Abstract
Microbial translocation from the gastrointestinal tract has been implicated in chronic activation of the immune system during progressive HIV-1 infection by ill-defined mechanisms. We recently identified a gene encoding syndecan-1 (SYN1) in microarray studies of HIV-1 infection in lymphatic tissues and show here that increased expression of SYN1 in the gut of HIV-1-infected individuals is associated with increased microbial translocation. We further show that: (i) microbial access to SYN1 in the intestinal epithelium could be mediated by compromised barrier function through the up-regulation of claudin-2; (ii) increases in SYN1 and microbial translocation are associated with systemic immune activation; and (iii) SYN1 expression and microbial translocation are inversely correlated with peripheral blood CD4+ T cell counts. We thus propose a new mechanism in which claudin-2 and SYN1 work in concert to enhance microbial translocation across the intestinal epithelial barrier to contribute to chronic immune activation and CD4+ T cell depletion.
Keywords: HIV-1, microbial translocation, syndecan-1, claudin-2, immune activation, intestinal epithelium
Introduction
Microbial translocation from the gastrointestinal tract has been proposed as a causal link to chronic immune activation during progressive HIV-1 infection1. Recent data has implicated the loss of immunological defenses2–4 and the physical breakdown of the intestinal barrier5–7 as mechanisms that enhance translocation of bacteria and their products into the gut lamina propria. Here we provide evidence of a new mechanism in HIV-1 infection in which a cell surface protein known as syndecan-1 (SYN1) facilitates microbial translocation across an intestinal epithelial barrier compromised via the aberrant expression of the pore-forming, tight junction (TJ) protein, claudin-2 (CLD2).
SYN1 is one of four members of a family of cell surface heparin-sulfate proteoglycans expressed with temporal and spatial specificities on various cell types within many different tissues and involved in a wide array of cellular processes8. SYN1 is primarily expressed on the basolateral surface of intestinal epithelium9 and also on plasma cells10 and plays a role in cell-cell and cell-matrix interactions, migration, proliferation, and cell differentiation8. Increasing in vitro evidence has linked SYN1 with gut microbial translocation through the ability of bacteria to bind SYN1’s heparin-sulfate (HS) motifs11–15. However, the ability of luminal bacteria to interact with SYN1 is likely limited in healthy individuals due to its basolateral location but can take on increasing importance in persons with increased intestinal permeability, such as those individuals infected with HIV-15–7.
Thus, when we identified SYN1 from a recent microarray analysis as a gene increased in expression during chronic HIV-1 infection16, we investigated a potential role for SYN1 in microbial translocation during HIV-1 infection. In this report, we indeed show that there is both an increase in intestinal permeability during HIV-1 infection through the up-regulation of CLD2 as well as a strong association between SYN1 expression in the intestinal epithelium and microbial translocation and systemic immune activation.
Methods
Gut and Lymph Node Tissue from HIV-1-infected Individuals
Ileal, rectal, and inguinal lymph node biopsies from 18 HIV-1-infected subjects at different clinical stages and 4 uninfected subjects were obtained for this University of Minnesota institutional review board-approved study. Tissues were fixed in paraformaldehyde or Streck’s Tissue Fixative before embedding in paraffin.
Microarray Analysis of Inguinal Lymph Nodes
RNA extractions, synthesis of biotin-labeled cRNA probes, and microarray hybridization followed previously published procedures16.
Immunohistochemistry/Immunofluorescence
Immunohistochemistry and immunofluorescence were performed as previously described17 using a biotin-free detection system on 5-µm tissue sections mounted on glass slides. Tissues were deparaffinized and rehydrated in deionized water. Heat-induced epitope retrieval was performed using either a water-bath (95–98°C for 10–20 min) or high-pressure cooker (120°C for 30 sec) in one of the following buffers: DiVA Decloaker (Biocare Medical), 10mM sodium citrate, pH 6.0, or 1mM EDTA, pH 8.0, followed by cooling to room temperature. Tissues sections were blocked with SNIPER Blocking Reagent (Biocare Medical) for 15 min at room temperature. Endogenous peroxidase was blocked with 3% (v/v) H2O2 in methanol. Primary antibodies were diluted in TNB (0.1M Tris-HCl, pH 7.5; 0.15M NaCl; 0.05% Tween 20 with Dupont blocking buffer) and incubated overnight at 4°C. After the primary antibody incubation, sections were washed with PBS. Sections for immunofluorescence were then incubated with rabbit or mouse fluorophore-cojugated secondary antibodies in 5% non-fat milk for 2 hr at room temperature. These sections were washed, nuclei counterstained blue with TOTO-3, and mounted using Aqua Poly/Mount (Polysciences Inc.). Sections for immunohostochemistry were incubated with mouse or rabbit polymer system reagents conjugated with horseradish peroxidase (Dako Cytomation) according to the manufacturer's instructions, developed with 3,3’-diaminobenzidine (Vector Laboratories), counterstained with Harris hematoxylin (Surgipath Medical), and mounted using Permount (Fisher Scientific). Stained sections were examined either by light microscopy or immunofluorescent confocal microscopy at ambient temperatures. Light micrographs were taken using an Olympus BX60 upright microscope with the following objectives: ×10 (0.3 NA), ×20 (0.5 NA), and ×40 (0.75 NA); images were acquired using a Spot color mosaic camera (model 11.2) and Spot acquisition software (version 4.5.9; Diagnostic Instruments). Immunofluorescent micrographs were taken using an Olympus BX61 Fluoview confocal microscope with the following objectives: ×20 (0.75 NA), ×40 (0.75 NA), and ×60 (1.42 NA); images were acquired using Olympus Fluoview software (version 1.7a).
To quantify levels of SYN1 expression in the gut, 16 randomly stained images from each specimen were captured and positive cells enumerated using Photoshop (CS2, version 9.0; Adobe Systems) with plug-ins from Reindeer Graphics. Specifically, this program utilizes a threshold tool to set a gray level that discriminates positively-stained cells from background and marks the positive signal with a red overlay. The program then measures the area of the signal above the threshold, expressed as a % of the total area. Images for SYN1 quantification focused on the gut epithelium to minimize the positive signal derived from SYN1 expression on plasma cells residing within the lamina propria. Data were expressed as % tissue area positive for SYN1.
A semi-quantitative approach was used to measure CLD2 expression. Stained images of the entire tissue section were captured from each specimen and CLD2 measured as a function of its expression in the luminal epithelial cells (the outer layer/perimeter of epithelium exposed to the intestinal lumen). The amount of CLD2 expression along the epithelial perimeter was determined and expressed as a range of % tissue area positive for CLD2 (e.g., > 90% indicates that CLD2 was expressed in over 90% of the luminal epithelial cells for a particular gut section).
To quantify numbers of translocated E. coli in the inguinal lymph node, 16 randomly stained images from each specimen were captured and positive cells enumerated by manually counting positively-stained bacteria in each image. The average # of bacteria per lymph node area was determined and log transformed. Data were expressed as log10 E. coli/mm2 of tissue. Isotype-matched IgG negative control antibodies in all instances yielded negative staining results (see Table, Supplemental Digital Content 1, which lists the primary antibodies and antigen retrieval methodologies).
Fluorescence in-situ Hybridization (FISH)
Cy3-conjugated FISH probes (EUB338: GCTGCCTCCCGTAGGAGT; NON338: ACTCCTACGGGAGGCAGC) were obtained from Eurofins MWG/Operon and used to detect bacterial 16s rRNA according to a procedure previously published18. Briefly, tissues were deparaffinized and rehydrated to 70% ethanol. Tissues were then immersed in 0.5M EDTA for 15 minutes at 37°C; in 5 µg/mL proteinase K (Boehringer Mannheim) in 100 mM Tris-HCl plus 50 mM EDTA, pH 7.5, for 30 minutes at 37°C; and in 70% ethanol for 1 minute. After dehydration in ethanol, tissue sections were air-dried, and a 13-µL probe mixture containing 1 µg/mL of the respective probe in hybridization mixture (0.45M NaCl; 45 mM sodium citrate, pH 7.0; 25% formamide; 10× Denhardt’s solution; 10% dextran sulfate; 1% Triton X-100; 0.1% sodium dodecyl sulfate) was applied to the sections overnight at 40°C in a humidified chamber. Sections were washed first in 0.1M Tris-HCl plus 0.9M NaCl, pH 7.5 for 20 minutes at 48°C, followed by PBS. Nuclei were counterstained blue with DAPI and mounted using Aqua Poly/Mount (Polysciences Inc.). Immunofluorescent micrographs were taken using an Olympus BX61 Fluoview confocal microscope.
