Abstract
All RNA species in yeast cells are subject to turnover. Work over the past 20 years has defined degradation mechanisms for messenger RNAs, transfer RNAs, ribosomal RNAs, and noncoding RNAs. In addition, numerous quality control mechanisms that target aberrant RNAs have been identified. Generally, each decay mechanism contains factors that funnel RNA substrates to abundant exo- and/or endonucleases. Key issues for future work include determining the mechanisms that control the specificity of RNA degradation and how RNA degradation processes interact with translation, RNA transport, and other cellular processes.
Keywords: RNA, decay, translation, quality control, decapping, deadenylation
ALL RNA species in eukaryotic cells are subject to turnover, which plays several roles in yeast cells. First, the differential degradation of messenger RNA (mRNAs) can play an important role in setting the basal level of mRNA expression and how that mRNA level is modulated by environmental stimuli. Second, numerous quality control systems degrade aberrant transfer RNA (tRNAs) and ribosomal RNA (rRNAs), as well as aberrant mRNAs, which might otherwise encode a defective protein product. Third, RNA degradation removes the by-products of gene expression, including excised introns and other RNA pieces released during RNA processing. Finally, RNA degradation mechanisms functions in removing intergenic, intragenic, promoter-associated, and antisense RNAs that arise either as regulatory RNAs or transcriptional noise.
Here I review our understanding of the pathways and nucleases of RNA turnover by considering the different classes of RNAs and how they are degraded. Three common themes emerge from this review. First, most RNA degradation mechanisms funnel RNAs to the cytoplasmic Xrn1 or nuclear Rat1 5′ to 3′ nucleases, or to the exosome, which is a conserved cytoplasmic and nuclear complex with both 3′ to 5′ exonuclease activities and an endonuclease cleavage site. Second, where examined, all RNAs are subject to quality control systems where nonfunctional RNAs are more rapidly degraded. Third, the RNA pathways are modulated by environmental inputs and interact with other cellular processes including translation, RNA processing, transcription, and stress responses.
Degradation of mRNA
Cytoplasmic turnover of mRNA
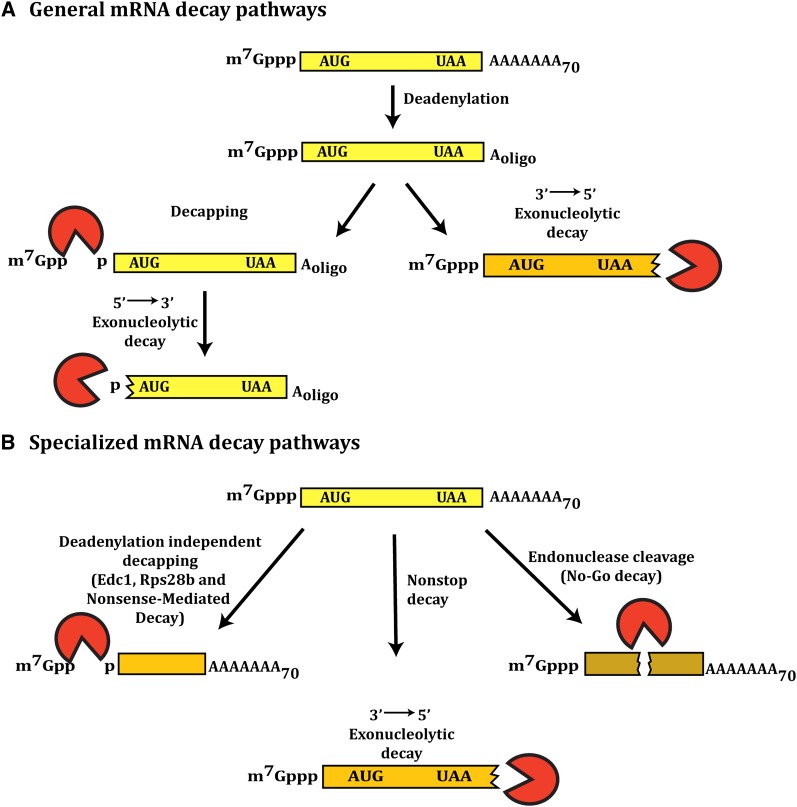
Cytoplasmic degradation of yeast mRNAs occurs by two general pathways, both of which are initiated by shortening of the 3′ poly(A) tail in a process referred to as deadenylation (Muhlrad and Parker 1992; Decker and Parker1993) (Figure 1A). Deadenylation in yeast is carried out by the Pan2/Pan3 complex as well as the by Ccr4/Pop2/Not complex (Brown and Sachs 1998; Tucker et al. 2001). Following deadenylation, mRNAs can be subjected to 3′ to 5′ degradation by the exosome (Anderson and Parker 1998). More commonly, mRNAs are decapped by the Dcp1/Dcp2 decapping enzyme and then subjected to 5′ to 3′ degradation by Xrn1 (Hsu and Stevens 1993; Muhlrad et al. 1994, 1995; Dunckley and Parker 1999; Van Dijk et al. 2002; Steiger et al. 2003).
