Abstract
This manuscript is part of a debate on the statement that “the use of short synthetic adhesion peptides, like RGD, is the best approach in the design of biomaterials that guide cell behavior for regenerative medicine and tissue engineering”. We take the position that although there are some acknowledged disadvantages of using short peptide ligands within biomaterials, it is not necessary to discard the notion of using peptides within biomaterials entirely, but rather to reinvent and evolve their use. Peptides possess advantageous chemical definition, access to non-native chemistries, amenability to de novo design, and applicability within parallel approaches. Biomaterials development programs that require such aspects may benefit from a peptide-based strategy.
1. Introduction
In this debate, our position is to emphasize the rationale for employing peptides, rather than whole proteins or biologically sourced materials, in the evolving design of biomaterials in the foreseeable future. We fully acknowledge that we are presenting one-sided arguments for the purposes of this debate, and the reality is that there is no reasonable up-or-down answer to this question, hence its appropriateness as a debate topic. Nevertheless, we will artificially polarize our stance in the discussion below, with the hope of illustrating the competing considerations that lead to the choice of one over the other. In reality, choosing between peptides and proteins in a given biomaterial setting usually requires the synthesis and consideration of many factors that often run counter to each other, including practicality, cost effectiveness, mechanistic clarity, biological specificity, immune responses, regulatory considerations, and others. It is our hope that the arguments that our opponents and we present here will effectively enumerate these competing considerations in order to facilitate the choice of a peptide-based approach or a protein-based approach in currently developing biomaterials strategies.
Our position is not to defend the use of adhesion peptides in isolation as a reasonable and complete approach for building biomaterials designed to act as surrogate ECMs. Recent work in Biomaterials has clearly indicated that such an approach falls considerably short of realizing the dynamic, context-specific, high-affinity, and multifunctional nature of whole proteins, native ECMs, or even protein domains. To us, defending this position directly would be untenable, and we believe that this paradigm will evolve. However, we do not believe that the use of peptides within biomaterials should be discounted entirely for the sake of using proteins or protein domains. So our position in this debate will be the following:
Although there are shortcomings of short peptide ligands within biomaterials, it is not necessary to discard the notion of using peptides within biomaterials entirely, but rather to reinvent their use.
It is our intent to illustrate some potential paths forward for the evolution of peptides within biomaterials that exploit some of peptides’ greatest advantages, including their chemical definition, their accessibility, their practicality, and their simplicity. We will highlight recent strategies for improving upon their shortcomings as biomaterials components, including their lack of specificity and their highly reduced functions.
In essence, the lack of overwhelming success of short ECM-derived ligands in biomaterials raises a simple question of how to proceed forward. We make arguments here for doing so using synthetic chemistry, including peptide chemistry. In the end, of course the best approaches may well include clever marriages of both protein-based and peptide-based strategies.
2. Advantages of using peptides
2.1 Peptides are chemically defined
Arguably the most advantageous property of peptides is that they are chemically defined. This in turn enables the systematic refinement of their structures, parallel experimental designs to discover novel peptides or combinations of them, and precise molecular manipulations required for mechanistic investigations. The definition of peptides arises from their chemical route of production, which is usually solid phase peptide synthesis (SPPS) [1, 2]. Using generally available resins, activating agents, amino acid derivatives, and solvents, peptides of up to 30–50 residues can be routinely prepared with good yields [2]. For such peptides, exceptional control over chemical identity can be achieved, even for “difficult to synthesize” sequences, which are becoming more accessible owing to the progressive introduction of newer and better reagents [3–6]. In contrast, proteins or tissue-derived matrices often lack such precise definition. When biologically sourced, biopolymers or ECMs can contain contaminating molecules that are difficult to define, much less remove. Troublesome contaminants like lipopolysaccharide can accompany proteins expressed in bacterial cultures, and tissue-derived matrices have a nonzero potential for harboring transmissible pathogens including viruses or prions. Although it can generally be assured in clinically developed ECM-based materials that such pathogens have been removed or neutralized, doing so requires significant attention, effort, and expense. More pertinent to the development of matrix-based biomaterials is that the incomplete molecular definition of ECMs can thwart efficient engineering of them. For example, the analysis of the glycoprotein content of a given natural ECM typically involves some combination of enzymatic digestion, solubilization, and extraction, through which the composition of the recovered materials and certainly the interconnectivity of them may change. This “compositional drift” can occur during the disassembly of protein matrices and also during the opposite process, matrix assembly. In this case, if one desires to produce a matrix with a defined ratio of proteins (e.g. 90% Collagen I and 10% laminin-111), the different components may not assemble or polymerize completely, making it uncertain to what extent the ratios between different matrix components may be maintained upon assembly. Compositional drift in analysis and production of protein-based ECMs in turn makes it difficult to study them and engineer them. Even in “pure” protein preparations, there may exist heterogeneity in folding, in essence producing a mix of biological activities, even though there may be homogeneity in the primary structure of the material. Peptide-based materials can circumvent these issues by being chemically defined.
2.2 Peptides can possess a diversity of functions
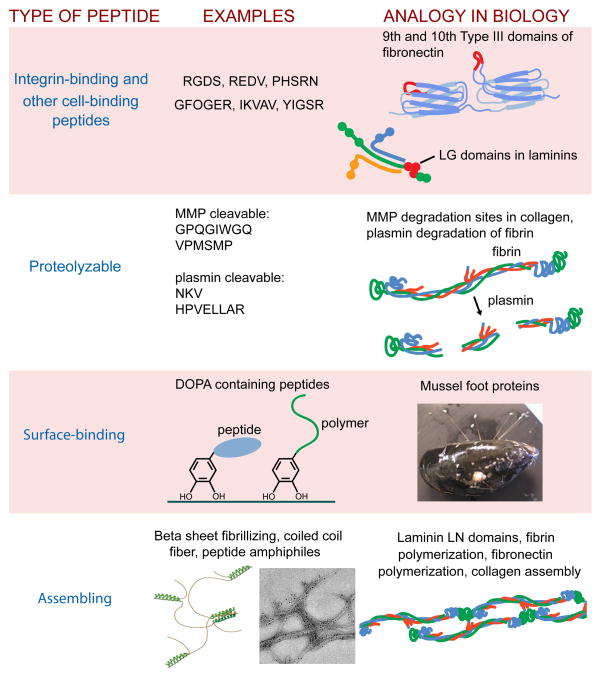
Although the debate statement mentions the integrin-binding peptide RGD as an example, peptides can be employed in biomaterials contexts for functions well beyond integrin binding. In fact, relegating peptides only to integrin binding drastically underutilizes their potential. Peptides used within biomaterials can possess a diversity of functions beyond cell binding, including specific proteolytic susceptibility, surface binding, matrix binding, self-assembly, growth factor binding, and combinations thereof (Box 1). Moreover, the tailorable nature of peptides leaves much room for the discovery and development of additional functions in the future.