Statistical Analysis
To test for comparisons of SYN1 across stage, we used a mixed effects model that has random intercepts specific to each subject. Parameter estimates are obtained using restricted maximum likelihood, and all p-values use a normal approximation to the sampling distribution of the Wald statistics specific for each model parameter. To test for an overall effect for stage, a contrast was formed that allowed for testing for a difference between all groups and uninfected subjects. The p-value for this overall effect is 0.0003. To test for differences between the 3 infected stages and uninfected individually, we use the Wald test statistic specific to the parameter for that stage. We did not detect statistically significant differences between any of the 3 positive stages using this model. To test for CLD2 differences across stage, the ANOVA method was used.
All associations were tested using a mixed effects model that has random intercepts specific to each subject to model correlation among measurements from the same subject. A normal approximation to the sampling distribution of the restricted maximum likelihood estimates was used to determine p-values. A logarithmic transformation was employed for analysis of all continuous variables to reduce the effect of potential influential observations and reduce dependence of variances on mean levels. All computations were conducted using S-PLUS Version 3.4 distributed by MathSoft, Inc.
Results
Gut Expression of SYN1 is Increased during HIV-1 Infection
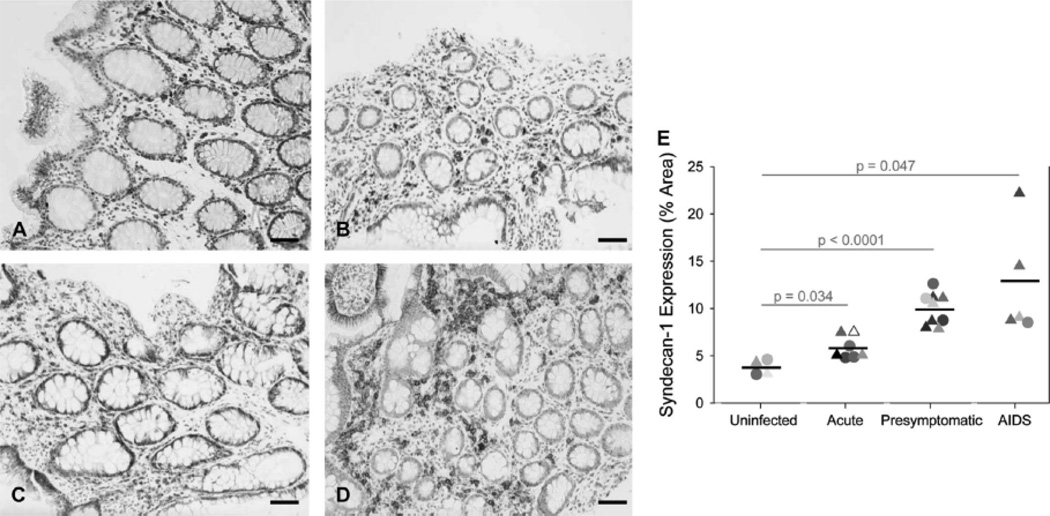
To establish a linkage between SYN1 and microbial translocation, we first determined the cellular/spatial localization and expression levels of SYN1 in the gut. We stained for SYN1 in ileal and rectal biopsies from uninfected and HIV-1-infected individuals in each clinical stage of disease—acute (defined as individuals infected within 4 months of documented seroconversion), presymptomatic (defined as individuals infected for at least 4 months with a CD4+ T cell count > 200 cells/µl), and AIDS (defined as infected individuals with a CD4+ T cell count < 200 cells/µl) (Table 1). SYN1 was present at basal levels in uninfected individuals (Fig. 1A and B) but steadily increased in the mucosal epithelium (Fig. 1C and D) of the large and small intestine during the progression of HIV-1 infection, with highest levels in the AIDS stage of disease (Fig. 1C and D). SYN1 was also present in leukocytes, primarily CD27+ plasma cells (see Figure, Supplemental Digital Content 2, which shows SYN1 and CD27 co-localization in leukocytes), residing within the lamina propria. The levels of SYN1 were quantified, revealing significant increases at all stages of HIV-1 infection: ~ 1.6-fold increase in the acute stage of disease and a larger 2.7–3.4 fold increase (p ≤ 0.047) in the presymptomatic and AIDS stages of disease (Fig. 1E).
Table 1.
Clinical Characteristics of Study Subjects
| Patient | Disease Stage |
Gender | Age | Race | Peripheral Blood CD4+ T Cell Count (Cells/µl) |
Plasma HIV-1 RNA Levels (Copies/ml) |
|---|---|---|---|---|---|---|
| 1425 | Uninfected | Male | 43 | Caucasian | 1,351 | Undetectable |
| 1442 | Uninfected | Female | 45 | Caucasian | 485 | Undetectable |
| 1001 | Uninfected | Female | 58 | Caucasian | 700 | Undetectable |
| 1003 | Uninfected | Female | 49 | Caucasian | 820 | Undetectable |
| 1329 | Acute | Male | 59 | Caucasian | 370 | 484,694 |
| 1391 | Acute | Male | 37 | African American | 234 | 24,718 |
| 1389 | Acute | Male | 32 | Caucasian | 824 | 32,173 |
| 1449 | Acute | Male | 30 | Caucasian | 333 | > 100,000 |
| 1435 | Acute | Male | 42 | Caucasian | 663 | > 100,000 |
| 1458 | Acute | Male | 51 | Caucasian | 400 | 439,000 |
| 1086 | Presymptomatic | Male | 30 | Caucasian | 512 | 20,562 |
| 1293 | Presymptomatic | Male | 36 | Caucasian | 905 | 14,225 |
| 1335 | Presymptomatic | Male | 32 | Caucasian | 400 | 15,284 |
| 1317 | Presymptomatic | Male | 31 | Caucasian | 399 | 120,469 |
| 1419 | Presymptomatic | Male | 37 | Caucasian | 245 | 61,432 |
| 1429 | Presymptomatic | Male | 27 | African American | 1,058 | 2,620 |
| 1428 | Presymptomatic | Male | 30 | Caucasian | 363 | 38,600 |
| 1413 | AIDS | Male | 50 | African American | 42 | 59,401 |
| 1438 | AIDS | Male | 49 | Caucasian | 147 | 4,960 |
| 1462 | AIDS | Male | 43 | Caucasian | 81 | 35,000 |
| 1327 | AIDS | Female | 40 | African American | 112 | 12,046 |
| 1446 | AIDS | Female | 45 | Caucasian | 200 | 150,500 |
Figure 1. SYN1 expression is significantly increased in the large and small intestine of the GI tract during HIV-1 infection.
Representative images of SYN1 expression in the gut of uninfected individuals (A, B) and HIV-1-infected persons in the presymptomatic stage of disease (C) and AIDS (D) were examined immunohistochemically (SYN1-positive cells appear brown while cell nuclei appear blue). (E) SYN1 expression was enumerated in each gut biopsy and reported as % tissue area positive for SYN1. The results are shown with significance where applicable (p < 0.05). Symbols: triangles and circles represent ileal and rectal biopsies, respectively, while the black bars denote the mean expression level of SYN1 in each stage of disease. Original magnifications: X200; scale bars: 50 µm.
