Abstract
Conventional genetics assumes common variance among alleles or genetic groups. However, evidence from vertebrate and invertebrate models suggests that residual genotypic variance may itself be under partial genetic control. Such a phenomenon would have great significance: high-variability alleles might confound the detection of “classically” acting genes or scatter predicted evolutionary outcomes among unpredicted trajectories. Of the few works on this phenomenon, many implicate sex in some aspect of its control. We found that female genetic hypercalciuric stone-forming (GHS) rats (Rattus norvegicus) had higher coefficients of variation (CVs) for urinary calcium (CV = 0.14) than GHS males (CV = 0.06), and the reverse in normocalciuric Wistar–Kyoto rats (WKY) (CV♂ = 0.14; CV♀ = 0.09), suggesting sex-by-genotype interaction on residual variance. We therefore investigated the effect of sex on absolute-transformed residuals in urinary calcium in an F2 GHS × WKY mapping cohort. Absolute residuals were associated with genotype at two microsatellites, D3Rat46 (RNO3, 33.9 Mb) and D4Mgh1 (RNO4, 84.8 MB) at Bonferroni thresholds across the entire cohort, and with the microsatellites D3Rat46, D9Mgh2 (RNO9, 84.4 Mb), and D12Rat25 (RNO12, 40.4 Mb) in females (P < 0.05) but not males. In GHS chromosome 1 congenic lines bred onto a WKY genomic background, we found that congenic males had significantly (P < 0.0001) higher CVs for urinary calcium (CV = 0.25) than females (CV = 0.15), supporting the hypothesis of the inheritance of sex-by-genotype interaction on this effect. Our findings suggest that genetic effects on residual variance are sex linked; heritable, sex-specific residuals might have great potential implications for evolution, adaptation, and genetic analysis.
Keywords: calcium, sex, phenotypic dispersion, QTL, rat
HOMOGENEITY of variance among genotypes, strains, or genetic groups has been a major assumption of quantitative genetic analysis and evolutionary theory (see Falconer and Mackay 1996; Lynch and Walsh 1998) for nearly 100 years (Fisher 1922). However, a number of vertebrate and invertebrate model systems suggest the possibility of genetic variance in residual variability or phenotypic heterogeneity: Drosophila experiments in the 1950s found twice the variance for body size in homozygote flies compared to heterozygotes (Reeve and Robertson 1953) and increasing phenotypic variance for both low- and high-selected lines selected for abdominal bristle number (Clayton and Robertson 1957). Reaction time in attention deficit/hyperactivity disorder (ADHD) (see Castellanos et al. 2005; Russell et al. 2006) shares familial effects with intra-individual variability (IIV) in reaction time, suggesting a genetic basis for residual variation (Andreou et al. 2007; Wood et al. 2009). Genetic effects on residual variance has been observed in several systems including rodent genetic models of ADHD (Perry et al. 2010a,b), associations of genotype with thermotolerance in fish (Perry et al. 2003), and abdominal bristles in Drosophila (Mackay and Lyman 2005). The basis of this phenomenon is unknown: genetic structure in residual variance might be from the inability of homozygotes to buffer their physiology against microfluctuations in external environment (genetic homeostasis) (Lerner 1977) or from genotype/allele-specific variance functions, much as allele-specific means (termed here allelic heterogeneity; see Hill and Zhang 2004). Numerous examples of this genetic phenomenon explicitly involve sex: operant behavior in rats (Perry et al. 2010a), sex-chromosomal × autosomal interaction for thermotolerance (Perry et al. 2003), clonal lines and bristle number in Drosophila (Clayton and Robertson 1957; Mackay and Lyman 2005), and gene expression in mice (Fraser and Schadt 2010). Sex may be a common element in this effect; buffering of environmental effects on genetic systems may be a predominantly female physiological process (Fraser and Schadt 2010).
Nephrolithiasis (kidney stones) is a common condition in industrialized countries, affecting up to 12% in men and 6% of women and having a recurrence of up to 10% (Devuyst and Pirson 2007). The most common risk factor for nephrolithiasis is hypercalciuria, the excess excretion of urinary calcium (Coe et al. 2005). We have used the genetic hypercalciuric stone-forming rat (GHS), a model of this phenotype developed from Sprague–Dawley (SD) rats selected for urinary calcium excretion (Li et al. 1993; Bushinsky et al. 1995, 2002, 2006; Levy et al. 1995; Krieger et al. 1996; Tsuruoka et al. 1997; Yao et al. 1998; Frick and Bushinsky 2003), to map two quantitative trait loci (QTL) for urinary calcium excretion using F2 intercrosses of GHS rats and normocalciuric Wistar–Kyoto (WKY) rats (Hoopes et al. 2008): HC1 (on rat chromosome (RNO) 1; (Hoopes et al. 2003), and HC2 (RNO4) (Scheinman et al. 2008), both of which were detected only in females. These genes are underlain by an array of genes associated with calcium physiology (Hoopes et al. 2006). However, further examination of Hoopes et al. (2003) revealed that female GHS rats also had a variance 40 times and a coefficient of variation (CV = σ/µ) 2.3 times that of male GHS rats and that nearly the reverse was true in male and female WKY progenitors, implicating genotype-by-sex interaction in the generation of random variation. Further, there was increasing generation-to-generation variability in urinary calcium over the selective development of the GHS line, a pattern suggestive of increasing variation in selection lines of Drosophila bred by Clayton and Robertson (1957) (see Hill and Bunger 2004; Hill and Zhang 2004). The architecture of urinary calcium and stone formation differs markedly between the sexes in mammals: males and females have different urinary calcium in rats (Hoopes et al. 2003) and humans (Coe 1988; Curhan et al. 1997), which have more stones than females (Monk and Bushinsky 2003; Alaya et al. 2010).
We hypothesized that sex would have a significant effect on genetic dispersion in calcium excretion in our rat model. Specifically, we hypothesized that: (i) such dispersion would be associated with particular microsatellite markers (individual locus model) rather than overall inbreeding coefficients (genetic homeostasis), (ii) sex would affect dispersion in calcium excretion, and (iii) the effects of QTL for individual phenotypic dispersion (PDi) in calcium excretion would be sex linked. We tested these hypotheses using residuals and heterozygosity analysis in 236 male and female F2 GHS × WKY rats genotyped at 176 microsatellites and annotational gene expression analysis in strain progenitors.
Materials and Methods
Husbandry and experimentation
Calcium excretion in the GHS, selectively bred from SDs over 70 generations (D. Bushinsky, University of Rochester), is eight times that of normocalciuric WKY and SD controls (Bushinsky and Favus 1988; Bushinsky et al. 2006). F1 GHS × WKY rats were bred from GHS females and WKY males, since mean urinary calcium and molecular differentiation were greatest between these groups (Hoopes et al. 2003). Male and female F2 rats were then bred from F1 rats for the mapping of QTL for calcium excretion. GHS, WKY, F1, and F2 rats were subjected to the same protocol for the assay of urinary calcium excretion: at 8 weeks of age, each rat was placed in a separate metabolic cage (Acme Metal Products, Chicago, IL) and fed 13 g/day of a normocalciuric rat diet (0.6% calcium, 0.65% phosphorus, 0.24% magnesium, 0.40% sodium, 0.43% potassium, 2.2 IU vitamin D3/g food) for 4 days. Rats consuming <12 g food or 15 ml water on any of the 4 days were removed from the experiment. Individual 24-hr urine collections on the fourth day of the protocol were stored in 12 M hydrochloric acid and urinary calcium assayed from photometric absorbance after reaction of the total sample with arsenazo III (Michalylova and Ilkova 1971) (see Bushinsky and Favus 1988; Kim et al. 1993; Li et al. 1993; Bushinsky et al. 1995, 2002; Tsuruoka et al. 1997). We produced three cohorts of F2 rats: March 1997, November 1998, and January 2001.
Genetic analysis
Whole-genomic DNA was extracted from frozen (−70°) liver tissue (Puregene DNA Isolation Kit, Gentra Laboratories, Minneapolis, MN). We PCR amplified 255 microsatellites on a PCT-200 (MJ Research, Waltham, MA) in the F2 rats using run parameters from Research Genetics (now Invitrogen). Initially, microsatellites were selectively genotyped within date-cohort (∼20% of the upper and lower tails for calcium excretion); those with interesting associations with urinary calcium were selected for further genotyping. DNA fragments were separated on either 6% acrylamide sequencing gels or 4% agarose gels (see Hoopes et al. 2003).
Residuals by sex and genotype
All statistics were run in SAS (2000). We calculated the mean (µ), standard deviation (σ), variance (σ2), and coefficient of variation (CV) within each sex in the F2 mapping cohort.