Figure 1 .
(A) General mRNA decay pathways. (B) Specialized mRNA decay pathways.
Yeast also contain specialized mRNA decay pathways that act in response to aberrancies in translation (Figure 1B). In these cases, mRNAs can be subject to either deadenylation independent decapping (Muhlrad and Parker 1994), rapid 3′ to 5′ degradation (Van Hoof et al. 2002), or endonuclease cleavage (Doma and Parker 2006). The available evidence suggests that these specialized mechanisms function primarily on aberrant mRNAs, although the nonsense-mediated decay (NMD) pathway does degrade a pool of “normal” mRNAs (see section on mRNA Quality Control Pathways).
Several observations suggest that, at least during mid-log growth phase, decapping is the predominant pathway of mRNA degradation. First, strains lacking the decapping enzyme grow very slowly or are lethal in some strain backgrounds (Beelman et al. 1996; Dunckley and Parker 1999; Giaever et al. 2002), while strains defective in cytoplasmic 3′ to 5′ mRNA degradation grow relatively normally (Anderson and Parker 1998; Giaever et al. 2002). Second, strains defective in decapping or 5′ to 3′ degradation show changes in both the steady-state levels and decay rates of many mRNAs (Beelman et al. 1996; He et al. 2003; Van Dijk et al. 2011). Finally, the genome-wide mapping of endonuclease sites in mRNAs has revealed that few yeast mRNAs are subject to endonucleolytic degradation (Y. Harigaya and R. Parker, unpublished data). However, it should be noted that 3′ to 5′ degradation of mRNAs is just slightly slower than decapping. For example, for the PGK1 and MFA2 mRNAs, computational analysis of experimental data has indicated that 3′ to 5′ decay is 1.5 and 6 times slower than decapping, respectively (Cao and Parker 2001).
As assessed by a variety of different methods (Passos and Parker 2008; Munchel et al. 2011), the degradation rates of individual mRNAs can vary by over an order of magnitude. This was first observed in decay rate measurements of groups of mRNAs (Herrick et al. 1990; Brown and Sachs 1998) and has now been confirmed by multiple genome-wide studies of mRNA decay rates (Wang et al. 2002; Grigull et al. 2004; Munchel et al. 2011). The decay rates of mRNAs are somewhat clustered by the function of the encoded protein (Herrick et al. 1990; Grigull et al. 2004; Wang et al. 2006; Beilharz and Preiss 2007). Differences in the decay rates of individual mRNAs can arise by differences in deadenylation rates, decapping rates, or the rates of 3′ to 5′ degradation (Cao and Parker 2001; Beilharz and Preiss 2007). For example, the MFA2 mRNA (t1/2 = 3′–4′) differs from the relatively stable PGK1 mRNA (t1/2 = 30′–45′) by having faster rates of deadenylation (15 adenosines/min compared to 3 adenosines/min), decapping (0.0077 sec−1 compared to 0.000462 sec−1) and 3′ to 5′ degradation (0.0012 sec−1 compared to 0.0003 sec−1) (Cao and Parker 2001). Thus, to understand differential control of mRNA decay rates, one must consider the processes of deadenylation, decapping, and 3′ to 5′ degradation and how they are regulated.