Box 1.
Peptides available for incorporation in biomaterials posess functions beyond cell binding
It is true that the one peptide that has been studied the most by far in biomaterials contexts has been RGD, originally developed as an easily produced small molecule capable of functioning analogously to the 10th type-III domain of fibronectin [7]. But since its discovery and exhaustive exploration within biomaterials, it is now known to engage many different integrin subtypes, and moreover it is found in many different ECM molecules beyond fibronectin [8]. In some applications, the broad integrin binding provided by RGD may be acceptable, but if cell-specific behaviors are required, or if a defined set of integrins are to be activated and bound, the information content of short integrin ligands like RGD will in most cases be insufficient (for expansion of this view, see the recent review by one of our opponents in this debate series [9]). The question arises, then, of how to provide for this missing information in biomaterials. There can be advantages of doing so in chemically precise ways, including the use of synthetic peptides, peptide derivatives, and peptidomimetics.
As one example of additional functions that can be installed within biomaterials through peptides, enzyme substrate sequences have received particular interest, especially within hydrogels. Using peptide sequences of varying proteolytic susceptibility, such matrices can be designed to degrade with controllable kinetics by a variety of different proteases including matrix metalloproteinases or plasmin [10–12], enabling the tailored degradation of the material or the release of a matrix-tethered payload via proteolysis [13, 14]. In contrast, controlling the degradation kinetics of protein matrices can be somewhat less systematic, given that the experimenter rarely has control over the precise arrangement and sequences of proteolytic cleavage sites within them. Further, protein matrices form insoluble networks through self-assembly, thus the degradation of one fiber may not lead to degradation of the matrix in a predictable manner and thus controlled degradation/release rates may not be easily achieved. Peptides can also be engineered to possess growth factor binding capabilities, important given the native capacity of the ECM for growth factor sequestration, presentation, activation, and release [15–20]. For example, heparin-mimicking sulfated peptides that can bind vascular endothelial growth factor have been identified using bead-peptide libraries of sulfated peptides [21] or through rational design [22]. Both of these examples illustrate how the chemical definition of peptides enables parallel approaches or de novo design, respectively.
Peptides can also be designed to self-assemble, achieving one of the basic properties of native ECMs, insoluble network formation, with chemically defined small molecules. Native ECMs self-polymerize through precise protein-protein interactions, as exemplified by the LN domains of laminins, the stretches of fibronectin involved in fibrillogenesis, and the “knobs and holes” that mediate fibrin polymerization. Analogously, several classes of peptides have been designed and exploited in biomaterials contexts for their self-assembling behaviors. These have included β-sheet fibrillizing peptides [23–27], peptide amphiphiles [28–30], coiled coil peptides [31–33], β-hairpins [34–37], short aromatic peptide derivatives [38–41], and others [42, 43]. In each case, design rules have been worked out in order to achieve predictable assembly into fibers, networks, and gels, and strategies for decorating these materials with functional peptide sequences continue to be reported [26–28, 33, 39, 44–46].
2.3 Non-native chemistries and functions are easily incorporated into peptides
Owing to peptides’ synthetic routes of production, opportunities abound for specifically installing structures not routinely accomplished in expressed proteins or tissue-derived matrices. A short list of these might include natural amino acids beyond the genetically encoded ones, D-amino acids, chemical functionalities for cross-linking or polymerization, fluorescent labels useful for structural analyses, polymer bioconjugates, amino acids with fluorinated side chains, post-translational modifications including hydroxylation, phosphorylation, glycosylation, lipidation, and so on. Access to these non-canonical structures can be used to improve upon biomolecules already found in nature or to design new molecules de novo. As an example, 3,4-dihydroxyphenyl-L-alanine (DOPA) is post-translationally produced from tyrosine in nature and is found in large proportion in certain marine biopolymers such as the adhesive plaques of mussels. Chemically derived DOPA-containing peptides, peptidomimetics, and peptide-polymers have been successfully engineered as coatings for modifying a range of synthetic materials [47–50], effectively installing the tenacious adhesive properties of mussel proteins into synthetic biomaterials. Other surface-binding and matrix-binding peptides have been identified through the use of peptide libraries [51, 52] or de novo design [53, 54], again highlighting peptides’ amenability to such approaches. Library screenings have also identified peptides that bind to the cartilage ECM [55], solid surfaces and cells [51, 56], bone-like minerals [57], and titanium implants [58], and that can promote the proliferation of pluripotent cells [59]. It is true that parallel investigations, de novo design, and non-canonical amino acids can be applied towards expressed proteins, but not as efficiently, directly, or cost-effectively as with peptides.
2.4 Peptides can be conjugated to biomaterials with precision
Conjugating peptides onto or within synthetic materials can be accomplished with more chemical precision than proteins because one generally has more control over the conjugation chemistry with peptides. For proteins that have more diversity in chemical properties, it can be difficult to orient the biomolecule such that it is correctly and uniformly positioned on a given surface, unless precise immobilization strategies are employed [60, 61]. With proteins, the exact region that interacts with the cell may only be partially known, so chemical modifications of proteins can also alter their biological activity in unpredictable ways. In contrast, peptides can be incorporated into biomaterials through specific chemistries, utilizing a range of options including commercially available crosslinkers [62], Michael addition of cysteine-containing peptides to vinyl sulfones, acrylate or maleimide conjugation [63–65], UV-initiated crosslinking [66], amine/carboxylic acid coupling [67], and chemoselective chemistries such as “click” chemistries [68, 69] and native chemical ligation [26, 70–72]. Each of these can be achieved with chemical specificity, whereas the covalent incorporation of proteins into biomaterials is typically achieved through non-specific amine/carboxylic acid coupling. These types of immobilization strategies can result in random modification of the protein, a heterogeneous mixture of conformations of attached proteins, and commonly a reduction of protein activity.