We estimated the residual deviation (r) of individual F2 rats from their predicted phenotype as defined by the independent variables in the general linear model
| (1) |
where was day 4 urinary calcium excretion, μ mean calcium excretion, αi microsatellite locus, rat litter, rearing date of the cohort, φs effect of sex, effect of rat weight, and the residual. These independent variables are associated with urinary calcium excretion in this F2 cohort (Hoopes et al. 2003). Residuals were absolute transformed for analysis; absolute residuals (|r|), as the absolute deviation from all independent variables, were considered to represent residual phenotypic instability (PDi). Since litter and rearing cohort were partially confounded, we used mean phenotype for each level of each term as a quantitative variable rather than a factorial. The locus term was included in the model as a control against mean urinary excretion among genotypes at known and unknown classically acting QTL for calcium excretion (Hoopes et al. 2008). Next, to determine the effect of sex on PDi, we used a reduced version of the above fitting no effect for sex, but modeling the effects of sex separately,
| (2) |
where was day 4 urinary calcium excretion, μ mean calcium excretion, αi microsatellite locus, ϕj rat litter, rearing date of the cohort, effect of rat weight, and the residual. Only loci for which at least 66% of all individuals had been genotyped were included in either model analysis to limit false positives from small n.
We then tested for significant differences among genotypes for PDi in model 1 and Model 2 using nonparametric Kruskal–Wallis (K-W) ranking via χ2 (SAS 2000) with microsatellite locus as the independent variable to avoid bias from the altered phenotypic structure; K-W ranking was preferred to avoid complications of the distribution under linear statistics. Mean PDi and CV and σ2 for untransformed calcium excretion were estimated by genotype within each microsatellite locus; t-tests with Bonferroni correction were used to separate differences in PDi among genotypes (SAS 2000). Mode of action (additivity, dominance, or over-/underdominance) at each putative QTL was estimated using general linear models with contrast statements in SAS (2000) based on heterozygote potence ratio (hp = Q/L) (Griffing 1990), where L was the linear difference between the parental (GHS, WKY) genotypes (L = |µP1 – µP2|) and Q the difference of twice the mean quadratic heterozygote effect (Q = 2F1 – L). Contrast vectors of −1 0 1 were fit for WKY, heterozygote and GHS genotypes to calculate L. Contrast vectors were +/−0.5 +/−0.5 −/+1 for positive/negative WKY allele dominance (Lynch and Walsh 1998), −/+0.5 −/+0.5 +/−1 for GHS dominance, and +/−0.5 −/+1 +/−0.5 for under-/overdominance, respectively, for WKY, heterozygote, and GHS genotypes.
P-values at or below the Bonferroni-adjusted α0.05 threshold were considered to be significant evidence for rejection of the null hypothesis, and those at or below the sequential false discovery rate (FDR) (Verhoeven et al. 2005) as suggestive. FDR is less stringent than Bonferroni for family-wise error rate (FWER) but has less incidence of type II error (Holm 1979; Moran 2003; Verhoeven et al. 2005). In the FDR, empirical P-values (Pemp) from the m independent tests were ranked from P1 … Pm. Pi for each locus (i – m) was then tested against the partial inequality Pi ≤ αi/m, where α = 0.05 and i was the rank of that test based on Pemp in ascending order. Where P(i=emp) ≤ αi/m, that null hypothesis and those with lower Pi (ranked from 1 to P(i=emp) were rejected (Benjamini and Hochberg 1995; Benjamini et al. 2001; Verhoeven et al. 2005). The significance of all loci was also tested using permutational thresholds created from 1000 bootstrapped values in which phenotype had been randomized against all explanatory variables, creating a distribution of values ranked . values for each test on the above criteria were compared against this distribution; those higher than the fifth percentile of the distribution were considered to support the null hypothesis (Churchill and Doerge 1994) after correction for multiple testing. We tested whether sexual differences in expression at QTL for PDi in urinary calcium were scalar (similar PDi means genotypic means for males and females, but nonsignificant in one sex) (see Lynch and Walsh 1998), or whether crossovers in mean PDi occurred for the same genotype in males and females. For this test, we calculated PDi as above and used a subsequent model with a main effect for the microsatellite locus (αi) and an interactive term for sex-by-locus interaction (αiλj) for those loci with significant (Bonferroni) or suggestive (FDR) associations with PDi in urinary calcium. FDR thresholds were calculated separately for tests using all individuals (model 1), males, and females (model 2).
General differences in PDi by sex were tested using nonparametric ranking on absolute residuals from a model as above but without terms for sex or microsatellite locus. Sexual differences in PDi overall were then tested using a one-way nonparametric model (SAS 2000).
Multilocus heterozygosity
We regressed multilocus heterozygosity (MLH = p(homozygous loci)) for each chromosome RNO1-X, and genome-wide MLH from loci on all chromosomes, on PDi from a general linear model including body weight, litter, sex, and cohort date as a rough test of the role of genetic homeostasis on dispersion (“heterozygosity–trait correlation” (Hansson and Westerberg 2002) in the complete F2 cohort. MLH-PDi regression was also modeled separately within males and females to determine whether MLH-PDi associations differed by sex for comparison to results from model 2, above. P-values by chromosome were Bonferroni corrected for multiple testing (Verhoeven et al. 2005).
Gene expression
We screened gene expression in kidney and duodenal tissue collected from three female GHS and three female SD rats; the latter strain was chosen to represent the unselected state of the GHS strain comparable to the GHS save for selective history. As the sites of calcium uptake and excretion (Moe and Bonny 2005), renal and duodenal tissue was sampled from GHS and SD rats immediately post-mortem. RNA analysis was carried out at the University of Rochester Functional Genomics Center (http://www.urmc.rochester.edu/fgc/index.cfm) according to Affymetrix protocols (Affymetrix 2009). RNA was extracted with Trizol (Invitrogen) and purified by column chromatography (Qiagen RNeasy kit) with DNase treatment. RNA quality was determined using an Agilent 2100 Bioanalyzer. Affymetrix kits were used to produce cleaned biotinylated complementary RNA (cRNA). cRNA was hybridized for 17 hr to Affymetrix Rat Genome 230 GeneChips (Affymetrix, Santa Clara, CA) in 300-µl cocktails at 42°. All R230A chips were washed in a GeneChip Fluidics Station 450 and subsequently scanned in a GeneChip Scanner 3000 7G. We ignored all fluorescence values below a signal of 50 fluorescence units as possibly erroneous.
To identify genes with high variance in expression, CV was calculated for each gene amplified in the R230A array panel within strain and the ratio of CVGHS/CVSD (= CVR) calculated to account for integral variance in gene expression using CV from the unselected SD strain. We tested for gene function enrichment by annotation for CVR at those genes with nonerroneous signal (as interpreted above) within a 20-Mb (10 Mb upstream and downstream) window around our significant and suggestive QTL. The SNP probe Affymetrix identifications of all genes within these windows were submitted to the Database for Annotation,Visualization and Integrated Discovery (DAVID v. 6.7) (Huang et al. 2009). This program groups genes by biological function and tests for significant enrichment according to functional annotation and physiological relationship to other genes on the basis of published gene annotation lists (i.e., Cicala et al. 2002). P-values from Fisher’s exact tests of enrichment proportion were Benjamini adjusted for multiple testing in DAVID to control family-wise error rate; an adjusted threshold of P0.05 was considered significant. “Fuzzy functional annotation clustering” in DAVID was also used to identify modular gene clusters with enrichment in variance for gene expression according to annotation (Huang et al. 2009).
Results
Genetic dispersion
Variance in urinary calcium excretion:
Female F2 rats had significantly higher PDi than males in the F2 cohort (χ2 = 5.23, P = 0.0221, µ♀ = 0.91 ± 0.12 mg/day, µ♂ = 0.70 ± 0.13 mg/day). Variance and coefficients of variance were considerably higher in the F2 rats (σ2 = 1.59, CV = 0.55) than in the WKY and GHS progenitors (see Hoopes et al. 2003).
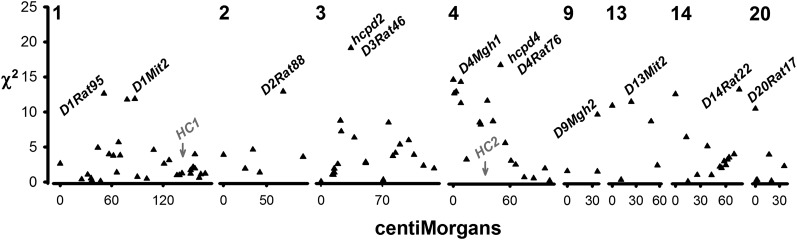
We identified significant genotypic differences (P < 0.05) in PDi at the microsatellites D3Rat46 (RNO3, 33.9 Mb) and D4Rat76 (RNO4, 84.8 Mb) over the entire population after Bonferroni correction (Table 1 and Figure 1). We considered these to represent QTL for PDi in urinary calcium excretion, here termed hcpd1 (linked to D3Rat46) and hcpd2 (linked to D4Rat76) (Table 1). Contrast analysis indicated that hcpd1 was underdominant (P < 0.001); σ2 in untransformed urinary calcium excretion in WKY homozygotes was three times that in WKY/GHS heterozygotes and twice that of GHS homozygotes (Table 1). Hcpd2 was additive (P < 0.001); PDi for urinary calcium excretion was significantly higher for WG heterozygotes than WW homozygotes (P < 0.01), and higher in GG homozygotes than WG heterozygotes (P < 0.05; Table 1). CVs for urinary calcium within genotypes strongly resembled results from PDi, although hcpd2 appeared more additive at D4Rat76 (Table 1). Hcpd2 was overlapped by the very wide (∼90 Mb) 95% Bayesian CI for the conventionally acting QTL for hypercalciuria HC2 (43.4 Mb) (Hoopes et al. 2003; Scheinman et al. 2008), although physically somewhat distant from it (D4Mgh1, 17.6 Mb; hcpd2, 84.8 Mb). No other locus associated with PDi in this study was linked to any classically acting QTL for hypercalciuria. PDi at each locus was statistically independent of all others, with the exception of D4Mgh1 and hcpd2 (P < 0.001, Spearman rank and Pearson tests) (SAS 2000), although the mode of action at these two loci differed (overdominant vs. additive; Table 1).
