Abstract
Notch is a receptor that mediates cell–cell interactions in animal development, and aberrations in Notch signal transduction can cause cancer and other human diseases. Here, I describe the major advances in the Notch field from the identification of the first mutant in Drosophila almost a century ago through the elucidation of the unusual mechanism of signal transduction a little over a decade ago. As an essay for the GENETICS Perspectives series, it is my personal and critical commentary as well as an historical account of discovery.
THE overarching theme of this essay is how genetic analysis illuminated the role of Notch in mediating cell–cell interactions during development, identified the core components of the signaling system, and elucidated the mechanism of signal transduction. However, I also found myself developing three other themes, which I will return to at the end in Concluding Remarks. One theme is the remarkable synergy that occurred between genetics and molecular biology—something that we take for granted today but had a revelatory feeling when the two approaches converged on animal development in the 1980s. Another theme is how scientific understanding is achieved. Finally, there is a coming-of-age theme about how model organisms came to occupy a prominent place in modern biology research, with an emphasis on Caenorhabditis elegans, my personal favorite.
1930s–1970s
The earliest alleles of Notch arose as spontaneous dominant mutations in fly stocks (see Mohr 1919). It was relatively easy to recover them because Notch is haploinsufficient in Drosophila: a deletion that removes Notch causes the eponymous notch-like indentations of the wing margin. Continuing work on Notch—beginning with one of the first characterized chromosomal deficiencies (Mohr 1919) through the 1970s—occurred primarily in the context of advancing concepts of the nature of genes. The many different kinds of alleles of Notch generated during this era became a treasure trove for molecular biologists when cloning and sequencing became possible in the 1980s.
Donald F. Poulson is generally regarded as the founding father of the Notch field in Drosophila, as he first described the hallmark phenotype of dying homozygous null Notch mutant embryos. These embryos display hypertrophy of the nervous system at the expense of ectoderm (later called the “neurogenic phenotype”; see Figure 1) as well as many abnormalities in non-neural tissues (Poulson 1939, 1940). It was a major advance of general significance when Poulson looked carefully at the anatomy of dead embryos and saw that smaller and smaller cytological deficiencies, and even mutations that did not result in cytological deficiency, caused discrete cell-fate transformations. Poulson may have been the first Drosophila geneticist forging important connections between genes and embryogenesis at a time when most Drosophila workers were focused on adult morphological mutants.
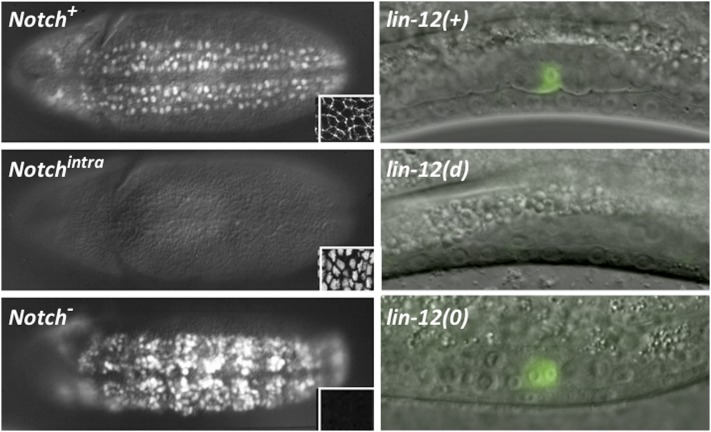
Figure 1 .
Photomicrographs showing wild-type and lin-12/Notch mutant phenotypes. (Left) Drosophila embryos. Neuroblasts are marked by anti-Hunchback staining, and insets show the results of staining with an antibody to the intracellular domain of Notch. (Top Left) A wild-type embryo showing the normal pattern of neuroblast segregation from the ventral ectoderm and Notch protein predominantly at the surface of ectodermal cells. (Middle Left) An embryo expressing Notchintra protein ubiquitously under heat-shock control: all ventral ectodermal cells remain ectodermal, and Notchintra protein accumulates predominantly in nuclei. (Bottom Left) A Notch− embryo: all ventral ectodermal cells segregate as neuroblasts, the classic “neurogenic phenotype.” (Right) C. elegans hermaphrodite gonads. Green fluorescent protein marks the anchor cell (AC) in a wild-type hermaphrodite, two ACs in a lin-12(0) hermaphrodite, and the lack of an AC in a lin-12(d) hermaphrodite. Drosophila photomicrographs courtesy of Gary Struhl; C. elegans photomicrographs courtesy of Maria Sallee.
Because effects on wing morphology and bristle number and spacing were an easy mark for early geneticists, many different kinds of alleles of Notch were identified, resulting in different effects on these adult traits. In the 1960s and 1970s, fly geneticists were focused on understanding the apparent complexity of the locus through studying the nature and interactions of different alleles, generally by describing the phenotypes of various trans-heterozygotes (Welshons and Von Halle 1962; Foster 1975; Portin 1975). These studies were more concerned with the genetic properties of alleles than with their normal roles in development, reflecting the prevalent preoccupation of that era—how the structure and organization of genes in animals could be related to those in microorganisms—at a time when the operon concept was still relatively new and enhancers and introns had not yet been discovered. One article from this era that stands out as more modern in its sensibility is that of Shellenbarger and Mohler (1975), who interpreted the results of temperature-shift experiments as indicating that some of the genetic complexity reflected different spatial and temporal functions for Notch during development.
1980s
The 1980s were shaped by the tremendous impact of molecular biology on the concept of the gene and how knowing gene products could lead to mechanistic insights. Some critical advances in the understanding of Notch that occurred during this decade were made using a new experimental organism, C. elegans. I will describe the major developments in the Notch story during this decade after a brief digression to introduce C. elegans and my own entry into the field during this period.
Enter C. elegans and the identification of lin-12/Notch
I was very fortunate to enter graduate school at the Massachusetts Institute of Technology (MIT) in 1977. Both the venue and timing were propitious. An MIT education meant being steeped in the classics of molecular biology. And, at that time, developmental genetics was beginning a remarkable and sustained log phase of growth and discovery: the impact of the instant-classic Drosophila articles such as Lewis (1978) and Nusslein-Volhard and Wieschaus (1980) was profound and increased immeasurably as the power of molecular biology began to be harnessed to genetics. And, fatefully, Bob Horvitz joined the MIT faculty during my first year, captivating me with the prospect of approaching developmental biology with the sensibility of a phage geneticist: C. elegans as an experimental system was expressly chosen as the metazoan analog of phage by Sydney Brenner (Brenner 1974), who was already a personal hero of mine from the classic molecular genetics articles that we read in class. Bob soon established an active group that included Victor Ambros, Edwin (Chip) Ferguson, Bill Fixsen, Paul Sternberg, and, a little later, Gary Ruvkun—fantastic colleagues as well as supportive friends.
I spent much of my graduate career honing my skills as a geneticist on intellectually pleasurable but rather arcane aspects of functional redundancy (Greenwald and Horvitz 1980, 1982, 1986). However, everyone else in the lab was studying cell-lineage mutants, and I wanted to do so, too.
At the time, the C. elegans larval lineage had been completed and found to be largely invariant (Sulston and Horvitz 1977; Kimble and Hirsh 1979). The lineage of the vulva, one of the main organs that develops in the larva, seemed to be exceptionally tractable to genetic analysis: the ability to cultivate C. elegans as a self-fertile hermaphrodite allowed mutants with vulval abnormalities or even lacking a vulva altogether to be readily obtained (Horvitz and Sulston 1980; see also the Perspectives by Horvitz and Sulston 1990). In addition, laser microbeam ablation experiments, using a system invented by John White, revealed that cell–cell interactions played a role in vulval development by showing that ablation of certain cells changed the fate of neighboring cells (Sulston and White 1980; Kimble 1981). These features made vulval development a powerful paradigm for genetic analysis of signaling systems—although I do not know if any of us realized just how spectacularly successful it would prove to be at the time.
The first alleles of lin-12 were dominant mutations that were isolated as part of Chip’s epic analysis of a large number of mutations affecting vulval development (Ferguson and Horvitz 1985; Ferguson et al. 1987). Chip had mapped several dominant mutations with different vulval phenotypes to a single chromosomal region. Provisionally, these mutations were assigned to a single locus, lin-12 (lin: abnormal cell lineage). When I was contemplating a lineage project, I was attracted to lin-12 because of its potential genetic complexity—like many of the fly geneticists who studied Notch.
Notch as a binary switch for cell-fate decisions mediated by cell–cell interactions
When I began to work on lin-12, I first generated null alleles [lin-12(0)] for phenotypic analysis as well as for classical gene dosage analysis to gain insight into the nature of the dominant mutations (Muller 1932). The gene dosage analysis revealed that the lin-12(d) mutations were hypermorphs, i.e., mutations that result in elevated gene activity. Thus, I had alleles in hand with opposite effects on gene activity to test the exciting prospect that lin-12 functioned as a “genetic switch” as did the genes that I had loved learning about for λ and the yeast mating type, and a paradigm that was also applied to the Drosophila homeotic genes (Lewis 1978; Struhl 1981).
The simple cellular anatomy and invariant cell lineage of wild-type hermaphrodites allows mutants to be understood in terms of altered cell-fate decisions by individual cells (Horvitz and Sulston 1980; Sulston and Horvitz 1981). When Paul Sternberg, with his encyclopedic knowledge of postembryonic worm anatomy and cell lineage, examined the different kinds of lin-12 mutants, he observed that the hypermorphic and null alleles had the opposite effect on cell fate in many different cell-fate decisions. Because opposite alterations in the level of lin-12 activity had opposite effects on these cell-fate decisions, we inferred that lin-12 indeed functions as a genetic switch (Greenwald et al. 1983)—the first switch gene described in the worm.
Paul identified many different cell-lineage alterations. Looking strictly at the lineage trees, it was interesting that lin-12 was acting late in a hierarchy to diversify it, making otherwise similar lineages different (Horvitz et al. 1983). However, in examining the lineage alterations of lin-12 mutants, what seemed most striking to me was that many of the cell-fate decisions altered in lin-12 mutants involved cell–cell interactions.