3. Addressing the shortcomings of peptide-based biomaterials
3.1 Chemical Access to Longer Peptides
Although peptides are practical molecules for conferring biological functions to synthetic materials, they do possess a number of shortcomings; however, many of their shortcomings can be improved upon and have been improved upon recently (Box 2). Many limitations of peptides are a product of their reduced length, which in turn contributes to their generally poor specificity, their conformational flexibility, and their tendency not to fold into stable secondary, tertiary, and quaternary structures. This is a significant hurdle to overcome, and it is pertinent not just for employing peptides within biomaterials, but within most biomedical contexts. As a result, there has been significant motivation to extend the length attainable for synthetic peptides in general, driving many recent innovations in peptide chemistry. Some of these strategies have been applied to biomaterials contexts, but most have been significantly underutilized, thus representing an extremely useful set of resources that remain relatively untapped within biomaterials research and development. For example, chemoselective ligation strategies enable the joining of unprotected peptides or peptide derivatives to make longer polyamino acids than could be produced directly using solid phase peptide synthesis [73]. Native chemical ligation was introduced in 1994 as a means for joining two peptides, one with a C-terminal thioester and one with an N-terminal cysteine residue, forming a new peptide bond between the two in the process [70, 74]. The technique is highly selective chemically, enabling the conjoining of peptides of almost any amino acid content in near quantitative yield, and it has been successful for chemically synthesizing well-folded proteins of over 200 amino acids in length [74, 75]. Given the technique’s importance in synthetic biology and in total protein synthesis, it is surprising that native chemical ligation has not been extensively utilized in biomaterials. A few reports have utilized native chemical ligation for the polymerization or cross-linking of biomaterials [26, 71, 72], and it has been utilized to chemically synthesize ECM domains such as type I thrombospondin repeats [76] but combining such approaches to provide for well-folded ligands in biomaterials contexts has been under-explored. Similarly, other chemoselective conjugation techniques have been applied towards biomaterials, including copper-catalyzed alkyne-azide cycloaddition [68, 69, 77], oxime ligation [78, 79], and intein-mediated ligation [80–82], but these techniques have yet to be fully embraced in biomaterials design despite their promise of overcoming the challenges associated with peptides’ short length. The example of intein-mediated ligation also illustrates a useful technique combining expressed proteins with chemoselective chemistry, despite the fact that we are focusing on purely synthetic approaches for the purpose of the debate format. In addition to significant progress in peptide conjugation, there has also been continual refinement in the resins, coupling reagents, and amino acid derivatives available, progressively lowering the cost of peptide synthesis, extending the length of sequences possible, improving purity, and simplifying work-up. Owing to such improvements in peptide synthesis, conjugation, and polymerization, it seems clear that longer and longer amino acid sequences will progressively become available in biomaterials contexts. It is up to the field to exploit these advancements.
Box 2.
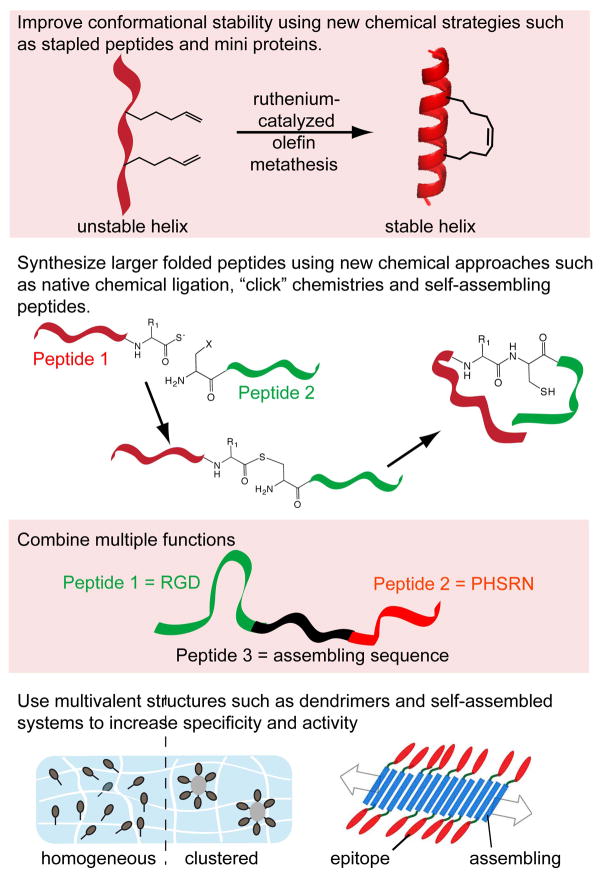
Addressing shortcomings of peptides in biomaterials. Problems of poor affinity could be addressed with covalent stabilization approaches such as “stapled” peptides (top). The short nature of synthetic peptides could be improved with chemoselective conjugation strategies such as native chemical ligation (middle top). Multiple functions can be combined, for example using self-assembling strategies (middle bottom), and materials with extreme multivalency can be produced, for example with dendrimers or self-assembly (bottom).
3.2 Strategies for improving peptide affinity
In general, peptides do not fold into defined or stable secondary, tertiary, or quaternary structures, and this drastically limits the affinities that can be achieved with them in comparison to well-folded proteins. Extending the synthetically accessible length of peptides will certainly improve the ability to directly synthesize folded domains, but even without lengthening peptides, many strategies currently exist for stabilizing particular folds through non-covalent or covalent means. Design rules have been developed for specifying certain folding in oligomerized structures, including β-sheets, α-helices [31, 33, 83], and collagen triple helices [84]. In addition, synthetic miniproteins constructed around motifs such as Trp cages and cystine knots [85, 86] are powerful scaffolds with which to engineer high-affinity biomaterials components. Cystine knots have been recently utilized to design peptide integrin ligands with low-nanomolar affinity [87], but many of these strategies have only cursorily been applied towards biomaterials. Analogously to the uses of chemical ligation described above, the predominant use of controlled secondary structural folding within biomaterials has been to polymerize or assemble the peptides into predictable networks, as has been achieved with β-sheet peptides [23, 25, 27, 42, 88, 89], coiled coil peptides [32], peptide amphiphiles [28], and collagen-like peptides [90]. In most of these strategies, the overarching concept is to utilize the secondary structure motifs to oligomerize other functional components, such as pendant functional amino acid sequences, into predictable assemblies. By comparison, using these design rules to stabilize or improve the affinity of the biofunctional components of the materials has received much less interest, leaving exciting opportunities to be explored.
For biomaterials containing peptides, many routes are available for installing considerable multivalency into the material, and this can be utilized to overcome shortcomings in peptide affinity. For example, dendrimers, multi-arm functionalized polymers, and self-assembled structures are all available to provide highly repetitive presentations of peptides. High-density ligand display has been shown to be important in the biological functionality of self-assembled materials, which are capable of displaying far higher densities of functional components on the surface of fibrils or oligomers than is possible for protein-based scaffolds [27, 30]. This may not be appropriate for all biomaterials strategies, but it can be advantageous for many.