Increased CLD2 Expression Begins in Early HIV-1 Infection and is Co-expressed with SYN1 in the Intestinal Epithelium
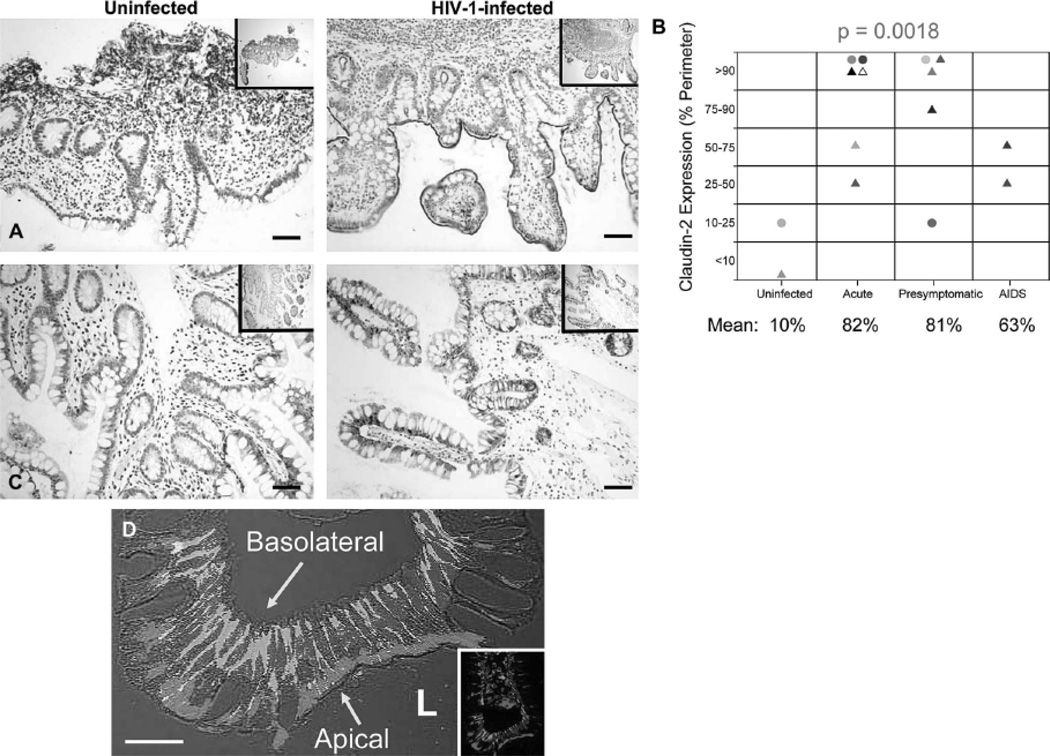
The maintenance and preservation of an intact intestinal epithelial barrier is important in limiting microbial access to the basolateral surface of epithelial cells as well as the host’s immune cells residing within the lamina propria. Key players in this process are the claudins, tight junction proteins involved in establishing the paracellular barrier that controls the flow of molecules in the intercellular space between epithelial cells19. Aberrant expression of TJ proteins and compromised barrier function have been reported for individuals in a number of different settings, such as inflammation19, 20 and virus infections21–25, including HIV-17. Since altered expression of claudins could potentially impair barrier protection and increase paracellular permeability, providing bacteria potential access to SYN1, a mechanism which could enhance microbial translocation, we investigated the expression of claudins (claudins-1, -2, and -4) in the gut of uninfected and HIV-1 infected individuals. We found a substantial up-regulation of the pore-forming junctional protein, CLD226, along the extracellular surface of the intestinal epithelium and crypts (Fig. 2A) beginning in early infection (acute stage) and continuing throughout disease progression (Fig. 2B) while expression of claudins-1 and -4 were essentially unchanged (Fig. 2C). Moreover, CLD2 often was co-expressed with SYN1, CLD2 along the apical surface of the epithelium and SYN1 along the basolateral surface (Fig. 2D). Thus, increased expression of CLD2, a junctional protein shown to increase paracellular permeability26 and promote TJ strand discontinuities27, during HIV-1 infection could compromise barrier protection, potentially allowing microbes access to the basolateral location of SYN1.
Figure 2. CLD2 is Upregulated during HIV-1 Infection and Co-expressed with SYN1 in Intestinal Epithelial Cells.
(A) Representative images of CLD2 expression in the ileum of an uninfected and HIV-1-infected individual in the acute stage of disease were examined immunohistochemically (CLD2-positive cells appear brown while cell nuclei appear blue). (B) CLD2 expression along the intestinal epithelium was quantified in each gut biopsy and reported as a range of % tissue perimeter positive for CLD2. Differences in CLD2 expression across stage were deemed significant (p = 0.0018). Symbols: triangles and circles represent ileal and rectal biopsies, respectively. (C) Representative images of claudin-1 expression in the ileum of an uninfected and HIV-1-infected individual in the presymptomatic stage of disease were examined immunohistochemically (Claudin-1-positive cells appear brown while cell nuclei appear blue). (D) Representative image of SYN1 and CLD2 expression in the rectum of an HIV-1-infected individual was examined immunofluorescently, showing the apical location of CLD2 (red) and basolateral location of SYN1 (green) (cell nuclei appear blue). Original magnifications: X200 (A and B), X600 (C); scale bars: 20 µm (C) and 50 µm (A and B).
SYN1 Expression is Associated with Increases in Microbial Translocation and Systemic Dissemination
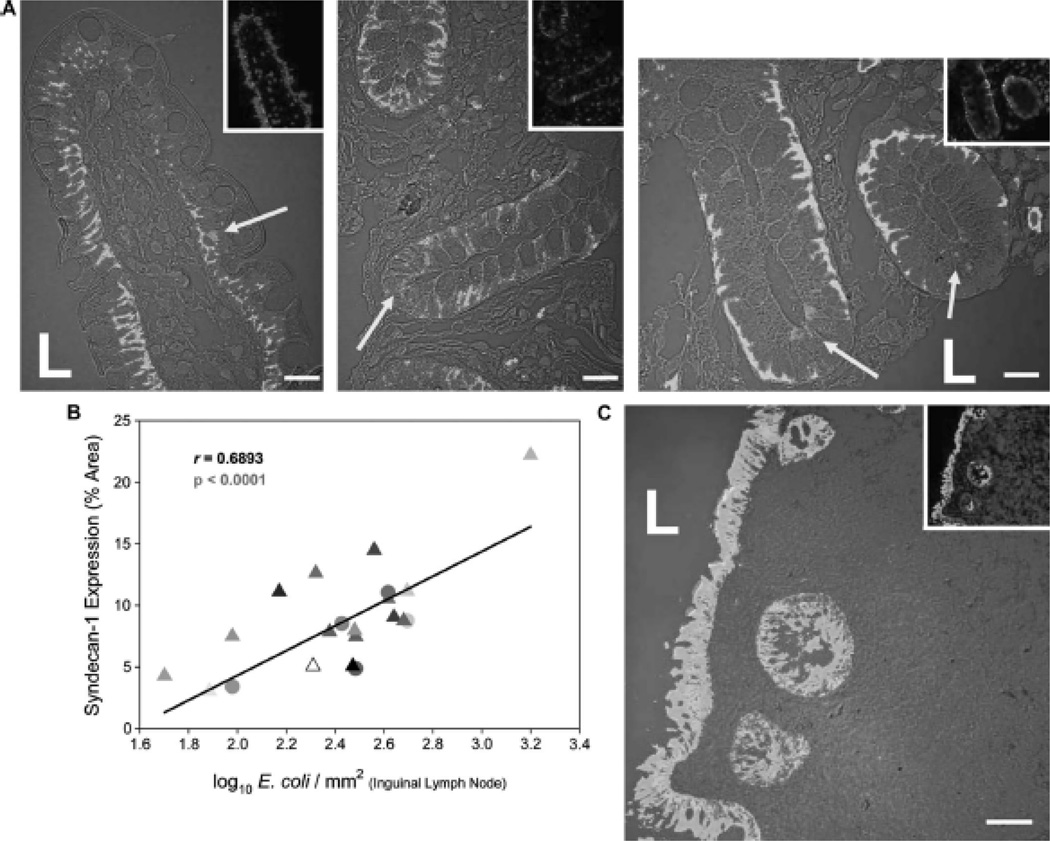
We tested the relationship between microbial translocation and SYN1 expression in vivo during HIV-1 infection by visualizing translocated bacteria (using an antibody recognizing E. coli) and SYN1-expressing cells in gut biopsies from HIV-1-infected persons. As shown in representative images in Fig. 3A, translocated E. coli often co-localized with SYN1-expressing epithelial cells in the gut of HIV-1-infected individuals. To confirm detection of translocated bacteria, we also employed fluorescence in situ hybridization (FISH) using a universal bacterial 16s ribosomal RNA probe and found general agreement between immunofluorescence and FISH detection methods (see Figure, Supplemental Digital Content 3, which shows translocated E. coli in the lamina propria of the gut). Isotype control antibodies yielded negative staining results (see Figure, Supplemental Digital Content 4, which shows negative staining results when isotype-matched antibodies are used in tissue specimens).