I will conclude this section by providing as an example a decision that I will come back to later. This decision occurs during hermaphrodite gonadogenesis and is the most striking exception to the general rule of the invariant lineage: in wild-type hermaphrodites, there are two cells in the hermaphrodite gonad, defined by their lineage history, with variable fates, suggesting that cell–cell interactions play a role in their specification (Kimble and Hirsh 1979). Each cell has the potential to be either an anchor cell (AC) or a ventral uterine precursor cell (VU); every wild-type hermaphrodite has a single AC (Kimble and Hirsh 1979) (Figure 1). When all other gonadal cells are ablated except for one of these two, the solitary cell always becomes an AC, indicating that cell–cell interactions are necessary for one of these cells to become a VU (Kimble 1981).
The role of the AC is to induce the vulva (Kimble 1981). In the lin-12(d) hypermorphic mutants, both of these cells become VUs, so the vulva is not induced. In null mutants, both become ACs (Figure 1). Similarly, many other cell-fate decisions displayed such reciprocal behavior, with the lin-12(d) hypermorphic and the lin-12(0) null alleles having opposite effects on cell fate (Greenwald et al. 1983) (Figure 1). Although we did not know at the time that LIN-12 was a Notch ortholog, these genetic data were the first demonstration of the binary nature of Notch-mediated decisions. Indeed, the equivalent experiment of oppositely altering Notch activity in Drosophila was achieved only when constitutively active forms could be engineered a decade later.
“Neurogenic genes” and a potential Notch pathway in Drosophila
While I was studying what turned out to be a C. elegans Notch, Jose Campos-Ortega and colleagues were screening the collection of Nusslein-Volhard and Wieschaus for other mutants with the classic Notch neurogenic phenotype. In a seminal article, Lehmann et al. (1983) described six other genes, including two that were critical to the elucidation of the Notch signal transduction pathway: Delta, subsequently shown to encode a ligand, and Enhancer of split [E(spl)], subsequently shown to be a direct transcriptional target.
Soon thereafter, landmark laser ablation experiments in the grasshopper demonstrated that cell–cell interactions influence neurogenesis in the embryonic ectoderm (Taghert et al. 1984; Doe and Goodman 1985). These observations, along with the molecular identification of Notch as a transmembrane protein (see below), suggested that the neurogenic genes together mediate these cell–cell interactions. However, because neuroblasts delaminate, another idea was that these genes mediate cell adhesion within the ectoderm, a view that I will return to below.
The screen of Lehmann et al. (1983) was the first of many other successful screens in Drosophila and C. elegans specifically for additional components of the Notch signaling system. I do not have enough space to describe them all, but suffice it to say that virtually every core component of the signaling system was first identified or first linked to Notch through powerful genetic screens in flies and worms, including, of course, Notch itself. This point will be apparent in the sections below on ligands, CSL proteins, and γ-secretase.
Linking lin-12 and Notch: the awesome power of molecular biology
Molecular biology revolutionized developmental genetics in the 1980s; the change was rapid and profound. Now we could learn the effect of mutation on the gene product itself and combine genetic approaches with the tools of molecular biology to test and understand mechanism as well as process. And only now would the extent of the conservation of developmental control genes become apparent.
The Drosophila Notch gene was a particularly attractive candidate for molecular analysis for both its genetic complexity and its involvement in neurogenesis. The cloning of Notch by “chromosomal walking” was reported by two groups (Artavanis-Tsakonas et al. 1983; Kidd et al. 1983) contemporaneously with the publication of cloning of members of the two most famous developmental gene complexes, the Antp and Ubx homeotic genes (Bender et al. 1983; Garber et al. 1983; Scott et al. 1983). The starting point for the walk (or “jump”) to Notch was an inversion breakpoint that had been characterized cytologically as juxtaposing Notch sequences with a previously cloned gene. Notch was subsequently sequenced independently by both groups and found to encode a transmembrane protein with repeated epidermal growth factor (EGF)-like motifs in the predicted extracellular domain as well as other repeated motifs (Wharton et al. 1985; Kidd et al. 1986).
As the fly people began succeeding in cloning their developmental genes, it became imperative that we C. elegans people had to clone ours, too. Victor Ambros and Gary Ruvkun encouraged me to try to clone lin-12 for my postdoctoral work at the Medical Research Council Laboratory of Molecular Biology in Cambridge, England. Again, I was fortunate in where I was training: there was no better place to do molecular biology, especially DNA sequencing and analysis. So, with the support and encouragement of my postdoctoral sponsor, Jonathan Hodgkin, and with a lot of help from many people, including my main molecular biology gurus, Bob Holmgren and Andy Fire, I began my quest to clone lin-12.
That I would succeed in cloning lin-12 was not a given. To give some context for the times: when I started my postdoctoral work in 1983, the remarkable C. elegans genome project was still at the stage of gridding the cosmid clones for physical mapping, and no one had yet cloned a C. elegans gene that had only been defined genetically.
Ideas of how to clone worm genes were widely being discussed in the field; most strategies were based on Tc1, an element with sequence features of a transposable element (Emmons et al. 1983; Liao et al. 1983). Tc1 is present in >300 copies in the Bergerac strain but in only ∼30 copies in the canonical Bristol strain; thus, one approach to cloning genes was to use Tc1 as a restriction-fragment length polymorphism (Files et al. 1983) to provide an entry point into a chromosomal walk. Another strategy was transposon tagging, based on the as-yet-unverified proposal that Tc1 transposition might be the molecular basis for a high spontaneous mutation rate in the Bergerac strain (Moerman and Waterston 1984). I tried both strategies in parallel; transposon-tagging, accomplished through the reversion of lin-12(d) by insertion of Tc1 into the locus, worked first. I decided not to publish the cloning on its own, instead waiting to see if sequence information might reveal something interesting. And it sure did—the presence of EGF-like motifs.
I reported the cloning and partial sequence analysis of lin-12 (Greenwald 1985) in the same issue of Cell as one of the two reports of the sequence of Notch (Wharton et al. 1985; Kidd et al. 1986). The copublication of sequence information about LIN-12 and Notch in Cell was no coincidence. As I recall, the sequence of Notch had been kept tightly under wraps, but the presence of EGF-like motifs in LIN-12 had been big news for several months and presented not just by me in several venues—I was on the job market!—but also by others at many meetings because it showed that worm developmental genes could be cloned and that “our” gene products had elements of homology to human proteins, too. I suppose that is how Benjamin Lewin, the founding editor of Cell, knew about my work and why he invited me to submit my article so as to come out back-to-back with Notch.
It is hard to capture the excitement that the EGF homology engendered, but it was truly thrilling. The homeobox had been identified only the year before (McGinnis et al. 1984; Scott and Weiner 1984), and the potential importance of vertebrate Hox genes for development was only just beginning to emerge through studying their expression (Carrasco et al. 1984; Hart et al. 1985). I think discovery of the EGF-like motifs in LIN-12/Notch was only the second time that sequence similarity had been reported between an important regulator of invertebrate development and a vertebrate gene. The fact that EGF had already been implicated as a key gene in mammalian development gave it particular resonance at a time when the meaning of the homeobox conservation was still mysterious.
The domain organization of Notch proteins in animals from hydra to human is shown in Figure 2. Fortunately, Kathleen Weston, a graduate student working on cytomegalovirus and a sequencer par excellence, became interested in lin-12 and rapidly sequenced and analyzed a draft sequence of much of the remaining coding region. From her work, we knew early on that LIN-12, like Notch, was a transmembrane protein as well and that the homology to Notch extended throughout the protein, even though we did not complete the full genomic and cDNA sequences until later (Yochem et al. 1988).
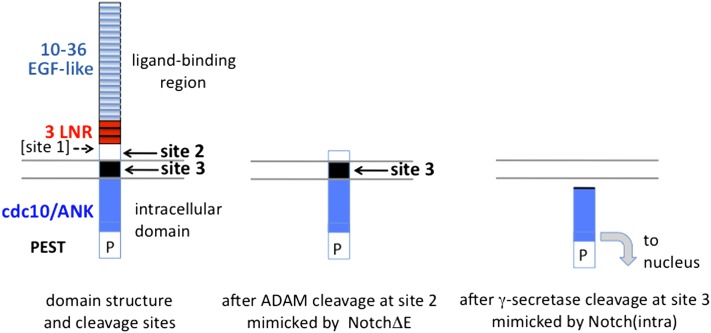
Figure 2 .
Notch domain organization, intermediates in signal transduction, and mimics that result in constitutive activity. Schematic domain structure of Notch proteins, showing epidermal growth factor (EGF)-like and LNR (LIN-12/Notch Repeat) motifs in the ectodomain and the cdc10/Ankyrin (ANK) repeat motifs in the intracellular domain. The PEST sequence influences the stability of the intracellular domain. The text details how the mechanism of signal transduction was determined. Cleavages at sites 2 and 3 are part of the mechanism of signal transduction. After ligand binding, exposure of site 2 allows for ADAM protease to cleave, resulting in ectodomain shedding. Site 2 cleavage can be mimicked by a truncated product that deletes much of the extracellular domain (center). Virtually any type 1 transmembrane protein with a short ectodomain can serve as a substrate for γ-secretase (Struhl and Adachi 2000). Thus, the cleavage of Notch at site 2 creates a substrate for γ-secretase cleavage at site 3, thereby releasing the intracellular domain for translocation to the nucleus and association with CSL for target gene activation. Site 3 cleavage can be mimicked by expression of the intracellular domain alone. Mammalian Notch is cleaved by Furin at site 1 during secretory trafficking, resulting in a heterodimer between the amino- and carboxy-terminal fragments (Blaumueller et al. 1997; Logeat et al. 1998). Drosophila Notch is not cleaved by Furin (Kidd and Lieber 2002), and it is not known whether the C. elegans Notch proteins LIN-12 and GLP-1 are cleaved. Site 1 cleavage is not regulated by ligand and hence is not part of the signal transduction mechanism per se.
Cell autonomy of lin-12 and Notch and feedback mechanisms in lateral “specification”
In 1986, I joined the Biology Department at Princeton, where I was treated as an honorary member of the incredible fly community; everyone, especially my senior colleagues, Eric Wieschaus, Tom Cline, and Paul Schedl, not only made me feel very welcome personally but also accepted the worm as a legitimate model organism without reservation and, in particular, accepted the relevance of my work on lin-12 to Notch. And compounding my great fortune, some outstanding students were willing to take the risk of joining my lab.