Covalent stabilization of peptides can also significantly improve their specificities. This has been well documented for cyclic RGD ligands [91], and peptide cyclization remains a simple, powerful method for ligand stabilization, especially for peptides that assume a looped conformation in their native structures, as RGD does [92]. For the covalent stabilization of more complex folds, other strategies have recently been developed and are poised to make a significant contribution to biomaterials. These strategies include the miniprotein scaffolds discussed above such as cystine-knot proteins, as well as stapled peptides, in which ruthenium-catalyzed olefin metathesis is used to place hydrocarbon bridges between specific side chains of peptides [93]. This approach has been used to stabilize the secondary structures of several different peptides, which in turn has greatly improved their affinities [94], made them resistant to proteolysis [95], and improved their ability to cross the cell membrane [96]. The stabilization of peptides with this technique is significant enough to even provide for oral bioavailability, which has been a longstanding shortcoming of peptide therapeutics owing to their susceptibility to proteolysis in the gastrointestinal tract [95]. Stapled peptides have been explored as therapeutics for cancer [94, 97], HIV fusion inhibitors [95], and to initiate and study apoptotic mechanisms [96, 98], but they have not yet to our knowledge been utilized in biomaterials applications. Doing so may open up new possibilities for biomaterials design by providing for more specific, stable, and of course chemically defined synthetic ligands.
3.3 Installing multifunctionality and modularity in peptides
One shortcoming of peptides is that they typically only possess one function, whether it is integrin binding, enzymatic susceptibility, growth factor binding, oligomerization, or another. In contrast, native ECM proteins are exceptionally more multifunctional, combining all of these functions elegantly within one molecule or a network of molecules. This multifunctional nature of ECM proteins is both their advantage over peptides and their disadvantage, as it contributes to the challenge of defining them, manipulating them, purifying them, and studying them. Even though peptides are extremely limited in terms of functionality, methods are being developed for combining them together in multifactorial combinations, in order to integrate multiple functions within synthetic matrices [99]. Recent examples include polymer hydrogels containing peptides of multiple different functions [100], self-assembling biomaterials in which multiple different peptides are co-assembled [32, 42, 99], and multidomain self-assembling peptides incorporating several functions, for example cell-adhesion, fibril formation, and proteolytic susceptibility [88, 89]. Unlike native ECMs, these materials have the advantage of being chemically defined, with known amounts of each functional component, so unlike native protein matrices their compositions can be systematically tailored to fit the needs of the application or experimental goal, as discussed above [99].
3.4 The cost of synthetic peptides
Peptides have sometimes been labeled as being prohibitively expensive, but peptide synthesis can be achieved on research scales with reasonable costs, and large-scale industrial production of peptides indicates their economic viability. On an industrial scale, commercialized pharmaceuticals such as enfuvirtide (a 36-amino acid synthetic peptide that acts as an HIV fusion inhibitor) and eptifibatide (an antiplatelet small cyclic peptide) illustrate peptides’ economic viability [101, 102]. On a research scale, peptide synthesis costs continue to decline as different coupling reagents, resins, amino acids, and solvents become increasingly available. Costs are especially attractive when considering all factors such as time for synthesis and simplicity of purification. In our laboratories, once the initial start cost is assumed, reasonably pure batches of a few hundred milligrams of average-length peptides can be produced in a few days with a cost of around two hundred US dollars. On a research scale, we find this to be not especially onerous. Peptides are also available through many companies specializing in custom synthesis, with reasonable pricing. For in vivo or immunological work, the purification requirements of peptides are especially attractive because they do not contain biological contaminants, especially troublesome ones like lipopolysaccharide. In contrast, proteins require the continuous optimization of their production for each recombinant protein, the purchase of large fermentors to be able to produce enough material for biomaterials applications, and the cloning of each different protein, which is not always straightforward.
4. Thoughts
In order to design biomaterials capable of driving specific cell phenotypes essential for proper development, homeostasis, repair, and potential regeneration of tissues, it will be necessary to design materials exhibiting complexity, coordinated molecular processes, and dynamic interactions with cells and tissues. As we have outlined above, many of these goals could be achieved with peptides and other chemically defined materials. A wealth of newly developed strategies in peptide design, chemistry, assembly, and materials science have yet to be exploited fully in biomaterials contexts, and more strategies are sure to be forthcoming as peptide science continues to progress. However, the evolution of peptides in biomaterials cannot simply be built on advancements in chemistry and peptide design, for they must also take advantage of new findings and understanding in ECM biology. New chemistries will only be effective if they engage relevant processes operating in tissues and native ECMs. In this regard, better communication between chemists, materials scientists, and ECM biologists would be a welcome development. Not only can chemically defined matrices be directed towards improved biomaterials, but they could also be extremely helpful for developing and testing specific hypotheses in ECM biology. Although peptides will surely not always be the most appropriate choice for all biomaterials research and development, many such efforts will benefit from peptide use.
Acknowledgments
The authors would like to thank Harry Bermudez, Josh Gasiorowski, and Greg Hudalla for suggestions on the manuscript, and Phil Messersmith for supplying the image of the mussel.
Contributor Information
Joel H. Collier, Email: collier@uchicago.edu.
Tatiana Segura, Email: tsegura@ucla.edu.
