Abstract
Background
Bone marrow stromal cells (BMSCs) are being used for immune modulatory, anti-inflammatory and tissue engineering applications, but the properties responsible for these effects are not completely understood. Human BMSCs were characterized to identify factors that might be responsible for their clinical effects and biomarkers for assessing their quality.
Methods
Early passage BMSCs prepared from marrow aspirates of 4 healthy subjects were compared to 3 human embryonic stem cell (hESC) samples, CD34+ cells from 3 healthy subjects and 3 fibroblast cell lines. The cells were analyzed with oligonucleotide expression microarrays with more than 35,000 probes.
Results
BMSC gene expression signatures of BMSCs differed from those of hematopoietic stem cells (HSCs), hESCs and fibroblasts. Genes up-regulated in BMSCs were involved with cell movement, cell-to-cell signaling and interaction and proliferation. The up-regulated genes most likely belonged to pathways for integrin signaling, integrin linked kinase (ILK) signaling, NFR2-mediated oxidative stress response, regulation of actin-based motility by Rho, actin cytoskeletal signaling, caveolar-mediated endocytosis, clathrin-mediated endocytosis and Wnt/β catenin signaling. Among the most highly up-regulated genes were structural extracellular (ECM) proteins:α5 and β 5 integrin chains, fibronectin, collagen type IIIα1 and Vα1; and functional EMC proteins: connective tissue growth factor (CTGF), transforming growth factor beta induced protein (TGFBI) and ADAM12.
Conclusions
Global analysis of human BMSCs suggests that they are mobile, metabolically active, proliferative and interactive cells that make use of integrins and integrin signaling. They produce abundant ECM proteins that may contribute to their clinical immune modulatory and anti-inflammatory effects.
Introduction
Bone Marrow Stromal Cells (BMSCs), also known as mesenchymal stem cells, are adherent, fibroblast-like cells found in the bone marrow. They can be isolated from bone marrow aspirates and bone/bone marrow biopsies by adherence to plastic. They are a heterogeneous population of cells which include a subset of stem cells (1). BMSCs can be expanded ex vivo, and the entire population can still maintain its capacity to differentiate into adipose tissue, cartilage, and bone. BMSCs have immunosuppressive and anti-inflammatory properties. They home to sites of tissue injury and have a beneficial effect when administered intravenously or directly to an injured tissue (2;3).
BMSCs have been used to treat Osteogenesis imperfecta and in-born errors of metabolism (4–6). Their use for bone regeneration is well established in experimental settings (7–10). Current evidence indicates that true trans-differentiation of BMSCs into phenotypes outside of their lineage is either very rare or non-existent (1;11). However, there is good evidence that BMSCs have a beneficial effect when administered directly to an injured tissue or intravenously (2;3;12). It is thought that the immunosuppressive, anti-inflammatory and tissue regenerative effects of BMSCs are due to the repertoire of cytokines and growth factors they secrete (1;13–16). BMSCs have been found to have a beneficial effects on acute GVHD following hematopoietic transplantation among HLA matched siblings and unrelated subjects (17;18). They have also been tested in the treatment of inflammatory bowel disease, ischemic heart disease (12), cirrhosis (19), autoimmune diseases (20;21), multiple sclerosis (22), stroke and traumatic brain injury (23).
The immunosuppressive effects of BMSCs have been well studied and they have been found to affect T cell, B cell, NK cell, monocyte and dendritic cell function (20;24;25). Factors implicated in the immunosuppressive properties of BMSCs include transforming growth factor beta-1 (TGFB1), interleukin-6 (IL-6), IL-8, prostaglandin E2, indoleamine 2,3-dioxygenase (IDO), hepatocyte growth factor (HGF), inducible nitric oxide synthase (iNOS), and insulin like growth factor (24). Paracrine effects are also thought to be responsible for many of the reparative and regenerative effects of BMSCs. BMSCs are an important component of the marrow niche, where they support and regulate hematopoietic stem cells and hematopoiesis. It may be that BMSCs are also able to “nurse” other stem cells by direct interaction or by paracrine stimuli (1).
A more precise characterization of the factors responsible for the clinical activity of BMSCs will likely lead to improved clinical therapy. We used global transcriptome analysis to characterize BMSCs. In addition, we made an attempt to identify biomarkers that could be used to assess the quality of clinical BMSC products and might be useful for assessing the potency of BMSC products. These biomarkers could be useful for assessing the stability, consistency, comparability and potency of clinical BMSCs products. BMSCs were compared to two types of stem cells, human embryonic stem cells (hESCs) and hematopoietic stem cells (HSCs), to identify genes expressed by the majority of BMSCs. They were also compared to fibroblasts to identify genes expressed by stem cells within the BMSC population.
Methods
Study Design
BMSCs were prepared using marrow aspirates collected from 7 healthy adults after obtaining informed consent. Human embryonic stem cells (hESCs) were obtained from NIH approved cell lines and were cultured as previously described (WA09 and WA01, WiCell Research Institute, Madison, WI and hESC BG01V, American Type Culture Collection (ATCC), Manassas, VA.) (26). Hematopoietic stem cells were obtained from G-CSF-mobilized peripheral blood stem cell (PBSC) concentrates. Three healthy subjects were given 5 days of G-CSF (10 micrograms/kg daily) and PBSCs were collected by apheresis. CD34+ cells were isolated from the PBSC concentrates using monoclonal antibodies and paramagnetic beads (CliniMacs, Miltenyi Biotec, Auburn, CA). The isolated cells were analyzed by flow cytometry and ≥ 95% expressed CD34 antigen. Three fibroblast cell lines were also tested: NuFF1 (Global Stem Inc, Rockville, MD), CCD-1112Sk and CCD 1108Sk (ATCC, Manassas, VA). All three were cultured according to manufacturer’s instructions. These studies were approved by an NIH institutional review board.
BMSC Culture
Approximately 10 to 15 mL of marrow was aspirated from the posterior iliac crest of 4 healthy subjects. The aspirates were collected into 3 mL syringes containing 1.5 of media (heparin, 150 units in 1.5 mL of DMEM with gentamicin). The cells were counted and plated in 75 cm2 flasks (culture flasks, canted neck, polystyrene, Corning Life Sciences, Corning, NY) at a cell density of 2 × 105/cm2 in complete media (alpha MEM, 2mM Glutamine, Gentamicin and 20% pre-selected Fetal Bovine Serum). Nonadherent cells were removed after 24 hours, and the adherent cells were cultured for approximately 13 days. During this period, media was changed every 3 days. The cells were harvested when the colonies reached 70–80% confluence using human recombinant trypsin (TrypLE Express, Invitrogen, Life Technologies, Grand Island, NY).
Cells from passage 1 were used for further expansion. Passage 1 BMSCs were plated at a cell density of 3–4 × 103/cm2 in a 2-layer cell factory (Nunc Cell Factory CF2, Thermo Fisher Scientific, Rochester, NY) in the presence of complete media. The cultures were observed everyday and media was changed every two to three days based on the growth rate of the BMSCs. When the cells reached 70–80% confluence (day 5 to day 7), the expanded, passage 2, the BMSCs were harvested using human recombinant trypsin (Invitrogen). The passage 2 BMSCs were transferred to a 4-layer cell factory (Nunc) and were processed, cultured and harvested as described for the 2-layer cell factory. If necessary, the passage 3 cells were expanded once more using a 10-layer cell factory (Nunc) (P4). Cells at each passage were harvested and tested for phenotype, viability, and gene expression analysis. For these studies passage 3 or 4 BMSCs were tested. These BMSCs were >70% viable and expressed CD73, CD90, CD105 and CD146 but not CD11b, CD14, CD19, CD34 or CD45,
Microarray Gene Expression Analysis
Total RNA was extracted from all cell types using Trizol (Invitrogen, Carlsbad, CA). RNA integrity was assessed using a 2100 Bioanalyzer (Agilent Technologies, Waldbronn, Germany). For each cell sample, total RNA (3 μg) was amplified into anti-sense RNA (aRNA). Total RNA from a pool of PBMCs from six healthy subjects was extracted and amplified into aRNA to serve as a reference. Both reference and test aRNA were directly labeled using a ULS aRNA Fluorescent Labeling Kit (Kreatech, Amsterdam, Netherlands). Cy3 was used for the reference samples and Cy5 for the test samples.
