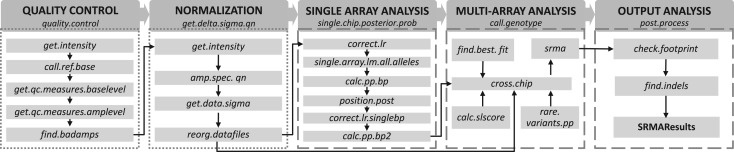
Fig. 1.
Workflow of SRMA. We provide wrapper functions for the five modules as indicated in the dark gray boxes. Under each module, we list the main functions that are called by the wrapper function. The functions in dotted boxes require aroma.affymetrix file structure and those in dashed boxes do not. The arrows suggest the order in which each function is used in the analysis pipeline. The main inputs of the first function ‘quality.control()’ are array related information and the data frame mapping amplicons to exons. The outputs are a Boolean matrix indicating the failed amplicons in samples and the chosen quality control metrics. The function ‘get.delta.sigma.qn()’ performs normalization. The main outputs of the function are arrays of normalized data organized by bases, RM/AM basepairs, samples and strands. The main outputs of the function ‘single.chip.posterior.prob()’ are the indices of the program-selected alternative alleles, the posterior probabilities of three variant classes. The main outputs of function ‘call.genotype()’ are the genotype calls, posterior probabilities and quality scores. The function ‘post.process()’ generates final insertion/deletions and SNV genotype calls with quality scores that are higher than given threshold.