References
- 1.Merrifield RB. Solid Phase Peptide Synthesis. I. The synthesis of a tetrapeptide. J Am Chem Soc. 1963;85:2149–2154. [Google Scholar]
- 2.Coin I, Beyermann M, Bienert M. Solid-phase peptide synthesis: from standard procedures to the synthesis of difficult sequences. Nat Protoc. 2007;2:3247–3256. doi: 10.1038/nprot.2007.454. [DOI] [PubMed] [Google Scholar]
- 3.Hyde C, Johnson T, Owen D, Quibell M, Sheppard RC. Some ‘difficult sequences’ made easy. A study of interchain association in solid-phase peptide synthesis. Int J Pept Protein Res. 1994;43:431–440. [PubMed] [Google Scholar]
- 4.Sohma Y, Sasaki M, Hayashi Y, Kimura T, Kiso Y. Novel and efficient synthesis of difficult sequence-containing peptides through O-N intramolecular acyl migration reaction of O-acyl isopeptides. Chem Commun (Camb) 2004;7:124–125. doi: 10.1039/b312129a. [DOI] [PubMed] [Google Scholar]
- 5.Carpino LA, Abdel-Maksoud AA, Mansour EM, Zewail MA. Segment coupling to a highly hindered N-terminal, alamethicin-related alpha-aminoisobutyric acid (Aib) residue. Tetrahedron Lett. 2007;48:7404–7407. doi: 10.1016/j.tetlet.2007.07.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mutter M, Nefzi A, Sato T, Sun X, Wahl F, Wohr T. Pseudo-prolines (psi Pro) for accessing “inaccessible” peptides. Pept Res. 1995;8:145–153. [PubMed] [Google Scholar]
- 7.Pierschbacher MD, Ruoslahti E. Cell attachment activity of fibronectin can be duplicated by small synthetic fragments of the molecule. Nature. 1984;309:30–33. doi: 10.1038/309030a0. [DOI] [PubMed] [Google Scholar]
- 8.Ruoslahti E. RGD and other recognition sequences for integrins. Annu Rev Cell Dev Biol. 1996;12:697–715. doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- 9.Carson AE, Barker TH. Emerging concepts in engineering extracellular matrix variants for directing cell phenotype. Regen Med. 2009;4:593–600. doi: 10.2217/rme.09.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lutolf MP, Lauer-Fields JL, Schmoekel HG, Metters AT, Weber FE, Fields GB, et al. Synthetic matrix metalloproteinase-sensitive hydrogels for the conduction of tissue regeneration: engineering cell-invasion characteristics. Proc Natl Acad Sci U S A. 2003;100:5413–5418. doi: 10.1073/pnas.0737381100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patterson J, Hubbell JA. SPARC-derived protease substrates to enhance the plasmin sensitivity of molecularly engineered PEG hydrogels. Biomaterials. doi: 10.1016/j.biomaterials.2010.10.016. [DOI] [PubMed] [Google Scholar]
- 12.Patterson J, Hubbell JA. Enhanced proteolytic degradation of molecularly engineered PEG hydrogels in response to MMP-1 and MMP-2. Biomaterials. 31:7836–7845. doi: 10.1016/j.biomaterials.2010.06.061. [DOI] [PubMed] [Google Scholar]
- 13.Tauro JR, Lee BS, Lateef SS, Gemeinhart RA. Matrix metalloprotease selective peptide substrates cleavage within hydrogel matrices for cancer chemotherapy activation. Peptides. 2008;29:1965–1973. doi: 10.1016/j.peptides.2008.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tokatlian T, Shrum CT, Kadoya WM, Segura T. Protease degradable tethers for controlled and cell-mediated release of nanoparticles in 2- and 3-dimensions. Biomaterials. 2010;31:8072–8080. doi: 10.1016/j.biomaterials.2010.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cochran S, Li CP, Ferro V. A surface plasmon resonance-based solution affinity assay for heparan sulfate-binding proteins. Glycoconj J. 2009;26:577–587. doi: 10.1007/s10719-008-9210-0. [DOI] [PubMed] [Google Scholar]
- 16.Xu Q, Yan B, Li S, Duan C. Fibronectin binds insulin-like growth factor-binding protein 5 and abolishes Its ligand-dependent action on cell migration. J Biol Chem. 2004;279:4269–4277. doi: 10.1074/jbc.M311586200. [DOI] [PubMed] [Google Scholar]
- 17.Rahman S, Patel Y, Murray J, Patel KV, Sumathipala R, Sobel M, et al. Novel hepatocyte growth factor (HGF) binding domains on fibronectin and vitronectin coordinate a distinct and amplified Met-integrin induced signalling pathway in endothelial cells. BMC Cell Biol. 2005;6:8. doi: 10.1186/1471-2121-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoshijima M, Hattori T, Inoue M, Araki D, Hanagata H, Miyauchi A, et al. CT domain of CCN2/CTGF directly interacts with fibronectin and enhances cell adhesion of chondrocytes through integrin alpha5beta1. FEBS Lett. 2006;580:1376–1382. doi: 10.1016/j.febslet.2006.01.061. [DOI] [PubMed] [Google Scholar]
- 19.Martino MM, Hubbell JA. The 12th–14th type III repeats of fibronectin function as a highly promiscuous growth factor-binding domain. FASEB J. 2010;24:4711–21. doi: 10.1096/fj.09-151282. [DOI] [PubMed] [Google Scholar]
- 20.Chen TT, Luque A, Lee S, Anderson SM, Segura T, Iruela-Arispe ML. Anchorage of VEGF to the extracellular matrix conveys differential signaling responses to endothelial cells. J Cell Biol. 188:595–609. doi: 10.1083/jcb.200906044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maynard HD, Hubbell JA. Discovery of a sulfated tetrapeptide that binds to vascular endothelial growth factor. Acta Biomater. 2005;1:451–459. doi: 10.1016/j.actbio.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 22.Kim SH, Kiick KL. Heparin-mimetic sulfated peptides with modulated affinities for heparin-binding peptides and growth factors. Peptides. 2007;28:2125–2136. doi: 10.1016/j.peptides.2007.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Genove E, Shen C, Zhang S, Semino CE. The effect of functionalized self-assembling peptide scaffolds on human aortic endothelial cell function. Biomaterials. 2005;26:3341–3351. doi: 10.1016/j.biomaterials.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 24.Horii A, Wang X, Gelain F, Zhang S. Biological designer self-assembling peptide nanofiber scaffolds significantly enhance osteoblast proliferation, differentiation and 3-D migration. PLoS One. 2007;2:e190. doi: 10.1371/journal.pone.0000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang S, Holmes T, Lockshin C, Rich A. Spontaneous assembly of a self-complementary oligopeptide to form a stable macroscopic membrane. Proc Natl Acad Sci U S A. 1993;90:3334–3338. doi: 10.1073/pnas.90.8.3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jung JP, Jones JL, Cronier SA, Collier JH. Modulating the mechanical properties of self-assembled peptide hydrogels via native chemical ligation. Biomaterials. 