Human oligonucleotide microarrays spanning the entire genome were printed in the Infectious Disease and Immunogenetics Section, DTM, Clinical Center, NIH using a commercial probe set containing 35,035 oligonucleotide probes, representing approximately 25,100 unique genes and 39,600 transcripts excluding control oligonucleotides (Operon Human Genome Array-Ready Oligo Set version 4.0, Huntsville, AL, USA). The design of the probe set was based on the Ensemble Human Database build (NCBI-35c), with full coverage of the NCBI human Reference sequence dataset (04/04/2005). The microarray was composed of 48 blocks with one spot printed per probe per slide. Hybridization was carried out in a water bath at 42°C for 18 to 24 hours. The arrays were washed and scanned using a GenePix scanner Pro 4.0 (Axon, Sunnyvale, CA) with a variable photomultiplier tube to obtain optimized signal intensities with minimum (<1% spots) intensity saturation.
Data Processing and Statistical Analysis
The raw data set was filtered according to a standard procedure to exclude spots below a minimum intensity that arbitrarily was set to an intensity parameter of 200 in both fluorescence channels. If the fluorescence intensity of one channel was greater than 200 for oligonuceotide array but the other channel was less than 200, the fluorescence of the low intensity channel was arbitrarily set to 200. Spots with diameters <20μm were excluded from the analysis. The filtered data was then normalized using the median over the entire array and was analyzed using BRB-ArrayTools (http://linus.nci.nih.gov/BRB-ArrayTools.html, National Cancer Institute (NCI), Biometric Research Branch, Division of Cancer Treatment and Diagnosis). Hierarchical cluster analysis and TreeView software were used to visualize the data (27;28). Gene annotation and functional pathway analysis was based on the Database for Annotation, Visualization and Integrated Discovery (DAVID) 2007 software (29) and GeneCards website (http://www.genecards.org/index.shtml).
Gene Expression Analysis by Quantitative PCR
To validate the results of the microarray analysis, 7 genes were selected for analysis by quantitative real-time/reverse-transcription polymerase chain reaction (RT-PCR). (Applied Biosystems). The expression of the 7 genes was analyzed according to manufacturers’ instructions and normalized using 18srRNA. Levels of expression were determined using the relative quantification method; the Ct values of the test genes were normalized to the Ct values of endogenous control 18srRNA. The fold change in gene expression or the relative quantity (RQ) was calculated based on RQ= 2−ΔCt, where ΔCt = average Ct of test sample-average Ct of endogenous control sample.
Results
Comparison of Genes Expressed by BMSCs, HSCs and hESCs
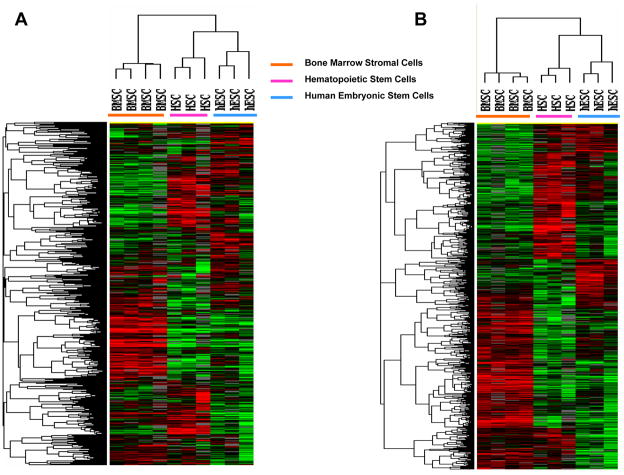
BMSCs produced from 4 healthy subjects and CD34+ cells from G-CSF-mobilized PBSC concentrates from 3 subjects and 3 hESC samples were analyzed by global transcriptome analysis. Among the more than 35,000 probes in the oligonucleotide microarray, the expression 4,475 probes were increased at least 2-fold in one sample and were expressed by 80% of the samples. Hierarchical clustering analysis of these 4,475 probes separated the samples in three clusters; one with all 4 BMSC samples, one with all 3 HSC samples and one with all 3 hESC samples (Figure 1, Panel A). Among the three types of cells, 2,800 of the 4,475 probes were differentially expressed (F-test, p ≤ 0.05) and hierarchical clustering analysis of the differentially expressed probes and 10 samples also separated the samples into the same 3 groups (Figure 1, Panel B). The expression of 2,538 genes was up-regulated in BMSCs compared to HSCs, and expression of 2,162 genes was down-regulated (t-tests; p<0.05) and the expression of 1,924 genes were up-regulated in BMSCs compared to hESCs and 1,248 were down-regulated (t-tests; p<0.05).
Figure 1. Global transcriptome analysis of BMSCs, HSCs and hESCs.
BMSCs from 4 subjects, CD34+ peripheral blood HSCs from 3 subjects and 3 hESC samples were analyzed with oligonucleotide microarray with >35,000 probes. Panel A shows the unsupervised hierarchical clustering analysis of the 4,475 probes that were increased at least 2-fold in one sample and expressed by 80% of the samples. Panel B shows the hierarchical clustering analysis of 2,800 differentially expressed genes (F-test, p<0.05).
A comparison of the molecular and cellular functions of genes up-regulated among these three cell types using Ingenuity software revealed that genes up-regulated in BMSCs compared to HSCs and hESCs were involved with cellular movement, cell-to-cell signaling and interaction, cell morphology, and cell death. In contrast, genes involved with RNA post-transcriptional modification, DNA replication, recombination and repair, RNA damage and repair and cell cycle were up-regulated in hESCs, and those involved with cellular development, gene expression, cell morphology and cell death were up-regulated in HSCs.
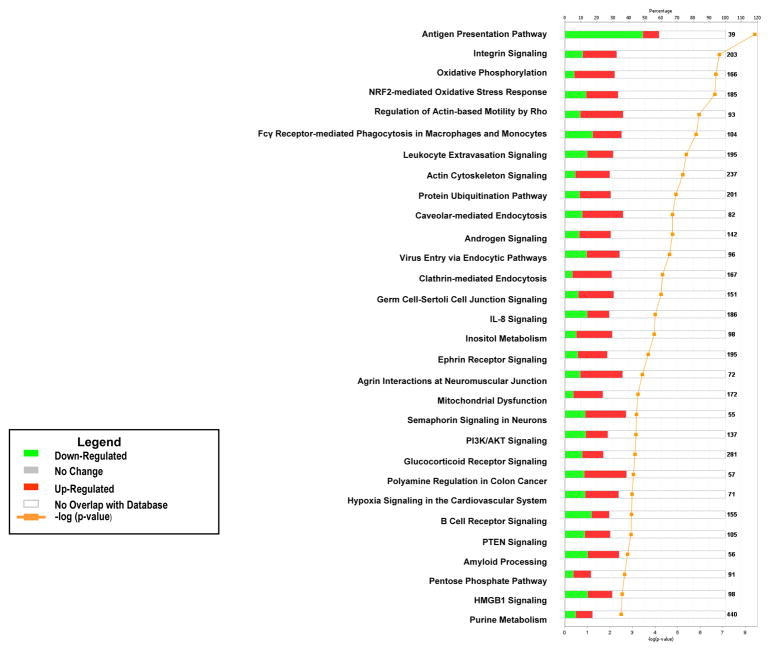
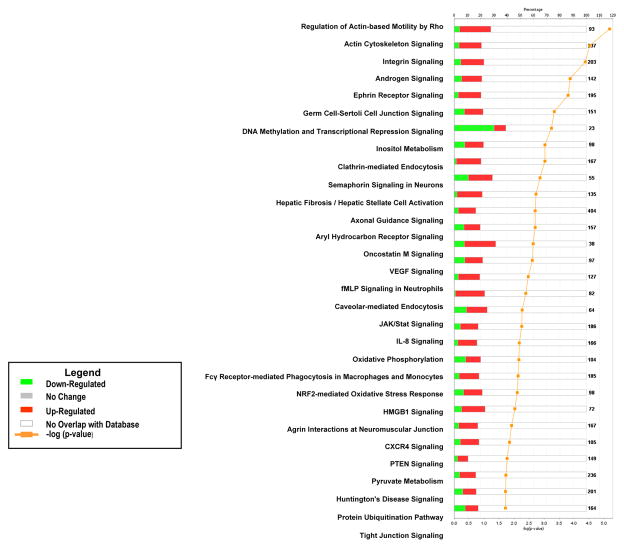
Pathway analysis of the genes differentially expressed between BMSCs and the two types of stem cells found that a significant number of BMSC up-regulated genes were involved with cellular growth, proliferation and development, organismal growth and development, cellular immune response, neurotransmitter and other nervous system signaling, cellular stress and injury, amino acid metabolism and carbohydrate metabolism (Supplemental Tables 1 and 2). A number of pathways included up-regulated BMSC genes compared to both HSCs and hESCs. These specific pathways include: integrin signaling, NFR2-mediated oxidative stress response, regulation of actin-based motility by Rho, actin cytoskeletal signaling, caveolar-mediated endocytosis, clathrin-mediated endocytosis, germ cell-sertoli cell junction signaling, IL-8 signaling, ephrin receptor signaling, HMGB1 signaling, PTEN signaling, androgen signaling, inositol metabolism and protein ubiquitination pathways (Figures 2 and 3).