Deadenylation
Two enzyme complexes catalyze poly(A) shortening (Table 1). The predominant deadenylase is the Ccr4/Pop2/Not complex (Daugeron et al. 2001; Tucker et al. 2001). This large complex consists of two active 3′ to 5′ exonucleases (Ccr4 and Pop2/Caf1) and includes the Not1, Not2, Not3, Not4, Not5, Caf40, and Caf130 proteins (Denis and Chen 2003). In yeast, at least during mid-log growth, the major deadenylase in this complex is the Ccr4 protein, a member of the ExoIII nuclease family, since mutations in the active site of this enzyme give defects in deadenylation similar to the ccr4Δ strain (Chen et al. 2002; Tucker et al. 2002). Ccr4 also interacts directly with the Pop2 protein through a leucine-rich-repeat region (Clark et al. 2004).
Table 1 . General factors involved in mRNA deadenylation.
| Factor | Function | References |
|---|---|---|
| Ccr4/Pop2/Not complex | Major mRNA deadenylaseCcr4 critical catalytic subunit, ExoIII family memberPop2: second catalytic subunit, RNaseD family memberNot1: large scaffolding proteinNot2–5, Caf130, Caf40: accessory proteins of unknown function | Daugeron et al. (2001); Tucker et al. (2001, 2002); Chen et al. (2002) |
| Pan2/Pan3 complex | Additional mRNA deadenylasePrimarily functions in initial trimming of poly(A) tailPan2: catalytic subunit; RNaseD family memberInteracts with and stimulated by Pab1Pan3: regulatory subunit | Brown et al. (1996); Boeck et al. (1996); Brown and Sachs (1998) |
| Tpa1 | Prolyl 4-hydroxylaseBinds poly(A)Interacts with eRF1 and eRF3Required for normal deadenylation and translation termination | Keeling et al. (2006); Henri et al. (2010); Kim et al. (2010) |
| Pab1 | Major poly(A)-binding proteinInhibits Ccr4 deadenylaseStimulates Pan2/Pan3 complexMay interact with eRF3 to affect deadenylationCouples deadenylation to decapping | Caponigro and Parker (1995); Boeck et al. (1996); Cosson et al. (2002); Tucker et al. (2002); Hosada et al. (2003) |
| eRF3 (Sup35) | Subunit of translation termination complex | Hosada et al. (2003); Funakoshi et al. (2007) |
| Interacts with Pab1 and thereby may influence deadenylation rates | ||
| Rpb4/Rpb7 | Two subunits of RNA polymerase II | Lotan et al. (2005, 2007) |
| Required for normal rates of deadenylation | ||
| May exit from nucleus as part of mRNP to affect cytoplasmic deadenylation |
The Pop2/Caf1 protein, a member of the RNaseD family, is a second exonuclease in the Ccr4/Pop2/Not complex (Thore et al. 2003). Despite the presence of noncanonical residues in its active site, one report describes Pop2 purified from bacteria as having 3′ exonuclease in vitro (Thore et al. 2003). However, all of the catalytic activity of Ccr4/Pop2 complexes purified from yeast is dependent on the Ccr4 active site, suggesting that Ccr4 is the critical active deadenylase (Goldstrohm et al. 2007). Strains lacking Pop2/Caf1 show a defect in deadenylation, but this is due to Pop2 interacting with and promoting Ccr4 function since mutations in the active site of Pop2 do not alter deadenylation of reporter mRNAs and overexpression of Ccr4 suppresses the deadenylation defects seen in a pop2Δ strain (Tucker et al. 2002; Viswanathan et al. 2004). Since Pop2/Caf1 has catalytic activity and Pop2 orthologs in other organisms play catalytic roles in deadenylation (Goldstrohm and Wickens 2008), it remains a formal possibility that Pop2 may function as a deadenylase under some conditions or for some mRNAs.
The roles of the Not, Caf40, and Caf130 proteins in deadenylation are not yet clear. One possibility is that they adapt the deadenylase complex to different mRNAs through the action of regulatory proteins. Consistent with this possibility, defects in some of the Not proteins can affect deadenylation of specific mRNAs (Tucker et al. 2002). An alternative is that the Not and Caf accessory proteins play roles in other functions of the Ccr4/Pop2/Not complex, which has been suggested to have roles in transcription initiation and elongation (Deluen et al. 2002; Swanson et al. 2003; Qiu et al. 2004; Kruk et al. 2011).