2008;29:2143–2151. doi: 10.1016/j.biomaterials.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rudra JS, Tian YF, Jung JP, Collier JH. A self-assembling peptide acting as an immune adjuvant. Proc Natl Acad Sci U S A. 2010;107:622–627. doi: 10.1073/pnas.0912124107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cui H, Webber MJ, Stupp SI. Self-assembly of peptide amphiphiles: from molecules to nanostructures to biomaterials. Biopolymers. 2010;94:1–18. doi: 10.1002/bip.21328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hartgerink JD, Beniash E, Stupp SI. Self-assembly and mineralization of peptide-amphiphile nanofibers. Science. 2001;294:1684–1688. doi: 10.1126/science.1063187. [DOI] [PubMed] [Google Scholar]
- 30.Silva GA, Czeisler C, Niece KL, Beniash E, Harrington DA, Kessler JA, et al. Selective differentiation of neural progenitor cells by high-epitope density nanofibers. Science. 2004;303:1352–1355. doi: 10.1126/science.1093783. [DOI] [PubMed] [Google Scholar]
- 31.Banwell EF, Abelardo ES, Adams DJ, Birchall MA, Corrigan A, Donald AM, et al. Rational design and application of responsive alpha-helical peptide hydrogels. Nat Mater. 2009;8:596–600. doi: 10.1038/nmat2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woolfson DN. Building fibrous biomaterials from alpha-helical and collagen-like coiled-coil peptides. Biopolymers. 2010;94:118–127. doi: 10.1002/bip.21345. [DOI] [PubMed] [Google Scholar]
- 33.Woolfson DN, Mahmoud ZN. More than just bare scaffolds: towards multi-component and decorated fibrous biomaterials. Chem Soc Rev. 2010;39:3464–3479. doi: 10.1039/c0cs00032a. [DOI] [PubMed] [Google Scholar]
- 34.Haines-Butterick L, Rajagopal K, Branco M, Salick D, Rughani R, Pilarz M, et al. Controlling hydrogelation kinetics by peptide design for three-dimensional encapsulation and injectable delivery of cells. Proc Natl Acad Sci U S A. 2007;104:7791–7796. doi: 10.1073/pnas.0701980104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kretsinger JK, Haines LA, Ozbas B, Pochan DJ, Schneider JP. Cytocompatibility of self-assembled beta-hairpin peptide hydrogel surfaces. Biomaterials. 2005;26:5177–5186. doi: 10.1016/j.biomaterials.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 36.Rajagopal K, Lamm MS, Haines-Butterick LA, Pochan DJ, Schneider JP. Tuning the pH responsiveness of beta-hairpin peptide folding, self-assembly, and hydrogel material formation. Biomacromolecules. 2009;10:2619–2625. doi: 10.1021/bm900544e. [DOI] [PubMed] [Google Scholar]
- 37.Schneider JP, Pochan DJ, Ozbas B, Rajagopal K, Pakstis L, Kretsinger J. Responsive hydrogels from the intramolecular folding and self-assembly of a designed peptide. J Am Chem Soc. 2002;124:15030–15037. doi: 10.1021/ja027993g. [DOI] [PubMed] [Google Scholar]
- 38.Jayawarna V, Richardson SM, Hirst AR, Hodson NW, Saiani A, Gough JE, et al. Introducing chemical functionality in Fmoc-peptide gels for cell culture. Acta Biomater. 2009;5:934–943. doi: 10.1016/j.actbio.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 39.Zhou M, Smith AM, Das AK, Hodson NW, Collins RF, Ulijn RV, et al. Self-assembled peptide-based hydrogels as scaffolds for anchorage-dependent cells. Biomaterials. 2009;30:2523–2530. doi: 10.1016/j.biomaterials.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 40.Yang Z, Liang G, Wang L, Xu B. Using a kinase/phosphatase switch to regulate a supramolecular hydrogel and forming the supramolecular hydrogel in vivo. J Am Chem Soc. 2006;128:3038–3043. doi: 10.1021/ja057412y. [DOI] [PubMed] [Google Scholar]
- 41.Yang Z, Liang G, Xu B. Enzymatic hydrogelation of small molecules. Acc Chem Res. 2008;41:315–326. doi: 10.1021/ar7001914. [DOI] [PubMed] [Google Scholar]
- 42.Collier JH, Rudra JS, Gasiorowski JZ, Jung JP. Multi-component extracellular matrices based on peptide self-assembly. Chem Soc Rev. 2010;39:3413–3424. doi: 10.1039/b914337h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jung JP, Gasiorowski JZ, Collier JH. Fibrillar peptide gels in biotechnology and biomedicine. Biopolymers. 2010;94:49–59. doi: 10.1002/bip.21326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mata A, Geng Y, Henrikson KJ, Aparicio C, Stock SR, Satcher RL, et al. Bone regeneration mediated by biomimetic mineralization of a nanofiber matrix. Biomaterials. 2010;31:6004–6012. doi: 10.1016/j.biomaterials.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Webber MJ, Tongers J, Renault MA, Roncalli JG, Losordo DW, Stupp SI. Development of bioactive peptide amphiphiles for therapeutic cell delivery. Acta Biomater. 2010;6:3–11. doi: 10.1016/j.actbio.2009.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jung JP, Nagaraj AK, Fox EK, Rudra JS, Devgun JM, Collier JH. Co-assembling peptides as defined matrices for endothelial cells. Biomaterials. 2009;30:2400–2410. doi: 10.1016/j.biomaterials.2009.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dalsin JL, Hu BH, Lee BP, Messersmith PB. Mussel adhesive protein mimetic polymers for the preparation of nonfouling surfaces. J Am Chem Soc. 2003;125:4253–4258. doi: 10.1021/ja0284963. [DOI] [PubMed] [Google Scholar]
- 48.Paunesku T, Rajh T, Wiederrecht G, Maser J, Vogt S, Stojicevic N, et al. Biology of TiO2-oligonucleotide nanocomposites. Nat Mater. 2003;2:343–346. doi: 10.1038/nmat875. [DOI] [PubMed] [Google Scholar]
- 49.Statz AR, Meagher RJ, Barron AE, Messersmith PB. New peptidomimetic polymers for antifouling surfaces. J Am Chem Soc. 2005;127:7972–7973. doi: 10.1021/ja0522534. [DOI] [PubMed] [Google Scholar]