Figure 2. Pathway analysis of differentially expressed genes in BMSCs compared to HSCs.
The canonical pathways significantly modulated by the genes differently expressed in BMSCs and HSCs are shown (Ingenuity Pathway Analysis, p < 0.05). The p value for each pathway is indicated by the bar and is expressed as −1 times the log of the p value. The line represents the ratio of the number of genes in a given pathway divided by the total number of genes that make up that pathway. The red bar indicated genes up-regulated in BMSCs and the green bar those up-regulated in HSCs. Only the 30 pathways with the most significant changes are shown.
Figure 3. Pathway analysis of differentially expressed genes in BMSCs compared to hESCs.
The canonical pathways significantly modulated by the genes differently expressed in BMSCs and hESCs are shown (Ingenuity Pathway Analysis, p < 0.05). The p value for each pathway is indicated by the bar and is expressed as −1 times the log of the p value. The line represents the ratio of the number of genes in a given pathway divided by the total number of genes that make up that pathway. The red bar indicated genes up-regulated in BMSCs and the green bar those up-regulated in hESCs. Only the 30 pathways with the most significant changes are shown.
Genes up-regulated in HSCs compared to BMSCs were most likely to be in immune pathways including, antigen presentation, B cell receptor signaling, activation of IRF by cytosolic pattern recognition receptors, primary immunodeficiency signaling, DC maturation, 4-1BB signaling in T lymphocytes, IL-8 signaling, and allograft rejection signaling (data not shown). Genes up-regulated in hESCs compared to BMSCs were most likely to be the DNA methylation and transcriptional repression signaling, role of Oct4 in mammalian embryonic stem cell pluripotency, RAN signaling and role of BRCA1 in DNA damage response pathways (data not shown).
The specific genes that were up- and down-regulated the greatest fold in BMSCs compared to HSCs are shown in Table 1 and compared to hESCs in Table 2. The most up-regulated genes in BMSCs compared to both HSCs and hESCs were transforming growth Factor beta induced (TGFBI) protein, ADAM12, Annexin 2 (ANXA2), and myoferlin (MYOF). The expression of these genes was up-regulated 20- to 347-fold in BMSCs compared to both HSCs and hESCs. Genes encoding several extracellular matrix proteins were also markedly up-regulated in either HSCs or hESCs including collagen, type III, alpha 1 (COL3A1), collagen, type V, alpha 1 (COL5A1), actin, alpha 1 (ACTA1), actin, alpha 2 (ACTA2), fibronectin (FN1), thrombospondin 1 and 2 and connective tissue growth factor (CTGF).
Table 1.
Genes differentially expressed between BMSC and CD34+ cells isolated from G-CSF-mobilized PBSCs.
| Genes up-regulated in BMSCs | Fold- Increase | Genes down-regulated in BMSCs | Fold- Decrease | ||
|---|---|---|---|---|---|
| TGFBI | transforming growth factor, beta-induced | 346 | LAPTM5 | lysosomal multispanning membrane protein 5 | 74.3 |
| COL3A1 | collagen, type III, alpha 1 | 153 | HLA-DRA | histocompatibility antigen HLA-DR | 54.1 |
| ADAM12 | ADAM metallopeptidase domain 12 | 90.7 | GNA15 | guanine nucleotide binding protein (G protein), alpha 15 (Gq class) | 43.5 |
| ACTA2 | actin, alpha 2, smooth muscle, aorta | 87.8 | MPO | myeloperoxidase | 35.3 |
| ANXA2 | annexin A2 | 65.5 | GMFG | glia maturation factor, gamma | 35.3 |
| CTGF | connective tissue growth factor | 64.2 | CCND2 | cyclin D2 | 34.3 |
| PENK | proenkephalin | 62.4 | PRSSL1 | protease, serine-like 1 | 33.2 |
| MXRA8 | matrix-remodelling associated 8 | 62.1 | ARHGDIB | Rho GDP dissociation inhibitor (GDI) beta | 31.96 |
| COL3A1 | collagen, type III, alpha 1 | 58.5 | ACSS1 | acyl-CoA synthetase short-chain family member 1 | 31.89 |
| MYOF | myoferlin | 57.8 | SYK | spleen tyrosine kinase, transcript variant 2 | 29.47 |
| FEZ1 | fasciculation and elongation protein zeta 1 | 57.7 | HLA-DPB1 | MHC, class II, DP beta 1 | 26.36 |
| NCKAP1 | NCK-associated protein 1 | 53.5 | GBP4 | guanylate binding protein 4 | 25.55 |
| NQO1 | NAD(P)H dehydrogenase, quinone 1 | 52.4 | IGHG1 | Immunoglobulin lambda heavy chain | 25.46 |
| FN1 | fibronectin 1 ( | 51.4 | ARHGDIB | Rho GDP dissociation inhibitor (GDI) beta | 25.36 |
| PCOLCE | procollagen C-endopeptidase enhancer | 47.5 | H1FX | H1 histone family, member X | 22.86 |
| AFAP1 | actin filament associated protein 1 | 46.7 | ICAM3 | intercellular adhesion molecule 3 | 22.06 |
| THBS1 | thrombospondin 1 | 43.4 | HLA-DRB3 | MHC, class II, DR beta 3 | 21.79 |
| AMOTL2 | angiomotin like 2 | 42.9 | HLA-DRA | MHC, class II, DR alpha | 21.71 |
| CTSL1 | cathepsin L1 | 42.3 | HLA-DRB4 | MHC, class II, DR beta 4 | 21.40 |
| LARP6 | La ribonucleoprotein domain family, member 6 | 37.9 | SELL | selectin L | 20.90 |
| MT2A | metallothionein 2A | 37.7 | HLA-DMA | MHC, class II, DM alpha | 20.84 |
| SLC17A8 | solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 8 | 37.4 | LOC100131564 | (xs92) mRNA, 445bp | 19.45 |
| ACTN1 | actinin, alpha 1 | 37.4 | STAG3L2 | PREDICTED: Homo sapiens hypothetical protein LOC100132740 | 18.48 |
| ITGB5 | integrin, beta 5 | 36.8 | CYTL1 | cytokine-like 1 | 18.05 |
| FN1 | fibronectin 1, transcript variant 5 | 36.1 | CHST13 | carbohydrate (chondroitin 4) sulfotransferase 13 | 17.88 |
p < 1 × 10−3 for all genes
Table 2.