Table 1 . QTL effects linked to simple sequence length polymorphisms (SSLP) for urinary calcium excretion residuals in a cohort of 236 F2 genetic hypercalciuric stone-forming (GHS) × Wistar–Kyoto (WKY) rats at the 5% (α0.05) and 10% (α0.10) false discovery rate (FDR; Verhoeven et al. 2005).
| Mbp (cM) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Locus | Region | n | () | () | () | P | r2 | Ar | Pcont |
| D1Rat95 | 66.8 (51) | 160 | 0.75 ± 0.19 | 0.60 ± 0.16a** | 0.98 ± 0.19a** | 0.0018 | 5.73 | UD | 0.0031 |
| 1p11 | 1.43 (52.1) | 1.03 (48.8) | 2.46 (61.3) | ||||||
| D1Mit2 | 135.0 (87) | 236 | 0.78 ± 0.18 | 0.70 ± 0.12a** | 1.05 ± 0.17a** | 0.0026 | 4.06 | DW | 0.0031 |
| 1q31 | 1.17 (47.7) | 1.18 (47.8) | 2.79 (61.2) | ||||||
| D2Rat88 | 222.3 (69) | 178 | 0.84 ± 0.21 | 1.08 ± 0.15a** | 0.67 ± 0.20a** | 0.0016 | 5.25 | OD | 0.0023 |
| 2q42 | 1.38 (47.9) | 2.69 (61.5) | 1.07 (49.1) | ||||||
| D3Rat46 | 33.9 (34) | 231 | 1.09 ± 0.18a** | 0.65 ± 0.12a** | 0.84 ± 0.17 | <0.0001 | 6.03 | UD | 0.0007 |
| 3q12 | 2.90 (63.8) | 0.95 (43.8) | 1.52 (53.4) | ||||||
| D4Mgh1 | 17.6 (8) | 236 | 0.58 ± 0.16a** | 0.94 ± 0.12a** | 0.73 ± 0.19 | 0.0008 | 6.01 | OD | 0.0012 |
| 4q12 | 0.75 (44.0) | 2.16 (56.8) | 0.96 (40.8) | ||||||
| D4Mit2 | 55.4 (36) | 236 | 0.58 ± 0.17a* | 0.82 ± 0.12 | 0.92 ± 0.17a* | 0.0029 | 3.59 | DG | 0.0073 |
| 4q22 | 0.83 (51.6) | 1.61 (52.5) | 2.06 (46.9) | ||||||
| D4Rat76 | 84.9 (50) | 231 | 0.55 ± 0.17a**b*** | 0.79 ± 0.12a**c* | 1.01 ± 0.17b***c* | 0.0002 | 5.36 | AD | 0.0004 |
| 4q24 | 0.89 (45.1) | 1.44 (53.0) | 2.24 (52.4) | ||||||
| D9Mgh2 | 84.4 (36) | 197 | 0.86 ± 0.20 | 0.82 ± 0.14a** | 1.09 ± 0.22a** | 0.0079 | 2.21 | DW | 0.0267 |
| 9q35 | 1.46 (49.8) | 1.97 (61.1) | 2.11 (56.0) | ||||||
| D13Mit2 | 62.1 (24) | 197 | 0.65 ± 0.18a* | 0.96 ± 0.14a* | 0.99 ± 0.23 | 0.0032 | 5.21 | DG | 0.0024 |
| 13q21 | 2.20 (44.2) | 2.35 (61.1) | 2.80 (56.9) | ||||||
| D14Rat22 | 102.5 (76) | 236 | 0.78 ± 0.18 | 0.73 ± 0.12a* | 1.04 ± 0.18a* | 0.0013 | 3.25 | DW | 0.0044 |
| 14q22 | 2.39 (43.7) | 2.30 (54.0) | 2.53 (58.7) | ||||||
| D20Rat17 | 3.4 (0) | 178 | 0.77 ± 0.20a* | 0.86 ± 0.15 | 1.16 ± 0.21a* | 0.0052 | 4.99 | DW | 0.0022 |
| 20q12 | 2.22 (49.8) | 2.43 (50.9) | 2.82 (67.9) | ||||||
Genomic location within the chromosome is indicated in megabase pairs and in centimorgans on our F2 GHS × WKY genomic map. Variance in untransformed urinary calcium excretion within genotypes (), coefficient of variation within genotypes (CVgeno), number of genotyped and phenotyped animals (n), adjusted P value (Padj) from the ratio of the empirical P value against the 5% FDR threshold, correlational coefficient (r2) from linear statistical analysis, and variance within genotype (WW = WKY homozygote, WG = WKY/GHS heterozygote, GG = GHS homozygote) for PDi in calcium excretion are given. Ar indicates the architecture (AD, additive; DW, WKY dominant; DG, GHS dominant; OD, overdominance; UD, underdominance) by SSLP locus and Pcont the significance of the contrast test associated with that architecture. All statistics were run in (SAS 2000). Means by genotype (µ) are given in milligrams per day. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Significant differences among genotypes for mean PDi.
Figure 1 .
scores from all tests of the association of PDi according to simple sequence length polymorphism (SSLP; microsatellite) locus position (cM). Names of microsatellite loci significantly associated with PDi at the 5% false discovery rate are indicated. The positions of the previously identified conventionally acting quantitative trait loci for urinary calcium excretion hypercalciuria 1 (HC1) and hypercalciuria 2 (HC2) (Scheinman et al. 2008) are indicated in gray type for comparison.
Genotype at six additional microsatellite loci was associated with PDi at the α0.05 FDR threshold: D1Mit2, D4Mgh1, D4Mit2, D9Mgh2, D13Mit2, and D14Rat22 (Table 1 and Figure 1). Several markers linked to D4Mgh1 spanning 8 cM ≈ 14.6 Mb (Scheinman et al. 2008) were associated with PDi at the 5% FDR; we consider our results to indicate eight loci for PDi at the 5% FDR (Table 1). PDi was not associated with rearing period, litter, weight, or sex at any microsatellite locus (Pavg > 0.4). At genotypes with high PDi (i.e., GHS homozygotes at D1Mit2, WKY/GHS at D4Mgh1) variance in untransformed calcium excretion was two to three times higher than that of the other genotypes.
Sex-limited QTL for dispersion:
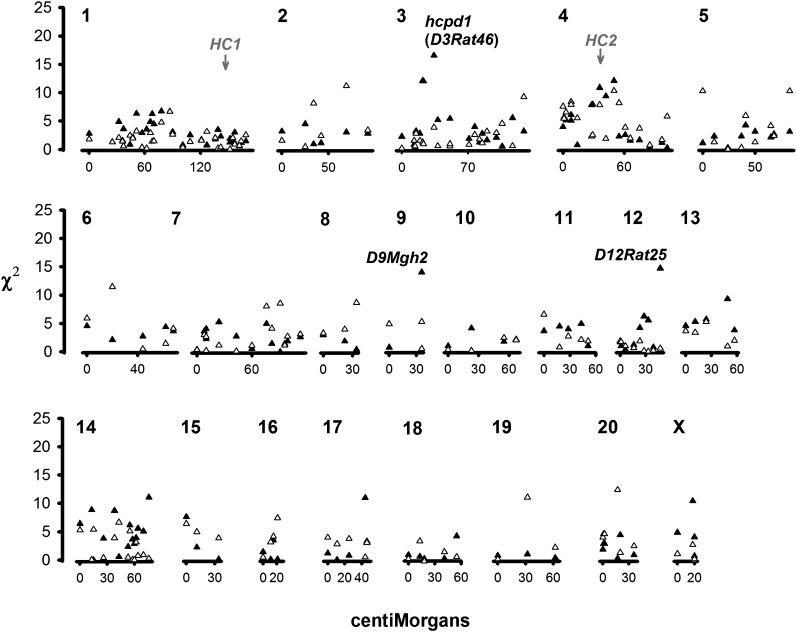
D3Rat46, which was linked to the QTL hcpd1 in the complete F2 cohort (above), was associated with PDi in urinary calcium excretion at the α0.05 Bonferroni threshold (Figure 2). At this position, female WKY homozygotes had significantly higher PDi than WKY/GHS heterozygotes after Bonferroni correction (P = 0.003; Figure 2 and Table 2). There was an a priori difference between GHS/WKY heterozygotes and WKY homozygotes in male F2s (P = 0.0486), but this locus was not significantly associated with PDi in males after multiple correction (P > 0.1). There was no evidence of significant interaction between D3Rat46 and the sex of the F2 rat after Bonferroni correction (P = 0.128). Contrast tests in SAS (2000) indicated that PDi at this locus was overdominant in females (F2, 125 = 8.62, P = 0.004).