Figure 3. Translocated E. coli localize spatially with SYN1-expressing cells in the gut of HIV-1-infected individuals and can be disseminated systemically.
(A) Representative optical images of E. coli (red fluorescence) and SYN1 expression (green fluorescence) in the gut of HIV-1-infected individuals in the presymptomatic and AIDS stages of disease were examined immunofluorescently. (B) SYN1-expressing cells within the gut were significantly correlated with numbers of translocated E. coli in the inguinal lymph node. (C) Representative image of cytokeratin expression (green fluorescence) and E. coli (red fluorescence) reveals translocation in the presence of intact epithelium in a local section of the ileum of an HIV-1-infected individual in the presymptomatic stage of disease (cell nuclei appear blue). Symbols: L represents the luminal space and yellow arrows denote translocated E. coli localized spatially with SYN1-expressing cells. Original magnifications: X600 (A), X200 (C); scale bars: 20 µm (A) and 50 µm (C).
Bacteria that translocate into the gut lamina propria can then spread systemically via the lymphatic system to distal organ sites. We found evidence that SYN1 expression not only was associated with enhanced microbial translocation in the gut but also with dissemination of bacteria. We discovered a significant positive correlation between gut expression of SYN1 and numbers of disseminated bacteria in inguinal lymph nodes (r = 0.6893, p < 0.0001) (Fig. 3B). As physical damage and breaches in the intestinal epithelium could be an alternative or additional mechanism of microbial translocation into the lamina propria, we used cytokeratin-specific antibodies to microscopically examine the structural integrity of the intestinal epithelium. We detected little morphological evidence of extensive epithelial cell damage or widespread breaches in the continuous layering of cytokeratin+ columnar epithelial cells (Fig. 3C). Moreover, in those areas of the gut with discrete, focal breaks, we found little accumulation of bacteria in the immediate vicinity. We thus conclude from these data and previous in vitro findings11–15 that increased SYN1 and CLD2 expression in the intestinal epithelium during HIV-1 infection could be one mechanism facilitating gut microbial translocation and subsequent spread systemically.
SYN1 Expression is Associated with Both Systemic Immune Activation and Anti-Bacterial Responses
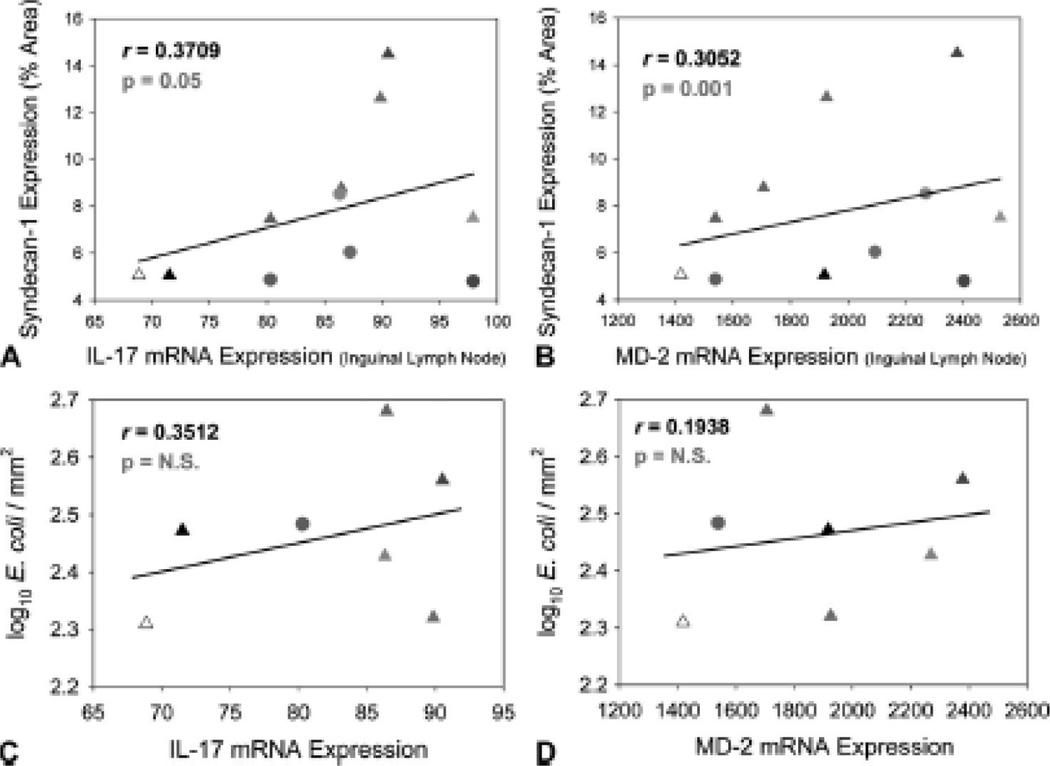
The host responds to increases in bacteria and microbial immunogens such as LPS and bacterial DNA through a number of regulatory mechanisms—IL-17 is an increasingly important anti-bacterial cytokine involved in inhibiting replication and dissemination of bacteria28 while Myeloid Differentiation Factor 2 (MD-2) is the essential sensor of LPS for Toll-like receptor 4 (TLR4) signaling, whose levels increase in response to bacterial infection29. We found a significant positive correlation between SYN1 expression in the gut and mRNA levels of both IL-17 and MD-2 in the inguinal lymph node (r = 0.3709 and 0.3052, respectively, p ≤ 0.05) (Fig. 4A and B). Additionally, we found a relationship between numbers of disseminated bacteria and mRNA levels of both IL-17 and MD-2 in the inguinal lymph node (Fig. 4C and D); however, the latter analysis was limited and not powered to detect significance. These data indicate an extensive antibacterial host response in the lymph nodes, providing supporting evidence of systemic microbial translocation via a SYN1-dependent pathway.
Figure 4. SYN1 expression in the gut of HIV-1-infected individuals and the extent of microbial translocation are associated with a systemic anti-bacterial response.
SYN1-expressing cells within the gut were significantly correlated with mRNA levels of (A) IL-17 and (B) MD-2 in the inguinal lymph node. Numbers of translocated E. coli within the lymph node were associated with mRNA levels (C) IL-17 and (D) MD-2.