Genes differentially expressed between BMSCs and hES
| Genes up-regulated in BMSCs | Fold- Increase | Genes down-regulated in BMSCs | Fold- Decrease | ||
|---|---|---|---|---|---|
| TGFBI | transforming growth factor, beta-induced, 68kDa | 298 | SLC7A3 | solute carrier family 7 (cationic amino acid transporter, y+ system), member 3 | 48.8 |
| ADAM12 | ADAM metallopeptidase domain 12 | 82.8 | TERF1 | telomeric repeat binding factor (NIMA-interacting) 1 | 32.6 |
| IGFBP7 | insulin-like growth factor binding protein 7 | 61.4 | psiTPTE22 | TPTE pseudogene (psiTPTE22) | 30.1 |
| CERCAM | cerebral endothelial cell adhesion molecule | 59.9 | psiTPTE22 | TPTE pseudogene (psiTPTE22) | 29.2 |
| COL5A1 | collagen, type V, alpha 1 | 56.3 | POU5F1B | POU class 5 homeobox 1B | 25.8 |
| ACTA2 | actin, alpha 2, smooth muscle, aorta | 51.3 | O00627 | Endogenous retrovirus H D1 leader region/integrase- derived ORF1 ORF2, and putative envelope protein | 22.8 |
| TSPO | translocator protein (18kDa), | 49.7 | CCND2 | cyclin D2 | 14.7 |
| CRIP2 | cysteine-rich protein 2 | 39.1 | LEFTY1 | left-right determination factor 1 | 13.8 |
| MYOF | myoferlin | 31.0 | FAM60A | family with sequence similarity 60, member A | 13.5 |
| COMT | catechol-O-methyltransferase | 30.0 | CDH3 | cadherin 3, type 1, P-cadherin | 13.4 |
| THBS2 | thrombospondin 2 | 30.0 | JARID2 | jumonji, AT rich interactive domain 2 | 13.2 |
| TNS1 | tensin 1 | 28.9 | TUBB2B | tubulin, beta 2B | 12.6 |
| IAH1 | isoamyl acetate-hydrolyzing esterase 1 homolog | 27.2 | HNRNPD | heterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) | 12.3 |
| TSPO | translocator protein (18kDa) | 27.3 | PLCXD1 | phosphatidylinositol-specific phospholipase C, X domain containing 1 | 11.4 |
| ALPK2 | alpha-kinase 2 | 27.1 | CD24 | CD24 molecule | 10.9 |
| EMILIN1 | elastin microfibril interfacer 1 | 25.4 | TRIM24 | tripartite motif-containing 24 | 10.8 |
| MVP | major vault protein | 24.7 | GARNL4 | GTPase activating Rap/RanGAP domain-like 4 | 10.5 |
| CCDC92 | coiled-coil domain containing 92 | 24.6 | FXYD6 | FXYD domain containing ion transport regulator 6 | 9.61 |
| ANXA2 | annexin A2 | 24.4 | ANKRD10 | Ankyrin repeat domain 10 | 9.59 |
| CAPN2 | calpain 2, (m/II) large subunit | 23.6 | AMT | aminomethyltransferase | 9.44 |
| ITGA5 | integrin, alpha 5 (fibronectin receptor, alpha polypeptide) | 22.2 | PABPN1 | poly(A) binding protein, nuclear 1 | 9.36 |
| GALNT1 | UDP-N-acetyl-alpha-D- galactosamine:polypeptide N- acetylgalactosaminyltransferase 1 (GalNAc-T1) | 22.1 | APOE | apolipoprotein E | 9.30 |
| CYP1B1 | cytochrome P450, family 1, subfamily B, polypeptide 1 | 21.4 | CDKN1C | cyclin-dependent kinase inhibitor 1C (p57, Kip2) | 9.28 |
| EFEMP2 | EGF-containing fibulin-like extracellular matrix protein 2 | 20.9 | CHD7 | chromodomain helicase DNA binding protein 7 | 9.10 |
| TRIB3 | tribbles homolog 3 (Drosophila) | 20.8 | FOXA3 | forkhead box A3 | 9.07 |
p < 1 × 10−2 for all genes
The most up-regulated genes in HSCs compared to BMSCs were distinct from those up-regulated in hESCs. Several MHC Class II genes were highly up-regulated in HSCs (Table 1), whereas OCT4 (POU5F1B), LEFTY1, and FOXA3 were some of the most up-regulated genes in hESCs compared to BMSCs (Table 2).
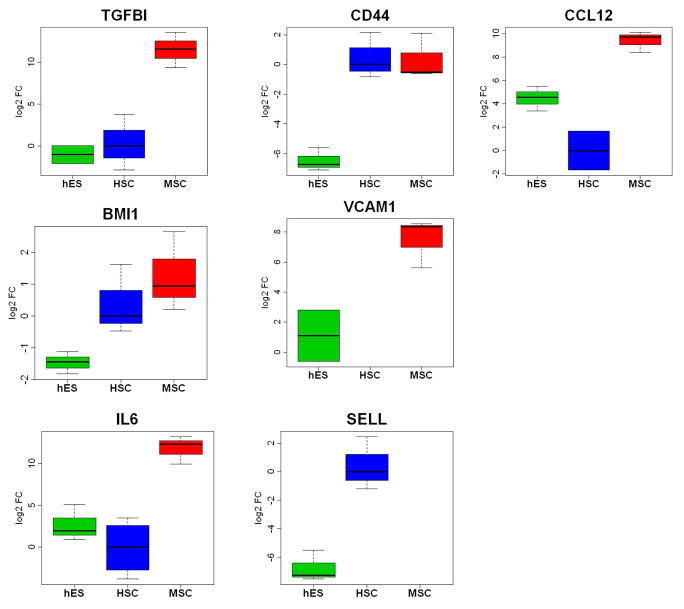
Quantitative real time PCR analysis confirmed that the expression of TGFBI, CCL12, IL-6 and VCAM-1 was greater in BMSCs than in HSCs or hESCs (Figure 4).
Figure 4. Comparison of the expression of selected gene between BMSCs, HSCs and hESCs.
Quantitative real time PCR was used to measure gene expression levels in the 4 BMSC, 3 HSC and 3 hESC samples of TGFBI, CD44, CCL12, BMI1, VCAM1, IL6 and SELL.
Comparison of Gene Expression in BMSCs and Fibroblasts
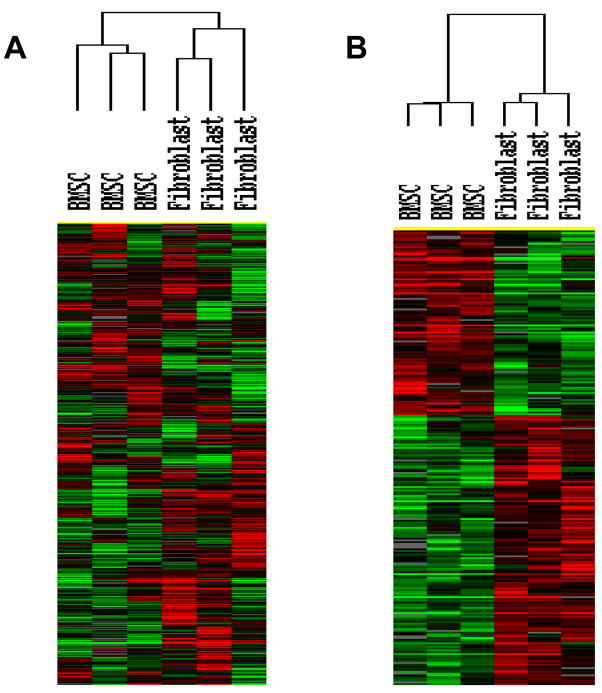
Global gene expression of the same 4 BMSC samples was compared with 3 fibroblasts samples. Both unsupervised and supervised hierarchical gene expression analysis separated the samples into two clusters, one with all 4 BMSC samples and one with the 3 fibroblast samples (Figure 5). Among the two groups, 1,991 genes were differentially expressed (F-test, p< 0.05).
Figure 5. Global transcriptome analysis of BMSCs and fibroblasts.
BMSCs from 3 subjects and 3 fibroblast cell lines were analyzed with oligonucleotide microarray with >35,000 probes. Panel A shows the unsupervised hierarchical clustering analysis of the 4,944 probes that were increased at least 2-fold in one sample and were expressed by 80% of the samples. Panel B shows that hierarchical clustering analysis of 1,991 differentially expressed genes (F-test, p<0.05).
The molecular and cellular function of genes up-regulated in BMSCs compared to fibroblasts included those involved in cell-to-cell signaling and interaction, cell compromise, and cellular growth and proliferation. The up-regulated genes in fibroblasts were involved protein synthesis, gene expression and cell morphology(Supplement Table 3).