EGF had been discovered in the 1960s as a signal important in development; molecular cloning revealed that it was generated from a transmembrane protein precursor that also included multiple EGF-like motifs as well as bona fide EGF (Gray et al. 1983; Scott et al. 1983a). Thus, the presence of multiple EGF-like motifs in the extracellular domain made it conceivable that Notch functioned as a signal between cells. Alternatively, Notch might function as a receptor with its large, conserved intracellular domain mediating signal transduction. In addition, because neuroblasts delaminate from an ectodermal monolayer, some in the Drosophila community favored a model for Notch as a cell adhesion molecule, with reduction in Notch activity precipitating extrusion of neural precursor cells to allow them to receive signals inducing neural differentiation. Determining whether Notch functions nonautonomously in the signaling cell, autonomously in the receiving cell, or possibly in both cells would help differentiate among these possibilities.
In the Drosophila community, the question of Notch autonomy was controversial. Before my arrival at Princeton, my colleagues Hoppe and Greenspan (1986) analyzed gynandromorphs and suggested that Notch acts cell-autonomously in the embryonic neurectoderm, but the resolution of their mosaic analysis was severely limited by the technical constraints of available markers at that time, particularly the reliance on cuticular markers that did not allow a clear conclusion at the level of individual cells and their immediate neighbors. In contrast, in what initially seemed to be a more definitive test offering single-cell resolution, Technau and Campos-Ortega (1987) performed transplantation experiments using markers for neural differentiation that could be scored on a cell-by-cell basis; they concluded that Notch function was nonautonomous, reporting that cells that were transplanted from the neurogenic ectoderm of mutant donor embryos into wild-type host embryos could give rise to either neurons or epidermis. However, they lacked an independent marker for the genotype of the Notch(−) donor embryos produced from crossing heterozygotes, which now appears to have led to errors in inferring the donor genotypes. At the time, the limitations of the two studies and their contradicting conclusions left the question of Notch autonomy unsettled.
The time was ripe for addressing this question in C. elegans. Bob Herman had recently developed an elegant method for genetic mosaic analysis based on the spontaneous loss of free duplications (Herman 1984); Ed Hedgecock had identified a mutation that altered nucleolar morphology and allowed the genotype of individual cells in mosaics to be deduced (Hedgecock and Herman 1995); and Judith Austin and Judith Kimble had generated a free duplication that contained wild-type sequences for both lin-12 and ncl-1 (Austin and Kimble 1987; see also below).
Thus, my student Geraldine Seydoux had the means to generate genetic mosaics affecting a pair of adjacent cells of the somatic gonad that have a lin-12-mediated choice between the AC and VU fates, with lin-12 activity promoting the VU fate as described above (Greenwald et al. 1983). These two cells can be distinguished on the basis of their lineage histories and positions in the gonad primordium; each has a 50% chance of becoming the AC, and signaling between them specifies their fates (Kimble and Hirsh 1979; Kimble 1981; Seydoux and Greenwald 1989). The “AC/VU decision” is therefore an example of a process that has been classically called “lateral inhibition,” originally the neuroscience term for when an excited neuron reduces the excitability of its neighbors (see Meinhardt and Gierer 2000).
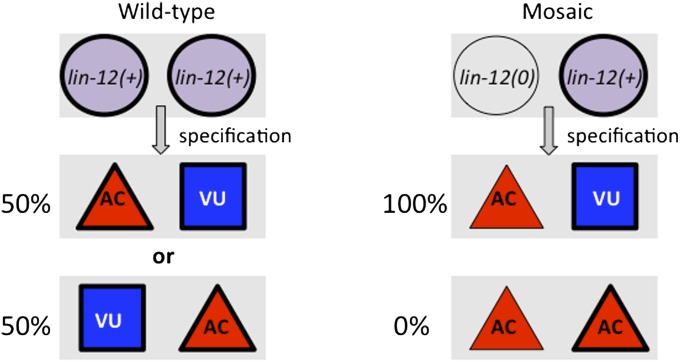
Geraldine screened for genetic mosaics in which one of the two cells was lin-12(0) and the other was lin-12(+) and examined how the fate of each cell correlated with its genotype. She found that the lin-12(0) cell always became an AC, indicating that lin-12 functions cell-autonomously to promote the VU fate (Figure 3).
Figure 3 .
The anchor cell (AC)/ventral uterine precursor cell (VU) decision, cell autonomy, and bias in cell-fate choice in genetic mosaics. See text for description.
With hindsight, it is clear that the more profound observation was that there is a nonautonomous aspect to lin-12 function as well. Geraldine found that in the mosaic situation the lin-12(+) cell no longer had a choice of fate and always became a VU (Figure 3). We were initially surprised by this observation because, in thinking of the AC/VU decision as lateral inhibition, we might have expected that 50% of the time the lin-12(+) cell would still become an AC through its intrinsic propensity to do so, regardless of its neighbor’s inability to be inhibited. However, the lin-12(+) cell always became a VU, suggesting that its fate was biased by the decision of its lin-12(0) neighbor, which would not be able to receive the VU-promoting signal and hence had no choice but to be an AC. From this bias in cell-fate choice of the lin-12(+) cell in these mosaics, we inferred the existence of a feedback mechanism that amplifies small stochastic initial differences in the level of lin-12 activity. Subsequently, my students Hilary Wilkinson and Kevin Fitzgerald found that the feedback mechanism involves transcriptional regulation of both lin-12 and the gene encoding its ligand in this decision, lag-2, in response to lin-12 activity, amplifying the small, stochastic initial difference (Wilkinson et al. 1994).
Although the “hot” issue at the time was the question of autonomy, the bias in cell-fate choice for the lin-12(+) cell, reflected in the nonautonomous effect on its fate, was more important in terms of a more general understanding of the decision-making process. Indeed, the key point is that the process is not simple “inhibition” because the cells communicate with each other to reach a consensus about which fates to adopt—the reason that I prefer the term lateral “specification” (Greenwald and Rubin 1992), although, alas, the term never caught on. The bias in cell-fate choice in mosaics also suggested that Notch functions in reception of a signal rather than as a passive cell adhesion molecule that facilitates other signaling events: if Notch were simply mediating adhesion, then the degree of adhesion between two cells should be reduced without causing a bias.
Findings in the worm were not regarded as general until Heitzler and Simpson (1991) published a beautiful mosaic analysis in flies that reached conclusions similar to ours. They analyzed mosaics affecting bristles on the notum, rather than the classic embryonic neurogenic phenotype. Bristle patterning was another classic paradigm for lateral inhibition: Curt Stern had shown that, when an achaete mutant clone eliminated a landmark bristle, if the clone border was near the bristle site, the nearby adjacent wild-type tissue could form a bristle, slightly displaced from the landmark site, suggesting that loss of the landmark bristle released a nearby epidermal cell from inhibition (Stern 1954). Subsequent studies led to the concept of a proneural cluster of cells, each having the potential to generate a sense organ precursor (SOP) for a bristle, with single SOPs specified as a result of lateral inhibition (Simpson 1990).
Heitzler and Simpson (1991) found that, in genetic mosaics containing Notch− and Notch+ cells within a proneural cluster, a Notch− cell always became an SOP—indicating autonomy of Notch function in epidermal specification—and a Notch+ cell always became epidermal, a bias in cell-fate choice similar to what we observed for the AC/VU decision. They went further by analyzing mosaic proneural clusters juxtaposing cells that differed only in the number of copies of Notch. They found that, when a cell with one copy of Notch+ and a cell with two copies of Notch+, or even two copies of Notch+ vs. three copies of Notch+, were juxtaposed, the cell with fewer copies generally became an SOP and the cell with more copies became epidermal. This result was quite astonishing and suggested that the feedback mechanism is both sensitive enough to detect small initial differences and accurate enough to ensure that the outcome invariably tracks with the relative level of Notch activity.
glp-1
During this decade, glp-1, a second C. elegans Notch gene, was also identified. glp-1 was defined genetically in two different screens: loss of zygotic function dramatically reduces germline proliferation and loss of maternal function alters early embryonic cell fate (Austin and Kimble 1987; Priess et al. 1987). These phenotypes, like the lin-12 phenotypes, reflect a failure of cell–cell interactions: germline proliferation depends on a signal from the soma (reviewed in Kimble and Crittenden 2005) and early embryonic development involves numerous successive glp-1-mediated inductive signaling events (reviewed in Priess 2005). Genetic mosaic analysis established that glp-1 functions autonomously in the germline to promote its proliferation (Austin and Kimble 1987), so when we identified another C. elegans Notch gene by low-stringency hybridization and found that it corresponded to glp-1, the interpretation that Notch functions in receiving cells was reinforced (Yochem and Greenwald 1989).
lin-12 and glp-1 appear to have arisen by a gene duplication event and are located very close together in the genome. When Lambie and Kimble (1991) made the heroic effort of constructing the lin-12 glp-1 double mutant, they found that concomitant loss of zygotic lin-12 and glp-1 activity causes larval arrest with novel cell-fate transformations, defining the “Lag” phenotype (for Lin-12 and Glp-1). Their results suggested that the two Notch proteins are functionally redundant, an inference supported by the finding that GLP-1 is able to substitute for LIN-12 in cell-fate decisions when expressed using lin-12 regulatory sequences (Fitzgerald et al. 1993). These studies together implied that different roles for Notch, as the mediator of inductive interactions or lateral specification, reflect different regulatory mechanisms rather than intrinsic differences in the Notch proteins themselves. The results of Lambie and Kimble (1991) also prefigured findings in vertebrates, where there is substantial functional redundancy among the four Notch proteins (e.g., Krebs et al. 2000).
Understanding that zygotic loss of both lin-12 and glp-1 causes a synthetic phenotype also allowed Lambie and Kimble (1991) to isolate strong alleles of two critical core components, lag-1 and lag-2 in a genetic screen analogous to the neurogenic screen of Lehmann et al. (1983). The Lag phenotype has also been important for assessing the roles of other potential core components in C. elegans, as it represents the loss of all zygotic Notch activity in C. elegans.
Ligands
Although much of the remainder of this essay concerns the elucidation of the mechanism of signal transduction by activated Notch, the process of signal transduction is initiated by the binding of a ligand. Thus, I will briefly describe another important development during this period—the genetic and molecular characterization of Delta, the first member of the DSL protein family of ligands for Notch.
Delta, like Notch, was defined in the early days of Drosophila genetics; it is also haploinsufficient, although deficiency heterozygotes show thickening of the wing veins, called “deltas,” rather than notching. Delta was clearly implicated in the Notch pathway via the neurogenic phenotype of homozygous null mutants (Lehmann et al. 1983). Cloning and sequence analysis revealed that Delta encodes a transmembrane protein with multiple EGF-like motifs (Vassin et al. 1987; Kopczynski et al. 1988); this basic structure, which also includes an N-terminal DSL domain, is the hallmark of this family. When Delta was expressed in cultured cells, it promoted aggregation with Notch-expressing cells, suggesting a physical interaction between the two proteins then mainly thought to be suggestive of a passive role in mediating cell–cell adhesion (Fehon et al. 1990).