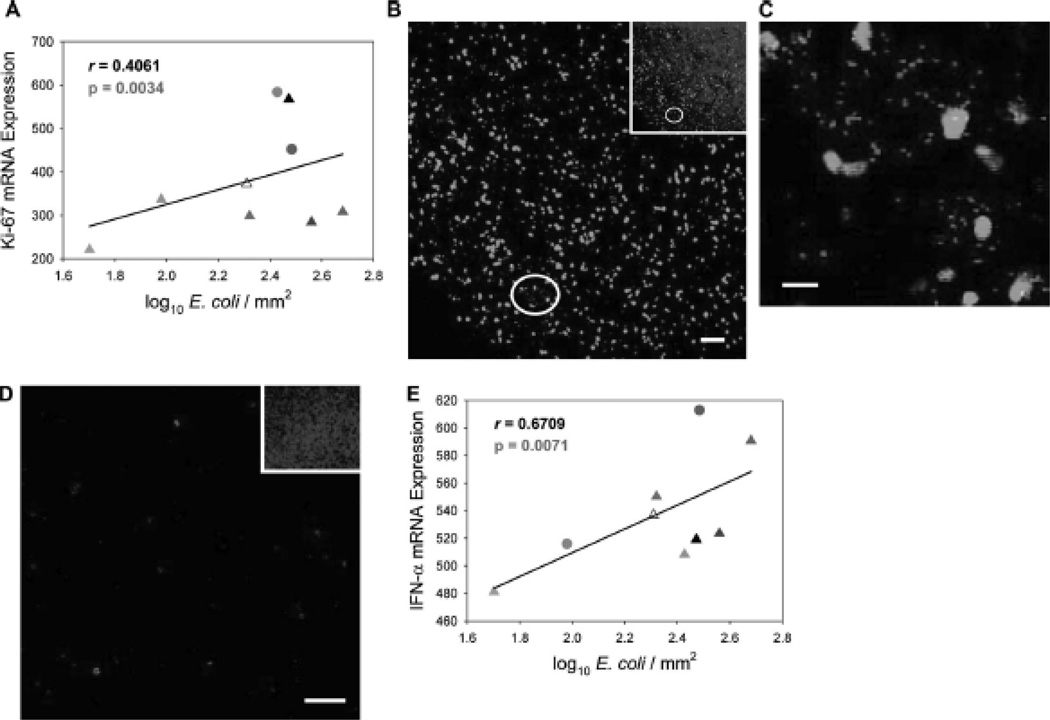
As previous studies have reported the ability of microbial products such as LPS and bacterial DNA to activate immune cells in vitro30 and in vivo1, 31–33, we wanted to determine whether there was a linkage between microbial translocation and systemic immune activation as measured in distal lymphoid tissue. First, we compared the extent of microbial translocation (numbers of translocated E. coli in lymph nodes) to the mRNA expression levels of the cell activation marker Ki-67 in inguinal lymph nodes and found a significant positive correlation between systemic microbial translocation and cell activation (r = 0.4061, p = 0.0034) (Fig. 5A). Second, we observed high levels of immune activation in lymph nodes in spatial conjunction with locally high numbers of translocated bacteria. In representative images in Fig. 5B and C, there are numerous E. coli within an inguinal lymph node associated with increased numbers of Ki-67-expressing cells in an HIV-1-infected person compared to uninfected individuals (Fig. 5D) in which translocated bacteria were not detected. Third, we found a significant positive correlation between IFN-α expression and numbers of translocated bacteria in lymph nodes (r = 0.6709, p = 0.0071), immune stimulation consistent with toll-like receptor recognition of microbial products34 (Fig. 5E). These data support a mechanism of widespread cell activation and anti-bacterial responses during HIV-1 infection in which SYN1 expression in the gut may be augmenting microbial translocation and systemic spread of bacteria.
Figure 5. The extent of microbial translocation is associated with immune activation in distal lymphatic tissue.
(A) Numbers of translocated E. coli were significantly correlated with mRNA levels of Ki-67 in the inguinal lymph node. (B) Representative images of E. coli (red fluorescence) and Ki-67 positive cells (green fluorescence) in the inguinal lymph node of an HIV-1-infected individual in the AIDS stage of disease (B and C) and inguinal lymph node of an uninfected individual (D) were examined immunofluorescently. The image in (C) is an enlarged view of the area circled in white. The insets show the total numbers of cells (cell nuclei appear blue) in each image. (E) Numbers of translocated E. coli were significantly correlated with mRNA levels of IFN-α in the inguinal lymph node. Original magnifications: X200 (B, D, and insets) and X600 (C); scale bars: 20 µm (C) and 50 µm (B and D).
Levels of SYN1 and Microbial Translocation are Inversely Correlated with CD4+ T Cell Survival
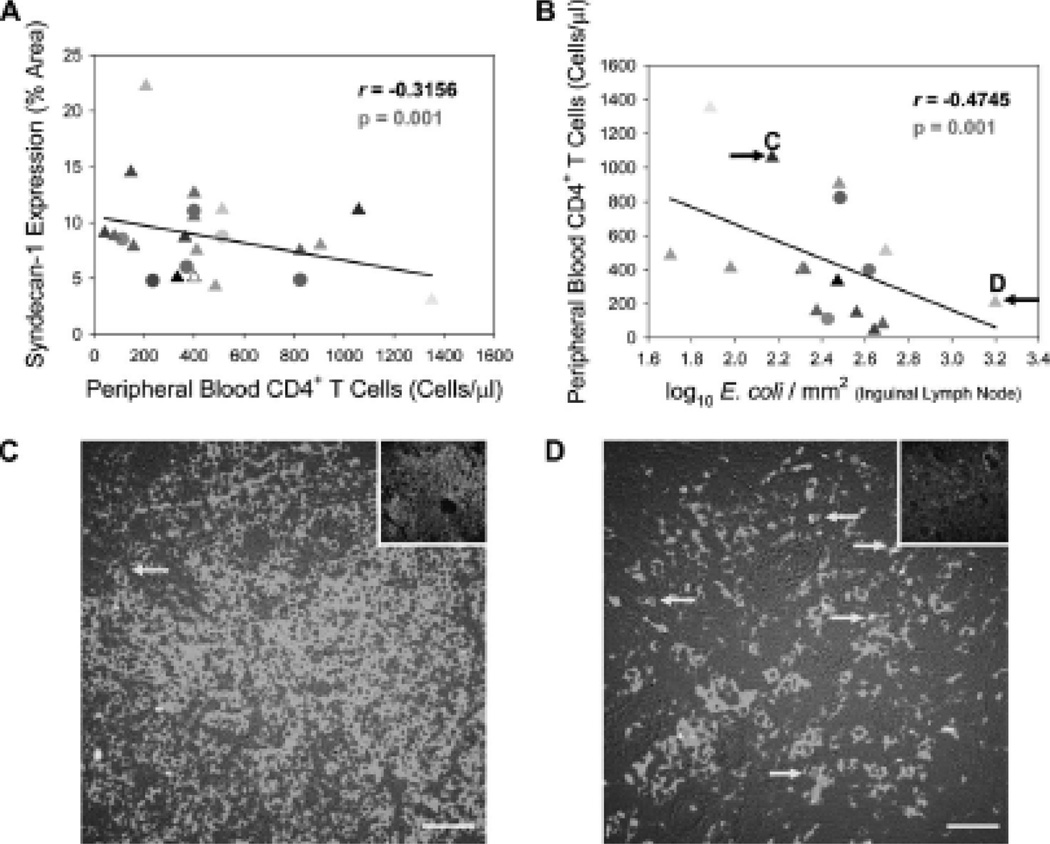
Microbial translocation has been purported as a contributing factor to CD4+ T cell loss during HIV-1 infection due to its role in chronic immune activation and reports showing an inverse correlation between the levels of microbial translocation and numbers of CD4+ T cells1, 33, 35, 36. We therefore wanted to determine whether SYN1 expression and augmented microbial translocation was linked to CD4+ T cell numbers in the peripheral blood. We indeed found inverse correlations between SYN1 expression in the gut and peripheral blood CD4+ T cells (r = −0.3156, p = 0.001) (Fig. 6A) and between the extent of microbial translocation (numbers of translocated E. coli in lymph nodes) and peripheral blood CD4+ T cells (r = −0.4745, p = 0.001) (Fig. 6B), consistent with SYN1-induced microbial translocation contributing directly or indirectly to CD4+ T cell depletion as measured in the peripheral blood.