Table 3.
Genes differentially expressed between BMSCs and fibroblasts.
| Genes up-regulated most in BMSCs | Fold Increase | Genes down-regulated most in BMSCs Name | Fold Decrease | ||
|---|---|---|---|---|---|
| HAPLN1 | hyaluronan and proteoglycan link protein 1 | 21.9 | MMP3 | matrix metallopeptidase 3 (stromelysin 1, progelatinase) | 51.0 |
| PCOLCE2 | procollagen C-endopeptidase enhancer 2 | 7.72 | CPZ | carboxypeptidase Z | 39.2 |
| HAS2 | hyaluronan synthase 2 | 7.46 | MASP1 | mannan-binding lectin serine peptidase 1 (C4/C2 activating component of Ra-reactive factor) | 18.1 |
| FOS | FBJ murine osteosarcoma viral oncogene homolog | 6.76 | PTGS1 | prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | 15.1 |
| FOSB | FBJ murine osteosarcoma viral oncogene homolog B | 6.71 | MME | membrane metallo-endopeptidase | 13.6 |
| GALNT1 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N- acetylgalactosaminyltransferase 1 (GalNAc-T1) | 6.65 | SVEP1 | sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 | 13.1 |
| ALDH1B1 | aldehyde dehydrogenase 1 family | 6.63 | TWIST2 | twist homolog 2 (Drosophila) | 12.7 |
| SIAE | sialic acid acetylesterase | 6.46 | SFRP1 | secreted frizzled-related protein 1 | 12.7 |
| SORCS2 | sortilin-related VPS10 domain containing receptor 2 | 6.18 | OSR2 | odd-skipped related 2 (Drosophila) | 12.3 |
| HOXB7 | homeobox B7 | 5.84 | CAMK2N1 | calcium/calmodulin-dependent protein kinase II inhibitor 1 | 10.4 |
| SMAD7 | SMAD family member 7 | 5.61 | CTSC | cathepsin C | 10.1 |
| LCTL | lactase-like | 5.59 | EMILIN2 | elastin microfibril interfacer 2 | 9.25 |
| ETV7 | ets variant 7 | 5.52 | HSPB3 | heat shock 27kDa protein 3 | 9.18 |
| ANKH | ankylosis | 5.49 | ADM | adrenomedullin | 8.79 |
| PASD1 | PAS domain containing 1 | 5.36 | FAM129A | family with sequence similarity 129, member A | 8.45 |
| P2RY6 | pyrimidinergic receptor P2Y | 5.26 | CXCR7 | Chemokine (C-X-C motif) receptor 7 | 8.41 |
| FN1 | fibronectin 1 | 5.07 | TSEN54 | tRNA splicing endonuclease 54 homolog (S. cerevisiae) | 8.16 |
| VCAN | versican | 4.95 | COL15A1 | Collagen, type XV, alpha 1 | 8.09 |
| KCNH2 | potassium voltage-gated channel | 4.90 | PTGFR | prostaglandin F receptor | 8.03 |
| VCAM1 | vascular cell adhesion molecule 1 | 4.88 | CD69 | CD69 molecule | 7.65 |
| ABI3BP | ABI family | 4.83 | WWOX | WW domain containing oxidoreductase | 7.47 |
| TTC3 | tetratricopeptide repeat domain 3 | 4.65 | IFIT1 | interferon-induced protein with tetratricopeptide repeats 1 | 7.08 |
| PDLIM5 | PDZ and LIM domain 5 | 4.63 | PDPN | podoplanin | 7.01 |
| SLC22A6 | solute carrier family 22 (organic anion transporter) | 4.56 | CTSC | cathepsin C | 6.61 |
| DACT1 | dapper | 4.52 | DPP4 | dipeptidyl-peptidase 4 | 6.58 |
P < 1 × 10−2 for all genes
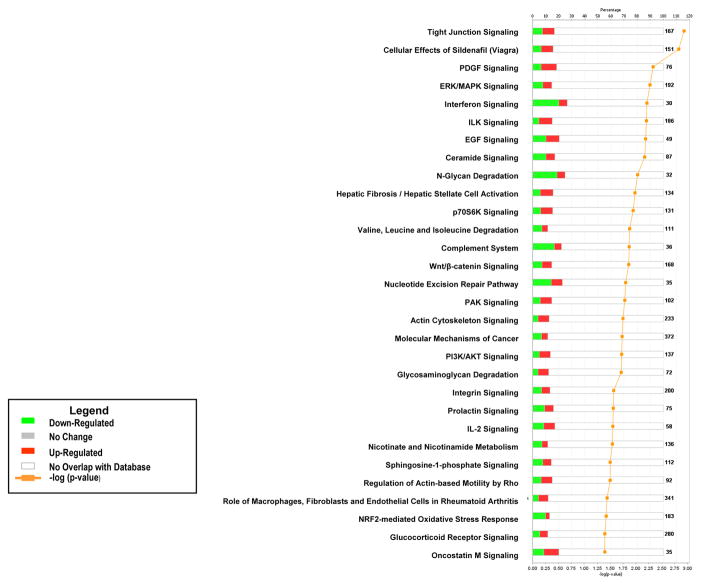
Pathway analysis of the genes differentially expressed between BMSCs and fibroblasts found that a significant number of up-regulated genes in the BMSCs were involved in cellular growth, proliferation and development, organismal growth and development, intracellular and second messenger signaling, cellular immune response, and neurotransmitters and other nervous system signaling
The specific pathways up-regulated in BMSCs included ILK signaling, integrin signaling, PDGF signaling, EGF signaling, Wnt/β-catenin signaling, PI3K/AKT signaling, PAK signaling, tight junction signaling, IL-2 signaling, and pathways responsible for the role of macrophages, fibroblasts and endothelial cells in rheumatoid arthritis (Figure 6). Up-regulated pathways in fibroblasts included interferon signaling, N-glycan degradation, and complement system pathways. Among the most up-regulated genes in BMSCs were FOS, FOSB, HOXB7, SMAD7, FN1 and VCAM1 (Table 3).
Figure 6. Pathway analysis of differentially expressed genes in BMSCs compared to Fibroblasts.
The canonical pathways significantly modulated by the genes differently expressed in BMSCs and fibroblasts are shown (Ingenuity Pathway Analysis p < 0.05). The p value for each pathway is indicated by the bar and is expressed as −1 times the log of the p value. The line represents the ratio of the number of genes in a given pathway divided by the total number of genes that make up that pathway. The red bar indicated genes up-regulated in BMSCs and the green bar those up-regulated in fibroblasts. Only the 30 pathways with the most significant changes are shown.
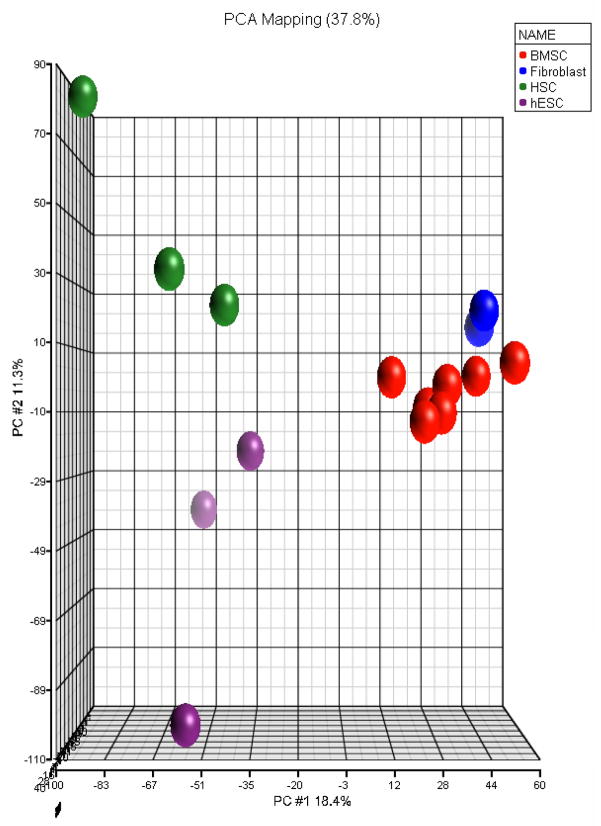
The expression of BMSC genes encoding functionally important proteins was compared to fibroblasts, HSC and hESCs. The up-regulated genes in BMSCs compared to HSCs and hESCs included HGF, IL6, VCAM1, CXCL12, PTGES, CD44 and CD59. When compared to HSCs, HGF was up-regulated 6.4-fold; IL-6 13.7-fold; PTGES 13.2-fold; and VCAM1 9.8-fold. In contrast, when BMSCs were compared to fibroblasts, only VCAM1 was up-regulated. This suggested that BMSCs are more similar to fibroblasts than HSCs or hESCs. To further compare BMSCs, fibroblasts, HSCs and hESCs, the gene expression data from all 16 samples was examined using multiple dimensional clustering (Figure 7). All samples of the same type clustered together. The HSCs and hESCs clustered more closely to each other than to BMSCs or fibroblasts, and BMSCs cluster more closely to fibroblasts than to either HSCs or hESCs.