Compelling evidence for a role of Delta in the signal-sending cell came from the mosaic analysis of Heitzler and Simpson (1991), who showed that Delta functions nonautonomously to promote the epidermal fate in neighboring cells, Furthermore, mosaics juxtaposing cells with different copy numbers of wild-type Delta genes displayed a bias in cell-fate choice, opposite to that displayed for Notch: the cell with the lower level of Delta activity was biased toward the epidermal fate, further underscoring the intimate relationship between Delta and Notch activity. Finally, the bias in cell-fate choice reveals that regulation of Delta activity is part of the feedback mechanism operating during lateral specification. Heitzler et al. (1996) subsequently found genetic circuitry consistent with this mechanism operating at the level of transcription of the Delta gene.
As noted above, the bias in cell-fate choice in lin-12 mosaics argued for a role of Notch as a receptor rather than a simple passive adhesion molecule. The finding that Delta mosaics displayed the opposite bias further supported the view that there is an instructive role, which, in view of the physical interaction between Delta and Notch, was consistent with its role as a ligand for Notch.
1990s
The basic Notch story as we think of it today emerged in the 1990s. The discovery of a new role for Notch as an oncogene in mammals started off the decade with éclat (Ellisen et al. 1991). This finding not only gave urgency to understanding more about this signaling system for potential insights into disease, but also provided the first clue as to the mechanism of signal transduction. By the end of a very active decade of research with parallel studies in C. elegans, Drosophila, and mammalian cultured cells, the key steps in the unusual mechanism of Notch signal transduction had been elucidated and accepted.
Notch as an oncogene and the first clue as to the mechanism of signal transduction
Ellisen et al. (1991) reported that three patients with T-cell acute lymphoblastic leukemia (T-ALL) had chromosomal translocations involving the Notch1 and the β-T-cell receptor gene. The breakpoints were all similar, resulting in a high level of expression of a message predicted to encode a truncated Notch1 product (then called “TAN1”) beginning near the last EGF-like motif.
This study was a major advance for two reasons. First, the association of T-ALL with similar translocations in different patients suggested a potential role for Notch as an oncogene. This possibility was supported by the finding that a mouse mammary tumor virus insertion called “int3” produced transcripts predicted to encode truncated Notch4 proteins and caused carcinomas and hyperplasia in mice (Gallahan et al. 1987; Jhappan et al. 1992; Robbins et al. 1992). It must be remembered that, at the time, forging connections between genes that control normal development and genes that contribute to cancer was still fairly new, and the extent to which development and oncogenesis were related mechanistically could only be conjectured. So at the time of publication the connection of Notch to cancer was very exciting news.
Second, for those of us puzzling over how Notch worked in development, the Ellisen et al. (1991) findings immediately suggested that analyzing the activity of engineered truncated forms of Notch would be a genetic approach to elucidating the mechanism of signal transduction and, by extension, cancer.
Notch and oncogenesis: Binary switch or differentiation block?
The exciting connection between activated Notch and cancer also began to influence how people thought about the normal role of Notch in vertebrate development. In particular, Jhappan et al. (1992) generated transgenic mice expressing the truncated form of Notch defined by the int-3 oncogenic insertion and found evidence of developmental arrest in mammary glands and other glands in which transcription of this form occurred. In addition, Coffman et al. (1993) expressed a truncated form of Notch in Xenopus and found that expression of some differentiation markers were delayed while animal caps displayed an extended period of competence to neural or mesodermal induction. From these studies, a view began to emerge that Notch activation delays or blocks differentiation, thereby maintaining the competence of cells to respond to other signaling events.
This view initially appeared to be supported by Drosophila studies in the Artavanis-Tsakonas lab when expression of activated Notch forms (see below) expressed under control of the sevenless promoter was interpreted as blocking the ability of the presumptive photoreceptor cells to differentiate until expression subsided, at which time the cells would choose an inappropriate fate because they were now exposed to inappropriate signals (Fortini et al. 1993). However, the experimental design, lacking mosaic analysis, did not allow the proper identification of which cells adopted which cell fates. Indeed, subsequent studies by several groups showed that Notch activation directly specifies the fates of the R3, R4, and R7 photoreceptors, whose differentiation was purportedly blocked, in lateral specification (R3/R4) and inductive signaling (R7) events (Cooper and Bray 1999, 2000; Fanto and Mlodzik 1999; Tomlinson and Struhl 1999, 2001).
At the time, I suggested that the binary decision of some cells was to choose between differentiating or remaining a stem cell and that Notch promoted the stem-cell choice rather than blocking differentiation per se (Greenwald 1994). My concern was essentially that thinking of a “block” would imply that the problem was in the execution of a fate, not at the level of a cell-fate decision. Now it is clear that Notch mediates many binary cell-fate decisions in mammalian development, including choices that promote maintenance of progenitor cells. Furthermore, altered cell-fate choices impacting progenitor cells caused by aberrations in Notch activity may contribute to its oncogenic effect. For example, Notch controls a binary decision between T- and B-cell progenitors (Tanigaki and Honjo 2007); one way that aberrant Notch activation contributes to cancer is to increase the number of T-cell progenitors (see Ferrando 2009). Notch activity also appears to control binary cell-fate decisions between stem cell and differentiated cell fate that go the “other way” in other cell types and thus can act as a tumor suppressor (see Lobry et al. 2011).
The cleavage model for Notch signal transduction
Many different groups recognized that the analysis of truncated forms of Notch might reveal the mechanism by which Notch transduces signals. At this time, in addition to studies in model organisms, studies of the Notch-signaling system in cell culture became an active area as well.
My own excitement about testing the effect of truncated forms, however, was tempered by the limitations of transgenic technology in C. elegans at the time. We simply did not know how to express proteins in the right time and place for our purposes. Fortunately—and not just for my work!—I am married to Gary Struhl, who was the first person to use the heat-shock promoter to create gain-of-function forms of developmental switch genes (Struhl 1985) and who had just developed “Flp-out” technology to express genes in specific tissues (Struhl and Basler 1993). Both of these technologies would prove to be useful for expressing truncated forms of Notch. Gary was attracted by what seemed likely to be an interesting mechanism for an important patterning gene and, for me, expressing a truncated form in the fly in parallel offered the promise of an answer even if the worm experiments did not work. And if we could show similar behavior of equivalent truncated forms in two systems, then it would underscore the universality of whatever we found individually.
It also was fun for us to be doing a project together while our ultimate collaboration, our daughter Abigail, was gestating. My student Kevin Fitzgerald graciously joined in while pursuing other valuable structure–function studies of Notch and DSL ligands (Fitzgerald et al. 1993; Wilkinson et al. 1994; Fitzgerald and Greenwald 1995), and after beginning in Princeton, we completed the project after moving to Columbia in 1993.
We reasoned that the predicted truncated protein associated with T-ALL would lack the signal sequence, suggesting that such a protein, if indeed produced and stable, would be cytosolic, so we decided to express just the intracellular domain (“intra”) as the most extreme case. Gary easily made flies expressing the Notch intracellular domain, and he and Kevin successfully implemented a novel expression strategy for worms to produce LIN-12(intra) using lin-12 regulatory sequences. As we had hoped, the results using the two systems were concordant: in worms, expression of LIN-12(intra) caused the phenotype associated with lin-12(d) alleles, and in flies, expression of Notch(intra) promoted epidermal differentiation opposite to the neurogenic phenotype of null alleles (Struhl et al. 1993). Further genetic analysis showed that the apparent signaling activity of the intracellular domain was constitutive and acting at the time of the respective cell fate decisions.
When Gary used an antibody to stain flies expressing Notch(intra), he had a big surprise: the protein was in the nucleus. This observation led us to formulate the cleavage model with a directness that surprises me now: “Our finding that Notch(intra) protein causes a gain-of-function phenotype and accumulates in the nucleus makes it worth considering the possibility that signal transduction mediated by the wild-type protein involves cleavage and transport of the intracellular domain to the nucleus and even the possibility that the intracellular domain of Notch may be directly involved in transcriptional regulation” (Struhl et al. 1993, p. 340).
Soon after our article was published, Toby Lieber, Simon Kidd, and Michael Young published a comprehensive study of many different truncated forms, including a comparable Notch(intra) form (Lieber et al. 1993). They also reported that Notch(intra) causes phenotypes consistent with Notch activation and localizes to the nucleus and considered a cleavage model and a potential role for Notch in transcriptional activation. However, they also generated other constitutively active truncated forms that could not be detected in the nucleus. Although Lieber et al. (1993) proposed reasonable ways to reconcile such observations with a cleavage model, the apparent lack of correlation between constitutive activity and nuclear localization in this study and several others became a major line of evidence advanced against the cleavage model by others. I will come back to this point below.
Several articles also described another informative truncated form, which lacked much of the extracellular domain yet contained a signal sequence, so that the resulting protein was targeted to the membrane (unlike the T-ALL leukemic truncation). This form, NotchΔE, was reported to have constitutive activity in Xenopus (Coffman et al. 1993), Drosophila (Lieber et al. 1993; Rebay et al. 1993), and C. elegans (referred to in Struhl et al. 1993), again underscoring that the mechanism of signal transduction was conserved in all animals.
Notch(intra) and NotchΔE, the key constitutively active forms used in subsequent genetic analysis, are diagrammed in Figure 2 as what we now know they are: mimics of the cleavage products of successive proteolytic processing events that ensue upon ligand binding. I note that the constitutive activities of these forms were observed to promote epidermal fate at the expense of neurogenesis, the opposite of the neurogenic phenotype caused by loss of Notch activity, thereby formally demonstrating the binary nature of Notch-mediated cell-fate choice in Drosophila (Lieber et al. 1993; Rebay et al. 1993; Struhl et al. 1993), as we had demonstrated using the lin-12(d) forms in C. elegans (Greenwald et al. 1983).
CSL: a sequence-specific DNA-binding protein and core component of the Notch-signaling system
Before discussing further why the cleavage model was initially resisted and the path by which it gained general acceptance, I must introduce a core component now generally called “CSL,” an acronym coined from some of the names it had been called in different systems: CBF1, Su(H), and LAG-1 (Christensen et al. 1996). CSL is now established as the sequence-specific DNA-binding protein with which Notch(intra) associates to promote target gene expression. Attaining an understanding of that role was also critical in the path to acceptance of the cleavage model and an important development in its own right during this time period as well.