Figure 6. SYN1 expression in the gut of HIV-1-infected individuals and microbial translocation are negative prognosticators for CD4+ T cell viability.
SYN1 expression in the gut (A) and numbers of translocated E. coli in the inguinal lymph node (B) were inversely correlated with numbers of peripheral blood CD4+ T cells. (C and D) Representative images of CD4+ T cells (green fluorescence) and cleaved caspase 3 (an indicator of apoptosis, red fluorescence) in the paracortical T cell zone of inguinal lymph nodes reveal higher amounts of apoptosis and CD4+ T cell loss coincident with higher levels of microbial translocation. The insets show the total numbers of cells (cell nuclei appear blue) in each image. The black arrows in (B) denote the HIV-1-infected individuals used for lymphatic tissue analysis in (C) and (D). The yellow arrows in (C) and (D) highlight CD4+ T cells undergoing apoptosis. Original magnification: X400; scale bars: 50 µm.
To examine this in the contextual environment of lymphatic tissue, we used immunofluorescence to observe CD4+ T cells present in inguinal lymph nodes from HIV-1-infected individuals with divergent levels of microbial translocation (Fig. 6B). We found a similar relationship between the extent of microbial translocation and lymph node CD4+ T cells, whereby greater loss of CD4+ T cells was detected in lymph nodes coincident with higher levels of microbial translocation (compare Figs. 6C and D). Furthermore, we found an increase in CD4+ T cell apoptosis in lymph nodes harboring greater numbers of translocated bacteria (compare Figs. 6C and D), consistent with previous reports showing LPS and other microbial products as inducers of activation-induced apoptosis in lymphocytes in vitro30 and in vivo32, 37. These data suggest that microbial translocation may be a contributing factor in chronic immune activation and CD4+ T cell loss both in the periphery and lymphatic tissue during HIV-1 infection.
Discussion
Increased levels of microbial translocation during HIV-1 infection have been well documented1, 31, 33, 35, 36, 38–40 and ascribed to such causes as the loss of immunological defenses2–4 and the physical breakdown of the intestinal barrier5–7. Here, we provide evidence of a new mechanism linking HIV-1 infection-associated increases in gut epithelial cell SYN1 and the pore-forming, TJ protein CLD2 to increased bacterial translocation in the gut and systemic spread into distal sites such as peripheral lymph nodes. This increase in microbial translocation is associated with anti-bacterial responses, measured as correlated increases in such mRNA markers (IL-17 and MD-2), which may explain why the presence of bacteria systemically is not associated with gram negative sepsis.
The mechanism by which SYN1 increases bacterial translocation has been reported to involve interactions between bacterial surface proteins and HS glycosaminoglycan motifs surfacing the extracellular domain of SYN1-expressing cells11–15. However, these HS motifs are likely shielded from luminal bacteria in healthy individuals due to the basolateral positioning of SYN1 along the intestinal epithelium. The dramatic increase in CLD2 during HIV-1 infection would provide an initial breach in barrier function, increasing permeability26, and allowing potential microbial access to the HS motifs of SYN1, which then can facilitate microbial translocation across the epithelial barrier and into the lamina propria.
The polarized epithelium of the host plays a significant role in resistance to bacterial infection, whereby disruption/alteration of the claudin composition of TJ’s can lead to imbalance of this equilibrium and increase paracellular permeability41. Bacteria have been shown to take advantage of “leaky” TJ’s, gaining access to the basolateral surfaces of the epithelium, ultimately increasing invasiveness and paracellular translocation42–48. Moreover, CLD2 has been implicated as one of the primary claudin determinants in epithelial barrier dysfunction and increased paracellular permeability in vivo49–51 by decreasing the “tightness” of individual claudin-based TJ strands27, 52; finally, the expression of CLD2 has also been associated in vivo with bacterial sepsis53.
In spite of CLD2’s role in increasing gut permeability and its initial up-regulation in acute/early infection, we did not find a direct relationship between CLD2 expression levels and microbial translocation, consistent with previous observations in which microbial translocation is not appreciable, as detected experimentally, until chronic HIV-1 infection1, 31, 36. The extent of microbial translocation is greatest in the presymptomatic/AIDS stages of disease when SYN1 levels are most upregulated (Fig. 3B). This then suggests that CLD2 may be required for SYN1 accessibility but SYN1 is the agent that actively facilitates microbial translocation. While this mechanism in the context of HIV-1 infection deserves further study, what is clear from our data is that SYN1-mediated microbial translocation differs markedly from other proposed mechanisms involving the loss of immunological defenses2–4 or physical breakdown of the intestinal barrier (increased apoptosis)5–7. We think that both the direct SYN1 pathway and these indirect mechanisms may work in concert to increase microbial translocation during HIV-1 infection.
We show that the extent of microbial translocation is associated with both systemic immune activation, measured as correlated increases in such mRNA markers (Ki-67 and IFN-α) as well as the close proximity of translocated E. coli to activated immune cells within inguinal lymph nodes, and apoptosis of CD4+ T cells in lymphatic tissue. While it remains difficult to dissect pathogenic contributions from the virus itself vs. bacteria and their immunogenic components, these data, nevertheless, suggest that microbial translocation may be a contributing factor in chronic immune activation and CD4+ T cell depletion.
While our study and previous reports suggest that SYN1-expressing cells can augment microbial translocation by enhancing bacterial binding and subsequent internalization into intestinal epithelial cells11–15, we suspect that this is an adverse off-target effect of SYN1’s physiological functions in HIV-1 infected lymph nodes where increased expression was first detected16. We further speculate, based on SYN1’s role in cell-cell and cell-matrix interactions8, that SYN1’s increased expression in the intestinal epithelium and infiltration of SYN1+ plasma cells into the gut during HIV-1 infection may be related to inflammation and/or tissue remodeling8, 54, 55. Because of the previously documented relationship between lymphatic tissue collagen deposition and CD4+ T cell depletion56, we will be particularly interested in future studies exploring the relationship of SYN1 to these immunopathological changes in which SYN1 might be both involved in the fibrotic changes and the chronic immune activation that drives them.
Supplementary Material
Representative images of SYN1 (green fluorescence) and CD27 (red fluorescence) expression in the rectum of an HIV-1-infected individual in the presymptomatic stage of disease were examined immunofluorescently (cell nuclei appear blue). Original magnification: X400; scale bars: 50 µm.
(A and B) Representative images of E. coli (red fluorescence) in the ileum of an HIV-1-infected individual in the presymptomatic stage of disease were examined using FISH (cell nuclei appear blue). Original magnifications: X600; scale bars: 20 µm.
(A) Representative optical images of E. coli (red fluorescence) and SYN1 expression (green fluorescence) from two identical sections of ileum from an HIV-1-infected individual in the presymptomatic stage of disease reveal minimal cross-reactivity with a matched isotype control antibody for E. coli (rabbit IgG). (B) Representative optical images of E. coli (red fluorescence) from a dry mount of purified E. coli reveal minimal cross-reactivity with a matched isotype control antibody. Original magnifications: X600; scale bars: 20 µm.
Acknowledgements
We thank all of the donor participants in this study. This work was supported by National Institute of Health (NIH) grant R01 AI056997 (A. T. H.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
This work was presented in part at the “Fourth International Workshop on HIV Persistence during Therapy,” St. Martin, West Indies, December 2009 (abstract # 44).
Author Contributions
A.J.S. designed and performed experiments; T.W.S. recruited subjects and procured biopsy samples; C.S.R. performed statistical analyses; A.J.S. and A.T.H. analyzed the data and wrote the manuscript.