Figure 2 .
χ2 scores from nonparametric testing of absolute residuals (phenotypic dispersion, PDi) for urinary calcium in male (solid triangles) and female (open triangles) female F2 GHS × WKY rats. The location of the significant quantitative trait locus (hcpd1) and the suggestively linked markers D9Mgh2 and D12Rat25 are indicated; also indicated are the significant QTL for classical mean differences in urinary calcium HC1 and HC2 (Scheinman et al. 2008).
Table 2 . Significant and suggestive QTL for PDi in urinary calcium excretion assayed at four microsatellites (Locus) in female F2 GHS × WKY rats.
| Locus | n | (CV) | (CV) | (CV) | P | Sig | r2 |
|---|---|---|---|---|---|---|---|
| D3Rat46 (hcpd1) | 125 | 3.76 (66.4) | 1.23 (45.8) | 1.58 (53.7) | 0.0002 | Bonf | 10.6% |
| D9Mgh2 | 105 | 1.15 (42.4) | 2.72 (68.5) | 2.61 (52.9) | 0.0009 | FDR | 10.7% |
| D12Rat25 | 101 | 3.49 (64.5) | 1.49 (55.0) | 0.983 (45.6) | 0.0007 | FDR | 16.9% |
Results significant at the α0.05 Bonferonni (Bonf) or FDR (Verhoeven et al. 2005) thresholds, the number of F2 rats (n), variances (σ2), coefficients of variance (CV) for each genotype, and empirical P-value are indicated. All statistical analysis was performed in SAS (2000). Proportions of PDi explained by microsatellite genotype (r2) were estimated using PROC VARCOMP (SAS 2000).
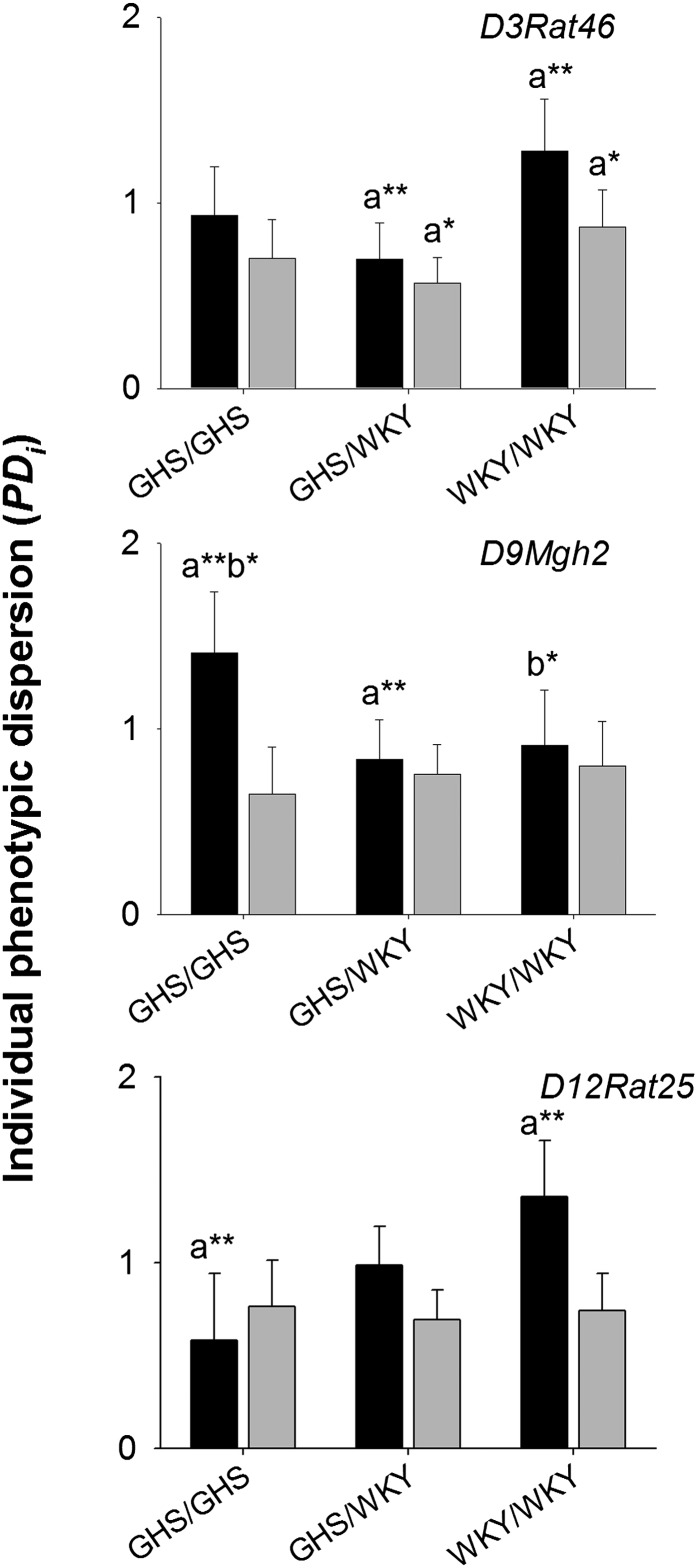
Two other loci—D9Mgh2, D12Rat25—were associated with PDi at the FDR in female F2 rats (P < 0.001; Figure 2 and Table 2). GHS homozygotes had significantly higher PDi than WKY/GHS heterozygotes (P = 0.0147) and WKY homozygotes lower PDi than GHS homozygotes in females at D9Mgh2 (P = 0.0873; Figure 3), which had negative dominance for the WKY allele (P = 0.0060). Contrast tests indicated additivity at D12Rat25 (P = 0.0018); WKY homozygotes had significantly higher PDi than GHS homozygotes (P = 0.0054; Figure 3 and Table 2). Neither of these loci were linked to PDi in male F2 rats (Figure 1). There was significant statistical interaction between sex and genotype at these microsatellite loci in the F2’s (D9Mgh2: F = 4.13, P = 0.0293; D12Rat25: F = 7.82, P = 0.0043). At D9Mgh2, female GHS homozygotes had significantly higher PDi than male GHS homozygotes (P = 0.012), while female WKY homozygotes had significantly higher PDi than male WKY homozygotes (P = 0.0225; see Figure 3). There was no significant sex–locus interaction for hcpd1 or hcpd2 (P > 0.3).
Figure 3 .
Mean individual phenotypic dispersion (PDi) in urinary calcium excretion at D3Rat46 (hcpd1), D9Mgh2, and D12Rat25 in female (solid bars) and male (shaded bars) F2 GHS × WKY intercrosses. Differences between means within sex are indicated as P < 0.05 (*), P < 0.01 (**).
Multilocus heterozygosity:
Whole-organismal multilocus heterozygosity (MLH) was not associated with residuals of calcium excretion (P > 0.8). MLH on RNO2 was marginally negatively association with residuals for calcium excretion (F = 3.16, P = 0.0766, β = -0.272 ± 0.300 |mg/day|) and MLH on RNO7 was marginally positively associated with calcium excretion residuals (F = 2.95, P = 0.0871, β = 0.251 ± 0.288 |mg/day|).
MLH was negatively associated with PDi in females on RNO3 (β = −0.639 ± 0.508, r2 = 0.0466, P = 0.0151), but positively associated with PDi at a priori significance in males on RNO15 (β = 0.303 ± 0.267, r2 = 0.0496, P = 0.0284) and 19 (β = 0.347 ± 0.276, r2 = 0.0534, P = 0.0157). None of these associations passed correction for multiple testing.
Gene expression
A total of 8846 renal genes and 9038 duodenal genes had an acceptable signal-to-noise ratio (>50) in the 230A Affymetrix R230A assay. Numerous genes with high CVs in expression in these regions were associated with acetylation, spliceosome formation, RNA modification, serine/threonine physiology, immunology, and metal-ion physiology (Supporting Information, File S1 and Table S1).
Renal genes with acceptable signal within 10 Mb of PDi QTL were enriched for acetylation (P < 0.05) and intracellular organelle lumen/organelle lumen (P < 0.05) functions by annotation (Table 3). Significant (α0.05) clusters of genes (EASE score (E) = geometric mean –log(P)) were identified for nuclear membranes/lumena (E = 2.91), mitochondrial (E = 1.87), protein catabolism (E = 1.52), and mitochondrial transit peptides (E = 1.39) using DAVID (Huang et al. 2009).