- 50.Lee H, Dellatore SM, Miller WM, Messersmith PB. Mussel-inspired surface chemistry for multifunctional coatings. Science. 2007;318:426–430. doi: 10.1126/science.1147241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baneyx F, Schwartz DT. Selection and analysis of solid-binding peptides. Curr Opin Biotechnol. 2007;18:312–317. doi: 10.1016/j.copbio.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 52.Daugherty PS. Protein engineering with bacterial display. Curr Opin Struct Biol. 2007;17:474–480. doi: 10.1016/j.sbi.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 53.Wang AY, Leong S, Liang YC, Huang RC, Chen CS, Yu SM. Immobilization of growth factors on collagen scaffolds mediated by polyanionic collagen mimetic peptides and its effect on endothelial cell morphogenesis. Biomacromolecules. 2008;9:2929–2936. doi: 10.1021/bm800727z. [DOI] [PubMed] [Google Scholar]
- 54.Wang AY, Mo X, Chen CS, Yu SM. Facile modification of collagen directed by collagen mimetic peptides. J Am Chem Soc. 2005;127:4130–4131. doi: 10.1021/ja0431915. [DOI] [PubMed] [Google Scholar]
- 55.Rothenfluh DA, Bermudez H, O’Neil CP, Hubbell JA. Biofunctional polymer nanoparticles for intra-articular targeting and retention in cartilage. Nat Mater. 2008;7:248–254. doi: 10.1038/nmat2116. [DOI] [PubMed] [Google Scholar]
- 56.Kato R, Kaga C, Kunimatsu M, Kobayashi T, Honda H. Peptide array-based interaction assay of solid-bound peptides and anchorage-dependant cells and its effectiveness in cell-adhesive peptide design. J Biosci Bioeng. 2006;101:485–495. doi: 10.1263/jbb.101.485. [DOI] [PubMed] [Google Scholar]
- 57.Segvich S, Biswas S, Becker U, Kohn DH. Identification of peptides with targeted adhesion to bone-like mineral via phage display and computational modeling. Cells Tissues Organs. 2009;189:245–251. doi: 10.1159/000151380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu Y, Mao J, Zhou B, Wei W, Gong S. Peptide aptamers against titanium-based implants identified through phage display. J Mater Sci Mater Med. 21:1103–1107. doi: 10.1007/s10856-009-3970-3. [DOI] [PubMed] [Google Scholar]
- 59.Derda R, Musah S, Orner BP, Klim JR, Li L, Kiessling LL. High-throughput discovery of synthetic surfaces that support proliferation of pluripotent cells. J Am Chem Soc. 132:1289–1295. doi: 10.1021/ja906089g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hodneland CD, Lee YS, Min DH, Mrksich M. Selective immobilization of proteins to self-assembled monolayers presenting active site-directed capture ligands. Proc Natl Acad Sci U S A. 2002;99:5048–5052. doi: 10.1073/pnas.072685299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eisenberg JL, Piper JL, Mrksich M. Using self-assembled monolayers to model cell adhesion to the 9th and 10th type III domains of fibronectin. Langmuir. 2009;25:13942–13951. doi: 10.1021/la901528c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hermanson GT. Bioconjugate techniques. San Diego: Academic Press; 1996. [Google Scholar]
- 63.Elbert DL, Hubbell JA. Conjugate addition reactions combined with free-radical cross-linking for the design of materials for tissue engineering. Biomacromolecules. 2001;2:430–441. doi: 10.1021/bm0056299. [DOI] [PubMed] [Google Scholar]
- 64.Elbert DL, Pratt AB, Lutolf MP, Halstenberg S, Hubbell JA. Protein delivery from materials formed by self-selective conjugate addition reactions. J Control Release. 2001;76:11–25. doi: 10.1016/s0168-3659(01)00398-4. [DOI] [PubMed] [Google Scholar]
- 65.Lutolf MP, Tirelli N, Cerritelli S, Cavalli L, Hubbell JA. Systematic modulation of Michael-type reactivity of thiols through the use of charged amino acids. Bioconjug Chem. 2001;12:1051–1056. doi: 10.1021/bc015519e. [DOI] [PubMed] [Google Scholar]
- 66.Khetan S, Katz JS, Burdick JA. Sequential crosslinking to control cellular spreading in 3-dimensional hydrogels. Soft Matter. 2009;5:1601–1606. [Google Scholar]
- 67.Rowley JA, Madlambayan G, Mooney DJ. Alginate hydrogels as synthetic extracellular matrix materials. Biomaterials. 1999;20:45–53. doi: 10.1016/s0142-9612(98)00107-0. [DOI] [PubMed] [Google Scholar]
- 68.Lutz JF, Zarafshani Z. Efficient construction of therapeutics, bioconjugates, biomaterials and bioactive surfaces using azide-alkyne “click” chemistry. Advanced drug delivery reviews. 2008;60:958–970. doi: 10.1016/j.addr.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 69.Parrish B, Breitenkamp RB, Emrick T. PEG- and peptide-grafted aliphatic polyesters by click chemistry. J Am Chem Soc. 2005;127:7404–7410. doi: 10.1021/ja050310n. [DOI] [PubMed] [Google Scholar]
- 70.Dawson PE, Muir TW, Clark-Lewis I, Kent SB. Synthesis of proteins by native chemical ligation. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 71.Hu BH, Su J, Messersmith PB. Hydrogels cross-linked by native chemical ligation. Biomacromolecules. 2009;10:2194–2200. doi: 10.1021/bm900366e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Paramonov SE, Gauba V, Hartgerink JD. Synthesis of collagen-like peptide polymers by native chemical ligation. Macromolecules. 2005;38:7555–7561. [Google Scholar]
- 73.Tiefenbrunn TK, Dawson PE. Chemoselective ligation techniques: modern applications of time-honored chemistry. Biopolymers. 2010;94:95–106. doi: 10.1002/bip.21337. [DOI] [PubMed] [Google Scholar]
- 74.Kent SB. Total chemical synthesis of proteins. Chemical Society reviews. 2009;38:338–351. doi: 10.1039/b700141j. [DOI] [PubMed] [Google Scholar]
- 75.Torbeev VY, Kent SB. Convergent chemical synthesis and crystal structure of a 203 amino acid “covalent dimer” HIV-1 protease enzyme molecule. Angewandte Chemie (International ed. 2007;46:1667–1670. doi: 10.1002/anie.200604087. [DOI] [PubMed] [Google Scholar]
- 76.Tiefenbrunn TK, Dawson PE. Chemical synthesis and biotinylation of the thrombospondin domain TSR2. Protein Sci. 2009;18:970–979. doi: 10.1002/pro.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hudalla GA, Murphy WL. Immobilization of peptides with distinct biological activities onto stem cell culture substrates using orthogonal chemistries. Langmuir. 2010;26:6449–6456. doi: 10.1021/la1008208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Christman KL, Broyer RM, Tolstyka ZP, Maynard HD. Site-specific protein immobilization through N-terminal oxime linkages. J Mater Chem. 2007;17:2021–2027. [Google Scholar]