The functional connection of CSL to Notch initially came from genetic studies of Drosophila Suppressor of Hairless [Su(H)], a classic modifier (see Nash 1970). Genetic analysis in several labs had connected Su(H) to the neurogenic genes and peripheral nervous system development. Molecular cloning established that Su(H) is the ortholog of CBF1 (also called RBP-J or KBF1) (Furukawa et al. 1992; Schweisguth and Posakony 1992). CBF1 had been found as a sequence-specific DNA-binding protein through association with many different cellular and viral promoters (Yano et al. 1987; Hamaguchi et al. 1989; Ling et al. 1993), so this orthology provided a crucial potential link between Notch signal transduction and transcription. This link was strengthened when Su(H) was found to associate physically with the intracellular domain of Notch (Fortini and Artavanis-Tsakonas 1994; Tamura et al. 1995) and by a combination of genetic and biochemical evidence establishing Su(H) as a direct transcriptional activator of genes of the Drosophila E(spl) complex (Jennings et al. 1994; Bailey and Posakony 1995; Lecourtois and Schweisguth 1995; Schweisguth 1995).
I note that, in the absence of the Drosophila genetic data, the broad expression and promiscuous binding of CSL would have obscured its critical role in Notch activity. Indeed, its various mammalian names underscore this point; e.g., CBF1 (C Binding Factor 1) refers to its being a cellular factor that binds to the Epstein-Barr virus “C” promoter, and “RBP-J” refers to its binding to the recombination signal sequence of immunoglobulin J κ gene.
Challenges to and acceptance of the cleavage model
A reasonable argument advanced against the cleavage model was that the intracellular domain of Notch could not be detected in nuclei in vivo in many contexts where Notch signal transduction was known to be active: nuclear Notch was not evident in wild-type animals or in transgenic animals carrying transmembrane constitutively active forms such as NotchΔE (e.g., Johansen et al. 1989; Fehon et al. 1990; Lieber et al. 1993; Roehl and Kimble 1993). Although plausible reasons for this lack were also postulated (see, e.g., Lieber et al. 1993), ultimately it would have to be reckoned with for the model to be validated and accepted.
Resistance to the cleavage model gained momentum when Fortini and Artavanis-Tsakonas (1994) claimed that, in Drosophila tissue culture, Su(H) protein is sequestered in the cytoplasm when coexpressed with Notch protein and is translocated to the nucleus when Notch binds to its ligand Delta. They proposed that the role of Notch was to sequester Su(H) from the nucleus in the absence of ligand binding. This simple tethering model was puzzling to Gary and me at the time because it did not seem to account for basic genetic observations in vivo, such as the fact that loss of Notch [which should release Su(H) from the proposed tether] had similar phenotypic effects as loss of Su(H). Nevertheless, the tethering model continued to exert a strong hold on the field, even after the central claim that the subcellular localization of Su(H) depends on the activity of Notch was shown to be wrong in vivo in Drosophila (Gho et al. 1996).
In contrast to the Drosophila cell culture findings, a growing body of work in mammalian cell culture supported the cleavage model. An important article by Jarriault et al. (1995) showed that CBF1 directly stimulates transcription of the Notch target gene HES1 in the presence of the ΔE truncated, constitutively active form of Notch, but not alone or in the presence of full-length (inactive) Notch. This cell culture study was possible only because both CBF1 and HES1 had been placed in the Notch pathway through Drosophila genetics by using Su(H), as described above, and the target gene E(spl) (Klambt et al. 1989; Jennings et al. 1994).
In addition, in compelling studies inspired by viral proteins that interact with the host CBF1 protein, essentially acting as Notch mimics, S. Diane Hayward and colleagues found that the Notch intracellular domain appears to abrogate repression by CBF1 and to recruit coactivators (Hsieh et al. 1996, 1997). Other contemporaneous studies reinforced these mechanistic insights.
Even though biochemists at the time were much more receptive than geneticists to the cleavage model, the experimental evidence available at that time was not yet compelling. There was no unequivocal evidence that proteolytic processing was necessary for transcriptional activation. Furthermore, the tissue culture work was performed using gain-of-function alleles and therefore could not address whether processing and transcriptional activation are normally regulated by ligand. Indeed, ligand inducibility is essential for a signaling system to have an instructive role.
Both Struhl and Adachi (1998) and Lecourtois and Schweisguth (1998) approached the question of ligand-dependent nuclear access by a similar approach to increase the sensitivity of detection of the Notch intracellular domain: they inserted a GAL4 DNA- binding domain moiety into the intracellular domain of an otherwise intact Notch protein and looked for transcriptional activation of a GAL4 target-lacZ reporter. Both groups found that β-galactosidase activity, which indicates nuclear access of the Notch intracellular domain, depends on ligand.
Struhl and Adachi (1998) also comprehensively assessed the relationship between nuclear access and signal transduction for constitutively active forms, providing compelling evidence that nuclear access is functionally relevant for signal transduction and transcriptional activation in vivo. In one set of experiments, they showed that signal transduction depended on nuclear access of the intracellular domain by targeting a derivative of Notch(intra) to the membrane using myristylation, thereby eliminating activity, or to the nucleus by adding nuclear localization sequences, thereby potentiating activity. In another set of experiments, they showed that signal transduction reflects transcriptional activation by the intracellular domain because adding a heterologous transcriptional activator further increased activity whereas adding a heterologous transcriptional repressor domain blocked signal transduction.
At the same time, Schroeter et al. (1998) assessed cleavage using transient transfection followed by immunoprecipitation in cultured mammalian cells. The immunoprecipitation step allowed them to obtain evidence for a scarce cleavage product, and two key experiments supported the hypothesis that this product was a bona fide intermediate in the signal transduction mechanism. First, Schroeter et al. (1998) identified and mutated a valine residue at the cleavage site in the transmembrane domain and reported that this mutation results in less cleavage product and reduced signaling ability in the NotchΔE but not Notch(intra) context, suggesting that cleavage and signaling are correlated. Second, they reported that ligand cotransfection was necessary for the detection of the cleavage product from the wild-type form, suggesting that cleavage was ligand-dependent.
Given the theme of this essay, I want to make a general comment about the contribution of in vivo approaches to assessing molecular mechanism, as there is a large community of investigators who put a premium on biochemical approaches in mammalian tissue culture over genetic experiments in model organisms. In particular, I want to note that there are virtues of in vivo genetic experiments that are difficult to achieve ex vivo. In the Drosophila experiments, transgenes were expressed at approximately endogenous levels; they were expressed in normal cells in their normal context, i.e., as part of epithelia, with their normal contacts, and receiving any other signals that they normally do; there were few experimental variables to control; and it was possible to remove endogenous components cleanly for assessing properties such as constitutive activity or ligand dependence, as well as the functional consequences of introducing tags and other probes of molecular function into the native ligands and receptors.
Indeed, these features of in vivo genetic analysis continue to be relevant for investigations into the mechanism of any biological process, and while ex vivo evidence has its own set of virtues, it is the combination of both approaches that is ultimately compelling.
For Notch, the two approaches synergized beautifully, and together the in vivo analysis of Struhl and Adachi (1998) and the ex vivo analysis of Schroeter et al. (1998) made a compelling case for the cleavage model, which became widely accepted at that time. However, there were still pockets of resistance, as can be seen from continued arguments against the cleavage model that were still being advanced afterward (Artavanis-Tsakonas et al. 1999). As described in the next section, elucidating the role of Presenilin in Notch signal transduction helped silence any remaining skepticism while adding a fascinating twist to the mechanism.
γ-Secretase and the release of the intracellular domain of Notch
All of the core components of the Notch signal transduction system were implicated as such through genetic analysis in worms and flies, and γ-secretase is no exception.
γ-Secretase was originally an inferred enzyme activity based on the proteolytic processing pattern of a transmembrane protein called β-APP. The peptide produced when β -APP is cleaved at the β site in its ectodomain and the γ site in its transmembrane domain can form β-amyloid plaques in the brain and cause Alzheimer’s disease. However, despite tremendous efforts in industry—where γ-secretase inhibitors were potentially valuable drugs—the identity of γ-secretase had been refractory to biochemical approaches. Genetic studies of familial early-onset Alzheimer’s disease (Sherrington et al. 1995) and of lin-12/Notch signaling in C. elegans (Levitan and Greenwald 1995) identified what proved to be the catalytic component of γ-secretase, Presenilin. Genetic analysis in C. elegans was later successful in identifying the other three core components of γ-secretase (Goutte et al. 2000, 2002; Francis et al. 2002).
My postdoctoral fellow Diane Levitan had been studying sel-12 (coincidentally, the 12th suppressor/enhancer of lin-12), a suppressor of the Multivulva phenotype caused by a lin-12(d) mutation. We were on the verge of submitting a manuscript when an astute colleague, Steve L’Hernault, called to tell us that SEL-12 was highly similar to the early-onset Alzheimer’s disease gene then called S182 (Sherrington et al. 1995) and now known as Presenilin (PS) 1. Although Sherrington et al. had not deposited the S182 sequence in public databases, the article included an alignment showing weak homology with the product of a C. elegans gene that Steve studied, SPE-4 (L’Hernault and Arduengo 1992). Aware that SEL-12 was also weakly homologous to SPE-4, Steve did the sequence analysis and discovered that SEL-12 and S182 are highly similar to each other, and we quickly retitled our manuscript to reflect that fact. Remarkably, Diane subsequently found that human PS1 could functionally replace SEL-12 in C. elegans (Levitan et al. 1996).
Although our genetic analysis indicated that Presenilin influences Notch signal transduction, it was not yet apparent that it was a core component of the signaling system, since sel-12 null mutants did not have the hallmark phenotypes associated with loss of Notch activity. However, the essential role of Presenilin became apparent when my student Xiajun Li identified a second C. elegans Presenilin gene, hop-1 (homolog of presenilin), and used RNA interference—still misunderstood at that time as antisense RNA—to demonstrate that depletion of hop-1 in a sel-12 null mutant background caused hallmark phenotypes associated with loss of Notch activity (Li and Greenwald 1997). Null alleles of the single Drosophila Presenilin gene were subsequently shown to cause Notch phenotypes, affirming that Presenilin is a conserved core component of the Notch signaling system (Struhl and Greenwald 1999; Ye et al. 1999).