Table 3 . Fold enrichment (fER), percentage hit rate by annotation term, and Benjamini-corrected significance (PBenj) for renal and duodenal genes with significant (>50 fluorescence units) mean expression within 10 Mb of microsatellite loci associated with PDi in urinary calcium.
| Kidney | Duodenum | |||||
|---|---|---|---|---|---|---|
| Pathway | fER | % | PBenj | fER | % | PBenj |
| Intracellular organelle lumena | 1.56 | 14.5 | 0.0089 | 1.54 | 12.1 | 0.3809 |
| Organelle lumena | 1.59 | 15.3 | 0.0098 | 1.54 | 12.6 | 0.4985 |
| Membrane-enclosed lumena | 1.54 | 15.3 | 0.0129 | 1.50 | 12.6 | 0.3379 |
| Acetylationb | 1.46 | 17.8 | 0.0374 | 1.63 | 17.3 | 0.0115 |
Fluorescence means were calculated from three female Genetic Hypercalciuric Stone-forming and three Sprague-Dawley rats. Database of origin are indicated in the footnotes.
SP_PIR_KEYWORDS.
GOTERM_CC_FAT.
Duodenal genes with acceptable signal linked to PDi QTL were enriched for acetylation (P < 0.05) genes according to annotation (Table 3). Hit rates were relatively high for each group (∼15%). Annotational clusters identified from duodenal samples included organellar lumena (E = 2.04), ribosomal translation (E = 1.78), intracellular protein transport (E = 1.57), and those located in the endoplasmic reticulum (E = 1.46).
Sex and CV in RNO1 congenic rat lines
Because of our findings of differences in dispersion between male and female F2 rats, we further tested for sex differences in residuals in our subcongenic RNO1 rat line (Hoopes et al. 2003). These lines are descended from a single GHS male (Hoopes et al. 2003). Congenic rats used in this analysis had been retrocrossed to WKY animals for 6–11 generations in the process of fixation of WKY genetic background, including RNO1 outside the 95% critical region for HC1 (220–239 Mbp). WKY genotype was determined during this selection using microsatellite genotyping (Hoopes et al. 2003). Four-day CVs in urinary calcium were calculated for each congenic rat. Given the higher CV for urinary calcium in WKY males, and the concentration of WKY genotype in the congenic lines via retrocross, we predicted higher CVs for male rats in the congenic lines. Congenic rats were fed a high-calcium diet (1.2% Ca, 0.65% PO4); TD.90312.PWD; Harlan Teklad, Indianapolis) for 10 days and urinary calcium was measured daily (as above). We calculated the CV for urinary calcium for each congenic individual; male–female differences in mean CV were compared for each sex within each line and generation using nonparametric ranking and t-tests.
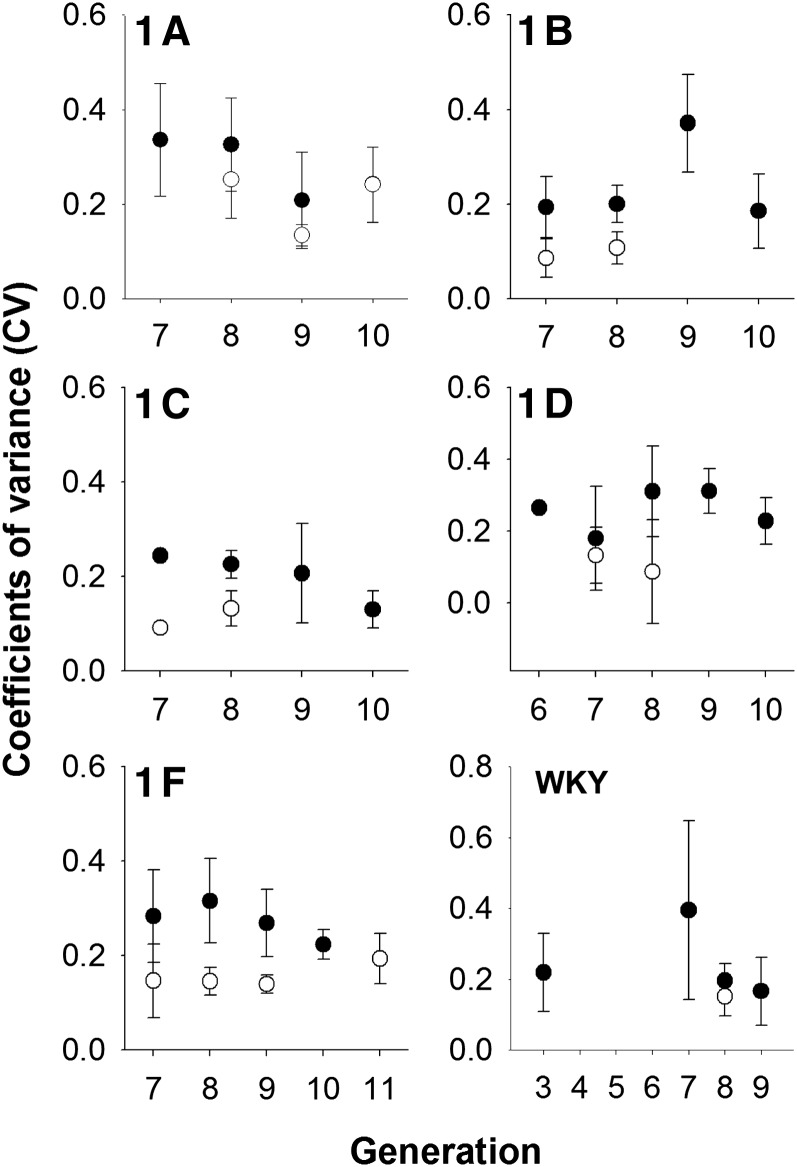
In 5 generations, 270 males and 61 female congenics were available. Males had significantly higher CV than females overall in the congenic population (F1,322 = 30.4, P < 0.0001, µM = 0.246 ± 0.0225, µF = 0.150 ± 0.0169) in multifactorial linear modeling and in nonparametric single-factor modeling (χ2 = 31.2, P < 0.0001). This effect was also detected in comparisons of CVs within subcongenic lines 1A–1F: of these lines, CV was significantly higher in males than in females in all lines (P < 0.05) except for line 1A, in which male CVs were only suggestively higher than those in females (P < 0.1) (Figure 4 and Table 4). There was no statistical difference between WKY males and females for urinary calcium excretion (P > 0.3) although urinary calcium was available only for a single generation of WKY females, and mean CVs for calcium excretion by sex in the WKYs were very similar to those in the other subcongenics, although not significantly different (µM = 0.238, µF = 0.158; Figure 4 and Table 4).
Figure 4 .
Coefficients of variation (CV) ± SE for 4-day urinary calcium excretion in male (●) and female (○) female chromosome 1 subcongenic lines 1A–1F and in WKY rats by generation.
Table 4 . Differences in CVs for urinary calcium excretion in male and female by line tested using Wilcoxon nonparametric one-way analysis.
| Line | N | χ2 | P | µM | µF |
|---|---|---|---|---|---|
| 1A | 39 | 2.84 | 0.0919 | 0.292 (0.0476) | 0.203 (0.0761) |
| 1B | 45 | 7.68 | 0.0056 | 0.217 (0.0331) | 0.106 (0.0853) |
| 1C | 42 | 5.82 | 0.0158 | 0.214 (0.0306) | 0.109 (0.0708) |
| 1D | 53 | 11.1 | 0.0008 | 0.266 (0.0478) | 0.123 (0.0911) |
| 1F | 66 | 8.87 | 0.0029 | 0.261 (0.0349) | 0.139 (0.0629) |
| WKY | 42 | 1.12 | 0.2905 | 0.238 (0.0670) | 0.158 (0.140) |
Mean CVs for calcium excretion in males (µM) and females (µF) (95% CI) are derived from calcium excretion across all generations within each subcongenic lines.
Discussion
Two microsatellite loci were significantly associated with absolute residual dispersion at Bonferroni-corrected significance thresholds across the entire population, which we consider evidence for QTL for residual variance in urinary calcium. The likelihood of all 11 tests significant at the 5% FDR being false positives was 9%, and 0.3% for all results significant a priori (see Moran 2003). MLH was not associated with dispersion. Allelic heterogeneity appears the most likely explanation for our findings (Hill and Bunger 2004; Hill and Zhang 2004). Our hypotheses of sex linkage on dispersion in urinary calcium are as follow: female F2 rats had higher dispersion in urinary calcium than males (P < 0.05), and QTL for phenotypic dispersion were detected only in female F2s (P < 0.05), while male WKY animals and male congenics in each RNO1 subcongenic line (being >1 – 0.56 ≈ 98% WKY genome) had significantly higher CVs for urinary calcium than females (P < 0.001), suggesting differences in sex linkage between GHS and WKY rather than sex limitation. Sex differences in the effect of hcpd1 (D3Rat46) and hcpd2 (D4Rat76) on dispersion were scalar, but there were significant male–female differences in dispersion patterns by genotype at D9Mgh2 and D12Rat25.