Figure 7. Comparison of BMSCs, fibroblasts, HSCs and hESCs.
Results of the gene expression analysis of 7 BMSCs, 3 fibroblast, 4 hESC and 4 HSCs samples were analyzed by multiple dimensional clustering (Partek Software). Since the transcriptome analysis of 3 fibroblast and 3 BMSC samples were perform at a different time than that of the 4 HSC, 4 hESC and 3 BMSC samples, genes whose expression differed due to a batch effect were removed by identifying genes whose expression differed between the two groups of BMSC samples and excluding them from the data set (p<0.05). A total of 4,300 genes were removed.
Discussion
An important step toward understanding the therapeutic effects of cultured BMSCs is their full characterization. Global gene expression analysis was used to compare BMSCs with HSCs, hESCs and fibroblasts. Analysis of the genes and pathways expressed by cultured BMSCs were consistent with a mobile, rapidly proliferative and metabolically active cell population capable of cell-to-cell signaling and interaction. Comparison of BMSCs with HSCs and hESCs confirmed that IL-6, HGF and PGE2 are likely important in the immune modulatory properties of BMSCs. Analysis of specific genes up-regulated in BMSC found that in addition to producing several cytokines and growth factors, they also produced large quantities of ECM proteins.
Pathway analysis of the genes expressed by BMSCs demonstrated that several pathways involved with cell adhesion and motility were up-regulated including integrin signaling, ILK signaling, actin cytoskeletal signaling and regulation of actin-based mobility by Rho pathways. Integrins are a heterodimeric superfamily of receptors involved with cell-cell and cell-matrix adhesion. They are involved with migration, proliferation, differentiation, gene expression, cell death and cell survival. They also appear to have a prominent role in BMSC function. Among the integrin genes markedly up-regulated in BMSCs were the α5 chain of the fibronectin receptor and the β5 chain of the vibronectin/osteopontin receptor. This is consistent with other studies that have found that the fibronectin and osteopontin receptors are highly expressed by BMSCs (30;31).
Integrins transmit signals through a variety of intracellular protein kinase and adaptor molecules such as ILK, FAK, Talin, paxillin, parvins, p130Cas, src-family kinases and Rho family GTPases. It appears that ILK and Rho family GTPases are important in integrin signaling in BMSCs.
Genes in both the ILK and regulation of actin-based mobility by Rho signaling pathways are up-regulated in BMSCs. ILK binds directly to the cytoplasmic domain of the β1 and β3 integrin chains and regulates a number of cellular processes and extracellular matrix accumulation (32). ILK links integrins to intracellular receptor kinases and actin.
The Rho family of GTPases link integrins and other cell surface receptors to the actin cytoskeleton (32). They are important regulators of the actin cytoskeleton and are involved with cell migration and adhesion. The Rho family of GTPases have been implicated in the proliferation and migration of HSCs and hematopoietic progenitor cells and some investigations have suggested that they are involved in BMSC migration and adhesion (33).
We found that genes encoding a number of factors found in the ECM are up-regulated in cultured BMSCs. The individual genes that were most up-regulated in BMSCs included several genes encoding ECM proteins including COL3A1, COL5A1, fibronectin hyaluronan and proteoglycan link protein 1 (HAPLN1), versican (VCAN), EGF-containing fibulin-like extracellular matrix protein 2 (EFEMP2), elastin microfibril intrerface 1 (EMILIN1), TGFBI, ADAM12, CTGF, and hyaluronan synthase (HAS2). Among these other have found that TGFBI and CTGF, COL3A1 are highly expressed in BMSCs (34;35).
Genes involved in the cavelolar-mediated and clathrin-mediated endocytosis pathways were up-regulated in BMSCs. Clathrin and cavelolar-mediated endocytosis are involved in the endocytosis of transmembrane receptors, which help regulate the sensitivity of cells to specific ligands. Endocytosis is also involved in regulating cell migration, mitosis and antigen presentation (36). Genes in these pathways have not previously been described to be expressed by BMSCs, and their role in BMSC function is not certain, but they may be involved with cell-to-cell interactions and signaling.
An important aspect of producing new clinical cellular therapies is to develope an assay that can be used to test the products at the end of manufacturing to ensure that they meet minimal criteria. One critical assessment made on all products is the measurement of its critical biological function(s) or potency. As new cellular therapies such as those involving BMSCs are developed, it is important to identify biomarkers that can be used to measure potency. Identifying potency biomarkers is critical, for without good biomarkers, it is not possible to determine whether a clinical trial fails due to inappropriate patient or disease selection or to a failure to consistently produce high quality products. Potency biomarkers are also important for testing product stability, consistency and comparability.
Measuring the potency of BMSCs is more difficult than measuring the potency other cellular therapies since they are being used for several diverse applications: bone and tissue regeneration, immune modulation and anti-inflammation. Furthermore, BMSCs may need to possess multiple critical functions for each of these applications. In addition, the properties and functions that are responsible for each of these therapies are not completely understood. Currently, there is no consensus concerning the specific biomarker(s) that should be used for BMSC potency testing. This study identified a number of genes expressed by BMSC that might be useful as biomarkers of BMSC anti-inflammatory, immune modulatory or the regeneration properties.
We found that BMSCs expressed IL-6, HGF and PGE2, all of which have been shown to play a role in the immune modulatory properties of BMSCs. These factors are potential BMSC potency biomarkers.
The chemokine ligand CXCL12 could also be a biomarker of immune modulation. CXCL12 is the ligand for an important hematopoietic stem cell adhesion molecule CXCR4. The interaction of CXCR4 and CXCL12 is thought to be critical for the homing of hematopoietic stem cells to their bone marrow niche (37). CXCR4 is also expressed by a number of immune cells and is also likely involved with the homing of these cells.
Many ECM proteins expressed by BMSCs have important functional properties that may contribute to the immune regulation and regenerative properties of BMSCs and could also be useful BMSC biomarkers. TGFβ I is a component of the extracellular matrix and is expressed ubiquitously by normal tissue. TGβ I mediates cell adhesion and migration through interactions with integrins and integrin receptors. Recent studies have suggested that TGFβ I inhibits tumorigenicity and tumor angiogenesis (38;39). Mice lacking TGFβ I are prone to spontaneous tumor development including lymphoma (39). The tumor suppressor effects of TGFβ I may be due to its suppression of CREB and cyclin D1. The anti-proliferative effects of TGFβ I may contribute to the ability of BMSCs to inhibit mixed lymphocyte reactions and modulate the immune response.
ADAM12 belongs to the ADAMs (a disintegrin and metalloproteases) family of proteins. It is a metalloprotease with it appears to regulated cell-cell and cell-extracellular matrix contacts through interaction with cell surface integrin and syndecan receptors and potentially influence the actin cytoskeleton (40). ADAM12 is expressed in muscle tissue and its expression is up-regulated during regeneration. Its expression is also increased in patients with hypertrophic myocardium (41). ADAM 12 may contribute to the tissue regenerative properties of BMSCs.