References
- 1.Brenchley JM, Price DA, Schacker TW, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12:1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- 2.Guadalupe M, Reay E, Sankaran S, et al. Severe CD4+ T-cell depletion in gut lymphoid tissue during primary HIV-1 infection and substantial delay in restoration following HAART. J Virol. 2003;77:11708–11717. doi: 10.1128/JVI.77.21.11708-11717.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mehandru S, Poles MA, Tenner-Racz K, et al. Primary HIV-1 infection is associated with preferential depletion of CD4+ T lymphocytes from effector sites in the gastrointestinal tract. J Exp Med. 2004;200:761–770. doi: 10.1084/jem.20041196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brenchley JM, Schacker TW, Ruff LE, et al. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J Exp Med. 2004;200:749–759. doi: 10.1084/jem.20040874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharpstone D, Neild P, Crane R, et al. Small intestinal transit, absorption, and permeability in patients with AIDS with and without diarrhea. Gut. 1999;45:70–76. doi: 10.1136/gut.45.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sankaran S, George MD, Reay E, et al. Rapid onset of intestinal epithelial barrier dysfunction in primary human immunodeficiency virus infection is driven by an imbalance between immune response and mucosal repair and regeneration. J Virol. 2008;82:538–545. doi: 10.1128/JVI.01449-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Epple H-J, Schneider T, Troeger H, et al. Impairment of the intestinal barrier is evident in untreated but absent in suppressively treated HIV-infected patients. Gut. 2009;58:220–227. doi: 10.1136/gut.2008.150425. [DOI] [PubMed] [Google Scholar]
- 8.Alexopoulou AN, Multhaupt HA, Couchman JR. Syndecans in wound healing, inflammation and vascular biology. Int J Biochem Cell Biol. 2007;39:505–528. doi: 10.1016/j.biocel.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 9.Bernfield M, Kokenyesi R, Kato M, et al. Biology of the syndecans: a family of transmembrane heparan sulfate proteoglycans. Annu Rev Cell Biol. 1992;8:365–393. doi: 10.1146/annurev.cb.08.110192.002053. [DOI] [PubMed] [Google Scholar]
- 10.Seftalioglu A, Karakus S. Syndecan-1/CD138 expression in normal myeloid, acute lymphoblastic and myeloblastic leukemia cells. Acta Histochem. 2003;105:213–221. doi: 10.1078/0065-1281-00706. [DOI] [PubMed] [Google Scholar]
- 11.van Putten JP, Paul SM. Binding of syndecan-like cell surface proteoglycan receptors is required for Neisseria gonorrhoeae entry into human mucosal cells. EMBO J. 1995;14:2144–2154. doi: 10.1002/j.1460-2075.1995.tb07208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freissler E, Meyer auf der Heyde A, David G, et al. Syndecan-1 and syndecan-4 can mediate the invasion of OpaHSPG-expressing Neisseria gonorrhoeae into epithelial cells. Cell Microbiol. 2000;2:69–82. doi: 10.1046/j.1462-5822.2000.00036.x. [DOI] [PubMed] [Google Scholar]
- 13.Park PW, Pier GB, Hinkes MT, et al. Exploitation of syndecan-1 shedding by Pseudomonas aeruginosa enhances virulence. Nature. 2001;411:98–102. doi: 10.1038/35075100. [DOI] [PubMed] [Google Scholar]
- 14.Henry-Stanley MJ, Hess DJ, Erickson EA, et al. Role of heparan sulfate in interactions of Listeria monocytogenes with enterocytes. Med Microbiol Immunol. 2003;192:107–115. doi: 10.1007/s00430-002-0165-7. [DOI] [PubMed] [Google Scholar]
- 15.Henry-Stanley MJ, Hess DJ, Erlandsen SL, et al. Ability of the heparan sulfate proteoglycan syndecan-1 to participate in bacterial translocation across the intestinal epithelial barrier. Shock. 2005;24:571–576. doi: 10.1097/01.shk.0000184286.95493.78. [DOI] [PubMed] [Google Scholar]
- 16.Li Q, Smith AJ, Schacker TW, et al. Microarray Analysis of Lymphatic Tissue Reveals Stage-Specific, Gene-Expression Signatures in HIV-1 Infection. J Immunol. 2009;183:1975–1982. doi: 10.4049/jimmunol.0803222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Q, Estes JD, Schlievert PM, et al. Glycerol monolaurate prevents mucosal SIV transmission. Nature. 2009;458:1034–1038. doi: 10.1038/nature07831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schultsz C, Van Den Berg FM, Ten Kate FW, et al. The intestinal mucus layer from patients with inflammatory bowel disease harbors high numbers of bacteria compared with controls. Gastroenterology. 1999;117:1089–1097. doi: 10.1016/s0016-5085(99)70393-8. [DOI] [PubMed] [Google Scholar]
- 19.Prasad S, Mingrino R, Kaukinen K, et al. Inflammatory processes have differential effects on claudins 2, 3 and 4 in colonic epithelial cells. Lab Invest. 2005;85:1139–1162. doi: 10.1038/labinvest.3700316. [DOI] [PubMed] [Google Scholar]
- 20.Zeissig S, Bürgel N, Günzel D, et al. Changes in expression and distribution of claudin 2, 5 and 8 lead to discontinuous tight junctions and barrier dysfunction in active Crohn's disease. Gut. 2007;56:61–72. doi: 10.1136/gut.2006.094375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dickman KG, Hempson SJ, Anderson J, et al. Rotavirus alters paracellular permeability and energy metabolism in Caco-2 cells. Am J Physiol Gastrointest Liver Physiol. 2000;279:G757–G766. doi: 10.1152/ajpgi.2000.279.4.G757. [DOI] [PubMed] [Google Scholar]
- 22.Nava P, López S, Arias CF, et al. The rotavirus surface protein VP8 modulates the gate and fence function of tight junctions in epithelial cells. J Cell Sci. 2004;117:5509–5519. doi: 10.1242/jcs.01425. [DOI] [PubMed] [Google Scholar]
- 23.Medigeshi GR, Hirsch AJ, Brien JD, et al. West nile virus capsid degradation of claudin proteins disrupts epithelial barrier function. J Virol. 2009;83:6125–6134. doi: 10.1128/JVI.02617-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Troeger H, Loddenkemper C, Schneider T, et al. Structural and functional changes of the duodenum in human norovirus infection. Gut. 2009;58:1070–1077. doi: 10.1136/gut.2008.160150. [DOI] [PubMed] [Google Scholar]
- 25.Yeo NK, Jang YJ. Rhinovirus infection-induced alteration of tight junction and adherens junction components in human nasal epithelial cells. Laryngoscope. 2010;120:346–352. doi: 10.1002/lary.20764. [DOI] [PubMed] [Google Scholar]
- 26.Amasheh S, Meiri N, Gitter AH, et al. Claudin-2 expression induces cation-selective channels in tight junctions of epithelial cells. J Cell Sci. 2002;115:4969–4976. doi: 10.1242/jcs.00165. [DOI] [PubMed] [Google Scholar]
- 27.Turksen K, Troy TC. Barriers built on claudins. J Cell Sci. 2004;117:2435–2447. doi: 10.1242/jcs.01235. [DOI] [PubMed] [Google Scholar]
- 28.Liu JZ, Pezeshki M, Raffatellu M. Th17 cytokines and host-pathogen interactions at the mucosa: dichotomies of help and harm. Cytokine. 2009;48:156–160. doi: 10.1016/j.cyto.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pugin J, Stern-Voeffray S, Daubeuf B, et al. MD-2 activity in plasma from patients with severe sepsis and septic shock. Blood. 2004;104:4071–4079. doi: 10.1182/blood-2003-04-1290. [DOI] [PubMed] [Google Scholar]