Sex may be the most fundamental contrast in biology (Andersson 1994; Gabory et al. 2009); there is widespread sexual dimorphism in gene expression (Waxman and Celenza 2003; Clodfelter et al. 2006; Yang et al. 2006; Wauthier and Waxman 2008) and intersexual genetic correlation is routinely less than unity (Chippindale et al. 2001; Stewart et al. 2010; van Doorn 2009). Previous work of ours indicates sex effects on the genetic structure of residuals for operant behavior in rats (Perry et al. 2010a) and sex chromosomal–autosomal interaction for thermotolerance in rainbow trout (Perry et al. 2003). Other work suggests the effects of sex on plasticity (Fernandez-Montraveta and Moya-Larano 2007; Stillwell et al. 2010) or on phenotypic residuals over environmental gradients (Mackay and Lyman 2005); however, we found no association of dispersion with rearing group or litter (P > 0.1). Various X-chromosomal peculiarities, including pseudoautosomal recombination and mutation (Lien et al. 2000; Lercher and Hurst 2002; Filatov and Gerrard 2003; Ross et al. 2005), variable X-chromosomal deactivation (Heard and Disteche 2006; Leeb and Wutz 2010) including among lineages (Patrat et al. 2009) might explain such effects (Charlesworth et al. 1987; Evans et al. 2007). Some molecular chaperones including hsp70, hsp90, and Piwi-interacting RNA canalize variability in morphological and over environmental and genetic axes (Gibert et al. 2007; Fernandino et al. 2011; Gangaraju et al. 2011). Work in mice suggests that environmental buffers of gene expression are sex specific (Fraser and Schadt 2010). Sex might also affect dispersion via simple hormonal fluctuations over estrus. All animals were used specifically at 8 weeks of age to minimize estral effects, which may have been common to litter and/or cohort. Other work of ours also indicates significant genetic effects on CVs for urinary calcium in males alone, suggesting a genetic basis for residual variability independent of estral cycling (Perry et al. 2011).
Gene expression
Enrichment for calcium excretion genes around QTL for untransformed urinary calcium (Hoopes et al. 2003) and for acetylation, luminal, and MHC II genes around our dispersion QTL suggests a firebreak between conventional genetic effects and those on variance; interesting, both conventionally acting QTL for calcium excretion in the F2 mapping cohort (HC1 and HC2) were detectable only in males (Scheinman et al. 2008; Perry et al. 2009). Lysine acetylation is a common (up to 90% of all Eukaryotic proteins (Glozak et al. 2005; Sadoul et al. 2008) modifier of histone and nonhistone proteins (Sadoul et al. 2008; Yang and Seto 2008) and may interact with other post-translation protein modification processes such as methylation and ubiquitination (Yang and Seto 2008). Such a ubiquitously active genetic pathway might explain the frequently encountered 10% CV in juvenile mammalian body size (see Hill and Zhang 2004). Several genes with high-expression CV linked to our dispersion loci are also known gene expression modifiers including several serine-rich protein genes, which are also involved in alternate splicing (Fu 1995). LSm5 is a member of the like-Smith protein gene family, ubiquitous in eukaryotes (Marz et al. 2008) and critically involved in spliceosome function by binding to U6 small nuclear RNA (snRNA) (Tkacz et al. 2008; Valadkhan 2010). Methylmalonic aciduria and homocystinuria type D protein (Mmadhc) is associated with methylation (Coelho et al. 2008; Radmanesh et al. 2008). Tra2a is critically involved with sexual differentiation in Drosophila (Hoshijima et al. 1991; Ryner and Baker 1991) and mammals (Tacke et al. 1998; Lieberman et al. 2001; Shiraishi et al. 2004) and promotes pre-mRNA splicing (Tacke et al. 1998).
Conclusions
Genes for residual variability might conceivably interfere with the detection, characterization, and localization of phenotype–genotype correlation by increasing type II error and reducing power due to increasing residual genetic variation. Sex-limited or sex-specific residual variance structures could create an entirely new level of analytical complexity, creating unsuspected genotypic architectures for seemingly classical phenotypes. Using our own example: hcpd2 is linked (16 cM; ∼40 Mb) to a classically acting QTL for hypercalciuria (HC2) in our study population (Perry et al. 2009). HC2 is partially sex limited to males and has an exceedingly wide 95% confidence interval (CI) (48 cM; ∼90 Mb (Scheinman et al. 2008); HC2 is also flanked by two positions associated with dispersion. HC1 was of equivalent effect but narrower CI (∼12 cM) and was not flanked by dispersion loci in our F2 population (Perry et al. 2009). Much of the theoretical correction for such effects might revolve around sex, requiring explicit inclusion of sex-specific phenotypic or genotypic heterogeneity. The fundamental construction of this phenomenon leaves no reason to believe that it should be confined to any particular system: such a phenomenon might theoretically affect any genetic analysis, although the risk would presumably be greatest for dispersion loci in linkage or linkage disequilibrium with more typical QTL. Practically, the identification of such effects would require either identification of genetic structure in residuals or the use of multiple measurements to account for genetic residuals. Dispersion genotypes could also promote escape from local fitness peaks by creating wider trait ranges over adaptive phenotypic landscapes (see Wright 1931), crossing of fitness troughs by liberating genotype–phenotype associations or even reducing sib competition by limiting overlap in phase space (Begon et al. 2006; Brigatti et al. 2007; Leone and Estevez 2008). Such a bet-hedging strategy might be more common in r-selected organisms; for example, juvenile coral growth rates are highly variable, but recruits to coral communities have stable growth functions (Vermeij 2006). Rapid evolution in changing environments is sometimes cited as an advantage of sex (Crow 1992); in addition to the exposure of deleterious mutations to selection (Maynard Smith 1978; Kondrashov 1988; Loewe and Hill 2010; Park et al. 2010), sex might also facilitate evolution by the liberation of residual variance. These suppositions remain largely untested: little is known of the potential statistical or natural state of such systems and further work is required to establish their role. Urinary calcium itself might have fitness relevance; calcium is a critical element in numerous physiological pathways including but not limited to signal transduction, membrane potential, cofactor activity, and bone physiology in addition to its function in muscular metabolism (Boron and Boulpaep 2011). Genetic dispersion in calcium metabolism might be a system for rapid reaction to changing ionic requirements, using bone as a stable reserve against physiological outlay; our findings suggest that dispersion systems might be common in evolutionary biology, active at many levels from large to small and integrally involved in the essentials of cellular physiology.