CTGF or CCN2 is a secreted protein that is strongly up regulated in fibrotic tissue. CTGF belongs to a group of secreted proteins called matricellular proteins. They are secreted and sequestered in the ECM, where they interact with cell surface receptors (42). CTGF interacts with a variety of receptors including integrin and growth factor receptors (42). CTGF functions to modulate the activity of other growth factors and it modulates adhesion, migration, and ECM production (42). It is also involved in the regulation of development, differentiation and tumorigenesis (43). CTGF production is induced in heart failure and the overexpression of CTGF in cardiomyocytes of transgenic mice exerts a prohypertrophic effect (44).
Among the most highly expressed BMSC genes were three related to bone development that could be biomarkers of the ability of BMSCs to produce bone: FOS, FOSB, and COL3A. One of these, FOSB, has been found to be highly expressed by another group (45). Fos proteins form heterodimers with Jun proteins to form AP-1 transcription factors which are expressed by osteoclasts and are involved with extracellular matrix gene expression (46;47). Type III collagen stimulates the growth of human osteoblastic cells (48).
In conclusion, global analysis of human BMSCs demonstrated that these cells are mobile, endocytotic, metabolically active and capable of producing ECM proteins. Integrins and integrin signaling and Wnt signaling appear to be important to these cells. The most highly expressed genes included those encoding for ECM proteins, some with important functional properties and may contribute to the clinical immune modulatory and anti-inflammatory effects of BMSCs. We identified some genes that might be useful biomarkers of BMSC quality. However, their value as possible potency biomarkers must be validated by comparing the expression of these biomarkers by clinical BMSC products with BMSC activity in animal models, biologic systems or in clinical trials.
Supplementary Material
Acknowledgments
This research was supported in part by the Intramural Research Program of the National Institute of Dental and Craniofacial Research, NIH and the Department of Transfusion Medicine, Clinical Center, NIH.
References
- 1.Bianco P, Robey PG, Simmons PJ. Mesenchymal stem cells: revisiting history, concepts, and assays. Cell Stem Cell. 2008 Apr 10;2(4):313–9. doi: 10.1016/j.stem.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen J, Li Y, Wang L, Zhang Z, Lu D, Lu M, et al. Therapeutic benefit of intravenous administration of bone marrow stromal cells after cerebral ischemia in rats. Stroke. 2001 Apr;32(4):1005–11. doi: 10.1161/01.str.32.4.1005. [DOI] [PubMed] [Google Scholar]
- 3.Mahmood A, Lu D, Chopp M. Marrow stromal cell transplantation after traumatic brain injury promotes cellular proliferation within the brain. Neurosurgery. 2004 Nov;55(5):1185–93. doi: 10.1227/01.neu.0000141042.14476.3c. [DOI] [PubMed] [Google Scholar]
- 4.Horwitz EM, Gordon PL, Koo WK, Marx JC, Neel MD, McNall RY, et al. Isolated allogeneic bone marrow-derived mesenchymal cells engraft and stimulate growth in children with osteogenesis imperfecta: Implications for cell therapy of bone. Proc Natl Acad Sci U S A. 2002 Jun 25;99(13):8932–7. doi: 10.1073/pnas.132252399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koc ON, Day J, Nieder M, Gerson SL, Lazarus HM, Krivit W. Allogeneic mesenchymal stem cell infusion for treatment of metachromatic leukodystrophy (MLD) and Hurler syndrome (MPS-IH) Bone Marrow Transplant. 2002 Aug;30(4):215–22. doi: 10.1038/sj.bmt.1703650. [DOI] [PubMed] [Google Scholar]
- 6.Meuleman N, Vanhaelen G, Tondreau T, Lewalle P, Kwan J, Bennani J, et al. Reduced intensity conditioning haematopoietic stem cell transplantation with mesenchymal stromal cells infusion for the treatment of metachromatic leukodystrophy: a case report. Haematologica. 2008 Jan;93(1):e11–e13. doi: 10.3324/haematol.11802. [DOI] [PubMed] [Google Scholar]
- 7.Mankani MH, Kuznetsov SA, Marshall GW, Robey PG. Creation of new bone by the percutaneous injection of human bone marrow stromal cell and HA/TCP suspensions. Tissue Eng Part A. 2008 Dec;14(12):1949–58. doi: 10.1089/ten.tea.2007.0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mankani MH, Kuznetsov SA, Shannon B, Nalla RK, Ritchie RO, Qin Y, et al. Canine cranial reconstruction using autologous bone marrow stromal cells. Am J Pathol. 2006 Feb;168(2):542–50. doi: 10.2353/ajpath.2006.050407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mankani MH, Kuznetsov SA, Wolfe RM, Marshall GW, Robey PG. In vivo bone formation by human bone marrow stromal cells: reconstruction of the mouse calvarium and mandible. Stem Cells. 2006 Sep;24(9):2140–9. doi: 10.1634/stemcells.2005-0567. [DOI] [PubMed] [Google Scholar]
- 10.Kuznetsov SA, Huang KE, Marshall GW, Robey PG, Mankani MH. Long-term stable canine mandibular augmentation using autologous bone marrow stromal cells and hydroxyapatite/tricalcium phosphate. Biomaterials. 2008 Nov;29(31):4211–6. doi: 10.1016/j.biomaterials.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Phinney DG, Prockop DJ. Concise review: mesenchymal stem/multipotent stromal cells: the state of transdifferentiation and modes of tissue repair--current views. Stem Cells. 2007 Nov;25(11):2896–902. doi: 10.1634/stemcells.2007-0637. [DOI] [PubMed] [Google Scholar]
- 12.Zhou Y, Wang S, Yu Z, Hoyt RF, Jr, Sachdev V, Vincent P, et al. Direct injection of autologous mesenchymal stromal cells improves myocardial function. Biochem Biophys Res Commun. 2009 Dec 18;390(3):902–7. doi: 10.1016/j.bbrc.2009.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caplan AI, Dennis JE. Mesenchymal stem cells as trophic mediators. J Cell Biochem. 2006 Aug 1;98(5):1076–84. doi: 10.1002/jcb.20886. [DOI] [PubMed] [Google Scholar]
- 14.Sadan O, Melamed E, Offen D. Bone-marrow-derived mesenchymal stem cell therapy for neurodegenerative diseases. Expert Opin Biol Ther. 2009 Dec;9(12):1487–97. doi: 10.1517/14712590903321439. [DOI] [PubMed] [Google Scholar]
- 15.Le BK, Ringden O. Immunobiology of human mesenchymal stem cells and future use in hematopoietic stem cell transplantation. Biol Blood Marrow Transplant. 2005 May;11(5):321–34. doi: 10.1016/j.bbmt.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 16.Horwitz EM, Dominici M. How do mesenchymal stromal cells exert their therapeutic benefit? Cytotherapy. 2008;10(8):771–4. doi: 10.1080/14653240802618085. [DOI] [PubMed] [Google Scholar]
- 17.Ball LM, Bernardo ME, Roelofs H, Lankester A, Cometa A, Egeler RM, et al. Cotransplantation of ex vivo expanded mesenchymal stem cells accelerates lymphocyte recovery and may reduce the risk of graft failure in haploidentical hematopoietic stem-cell transplantation. Blood. 2007 Oct 1;110(7):2764–7. doi: 10.1182/blood-2007-04-087056. [DOI] [PubMed] [Google Scholar]
- 18.Le BK, Frassoni F, Ball L, Locatelli F, Roelofs H, Lewis I, et al. Mesenchymal stem cells for treatment of steroid-resistant, severe, acute graft-versus-host disease: a phase II study. Lancet. 2008 May 10;371(9624):1579–86. doi: 10.1016/S0140-6736(08)60690-X. [DOI] [PubMed] [Google Scholar]
- 19.Gilchrist ES, Plevris JN. Bone marrow-derived stem cells in liver repair: 10 years down the line. Liver Transpl. 2010 Feb;16(2):118–29. doi: 10.1002/lt.21965. [DOI] [PubMed] [Google Scholar]