A second deadenylase complex consists of the Pan2 and Pan3 proteins, with Pan2, a RNaseD family member, being the catalytic subunit (Boeck et al. 1996). Pan2 and Ccr4 appear to be the only major deadenylases since pan2Δ ccr4Δ strains are slow growing and show no deadenylation of reporter mRNAs (Tucker et al. 2001). The activity of Pan2 is promoted by Pab1 (Boeck et al. 1996), while Pab1 appears to inhibit the action of the Ccr4 complex (Tucker et al. 2002). This suggests that mRNAs with Pab1 bound to the poly(A) tail will be resistant to deadenylation by Ccr4/Pop2 but will be substrates for Pan2/Pan3. Thus, the specific deadenylase active on an mRNA will be influenced by the nature of the protein complex on its 3′ poly(A) tail.
The Pan2/Pan3 and Ccr4/Pop2/Not complexes appear to function in a temporal manner on most mRNAs with Pan2/Pan3 first acting to shorten the nascent poly(A) tail from ∼90 residues to ∼65, although this can vary a bit between different mRNAs (Brown and Sachs 1998). This step appears to happen quickly since the poly(A) tail lengths longer than 70 residues are typically not observed in yeast cells unless Pan2 is inactive (Brown and Sachs 1998). This implies that there is a difference between the accessibility of the 0–65 A residues of the poly(A) tail and the 3′-most 25 nucleotides. Since Pab1 promotes Pan2 activity, one model is that this deadenylation reflects Pab1 bound to the first ∼65 residues of the A tail, but the 3′ most region is exposed and thereby rapidly deadenylated by Pan2. Since pan2Δ strains show relatively normal deadenylation of reporter mRNAs (Tucker et al. 2001), Ccr4 then appears to be responsible for the continued deadenylation of the mRNA. Since the Ccr4 complex is inhibited by Pab1 (Tucker et al. 2002), this phase of deadenylation implies that Pab1 is at least partially dissociating from the poly(A) tail. However, in strains lacking Ccr4 activity, the Pan2 complex can continue to deadenylate mRNAs, although at a slower rate than Ccr4 (Tucker et al. 2001). Interestingly, the Pan2 complex stalls at an A tail of ∼20–25 residues (Daugeron et al. 2001; Tucker et al. 2001), which might be a length at which the Pab1 can no longer associate with the mRNA, and therefore Pan2 activity becomes limited (Tucker et al. 2001). Interestingly, once the poly(A) tail reaches an oligo(A) length of 10–12 residues, the mRNA can become a substrate for decapping and for binding of the Pat1/Lsm1–7 complex at the 3′ end (Tharun and Parker 2001; Chowdhury et al. 2007), which enhances the rate of decapping. This exchange of the Pab1 protein for the Pat1/Lsm1–7 complex is part of the mechanism that allows decapping to be promoted following deadenylation (see below).
Control of deadenylation:
Three types of interactions are known to modulate deadenylation rate, either generally or on specific mRNAs. First, because the Ccr4 and Pan2 deadenylases are influenced by the binding of Pab1 to the poly(A) tail, the rate of deadenylation is influenced by features of the Pab1–poly(A) interaction and its dynamics. Because Pab1–mRNA interactions are influenced by translation per se, this leads to deadenylation being coupled to aspects of translation. Second, key regulators of deadenylation on specific mRNAs are sequence-specific binding proteins that either directly, or indirectly, recruit the deadenylases to the mRNA to accelerate deadenylation. Finally, deadenylation is regulated in response to environmental cues, including stress and nutrient limitations.
On the basis of the biochemical analyses of deadenylases, a working model for understanding how deadenylation is affected by mRNP dynamics is that when Pab1 is present on the poly(A) tail, the Ccr4 deadenylase is inhibited and Pan2 is stimulated, whereas, when Pab1 dissociates, Ccr4 deadenylation is accelerated and Pan2 deadenylation is inhibited. Consistent with this view, self-association of Pab1 limits its binding to poly(A) and increases Ccr4-dependent deadenylation (Simon and Seraphin 2007; Yao et al. 2007). Moreover, strains defective in Pab1 show a defect in the initial rapid Pan2-dependent poly(A) shortening (Caponigro and Parker 1995; Morrissey et al. 1999; Simon and Seraphin 2007).