Supplementary Material
Footnotes
Communicating editor: D. W. Threadgill
Literature Cited
- Affymetrix2009. GeneChip® Expression Analysis Technical Manual: With Specific Protocols for Using the GeneChip® Hybridization, Wash, and Stain Kit. P/N 702232 Rev. 3. 180 pp. Available at: http://media.affymetrix.com/support/downloads/manuals/expression_analysis_technical_manual.pdf [Google Scholar]
- Alaya A., Nouri A., Najjar M., 2010. Changes in stone composition according to age and gender in Tunisian children. J. Pediatr. Urol. 6: 364–371 [DOI] [PubMed] [Google Scholar]
- Andersson M., 1994. Sexual Selection. Princeton University Press, Princeton, NJ [Google Scholar]
- Andreou P., Neale B., Chen W., Christiansen H., Gabriels I., et al. , 2007. Reaction time performance in ADHD: improvement under fast-incentive condition and familial effects. Psychol. Med. 37: 1703–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begon M., Townsend C., Harper J., 2006. Ecology: From Individuals to Ecosystems. Blackwell, Malden, MA [Google Scholar]
- Benjamini Y., Hochberg Y., 1995. Controlling the false discovery rate: a practical and powerful approch to multiple testing. J. R. Stat. Soc. Ser. A Stat. Soc. 57: 289–300 [Google Scholar]
- Benjamini Y., Drai D., Elmer G., Kafkafi N., Golani I., 2001. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 125: 279–284 [DOI] [PubMed] [Google Scholar]
- Boron W., Boulpaep E., 2011. Medical Physiology: A Cellular and Molecular Approach. Saunders, Philadelphia [Google Scholar]
- Brigatti E., Sa Martins J., Roditi I., 2007. Evolution of biodiversity and sympatric speciation through competition in a unimodal distribution of resources. Physica A 376: 378–386 [Google Scholar]
- Bushinsky D., Favus M., 1988. Mechanism of hypercalciuria in genetic hypercalciuric rats: inherited defect in intestinal calcium transport. J. Clin. Invest. 82: 1585–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushinsky D., Grynpas M., Nilsson E., Nakagawa Y., Coe F., 1995. Stone formation in genetic hypercalciuric rats. Kidney Int. 48: 1705–1713 [DOI] [PubMed] [Google Scholar]
- Bushinsky D., Asplin J., Grynpas M., Evan A., Parker W., et al. , 2002. Calcium oxalate stone formation in genetic hypercalciuric stone-forming rats. Kidney Int. 61: 975–987 [DOI] [PubMed] [Google Scholar]
- Bushinsky D., Frick K., Nehrke K., 2006. Genetic hypercalciuric stone-forming rats. Curr. Opin. Nephrol. Hypertens. 15: 403–418 [DOI] [PubMed] [Google Scholar]
- Castellanos F. X., Sonuga-Barke E., Scheres A., DiMartino A., Hyde C., et al. , 2005. Varieties of attention-deficit/hyperactivity disorder-related to intra-individual variability. Biol. Psychiatry 57: 1416–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., Coyne J., Barton N., 1987. The relative rates of evolution of sex chromosomes and autosomes. Am. Nat. 130: 113–146 [Google Scholar]
- Chippindale A., Gibson J., Rice W., 2001. Negative genetic correlation for adult fitness between sexes reveals ontogenetic conflict in Drosophila. Proc. Natl. Acad. Sci. USA 98: 1671–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill G., Doerge R., 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicala C., Arthos J., Selig S. M., Dennis G., Jr, Hosack D. A., et al. , 2002. HIV envelope induces a cascade of cell signals in non-proliferating target cells that favor virus replication. Proc. Natl. Acad. Sci. USA 99: 9380–9385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton G., Robertson A., 1957. An experimental check on quantitative genetical theory. II. The long-term effects of selection. J. Genet. 55: 152–170 [Google Scholar]
- Clodfelter K., Holloway M., Hodor P., Park S., Ray W., et al. , 2006. Sex-dependent liver gene expression is extensive and largely dependent upon signal transducer and activator of transcription 5b (STAT5b): STAT5b-dependent activation of male genes and repression of female genes revealed by microarray analysis. Mol. Endocrinol. 20: 1333–1351 [DOI] [PubMed] [Google Scholar]
- Coe F., 1988. Prevention and Treatment of Kidney Stones. U.S. Public Health Service, Bethesda, MD [Google Scholar]
- Coe F., Evan A., Worchester E., 2005. Kidney stone disease. J. Clin. Invest. 115: 2598–2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho D., Suormala T., Stucki M., Lerner-Ellis J. P., Rosenblatt D. S., et al. , 2008. Gene identification for the cblD defect of vitamin B12 metabolism. N. Engl. J. Med. 358: 1454–1464 [DOI] [PubMed] [Google Scholar]
- Crow J., 1992. An advantage of sexual reproduction in a rapidly changing environment. J. Hered. 83: 169–173 [DOI] [PubMed] [Google Scholar]
- Curhan G., Willett W., Rimm E., Stampfer M., 1997. Family history and risk of kidney stones. J. Am. Soc. Nephrol. 8: 1568–1573 [DOI] [PubMed] [Google Scholar]
- Devuyst O., Pirson Y., 2007. Genetics of hypercalciuric stone forming diseases. Kidney Int. 72: 1065–1072 [DOI] [PubMed] [Google Scholar]
- Evans A., Mena P., McAllister B., 2007. Positive selection near an inversion breakpoint on the neo-X chromosome of Drosophila americana. Genetics 177: 1303–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer D. S., Mackay T. F. C., 1996. Introduction to Quantitative Genetics. Longman Group LTD, Essex, England. [Google Scholar]
- Fernandez-Montraveta C., Moya-Larano J., 2007. Sex-specific plasticity of growth and maturation size in a spider: implications for sexual size dimorphism. J. Evol. Biol. 20: 1689–1699 [DOI] [PubMed] [Google Scholar]
- Fernandino J., Popesku J., Paul-Prasanth B., Xiong H., Hattori R., et al. , 2011. Analysis of sexually dimorphic expression of genes at early gonadogenesis of pejerrey Odontesthes bonariensis using a heterologous microarray. Sex Dev. 5: 89–101 [DOI] [PubMed] [Google Scholar]
- Filatov D., Gerrard D., 2003. High mutation rates in human and ape pseudoautosomal genes. Gene 317: 67–77 [DOI] [PubMed] [Google Scholar]
- Fisher R., 1922. On the mathematical foundations of theoretical statistics. Philos. Trans. R. Soc. A 222: 309–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser H. B., Schadt E. E., 2010. The quantitative genetics of phenotypic robustness. PLoS ONE 5: e8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick K., Bushinsky D., 2003. Molecular mechanisms of primary hypercalciuria. J. Am. Soc. Nephrol. 14: 1082–1095 [DOI] [PubMed] [Google Scholar]
- Fu X., 1995. The superfamily of arginine/serine-rich splicing factors. RNA 1: 663–680 [PMC free article] [PubMed] [Google Scholar]
- Gabory A., Attig L., Junien C., 2009. Sexual dimorphism in environmental epigenetic programming. Mol. Cell. Endocrinol. 304: 8–18 [DOI] [PubMed] [Google Scholar]
- Gangaraju V. K., Yin H., Weiner M. M., Wang J., Huang X. A., et al. , 2011. Drosophila Piwi functions in Hsp90-mediated suppression of phenotypic variation. Nat. Genet. 43: 153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibert J. M., Peronnet F., Schlotterer C., 2007. Phenotypic plasticity in Drosophila pigmentation caused by temperature sensitivity of a chromatin regulator network. PLoS Genet. 3: e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glozak M., Sengupta N., Zhang X., Seto E., 2005. Acetylation and deacetylation of non-histone proteins. Gene 363: 15–23 [DOI] [PubMed] [Google Scholar]
- Griffing B., 1990. Use of a nutrient-controlled experiment to test heterosis hypotheses. Genetics 126: 753–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson B., Westerberg L., 2002. On the correlation between heterozygosity and fitness in natural populations. Mol. Ecol. 11: 2467–2474 [DOI] [PubMed] [Google Scholar]
- Heard E., Disteche C., 2006. Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev. 20: 1848–1867 [DOI] [PubMed] [Google Scholar]
- Hill W., Bunger L., 2004. Inferences on the genetics of quantitative traits from long-term selection in laboratory or farm animals. Plant Breed. Rev. 24: 169–210 [Google Scholar]
- Hill W., Zhang X.-S., 2004. Effects on phenotypic variability of directional selection arising through genetic differences in residual variability. Genet. Res. 83: 121–132 [DOI] [PubMed] [Google Scholar]
- Holm S., 1979. A simple sequentially rejective multiple test procedure. Scand. J. Stat. 6: 65–70 [Google Scholar]
- Hoopes R., Jr, Reid R., Sen S., Szpirer C., Dixon P., et al. , 2003. Quantitative trait loci for hypercalciuria in a rat model of kidney stone disease. J. Am. Soc. Nephrol. 14: 1844–1850 [DOI] [PubMed] [Google Scholar]
- Hoopes R., Jr, Middleton F., Sen S., Hueber P., Reid R., et al. , 2006. Isolation and confirmation of a calcium excretion quantitative trait locus on chromsome 1 in genetic hypercalciuric stone-forming rats. J. Am. Soc. Nephrol. 17: 1292–1304 [DOI] [PubMed] [Google Scholar]
- Hoopes R. J., Perry G., Reid R., Hueber P., Sen S., et al. , 2008. Novel sex-specific QTL effects on hypercalciuria in the genetic stone-forming rat map to chromosomes 1 and 4. Renal Week 2008, Philadelphia [Google Scholar]
- Hoshijima K., Inoue K., Higuchi I., Sakamoto H., Shimura Y., 1991. Control of doublesex alternative splicing by transformer and transformer-2 in Drosophila. Science 252: 833–836 [DOI] [PubMed] [Google Scholar]
- Huang D., Sherman B., Lempicki R., 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Kim M., Sessler N. E., Tembe V., Favus M. J., Bushinsky D. A., 1993. Response of genetic hypercalciuric rats to a low calcium diet. Kidney Int. 43: 189–196 [DOI] [PubMed] [Google Scholar]
- Kondrashov A., 1988. Deleterious mutations and the evolution of sexual reproduction. Nature 336: 435–440 [DOI] [PubMed] [Google Scholar]