In the intervening time, Schroeter et al. (1998) and Struhl and Adachi (1998) had provided compelling evidence for the cleavage model, and De Strooper et al. (1998) had found that Presenilin was required for transmembrane cleavage of β-APP. Given the parallels between β-APP processing and Notch processing, the next step was to assess the role of Presenilin in Notch cleavage. Three articles published together in Nature in 1999 did just that.
De Strooper et al. (1999) found that processing of NotchΔE was reduced in PS1-null cells; Gary and I found that nuclear access of NotchΔE-GAL-4 was blocked in PS null mutant clones in Drosophila (Struhl and Greenwald 1999). Both studies concluded that Presenilin promoted the transmembrane cleavage of Notch, consistent with a function as a protease itself or in facilitating protease function.
In contrast, Ye et al. (1999) examined processing and signal transduction of Notch in PS null mutant Drosophila embryos and reached different conclusions. The key genetic result that led to considerable commentary at the time was that Ye et al. (1999) claimed that NotchΔE signal transduction, as assessed by suppression of neurogenesis, was not affected in the absence of PS. Taken at face value, their results would indicate that signal transduction is not correlated with transmembrane cleavage (the assay used in the other two studies), thus challenging the cleavage model. However, Gary and I immediately recognized that Ye et al. (1999) had used a phenotypic assay for signal transduction that depended on the imaginal disks being “old enough” for neurogenesis to have occurred, but without independent markers for timing. Therefore, the apparent lack of neurogenesis might instead have been observed if the disks examined were too young or developmentally delayed by the manipulations used to express NotchΔE.
To address this direct challenge to the cleavage model, we re-examined the relationship between transmembrane cleavage and signal transduction in the same cellular context using internal controls for timing and found that they were strictly correlated, again validating the cleavage model and assigning Presenilin to the transmembrane cleavage step (Struhl and Greenwald 2001). There were no significant challenges to the cleavage model after that.
Concluding remarks
I think it is timely to remember that the history of Notch shows that molecular biology synergized with genetics but did not replace it. The current gadarene rush to “systems biology” has created a sense that classical genetics is being superseded. There was a similar feeling when molecular biology was first becoming a powerful force in fields that had previously been accessible only through genetics. While it remains to be seen whether “systems biology” will have a high and lasting impact, I think for it to arrive at its full potential, it will need to achieve some synergy with traditional genetics—at a minimum, for testing in vivo the models that emerge from genome-wide approaches.
I also think that it is interesting to see how the Notch story, as has been true for most scientific advances, did not develop as a neat linear narrative, but instead had its share of wrong directions and parallel paths, synergies and antagonisms, and restriction by and liberation from trends and expectations. With the intense pressure these days to make every story seem simple and tidy, I think it is important to remember (and for students to learn) that scientific understanding does not usually happen that way.
Finally, I think the Notch story offers a prime example of how and why flies and worms became such incredible systems for studying animal development. The major credit for these model systems, of course, belongs to others. But, with distance, I see that my work had some role in the acceptance of the worm, at least, as there were several “firsts” in the Notch field that came from studies in the worm, and first developments in the worm field that came from studies of Notch. I had no idea at the time that I was part of this larger story. I was just having a great time.
Acknowledgments
I am grateful to Tom Cline, David Hirsh, Oliver Hobert, Sophie Jarriault, Paul Sternberg, Gary Struhl, and Adam Wilkins for comments on this manuscript and to the many friends, students, and colleagues who have enlightened me in my career. Current work in my lab is supported by National Institutes of Health grant R01 095389 and the Howard Hughes Medical Institute.
Literature Cited
- Artavanis-Tsakonas S., Muskavitch M. A., Yedvobnick B., 1983. Molecular cloning of Notch, a locus affecting neurogenesis in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 80: 1977–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S., Rand M. D., Lake R. J., 1999. Notch signaling: cell fate control and signal integration in development. Science 284: 770–776 [DOI] [PubMed] [Google Scholar]
- Austin J., Kimble J., 1987. glp-1 is required in the germ line for regulation of the decision between mitosis and meiosis in C. elegans. Cell 51: 589–599 [DOI] [PubMed] [Google Scholar]
- Bailey A. M., Posakony J. W., 1995. Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes Dev. 9: 2609–2622 [DOI] [PubMed] [Google Scholar]
- Bender W., Akam M., Karch F., Beachy P. A., Peifer M., et al. , 1983. Molecular genetics of the bithorax complex in Drosophila melanogaster. Science 221: 23–29 [DOI] [PubMed] [Google Scholar]
- Blaumueller C. M., Qi H., Zagouras P., Artavanis-Tsakonas S., 1997. Intracellular cleavage of Notch leads to a heterodimeric receptor on the plasma membrane. Cell 90: 281–291 [DOI] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco A. E., McGinnis W., Gehring W. J., De Robertis E. M., 1984. Cloning of an X. laevis gene expressed during early embryogenesis coding for a peptide region homologous to Drosophila homeotic genes. Cell 37: 409–414 [DOI] [PubMed] [Google Scholar]
- Christensen S., Kodoyianni V., Bosenberg M., Friedman L., Kimble J., 1996. lag-1, a gene required for lin-12 and glp-1 signaling in Caenorhabditis elegans, is homologous to human CBF1 and Drosophila Su(H). Development 122: 1373–1383 [DOI] [PubMed] [Google Scholar]
- Coffman C. R., Skoglund P., Harris W. A., Kintner C. R., 1993. Expression of an extracellular deletion of Xotch diverts cell fate in Xenopus embryos. Cell 73: 659–671 [DOI] [PubMed] [Google Scholar]
- Cooper M. T., Bray S. J., 1999. Frizzled regulation of Notch signalling polarizes cell fate in the Drosophila eye. Nature 397: 526–530 [DOI] [PubMed] [Google Scholar]
- Cooper M. T., Bray S. J., 2000. R7 photoreceptor specification requires Notch activity. Curr. Biol. 10: 1507–1510 [DOI] [PubMed] [Google Scholar]
- Hart C. P., Awgulewitsch A., Fainsod A., McGinnis W., Ruddle F. H., 1985. Homeo box gene complex on mouse chromosome 11: molecular cloning, expression in embryogenesis, and homology to a human homeo box locus. Cell 43: 9–18 [DOI] [PubMed] [Google Scholar]
- De Strooper B., Saftig P., Craessaerts K., Vanderstichele H., Guhde G., et al. , 1998. Deficiency of presenilin-1 inhibits the normal cleavage of amyloid precursor protein. Nature 391: 387–390 [DOI] [PubMed] [Google Scholar]
- De Strooper B., Annaert W., Cupers P., Saftig P., Craessaerts K., et al. , 1999. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature 398: 518–522 [DOI] [PubMed] [Google Scholar]
- Doe C. Q., Goodman C. S., 1985. Early events in insect neurogenesis. II. The role of cell interactions and cell lineage in the determination of neuronal precursor cells. Dev. Biol. 111: 206–219 [DOI] [PubMed] [Google Scholar]
- Ellisen L. W., Bird J., West D. C., Soreng A. L., Reynolds T. C., et al. , 1991. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell 66: 649–661 [DOI] [PubMed] [Google Scholar]
- Emmons S. W., Yesner L., Ruan K. S., Katzenberg D., 1983. Evidence for a transposon in Caenorhabditis elegans. Cell 32: 55–65 [DOI] [PubMed] [Google Scholar]
- Fanto M., Mlodzik M., 1999. Asymmetric Notch activation specifies photoreceptors R3 and R4 and planar polarity in the Drosophila eye. Nature 397: 523–526 [DOI] [PubMed] [Google Scholar]
- Fehon R. G., Kooh P. J., Rebay I., Regan C. L., Xu T., et al. , 1990. Molecular interactions between the protein products of the neurogenic loci Notch and Delta, two EGF-homologous genes in Drosophila. Cell 61: 523–534 [DOI] [PubMed] [Google Scholar]
- Ferguson E. L., Horvitz H. R., 1985. Identification and characterization of 22 genes that affect the vulval cell lineages of the nematode Caenorhabditis elegans. Genetics 110: 17–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson E. L., Sternberg P. W., Horvitz H. R., 1987. A genetic pathway for the specification of the vulval cell lineages of Caenorhabditis elegans. Nature 326: 259–267 [DOI] [PubMed] [Google Scholar]
- Ferrando A. A., 2009. The role of NOTCH1 signaling in T-ALL. Hematology Am. Soc. Hematol. Educ. Program, pp 353–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Files J. G., Carr S., Hirsh D., 1983. Actin gene family of Caenorhabditis elegans. J. Mol. Biol. 164: 355–375 [DOI] [PubMed] [Google Scholar]
- Fitzgerald K., Greenwald I., 1995. Interchangeability of Caenorhabditis elegans DSL proteins and intrinsic signalling activity of their extracellular domains in vivo. Development 121: 4275–4282 [DOI] [PubMed] [Google Scholar]
- Fitzgerald K., Wilkinson H. A., Greenwald I., 1993. glp-1 can substitute for lin-12 in specifying cell fate decisions in Caenorhabditis elegans. Development 119: 1019–1027 [DOI] [PubMed] [Google Scholar]
- Fortini M. E., Artavanis-Tsakonas S., 1994. The suppressor of hairless protein participates in notch receptor signaling. Cell 79: 273–282 [DOI] [PubMed] [Google Scholar]
- Fortini M. E., Rebay I., Caron L. A., Artavanis-Tsakonas S., 1993. An activated Notch receptor blocks cell-fate commitment in the developing Drosophila eye. Nature 365: 555–557 [DOI] [PubMed] [Google Scholar]
- Foster G. G., 1975. Negative complementation at the notch locus of Drosophila melanogaster. Genetics 81: 99–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis R., McGrath G., Zhang J., Ruddy D. A., Sym M., et al. , 2002. aph-1 and pen-2 are required for Notch pathway signaling, gamma-secretase cleavage of betaAPP, and presenilin protein accumulation. Dev. Cell 3: 85–97 [DOI] [PubMed] [Google Scholar]