- 79.Taniguchi I, Kuhlman WA, Mayes AM, Griffith LG. Functional modification of biodegradable polyesters through a chemoselective approach: application to biomaterial surfaces. Polymer International. 2006;55:1385–1397. [Google Scholar]
- 80.Gao W, Liu W, Christensen T, Zalutsky MR, Chilkoti A. In situ growth of a PEG-like polymer from the C terminus of an intein fusion protein improves pharmacokinetics and tumor accumulation. Proceedings of the National Academy of Sciences of the United States of America. 107:16432–16437. doi: 10.1073/pnas.1006044107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Muir TW. Semisynthesis of proteins by expressed protein ligation. Annual review of biochemistry. 2003;72:249–289. doi: 10.1146/annurev.biochem.72.121801.161900. [DOI] [PubMed] [Google Scholar]
- 82.Evans TC, Jr, Benner J, Xu MQ. The cyclization and polymerization of bacterially expressed proteins using modified self-splicing inteins. The Journal of biological chemistry. 1999;274:18359–18363. doi: 10.1074/jbc.274.26.18359. [DOI] [PubMed] [Google Scholar]
- 83.Armstrong CT, Boyle AL, Bromley EH, Mahmoud ZN, Smith L, Thomson AR, et al. Rational design of peptide-based building blocks for nanoscience and synthetic biology. Faraday Discuss. 2009;143:305–317. doi: 10.1039/b901610d. discussion 359–372. [DOI] [PubMed] [Google Scholar]
- 84.Shoulders MD, Raines RT. Collagen structure and stability. Annu Rev Biochem. 2009;78:929–958. doi: 10.1146/annurev.biochem.77.032207.120833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kolmar H. Alternative binding proteins: biological activity and therapeutic potential of cystine-knot miniproteins. Febs J. 2008;275:2684–2690. doi: 10.1111/j.1742-4658.2008.06440.x. [DOI] [PubMed] [Google Scholar]
- 86.Daly NL, Rosengren KJ, Craik DJ. Discovery, structure and biological activities of cyclotides. Adv Drug Deliv Rev. 2009;61:918–930. doi: 10.1016/j.addr.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 87.Kimura RH, Levin AM, Cochran FV, Cochran JR. Engineered cystine knot peptides that bind alphavbeta3, alphavbeta5, and alpha5beta1 integrins with low-nanomolar affinity. Proteins. 2009;77:359–369. doi: 10.1002/prot.22441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Galler KM, Aulisa L, Regan KR, D’Souza RN, Hartgerink JD. Self-assembling multidomain peptide hydrogels: designed susceptibility to enzymatic cleavage allows enhanced cell migration and spreading. J Am Chem Soc. 2010;132:3217–3223. doi: 10.1021/ja910481t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Aulisa L, Dong H, Hartgerink JD. Self-assembly of multidomain peptides: sequence variation allows control over cross-linking and viscoelasticity. Biomacromolecules. 2009;10:2694–2698. doi: 10.1021/bm900634x. [DOI] [PubMed] [Google Scholar]
- 90.Fallas JA, O’Leary LE, Hartgerink JD. Synthetic collagen mimics: self-assembly of homotrimers, heterotrimers and higher order structures. Chem Soc Rev. 2010;39:3510–3527. doi: 10.1039/b919455j. [DOI] [PubMed] [Google Scholar]
- 91.Aumailley M, Gurrath M, Muller G, Calvete J, Timpl R, Kessler H. Arg-Gly-Asp constrained within cyclic pentapeptides. Strong and selective inhibitors of cell adhesion to vitronectin and laminin fragment P1. FEBS Lett. 1991;291:50–54. doi: 10.1016/0014-5793(91)81101-d. [DOI] [PubMed] [Google Scholar]
- 92.Pankov R, Yamada KM. Fibronectin at a glance. J Cell Sci. 2002;115:3861–3863. doi: 10.1242/jcs.00059. [DOI] [PubMed] [Google Scholar]
- 93.Schafmeister CE, Po J, Verdine GL. An all-hydrocarbon cross-linking system for enhancing the helicity and metabolic stability of peptides. J Am Chem Soc. 2000;122:5891–5892. [Google Scholar]
- 94.Bernal F, Tyler AF, Korsmeyer SJ, Walensky LD, Verdine GL. Reactivation of the p53 tumor suppressor pathway by a stapled p53 peptide. J Am Chem Soc. 2007;129:2456–2457. doi: 10.1021/ja0693587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bird GH, Madani N, Perry AF, Princiotto AM, Supko JG, He X, et al. Hydrocarbon double-stapling remedies the proteolytic instability of a lengthy peptide therapeutic. Proc Natl Acad Sci U S A. 2010;107:14093–14098. doi: 10.1073/pnas.1002713107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Walensky LD, Kung AL, Escher I, Malia TJ, Barbuto S, Wright RD, et al. Activation of apoptosis in vivo by a hydrocarbon-stapled BH3 helix. Science. 2004;305:1466–1470. doi: 10.1126/science.1099191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Moellering RE, Cornejo M, Davis TN, Del Bianco C, Aster JC, Blacklow SC, et al. Direct inhibition of the NOTCH transcription factor complex. Nature. 2009;462:182–188. doi: 10.1038/nature08543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Walensky LD, Pitter K, Morash J, Oh KJ, Barbuto S, Fisher J, et al. A stapled BID BH3 helix directly binds and activates BAX. Mol Cell. 2006;24:199–210. doi: 10.1016/j.molcel.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 99.Jung JP, Moyano JV, Collier JH. Multifactorial optimization of endothelial cell growth using modular synthetic extracellular matrices. Integr Biol. 2011 doi: 10.1039/c0ib00112k. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jia X, Kiick KL. Hybrid multicomponent hydrogels for tissue engineering. Macromol Biosci. 2009;9:140–156. doi: 10.1002/mabi.200800284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Matthews T, Salgo M, Greenberg M, Chung J, DeMasi R, Bolognesi D. Enfuvirtide: the first therapy to inhibit the entry of HIV-1 into host CD4 lymphocytes. Nat Rev Drug Discov. 2004;3:215–225. doi: 10.1038/nrd1331. [DOI] [PubMed] [Google Scholar]
- 102.Topol E, et al. Inhibition of platelet glycoprotein IIb/IIIa with eptifibatide in patients with acute coronary syndromes. The PURSUIT trial investigators. Platelet Glycoprotein IIb/IIIa in Unstable Angina: Receptor Suppression Using Integrilin Therapy. N Engl J Med. 1998;339:436–443. [Google Scholar]