main outputs of the function ‘single.chip.posterior.prob()’ are the indices of the program-selected alternative alleles, the posterior probabilities of three variant classes. The main outputs of function ‘call.genotype()’ are the genotype calls, posterior probabilities and quality scores. The function ‘post.process()’ generates final insertion/deletions and SNV genotype calls with quality scores that are higher than given threshold.
2.1 Preprocessing of resequencing data
Preprocessing of resequencing data includes two modules: Quality Control and Normalization, and is contingent on the R package ‘aroma.affymetrix’ (Bengtsson et al., 2008) for extracting probe intensities. This step requires raw CEL files and annotation files: a chip description file (CDF) organized by exon units and an aroma cell sequence (ACS) file. In addition, we also require a data frame, mapping amplicons (i.e. fragments generated during amplification experiments) to exons, and a data frame with information of reference alleles for all bases.
2.1.1 Quality control of resequencing data
The Quality Control module identifies amplicons that are not suitable for base-calling due to failure in target amplification and hybridization. The ‘aroma.affymetrix’ reads in raw intensities and allows users to perform strand-specific base position normalization within each array. We then calculate three metrics, including the median of the average (log2) probe intensity, median of the log ratios and reference call rate for each amplicon to evaluate the quality of the amplified targets. We use a criterion of R<0.9 (Wang et al., 2011) to identify failed amplicons. This criterion can be modified by users.
2.1.2 Data normalization
We exclude failed amplicons from normalization by changing the corresponding probe intensities to NAs. After quantile normalization at amplicon level across all samples, we calculate the differences (δ) and averages (σ) of log2 transformed intensities of reference match (RM) and alternative match (AM) probes; and record the base pair (reference versus alternative alleles) information. The GC content and the length of exons are also calculated here and stored in a data frame.
2.2 Single array and multi-array analysis
Single array and multi-array analysis of resequencing data are the core components of the SRMA package. Under the assumption that all variants are bi-allelic, we assign one alternative allele to each position and calculate the posterior probabilities of each position for three variant classes: containing no (SS), one (RS) and two (RR) copies of the reference allele. Multi-array analysis then determines the genotype for each position in each sample and calculates quality scores for the genotype calls.
2.2.1 Single array analysis
For each sample, a linear model is used to adjust the log ratios δ for a set of explainable variables, including average intensity σ, amplicon length, amplicon GC content and central base pair composition. We assume a Gaussian distribution for adjusted δ given the allele variant class, identical and independent distributions for each strand. We then choose one alternative allele using the single array posterior probabilities calculated for all samples at a base position (Wang et al., 2011). We then focus on the subset of data with the chosen alternative alleles and perform another iteration of linear regression and recalculate the single array posterior probabilities.
2.2.2 Multi-array analysis
This module starts from the initial genotype assignments based on single array posterior probabilities. The positions where all samples were designated as RR with the corresponding posterior probabilities >0.999 are considered to be reference-only positions. For the other positions that potentially contain variants, at each position, we first use k-means clustering to designate initial genotypes, and calculate a minor allele count (MAC) as the total number of alternative alleles across all samples. For common variant positions with MAC ≥ 4, we perform clustering on δ using EM algorithms as implemented in R package ‘mclust’ (Fraley and Raftery, 2002). For rare variant positions with MAC < 4, we classify genotypes on δ assuming known parameters for non-reference clusters (Wang et al., 2011). The genotype class with the highest posterior probability among all classes is assigned to each position for each sample. A sample-specific quality score q evaluates clustering quality based on silhouette width (Rousseeuw, 1987) and a position-specific quality score Q evaluates probe quality as a sum of the q scores across samples.
2.3 Output analysis
Output analysis includes detection of technical artifacts and identification of reliable rare single nucleotide variations (SNVs) and indels. We detect and eliminate the heterozygous call from footprint effect artifacts, low-homology regions and technical defects. We take known dbSNP positions mapped to the candidate genes and preserve all variant calls at these positions. To balance between FPR and FNR, we choose a threshold of 0.67 for both quality scores to exclude the low-quality genotype calls based on our validation data (Wang et al., 2011). This threshold can be modified by users. We provide the list of SNVs for all samples in the VCF4.0 format (Danecek et al., 2011).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Henrik Bengtsson for support with aroma.affymetrix, LeeAnn Chastain for copyediting.
Funding: Michael & Susan Dell Foundation (to N.Z.); U.S. National Institutes of Health through the following grants: P30 CA016672 (to N.Z. and W.W.), R01 EY016240 (to Y.X. and C.S.), 5R01 GM083084-03 (to T.P.S.). M.O'H and T.P.S acknowledge the NHMRC for support through an Australia Fellowship. P30 CA016672 (to N.Z. and W.W.), and R01 EY016240 (to T.P.S. and C.S.).
Conflict of Interest: none declared.
REFERENCES
- Bengtsson H., et al. aroma.affymetrix: A generic framework in R for analyzing small to very large Affymetrix data sets in bounded memory. Berkeley: Department of Statistics, University of California; 2008. Tech Report #745. [Google Scholar]
- Bodmer W., Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat. Genet. 2008;40:695–701. doi: 10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P., et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraley C., Raftery A.E. Model-based clustering, discriminant analysis, and density estimation. J. Am. Stat. Assoc. 2002;97:611–631. [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Vienna, Austria: 2010. ISBN 3-900051-07-0. [Google Scholar]
- Rousseeuw P.J. Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 1987;20:53–65. [Google Scholar]
- Shen P., et al. High-quality DNA sequence capture of 524 disease candidate genes. Proc. Natl Acad. Sci. USA. 2011;108:6549–6554. doi: 10.1073/pnas.1018981108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., et al. Identification of rare DNA variants in mitochondrial disorders with improved array-based sequencing. Nucleic Acids Res. 2011;39:44–58. doi: 10.1093/nar/gkq750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins E.J., et al. A DNA Resequencing Array for Genes Involved in Parkinson's Disease. Parkinsonism Rel. Disord. 2012 doi: 10.1016/j.parkreldis.2011.12.012. (in press) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.