- Furukawa T., Maruyama S., Kawaichi M., Honjo T., 1992. The Drosophila homolog of the immunoglobulin recombination signal-binding protein regulates peripheral nervous system development. Cell 69: 1191–1197 [DOI] [PubMed] [Google Scholar]
- Gallahan D., Kozak C., Callahan R., 1987. A new common integration region (int-3) for mouse mammary tumor virus on mouse chromosome 17. J. Virol. 61: 218–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber R. L., Kuroiwa A., Gehring W. J., 1983. Genomic and cDNA clones of the homeotic locus Antennapedia in Drosophila. EMBO J. 2: 2027–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gho M., Lecourtois M., Geraud G., Posakony J. W., Schweisguth F., 1996. Subcellular localization of Suppressor of Hairless in Drosophila sense organ cells during Notch signalling. Development 122: 1673–1682 [DOI] [PubMed] [Google Scholar]
- Goutte C., Hepler W., Mickey K. M., Priess J. R., 2000. aph-2 encodes a novel extracellular protein required for GLP-1-mediated signaling. Development 127: 2481–2492 [DOI] [PubMed] [Google Scholar]
- Goutte C., Tsunozaki M., Hale V. A., Priess J. R., 2002. APH-1 is a multipass membrane protein essential for the Notch signaling pathway in Caenorhabditis elegans embryos. Proc. Natl. Acad. Sci. USA 99: 775–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray A., Dull T. J., Ullrich A., 1983. Nucleotide sequence of epidermal growth factor cDNA predicts a 128,000-molecular weight protein precursor. Nature 303: 722–725 [DOI] [PubMed] [Google Scholar]
- Greenwald I., 1985. lin-12, a nematode homeotic gene, is homologous to a set of mammalian proteins that includes epidermal growth factor. Cell 43: 583–590 [DOI] [PubMed] [Google Scholar]
- Greenwald I., 1994. Structure/function studies of lin-12/Notch proteins. Curr. Opin. Genet. Dev. 4: 556–562 [DOI] [PubMed] [Google Scholar]
- Greenwald I., Horvitz H. R., 1986. A visible allele of the muscle gene sup-10X of C. elegans. Genetics 113: 63–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald I., Rubin G. M., 1992. Making a difference: the role of cell-cell interactions in establishing separate identities for equivalent cells. Cell 68: 271–281 [DOI] [PubMed] [Google Scholar]
- Greenwald I. S., Horvitz H. R., 1980. unc-93(e1500): a behavioral mutant of Caenorhabditis elegans that defines a gene with a wild-type null phenotype. Genetics 96: 147–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald I. S., Horvitz H. R., 1982. Dominant suppressors of a muscle mutant define an essential gene of Caenorhabditis elegans. Genetics 101: 211–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald I. S., Sternberg P. W., Horvitz H. R., 1983. The lin-12 locus specifies cell fates in Caenorhabditis elegans. Cell 34: 435–444 [DOI] [PubMed] [Google Scholar]
- Hamaguchi Y., Matsunami N., Yamamoto Y., Honjo T., 1989. Purification and characterization of a protein that binds to the recombination signal sequence of the immunoglobulin J kappa segment. Nucleic Acids Res. 17: 9015–9026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedgecock E. M., Herman R. K., 1995. The ncl-1 gene and genetic mosaics of Caenorhabditis elegans. Genetics 141: 989–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitzler P., Simpson P., 1991. The choice of cell fate in the epidermis of Drosophila. Cell 64: 1083–1092 [DOI] [PubMed] [Google Scholar]
- Heitzler P., Bourouis M., Ruel L., Carteret C., Simpson P., 1996. Genes of the Enhancer of split and achaete-scute complexes are required for a regulatory loop between Notch and Delta during lateral signalling in Drosophila. Development 122: 161–171 [DOI] [PubMed] [Google Scholar]
- Herman R. K., 1984. Analysis of genetic mosaics of the nematode Caneorhabditis elegans. Genetics 108: 165–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe P. E., Greenspan R. J., 1986. Local function of the Notch gene for embryonic ectodermal pathway choice in Drosophila. Cell 46: 773–783 [DOI] [PubMed] [Google Scholar]
- Horvitz H. R., Sulston J. E., 1980. Isolation and genetic characterization of cell-lineage mutants of the nematode Caenorhabditis elegans. Genetics 96: 435–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvitz H. R., Sulston J. E., 1990. Joy of the worm. Genetics 126: 287–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvitz H. R., Sternberg P. W., Greenwald I. S., Fixsen W., Ellis H. M., 1983. Mutations that affect neural cell lineages and cell fates during the development of the nematode Caenorhabditis elegans. Cold Spring Harb. Symp. Quant. Biol. 48(Pt. 2): 453–463 [DOI] [PubMed] [Google Scholar]
- Hsieh J. J., Henkel T., Salmon P., Robey E., Peterson M. G., et al. , 1996. Truncated mammalian Notch1 activates CBF1/RBPJk-repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA2. Mol. Cell. Biol. 16: 952–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh J. J., Nofziger D. E., Weinmaster G., Hayward S. D., 1997. Epstein-Barr virus immortalization: Notch2 interacts with CBF1 and blocks differentiation. J. Virol. 71: 1938–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarriault S., Brou C., Logeat F., Schroeter E. H., Kopan R., et al. , 1995. Signalling downstream of activated mammalian Notch. Nature 377: 355–358 [DOI] [PubMed] [Google Scholar]
- Jennings B., Preiss A., Delidakis C., Bray S., 1994. The Notch signalling pathway is required for Enhancer of split bHLH protein expression during neurogenesis in the Drosophila embryo. Development 120: 3537–3548 [DOI] [PubMed] [Google Scholar]
- Jhappan C., Gallahan D., Stahle C., Chu E., Smith G. H., et al. , 1992. Expression of an activated Notch-related int-3 transgene interferes with cell differentiation and induces neoplastic transformation in mammary and salivary glands. Genes Dev. 6: 345–355 [DOI] [PubMed] [Google Scholar]
- Johansen K. M., Fehon R. G., Artavanis-Tsakonas S., 1989. The notch gene product is a glycoprotein expressed on the cell surface of both epidermal and neuronal precursor cells during Drosophila development. J. Cell Biol. 109: 2427–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd S., Lieber T., 2002. Furin cleavage is not a requirement for Drosophila Notch function. Mech. Dev. 115: 41–51 [DOI] [PubMed] [Google Scholar]
- Kidd S., Lockett T. J., Young M. W., 1983. The Notch locus of Drosophila melanogaster. Cell 34: 421–433 [DOI] [PubMed] [Google Scholar]
- Kidd S., Kelley M. R., Young M. W., 1986. Sequence of the notch locus of Drosophila melanogaster: relationship of the encoded protein to mammalian clotting and growth factors. Mol. Cell. Biol. 6: 3094–3108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J., 1981. Alterations in cell lineage following laser ablation of cells in the somatic gonad of Caenorhabditis elegans. Dev. Biol. 87: 286–300 [DOI] [PubMed] [Google Scholar]
- Kimble J., Crittenden S. L., 2005. Germline proliferation and its control (August 15, 2005), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.13.1, http://www.wormbook.org [Google Scholar]
- Kimble J., Hirsh D., 1979. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev. Biol. 70: 396–417 [DOI] [PubMed] [Google Scholar]
- Klambt C., Knust E., Tietze K., Campos-Ortega J. A., 1989. Closely related transcripts encoded by the neurogenic gene complex enhancer of split of Drosophila melanogaster. EMBO J. 8: 203–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopczynski C. C., Alton A. K., Fechtel K., Kooh P. J., Muskavitch M. A., 1988. Delta, a Drosophila neurogenic gene, is transcriptionally complex and encodes a protein related to blood coagulation factors and epidermal growth factor of vertebrates. Genes Dev. 2: 1723–1735 [DOI] [PubMed] [Google Scholar]
- Krebs L. T., Xue Y., Norton C. R., Shutter J. R., Maguire M., et al. , 2000. Notch signaling is essential for vascular morphogenesis in mice. Genes Dev. 14: 1343–1352 [PMC free article] [PubMed] [Google Scholar]
- Lambie E. J., Kimble J., 1991. Two homologous regulatory genes, lin-12 and glp-1, have overlapping functions. Development 112: 231–240 [DOI] [PubMed] [Google Scholar]
- Lecourtois M., Schweisguth F., 1995. The neurogenic suppressor of hairless DNA-binding protein mediates the transcriptional activation of the enhancer of split complex genes triggered by Notch signaling. Genes Dev. 9: 2598–2608 [DOI] [PubMed] [Google Scholar]
- Lecourtois M., Schweisguth F., 1998. Indirect evidence for Delta-dependent intracellular processing of notch in Drosophila embryos. Curr. Biol. 8: 771–774 [DOI] [PubMed] [Google Scholar]
- Lehmann R., Jiménez F., Dietrich U., Campos-Ortega J. A., 1983. On the phenotype and development of mutants of early neurogenesis in Drosophila melanogaster. Dev. Genes Evol. 192: 63–74 [DOI] [PubMed] [Google Scholar]
- Levitan D., Greenwald I., 1995. Facilitation of lin-12-mediated signalling by sel-12, a Caenorhabitis elegans S182 Alzheimer's disease gene. Nature 77: 351–354 [DOI] [PubMed] [Google Scholar]
- Levitan D., Doyle T. G., Brousseau D., Lee M. K., Thinakaran G., et al. , 1996. Assessment of normal and mutant human presenilin function in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 93: 14940–14944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis E. B., 1978. A gene complex controlling segmentation in Drosophila. Nature 276: 565–570 [DOI] [PubMed] [Google Scholar]
- L’Hernault S. W., Arduengo P. M., 1992. Mutation of a putative sperm membrane protein in Caenorhabditis elegans prevents sperm differentiation but not its associated meiotic divisions. J. Cell Biol. 119: 55–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Greenwald I., 1997. HOP-1, a Caenorhabditis elegans presenilin, appears to be functionally redundant with SEL-12 presenilin and to facilitate LIN-12 and GLP-1 signaling. Proc. Natl. Acad. Sci. USA 94: 12204–12209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao L. W., Rosenzweig B., Hirsh D., 1983. Analysis of a transposable element in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 80: 3585–3589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber T., Kidd S., Alcamo E., Corbin V., Young M. W., 1993. Antineurogenic phenotypes induced by truncated Notch proteins indicate a role in signal transduction and may point to a novel function for Notch in nuclei. Genes Dev. 7: 1949–1965 [DOI] [PubMed] [Google Scholar]