- 20.Le BK, Ringden O. Immunomodulation by mesenchymal stem cells and clinical experience. J Intern Med. 2007 Nov;262(5):509–25. doi: 10.1111/j.1365-2796.2007.01844.x. [DOI] [PubMed] [Google Scholar]
- 21.Pistoia V, Raffaghello L. Potential of mesenchymal stem cells for the therapy of autoimmune diseases. Expert Rev Clin Immunol. 2010 Mar;6(2):211–8. doi: 10.1586/eci.09.86. [DOI] [PubMed] [Google Scholar]
- 22.Freedman MS, Bar-Or A, Atkins HL, Karussis D, Frassoni F, Lazarus H, et al. The therapeutic potential of mesenchymal stem cell transplantation as a treatment for multiple sclerosis: consensus report of the International MSCT Study Group. Mult Scler. 2010 Apr;16(4):503–10. doi: 10.1177/1352458509359727. [DOI] [PubMed] [Google Scholar]
- 23.Li Y, Chopp M. Marrow stromal cell transplantation in stroke and traumatic brain injury. Neurosci Lett. 2009 Jun 12;456(3):120–3. doi: 10.1016/j.neulet.2008.03.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kode JA, Mukherjee S, Joglekar MV, Hardikar AA. Mesenchymal stem cells: immunobiology and role in immunomodulation and tissue regeneration. Cytotherapy. 2009;11(4):377–91. doi: 10.1080/14653240903080367. [DOI] [PubMed] [Google Scholar]
- 25.van den Berk LC, Figdor CG, Torensma R. Mesenchymal stromal cells: tissue engineers and immune response modulators. Arch Immunol Ther Exp (Warsz) 2008 Sep;56(5):325–9. doi: 10.1007/s00005-008-0036-z. [DOI] [PubMed] [Google Scholar]
- 26.Ren J, Jin P, Wang E, Marincola FM, Stroncek DF. MicroRNA and gene expression patterns in the differentiation of human embryonic stem cells. J Transl Med. 2009;7:20. doi: 10.1186/1479-5876-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang E, Miller LD, Ohnmacht GA, Mocellin S, Perez-Diez A, Petersen D, et al. Prospective molecular profiling of melanoma metastases suggests classifiers of immune responsiveness. Cancer Res. 2002 Jul 1;62(13):3581–6. [PMC free article] [PubMed] [Google Scholar]
- 28.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998 Dec 8;95(25):14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(5):3. [PubMed] [Google Scholar]
- 30.Goessler UR, Bugert P, Bieback K, Stern-Straeter J, Bran G, Sadick H, et al. In vitro-Analysis of Integrin-Expression in Stem-Cells from Bone Marrow and Cord Blood during Chondrogenic Differentiation. J Cell Mol Med. 2008 Aug 4; doi: 10.1111/j.1582-4934.2008.00451.x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Goessler UR, Bugert P, Bieback K, Stern-Straeter J, Bran G, Hormann K, et al. Integrin expression in stem cells from bone marrow and adipose tissue during chondrogenic differentiation. Int J Mol Med. 2008 Mar;21(3):271–9. [PubMed] [Google Scholar]
- 32.Lal H, Verma SK, Foster DM, Golden HB, Reneau JC, Watson LE, et al. Integrins and proximal signaling mechanisms in cardiovascular disease. Front Biosci. 2009;14:2307–34. doi: 10.2741/3381. [DOI] [PubMed] [Google Scholar]
- 33.Jaganathan BG, Ruester B, Dressel L, Stein S, Grez M, Seifried E, et al. Rho inhibition induces migration of mesenchymal stromal cells. Stem Cells. 2007 Aug;25(8):1966–74. doi: 10.1634/stemcells.2007-0167. [DOI] [PubMed] [Google Scholar]
- 34.Markov V, Kusumi K, Tadesse MG, William DA, Hall DM, Lounev V, et al. Identification of cord blood-derived mesenchymal stem/stromal cell populations with distinct growth kinetics, differentiation potentials, and gene expression profiles. Stem Cells Dev. 2007 Feb;16(1):53–73. doi: 10.1089/scd.2006.0660. [DOI] [PubMed] [Google Scholar]
- 35.Jeong JA, Hong SH, Gang EJ, Ahn C, Hwang SH, Yang IH, et al. Differential gene expression profiling of human umbilical cord blood-derived mesenchymal stem cells by DNA microarray. Stem Cells. 2005 Apr;23(4):584–93. doi: 10.1634/stemcells.2004-0304. [DOI] [PubMed] [Google Scholar]
- 36.Doherty GJ, McMahon HT. Mechanisms of endocytosis. Annu Rev Biochem. 2009;78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- 37.Magnon C, Frenette PS. Hematopoietic stem cell trafficking. 2008 [PubMed] [Google Scholar]
- 38.Casey TM, Eneman J, Crocker A, White J, Tessitore J, Stanley M, et al. Cancer associated fibroblasts stimulated by transforming growth factor beta1 (TGF-beta 1) increase invasion rate of tumor cells: a population study. Breast Cancer Res Treat. 2008 Jul;110(1):39–49. doi: 10.1007/s10549-007-9684-7. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y, Wen G, Shao G, Wang C, Lin C, Fang H, et al. TGFBI deficiency predisposes mice to spontaneous tumor development. Cancer Res. 2009 Jan 1;69(1):37–44. doi: 10.1158/0008-5472.CAN-08-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kveiborg M, Albrechtsen R, Couchman JR, Wewer UM. Cellular roles of ADAM12 in health and disease. Int J Biochem Cell Biol. 2008;40(9):1685–702. doi: 10.1016/j.biocel.2008.01.025. [DOI] [PubMed] [Google Scholar]
- 41.Fedak PW, Moravec CS, McCarthy PM, Altamentova SM, Wong AP, Skrtic M, et al. Altered expression of disintegrin metalloproteinases and their inhibitor in human dilated cardiomyopathy. Circulation. 2006 Jan 17;113(2):238–45. doi: 10.1161/CIRCULATIONAHA.105.571414. [DOI] [PubMed] [Google Scholar]
- 42.Cicha I, Goppelt-Struebe M. Connective tissue growth factor: context-dependent functions and mechanisms of regulation. Biofactors. 2009 Mar;35(2):200–8. doi: 10.1002/biof.30. [DOI] [PubMed] [Google Scholar]
- 43.Rachfal AW, Brigstock DR. Structural and functional properties of CCN proteins. Vitam Horm. 2005;70:69–103. doi: 10.1016/S0083-6729(05)70003-0. [DOI] [PubMed] [Google Scholar]
- 44.Panek AN, Posch MG, Alenina N, Ghadge SK, Erdmann B, Popova E, et al. Connective tissue growth factor overexpression in cardiomyocytes promotes cardiac hypertrophy and protection against pressure overload. PLoS ONE. 2009;4(8):e6743. doi: 10.1371/journal.pone.0006743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ng F, Boucher S, Koh S, Sastry KS, Chase L, Lakshmipathy U, et al. PDGF, TGF-beta, and FGF signaling is important for differentiation and growth of mesenchymal stem cells (MSCs): transcriptional profiling can identify markers and signaling pathways important in differentiation of MSCs into adipogenic, chondrogenic, and osteogenic lineages. Blood. 2008 Jul 15;112(2):295–307. doi: 10.1182/blood-2007-07-103697. [DOI] [PubMed] [Google Scholar]
- 46.Wagner EF. Bone development and inflammatory disease is regulated by AP-1 (Fos/Jun) Ann Rheum Dis. 2010 Jan;69(Suppl 1):i86–i88. doi: 10.1136/ard.2009.119396. [DOI] [PubMed] [Google Scholar]
- 47.Wagner EF, Eferl R. Fos/AP-1 proteins in bone and the immune system. Immunol Rev. 2005 Dec;208:126–40. doi: 10.1111/j.0105-2896.2005.00332.x. [DOI] [PubMed] [Google Scholar]
- 48.Maehata Y, Takamizawa S, Ozawa S, Izukuri K, Kato Y, Sato S, et al. Type III collagen is essential for growth acceleration of human osteoblastic cells by ascorbic acid 2-phosphate, a long-acting vitamin C derivative. Matrix Biol. 2007 Jun;26(5):371–81. doi: 10.1016/j.matbio.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 49.Phinney DG, Gray AJ, Hill K, Pandey A. Murine mesenchymal and embryonic stem cells express a similar Hox gene profile. Biochem Biophys Res Commun. 2005 Dec 30;338(4):1759–65. doi: 10.1016/j.bbrc.2005.10.140. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.