- Krieger N., Stathopoulos V., Bushinsky D., 1996. Increased sensitivity to 1,25(OH)2D3 in bone from genetic hypercalciuric rats. Am. J. Physiol. 271: C130–C135 [DOI] [PubMed] [Google Scholar]
- Leeb M., Wutz A., 2010. Mechanistic concepts in X inactivation underlying dosage compensation in mammals. Heredity 105: 64–70 [DOI] [PubMed] [Google Scholar]
- Leone E., Estevez I., 2008. Space use according to the distribution of resources and level of competition. Poult. Sci. 87: 3–13 [DOI] [PubMed] [Google Scholar]
- Lercher M., Hurst L., 2002. Human SNP variability and mutation rate are higher in regions of high recombination. Trends Genet. 18: 337–340 [DOI] [PubMed] [Google Scholar]
- Lerner I., 1977. Genetic Homeostasis. Oliver & Boyd, London [Google Scholar]
- Levy F., Adams-Huet B., Pak C., 1995. Ambulatory evaluation of nephrolithiasis: an update of a 1980 protocol. Am. J. Med. 98: 50–59 [DOI] [PubMed] [Google Scholar]
- Li X., Tembe V., Horwitz G., Bushinsky D., Favus M., 1993. Increased intestinal vitamin D receptor in genetic hypercalciuric rats: a cause of intestinal calcium hyperabsorption. J. Clin. Invest. 91: 661–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman A., Friedlich D., Harmison G., Howell B., Jordan C., et al. , 2001. Androgens regulate the mammalian homologues of invertebrate sex determination genes tra-1 and fox-1. Biochem. Biophys. Res. Commun. 282: 499–506 [DOI] [PubMed] [Google Scholar]
- Lien S., Szyda J., Schechinger B., Rappold G., Arnheim N., 2000. Evidence for heterogeneity in recombination in the human pseudoautosomal region: high resolution analysis by sperm typing and radiation hybrid mapping. Am. J. Hum. Genet. 66: 557–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewe L., Hill W., 2010. The population genetics of mutations: good, bad and indifferent. Philos. Trans. R. Soc. B Biol. Sci. 365: 1153–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Walsh B., 1998. Genetics and Analysis of Quantitative Traits. Sinauer Associates, Sunderland, MA [Google Scholar]
- Mackay T., Lyman R., 2005. Drosophila bristles and the nature of quantitative genetic variation. Philos. Trans. R. Soc. B Biol. Sci. 360: 1513–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marz M., Kristen T., Stadler P., 2008. Evolution of spliceosomal snRNA genes in metazoan animals. J. Mol. Evol. 67: 594–607 [DOI] [PubMed] [Google Scholar]
- Maynard Smith J., 1978. The Evolution of Sex. Cambridge University Press, Cambridge, UK [Google Scholar]
- Michalylova V., Ilkova P., 1971. Photometric determination of micro-amounts of calcium with arsenazo III. Anal. Chim. Acta 53: 194–198 [Google Scholar]
- Moe O., Bonny O., 2005. Genetic hypercalciuria. Am. Soc. Nephrol. 16: 729–745 [DOI] [PubMed] [Google Scholar]
- Monk R., Bushinsky D., 2003. Nephrolithiasis, pp. 1411–1425 Williams’ Endocrinology, edited by Larsen P., Melmed S., Polonsky K. WB Saunders, Philadelphia [Google Scholar]
- Moran M., 2003. Arguments for rejecting the sequential Bonferroni in ecological studies. Oikos 100: 403–405 [Google Scholar]
- Park A., Jokela J., Michalakis Y., 2010. Parasites and deleterious mutations: interactions influencing the evolutionary maintenance of sex. J. Evol. Biol. 23: 1013–1023 [DOI] [PubMed] [Google Scholar]
- Patrat C., Okamoto I., Diabangouaya P., Vialon V., Le Baccon P., et al. , 2009. Dynamic changes in paternal X-chromosome activity during imprinted X-chromosome inactivation in mice. Proc. Natl. Acad. Sci. USA 106: 5198–5203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry G., Ferguson M., Danzmann R., 2003. Effects of genetic sex and genomic background on epistasis in rainbow trout (Oncorhynchus mykiss). Genetica 119: 35–50 [DOI] [PubMed] [Google Scholar]
- Perry G., Bushinsky D., Hoopes R. J., Reid R., Hueber P., et al. , 2009. Genetic effects on residual variance for calcium excretion in a rodent mapping panel for hypercalciuria. American Society of Nephrology Renal Week, San Diego [Google Scholar]
- Perry G., Sagvolden T., Faraone S., 2010a Intra-individual variability in genetic and environmental models of attention-deficit/hyperactivity disorder. Am. J. Med. Genet. B 153B: 1094–1101 [DOI] [PubMed] [Google Scholar]
- Perry G., Sagvolden T., Faraone S., 2010b Intraindividual variability (IIV) in a rat model of ADHD: the spontaneously hypertensive rat. Behav. Brain Funct. 6: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry G., Nehrke K., Bushinsky D., Nandi J., Lewandowski K., et al. , 2011. Heritable residual variation for renal physiology?: a QTL for the coefficient of variation in urinary calcium, p. 97 Rat Genomics and Models, edited by Cuppen C., Hubner N., Kwitek A., Gould M. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- Radmanesh A., Zaman T., Ghanaati H., Molaei S., Robertson R., et al. , 2008. Methylmalonic acidemia: brain imagings in 52 children and a review of the literature. Pediatr. Radiol. 38: 1054–1061 [DOI] [PubMed] [Google Scholar]
- Reeve E., Robertson F., 1953. Analysis of environmental variability in quantitative inheritance. Nature 171: 874–875 [DOI] [PubMed] [Google Scholar]
- Ross M. T., Grafham D. V., Coffey A. J., Scherer S., McLay K., et al. , 2005. The DNA sequence of the human X chromosome. Nature 434: 325–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell V., Oades R., Tannock R., Killeen P., Auerbach J., et al. , 2006. Response variability in attention-deficit/hyperactivity disorder: a neuronal and glial energetics hypothesis. Behav. Brain Funct. 2: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryner L., Baker B., 1991. Regulation of doublesex pre-mRNA processing by 3′-splice site activation. Genes Dev. 5: 2071–2085 [DOI] [PubMed] [Google Scholar]
- Sadoul K., Boyault C., Pabion M., Khochbin S., 2008. Regulation of protein turnover by acetyltransferases and deacetylases. Biochimie 90: 306–312 [DOI] [PubMed] [Google Scholar]
- SAS, 2000 Statistical Analysis Software. SAS Institute, Cary, NC.
- Scheinman S., Perry G., Reid R., Hueber P., Sen S., et al. , 2008. Novel sex-specific effects on hypercalciuria in the genetic hypercalciuric stone forming rat map to chromosomes 1 and 4. Renal Week 2008 Philadelphia [Google Scholar]
- Shiraishi E., Imazato H., Yamamoto T., Yokoi H., Abe S., et al. , 2004. Identification of two teleost homologs of the Drosophila sex determination factor, transformer-2 in medaka (Oryzias latipes). Mech. Dev. 121: 991–996 [DOI] [PubMed] [Google Scholar]
- Stewart A., Pischedda A., Rice W., 2010. Resolving intralocus sexual conflict: genetic mechanisms and time frame. J. Hered. 101(Suppl. 1): S94–S99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillwell R. C., Blanckenhorn W. U., Teder T., Davidowitz G., Fox C. W., 2010. Sex differences in phenotypic plasticity affect variation in sexual size dimorphism in insects: from physiology to evolution. Annu. Rev. Entomol. 55: 227–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacke R., Tohyama M., Ogawa S., Manley J., 1998. Human Tra2 proteins are sequence-specific activators of pre-mRNA splicing. Cell 93: 139–148 [DOI] [PubMed] [Google Scholar]
- Tkacz I. D., Cohen S., Salmon-Divon M., Michaeli S., 2008. Identification of the heptameric Lsm complex that binds U6 snRNA in Trypanosoma brucei. Mol. Biochem. Parasitol. 160: 22–31 [DOI] [PubMed] [Google Scholar]
- Tsuruoka S., Bushinsky D., Schwartz G., 1997. Defective renal calcium reabsorption in genetic hypercalciuric rats. Kidney Int. 51: 1540–1547 [DOI] [PubMed] [Google Scholar]
- Valadkhan S., 2010. Role of the snRNAs in spliceosomal active site. RNA Biol. 7: 345–353 [DOI] [PubMed] [Google Scholar]
- van Doorn G., 2009 Intralocus sexual conflict. Ann. N. Y. Acad. Sci. 1168: 52–71 [DOI] [PubMed] [Google Scholar]
- Verhoeven K., Simonsen K., McIntyre L., 2005. Implementing false discovery control: increasing your power. Oikos 108: 643–647 [Google Scholar]
- Vermeij M., 2006. Early life-history dynamics of Caribbean coral species on artificial substratum: the importance of competition, growth and variation in life-history strategy. Coral Reefs 25: 59–71 [Google Scholar]
- Wauthier V., Waxman D., 2008. Sex-specific early growth response genes in rat liver. Mol. Endocrinol. 22: 1962–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman D., Celenza J., 2003. Sexual dimorphism of hepatic gene expression: novel biological role of KRAB zinc finger repressors revealed. Genes Dev. 17: 2607–2613 [DOI] [PubMed] [Google Scholar]
- Wood A., Asherson P., Rijsdijk F., Kuntsi J., 2009. Is overactivity a core feature in ADHD?: familial and receiver operating characteristic curve analysis of mechanically assessed activity level. J. Am. Acad. Child Adolesc. Psychiatry 10: 1023–1030 [DOI] [PubMed] [Google Scholar]
- Wright S., 1931. Evolution in Mendelian populations. Genetics 16: 97–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Seto E., 2008. Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol. Cell 31: 449–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Schadt E., Wang S., Wang H., Arnold A., et al. , 2006. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 16: 995–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J., Kathpalia P., Bushinsky D. A., Favus M. J., 1998. Hyperresponsiveness of vitamin D receptor gene expression to 1,25-dihydroxyvitamin D3: a new characteristic of genetic hypercalciuric stone-forming rats. J. Clin. Invest. 101: 2223–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.