- Ling P. D., Rawlins D. R., Hayward S. D., 1993. The Epstein-Barr virus immortalizing protein EBNA-2 is targeted to DNA by a cellular enhancer-binding protein. Proc. Natl. Acad. Sci. USA 90: 9237–9241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobry C., Oh P., Aifantis I., 2011. Oncogenic and tumor suppressor functions of Notch in cancer: it’s NOTCH what you think. J. Exp. Med. 208: 1931–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logeat F., Bessia C., Brou C., LeBail O., Jarriault S., et al. , 1998. The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc. Natl. Acad. Sci. USA 95: 8108–8112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis W., Levine M. S., Hafen E., Kuroiwa A., Gehring W. J., 1984. A conserved DNA sequence in homoeotic genes of the Drosophila Antennapedia and bithorax complexes. Nature 308: 428–433 [DOI] [PubMed] [Google Scholar]
- Meinhardt H., Gierer A., 2000. Pattern formation by local self-activation and lateral inhibition. Bioessays 22: 753–760 [DOI] [PubMed] [Google Scholar]
- Moerman D. G., Waterston R. H., 1984. Spontaneous unstable unc-22 IV mutations in C. elegans var. Bergerac. Genetics 108: 859–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr O. L., 1919. Character changes caused by mutation of an entire region of a chromosome in Drosophila. Genetics 4: 275–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H. J., 1932. Further studies on the nature and causes of gene mutations. Proceedings of the 6th International Congress of Genetics, Ithaca, NY, pp. 213–255
- Nash D., 1970. The mutational basis for the “allelic” modifier mutants, ENHANCER and SUPPRESSOR OF HAIRLESS, of Drosophila melanogaster. Genetics 64: 471–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusslein-Volhard C., Wieschaus E., 1980. Mutations affecting segment number and polarity in Drosophila. Nature 287: 795–801 [DOI] [PubMed] [Google Scholar]
- Portin P., 1975. Allelic negative complementation at the Abruptex locus of Drosophila melanogaster. Genetics 81: 121–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulson D., 1940. The effects of certain X-chromosome deficiencies on the embryonic development of Drosophila melanogaster. J. Exp. Zool. 83: 271–335 [Google Scholar]
- Poulson D. F., 1939. Effects of Notch deficiencies. Drosoph. Inf. Serv. 12: 64 [Google Scholar]
- Priess J. R., 2005. Notch signaling in the C. elegans embryo (June 25, 2005), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.4.1, http://www.wormbook.org [Google Scholar]
- Priess J. R., Schnabel H., Schnabel R., 1987. The glp-1 locus and cellular interactions in early C. elegans embryos. Cell 51: 601–611 [DOI] [PubMed] [Google Scholar]
- Rebay I., Fehon R. G., Artavanis-Tsakonas S., 1993. Specific truncations of Drosophila Notch define dominant activated and dominant negative forms of the receptor. Cell 74: 319–329 [DOI] [PubMed] [Google Scholar]
- Robbins J., Blondel B. J., Gallahan D., Callahan R., 1992. Mouse mammary tumor gene int-3: a member of the notch gene family transforms mammary epithelial cells. J. Virol. 66: 2594–2599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roehl H., Kimble J., 1993. Control of cell fate in C. elegans by a GLP-1 peptide consisting primarily of ankyrin repeats. Nature 364: 632–635 [DOI] [PubMed] [Google Scholar]
- Schroeter E. H., Kisslinger J. A., Kopan R., 1998. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature 393: 382–386 [DOI] [PubMed] [Google Scholar]
- Schweisguth F., 1995. Suppressor of Hairless is required for signal reception during lateral inhibition in the Drosophila pupal notum. Development 121: 1875–1884 [DOI] [PubMed] [Google Scholar]
- Schweisguth F., Posakony J. W., 1992. Suppressor of Hairless, the Drosophila homolog of the mouse recombination signal-binding protein gene, controls sensory organ cell fates. Cell 69: 1199–1212 [DOI] [PubMed] [Google Scholar]
- Scott J., Urdea M., Quiroga M., Sanchez-Pescador R., Fong N., et al. , 1983a Structure of a mouse submaxillary messenger RNA encoding epidermal growth factor and seven related proteins. Science 221: 236–240 [DOI] [PubMed] [Google Scholar]
- Scott M. P., Weiner A. J., 1984. Structural relationships among genes that control development: sequence homology between the Antennapedia, Ultrabithorax, and fushi tarazu loci of Drosophila. Proc. Natl. Acad. Sci. USA 81: 4115–4119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott M. P., Weiner A. J., Hazelrigg T. I., Polisky B. A., Pirrotta V., et al. , 1983. The molecular organization of the Antennapedia locus of Drosophila. Cell 35: 763–776 [DOI] [PubMed] [Google Scholar]
- Seydoux G., Greenwald I., 1989. Cell autonomy of lin-12 function in a cell fate decision in C. elegans. Cell 57: 1237–1245 [DOI] [PubMed] [Google Scholar]
- Shellenbarger D. L., Mohler J. D., 1975. Temperature-sensitive mutations of the notch locus in Drosophila melanogaster. Genetics 81: 143–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherrington R., Rogaev E. I., Liang Y., Rogaeva E. A., Levesque G., et al. , 1995. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer’s disease. Nature 375: 754–760 [DOI] [PubMed] [Google Scholar]
- Simpson P., 1990. Lateral inhibition and the development of the sensory bristles of the adult peripheral nervous system of Drosophila. Development 109: 509–519 [DOI] [PubMed] [Google Scholar]
- Stern C., 1954. Two or three bristles. Am. Sci. 42: 213–247 [Google Scholar]
- Struhl G., 1981. A homoeotic mutation transforming leg to antenna in Drosophila. Nature 292: 635–638 [DOI] [PubMed] [Google Scholar]
- Struhl G., 1985. Near-reciprocal phenotypes caused by inactivation or indiscriminate expression of the Drosophila segmentation gene ftz. Nature 318: 677–680 [DOI] [PubMed] [Google Scholar]
- Struhl G., Adachi A., 1998. Nuclear access and action of notch in vivo. Cell 93: 649–660 [DOI] [PubMed] [Google Scholar]
- Struhl G., Adachi A., 2000. Requirements for presenilin-dependent cleavage of notch and other transmembrane proteins. Mol. Cell 6: 625–636 [DOI] [PubMed] [Google Scholar]
- Struhl G., Basler K., 1993. Organizing activity of wingless protein in Drosophila. Cell 72: 527–540 [DOI] [PubMed] [Google Scholar]
- Struhl G., Greenwald I., 1999. Presenilin is required for activity and nuclear access of Notch in Drosophila. Nature 398: 522–525 [DOI] [PubMed] [Google Scholar]
- Struhl G., Greenwald I., 2001. Presenilin-mediated transmembrane cleavage is required for Notch signal transduction in Drosophila. Proc. Natl. Acad. Sci. USA 98: 229–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G., Fitzgerald K., Greenwald I., 1993. Intrinsic activity of the Lin-12 and Notch intracellular domains in vivo. Cell 74: 331–345 [DOI] [PubMed] [Google Scholar]
- Sulston J. E., Horvitz H. R., 1977. Post-embryonic lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56: 110–156 [DOI] [PubMed] [Google Scholar]
- Sulston J. E., Horvitz H. R., 1981. Abnormal cell lineages in mutants of the nematode Caenorhabditis elegans. Dev. Biol. 82: 41–55 [DOI] [PubMed] [Google Scholar]
- Sulston J. E., White J. G., 1980. Regulation and cell autonomy during postembryonic development of Caenorhabditis elegans. Dev. Biol. 78: 577–597 [DOI] [PubMed] [Google Scholar]
- Taghert P. H., Doe C. Q., Goodman C. S., 1984. Cell determination and regulation during development of neuroblasts and neurones in grasshopper embryo. Nature 307: 163–165 [DOI] [PubMed] [Google Scholar]
- Tamura K., Taniguchi Y., Minoguchi S., Sakai T., Tun T., et al. , 1995. Physical interaction between a novel domain of the receptor Notch and the transcription factor RBP-J kappa/Su(H). Curr. Biol. 5: 1416–1423 [DOI] [PubMed] [Google Scholar]
- Tanigaki K., Honjo T., 2007. Regulation of lymphocyte development by Notch signaling. Nat. Immunol. 8: 451–456 [DOI] [PubMed] [Google Scholar]
- Technau G. M., Campos-Ortega J. A., 1987. Cell autonomy of expression of neurogenic genes of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 84: 4500–4504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson A., Struhl G., 1999. Decoding vectorial information from a gradient: sequential roles of the receptors Frizzled and Notch in establishing planar polarity in the Drosophila eye. Development 126: 5725–5738 [DOI] [PubMed] [Google Scholar]
- Tomlinson A., Struhl G., 2001. Delta/Notch and Boss/Sevenless signals act combinatorially to specify the Drosophila R7 photoreceptor. Mol. Cell 7: 487–495 [DOI] [PubMed] [Google Scholar]
- Vassin H., Bremer K. A., Knust E., Campos-Ortega J. A., 1987. The neurogenic gene Delta of Drosophila melanogaster is expressed in neurogenic territories and encodes a putative transmembrane protein with EGF-like repeats. EMBO J. 6: 3431–3440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welshons W. J., Von Halle E. S., 1962. Pseudoallelism at the notch locus in Drosophila. Genetics 47: 743–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton K. A., Johansen K. M., Xu T., Artavanis-Tsakonas S., 1985. Nucleotide sequence from the neurogenic locus notch implies a gene product that shares homology with proteins containing EGF-like repeats. Cell 43: 567–581 [DOI] [PubMed] [Google Scholar]
- Wilkinson H. A., Fitzgerald K., Greenwald I., 1994. Reciprocal changes in expression of the receptor lin-12 and its ligand lag-2 prior to commitment in a C. elegans cell fate decision. Cell 79: 1187–1198 [DOI] [PubMed] [Google Scholar]
- Yano O., Kanellopoulos J., Kieran M., Le Bail O., Israel A., et al. , 1987. Purification of KBF1, a common factor binding to both H-2 and beta 2-microglobulin enhancers. EMBO J. 6: 3317–3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Lukinova N., Fortini M. E., 1999. Neurogenic phenotypes and altered Notch processing in Drosophila Presenilin mutants. Nature 398: 525–529 [DOI] [PubMed] [Google Scholar]
- Yochem J., Greenwald I., 1989. glp-1 and lin-12, genes implicated in distinct cell-cell interactions in C. elegans, encode similar transmembrane proteins. Cell 58: 553–563 [DOI] [PubMed] [Google Scholar]
- Yochem J., Weston K., Greenwald I., 1988. The Caenorhabditis elegans lin-12 gene encodes a transmembrane protein with overall similarity to Drosophila Notch. Nature 335: 547–550 [DOI] [PubMed] [Google Scholar]