- 30.Funderburg N, Luciano AA, Jiang W, et al. Toll-like receptor ligands induce human T cell activation and death, a model for HIV pathogenesis. PLoS One. 2008;3:e1915. doi: 10.1371/journal.pone.0001915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ancuta P, Kamat A, Kunstman KJ, et al. Microbial translocation is associated with increased monocyte activation and dementia in AIDS patients. PLos One. 2008;3:e2516. doi: 10.1371/journal.pone.0002516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pandrea I, Gaufin T, Brenchley JM, et al. Cutting edge: Experimentally induced immune activation in natural hosts of simian immunodeficiency virus induces significant increases in viral replication and CD4+ T cell depletion. J Immunol. 2008;181:6687–6691. doi: 10.4049/jimmunol.181.10.6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang W, Lederman MM, Hunt P, et al. Plasma levels of bacterial DNA correlate with immune activation and the magnitude of immune restoration in persons with antiretroviral-treated HIV infection. J Infect Dis. 2009;199:1177–1185. doi: 10.1086/597476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heeg K, Dalpke A, Peter M, et al. Structural requirements for uptake and recognition of CpG oligonucleotides. Int J Med Microbiol. 2008;298:33–38. doi: 10.1016/j.ijmm.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 35.Baroncelli S, Galluzzo CM, Pirillo MF, et al. Microbial translocation is associated with residual viral replication in HAART-treated HIV+ subjects with <50copies/ml HIV-1 RNA. J Clin Virol. 2009;46:367–370. doi: 10.1016/j.jcv.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 36.Nowroozalizadeh S, Månsson F, da Silva Z, et al. Microbial translocation correlates with the severity of both HIV-1 and HIV-2 infections. J Infect Dis. 2010;201:1150–1154. doi: 10.1086/651430. [DOI] [PubMed] [Google Scholar]
- 37.Manhart N, Vierlinger K, Habel O, et al. Lipopolysaccharide causes atrophy of Peyer's patches and an increased expression of CD28 and B7 costimulatory ligands. Shock. 2000;14:478–483. doi: 10.1097/00024382-200014040-00010. [DOI] [PubMed] [Google Scholar]
- 38.Balagopal A, Philp FH, Astemborski J, et al. Human immunodeficiency virus-related microbial translocation and progression of hepatitis C. Gastroenterology. 2008;135:226–233. doi: 10.1053/j.gastro.2008.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lester RT, Yao XD, Ball TB, et al. HIV-1 RNA dysregulates the natural TLR response to subclinical endotoxemia in Kenyan female sex-workers. PLoS One. 2009;4:e5644. doi: 10.1371/journal.pone.0005644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Papasavvas E, Pistilli M, Reynolds G, et al. Delayed loss of control of plasma lipopolysaccharide levels after therapy interruption in chronically HIV-1-infected patients. AIDS. 2009;23:369–375. doi: 10.1097/QAD.0b013e32831e9c76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sousa S, Lecuit M, Cossart P. Microbial strategies to target, cross or disrupt epithelia. Curr Opin Cell Biol. 2005;17:489–498. doi: 10.1016/j.ceb.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 42.Mounier J, Vasselon T, Hellio R, et al. Shigella flexneri enters human colonic Caco-2 epithelial cells through the basolateral pole. Infect Immun. 1992;60:237–248. doi: 10.1128/iai.60.1.237-248.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fleiszig SM, Evans DJ, Do N, et al. Epithelial cell polarity affects susceptibility to Pseudomonas aeruginosa invasion and cytotoxicity. Infect Immun. 1997;65:2861–2867. doi: 10.1128/iai.65.7.2861-2867.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCormick BA. The use of transepithelial models to examine host-pathogen interactions. Curr Opin Microbiol. 2003;6:77–81. doi: 10.1016/s1369-5274(02)00003-6. [DOI] [PubMed] [Google Scholar]
- 45.Balkovetz DF, Katz J. Bacterial invasion by a paracellular route: divide and conquer. Microbes Infect. 2003;5:613–619. doi: 10.1016/s1286-4579(03)00089-3. [DOI] [PubMed] [Google Scholar]
- 46.Fedwick JP, Lapointe TK, Meddings JB, et al. Helicobacter pylori activates myosin light-chain kinase to disrupt claudin-4 and claudin-5 and increase epithelial permeability. Infect Immun. 2005;73:7844–7852. doi: 10.1128/IAI.73.12.7844-7852.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Köhler H, Sakaguchi T, Hurley BP, et al. Salmonella enterica serovar Typhimurium regulates intercellular junction proteins and facilitates transepithelial neutrophil and bacterial passage. Am J Physiol Gastrointest Liver Physiol. 2007;293:G178–G187. doi: 10.1152/ajpgi.00535.2006. [DOI] [PubMed] [Google Scholar]
- 48.Li Q, Zhang Q, Wang C, et al. Invasion of enteropathogenic Escherichia coli into host cells through epithelial tight junctions. FEBS J. 2008;275:6022–6032. doi: 10.1111/j.1742-4658.2008.06731.x. [DOI] [PubMed] [Google Scholar]
- 49.Zeissig S, Bürgel N, Günzel D, et al. Changes in expression and distribution of claudin 2, 5 and 8 lead to discontinuous tight junctions and barrier dysfunction in active Crohn's disease. Gut. 2007;56:61–72. doi: 10.1136/gut.2006.094375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reuter BK, Pizarro TT. Mechanisms of tight junction dysregulation in the SAMP1/YitFc model of Crohn's disease-like ileitis. Ann N Y Acad Sci. 2009;1165:301–307. doi: 10.1111/j.1749-6632.2009.04035.x. [DOI] [PubMed] [Google Scholar]
- 51.Clark JA, Gan H, Samocha AJ, et al. Enterocyte-specific epidermal growth factor prevents barrier dysfunction and improves mortality in murine peritonitis. Am J Physiol Gastrointest Liver Physiol. 2009;297:G471–G479. doi: 10.1152/ajpgi.00012.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Furuse M, Furuse K, Sasaki H, et al. Conversion of zonulae occludentes from tight to leaky strand type by introducing claudin-2 into Madin-Darby canine kidney I cells. J Cell Biol. 2001;153:263–272. doi: 10.1083/jcb.153.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Q, Zhang Q, Wang C, et al. Disruption of tight junctions during polymicrobial sepsis in vivo. J Pathol. 2009;218:210–221. doi: 10.1002/path.2525. [DOI] [PubMed] [Google Scholar]
- 54.Kliment CR, Englert JM, Gochuico BR, et al. Oxidative stress alters syndecan-1 distribution in lungs with pulmonary fibrosis. J Biol Chem. 2009;284:3537–3545. doi: 10.1074/jbc.M807001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zvibel I, Halfon P, Fishman S, et al. Syndecan 1 (CD138) serum levels: a novel biomarker in predicting liver fibrosis stage in patients with hepatitis C. Liver Int. 2009;29:208–212. doi: 10.1111/j.1478-3231.2008.01830.x. [DOI] [PubMed] [Google Scholar]
- 56.Estes JD, Haase AT, Schacker TW. The role of collagen deposition in depleting CD4+ T cells and limiting reconstitution in HIV-1 and SIV infections through damage to the secondary lymphoid organ niche. Semin Immunol. 2008;20:181–186. doi: 10.1016/j.smim.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representative images of SYN1 (green fluorescence) and CD27 (red fluorescence) expression in the rectum of an HIV-1-infected individual in the presymptomatic stage of disease were examined immunofluorescently (cell nuclei appear blue). Original magnification: X400; scale bars: 50 µm.
(A and B) Representative images of E. coli (red fluorescence) in the ileum of an HIV-1-infected individual in the presymptomatic stage of disease were examined using FISH (cell nuclei appear blue). Original magnifications: X600; scale bars: 20 µm.
(A) Representative optical images of E. coli (red fluorescence) and SYN1 expression (green fluorescence) from two identical sections of ileum from an HIV-1-infected individual in the presymptomatic stage of disease reveal minimal cross-reactivity with a matched isotype control antibody for E. coli (rabbit IgG). (B) Representative optical images of E. coli (red fluorescence) from a dry mount of purified E. coli reveal minimal cross-reactivity with a matched isotype control antibody. Original magnifications: X600; scale bars: 20 µm.