This model suggests that some of the effects of translation on deadenylation can be understood by their effects on Pab1 binding the poly(A) tail. For example, defects in translation initiation caused by a poor AUG context, a stem loop in the 5′ UTR, or mutations in translation initiation factors can increase the rates of deadenylation of yeast mRNAs (Muhlrad et al. 1995; Lagrandeur and Parker 1999; Schwartz and Parker 1999). One possibility is that defects in translation initiation either directly or indirectly destabilize Pab1 binding the poly(A) tail. Note that this model also predicts that Pan2-mediated deadenylation would be compromised by decreases in translation initiation. Surprisingly, in temperature-sensitive eIF4E strains, deadenylation of the Gal1 mRNA increases even in a ccr4Δ strain, which has been interpreted to suggest that eIF4E can also inhibit Pan2-based deadenylation (Lee et al. 2010). However, another possibility is that the “deadenylation” seen in a cdc33-1 ccr4Δ strain is not due to Pan2, but may be due to the cytoplasmic exosome (or to an unknown additional deadenylases), which is suggested by the observation that deadenylation is restored in a ccr4Δ pan2Δ strain when pab1 is mutated (M. Tucker and R. Parker, unpublished observation).
Deadenylation is also affected by aspects of translation termination. For example, premature translation termination accelerates poly(A) shortening as part of the process of NMD (see below and Cao and Parker 2003; Mitchell et al. 2003). This accelerated deadenylation may be a consequence of NMD leading to repression of translation and/or to dissociation of Pab1 from the mRNA, since decapping triggered by NMD is independent of the poly(A) tail (Muhlrad and Parker 1994). Similarly, the Tpa1 protein, a proline hydroxylase that binds poly(A) and interacts with translation termination factors, can influence the rate of deadenylation (Keeling et al. 2006; Henri et al. 2010).
The coupling of translation termination to deadenylation has been suggested to occur through direct interactions of the translation termination factor eRF3 with Pab1 (Cosson et al. 2002). This interaction appears to influence mRNA deadenylation since overexpression or deletion of the N-terminal domain of eRF3, where Pab1 interacts, leads to defects in deadenylation and mRNA decay (Kobayashi et al. 2004; Funakoshi et al. 2007). Since this effect seems to be primarily on the Ccr4 deadenylase (Funakoshi et al. 2007), one possibility is that eRF3–Pab1 interactions during translation termination transiently dissociate Pab1 from the poly(A) tail and increase deadenylation. However, it is important to note that translation termination is not required for deadenylation since mRNAs that never initiate translation due to stem-loop structures in their 5′ UTRs still deadenylate rapidly (Beelman and Parker 1994; Muhlrad et al. 1995).
Deadenylation also appears to be coupled to the process of transcription. The Rpb4 and Rpb7 subunits of RNA polymerase II are required for optimal deadenylation rates of yeast mRNAs (Lotan et al. 2005, 2007), and this has been proposed to occur by Rpb4 and Rbp7 loading on the mRNA during transcription to regulate cytoplasmic function (see Future Perspectives).
Control of deadenylation by mRNA-specific features:
There are now several examples of specific proteins that bind mRNAs in a sequence-specific manner to control deadenylation. Moreover, in many other cases, 3′ UTR elements modulate the poly(A) tail length, presumably by deadenylation, and identify a broad role of deadenylation regulation in gene expression (Beilharz and Preiss 2007). For example, the six members of the Puf protein family bind to specific-sequence 3′ UTR elements and regulate ∼10% of the yeast transcriptome (Olivas and Parker 2000; Gerber et al. 2004; Yosefzon et al. 2011). In yeast, the Puf1, Puf3, Puf4, and Puf5 proteins have all been shown to promote deadenylation and degradation of specific subsets of yeast mRNAs (Olivas and Parker 2000; Tadauchi et al. 2001; Hook et al. 2007; Ulbricht and Olivas 2008). Mechanistic studies have demonstrated that Puf5 promotes deadenylation at least in part by direct interaction with Pop2 and thereby recruitment of the Ccr4 deadenylase (Goldstrohm et al. 2006, 2007), although Puf proteins may also recruit the deadenylase complexes through other interactions. In addition, Puf proteins can also repress translation independently of deadenylation and therefore might also promote deadenylation indirectly (Chritton and Wickens 2011).
Other sequence-specific regulators of deadenylation include the Vts1 protein, which binds to a subset of yeast mRNAs through a specific stem-loop structure (Aviv et al. 2006) and recruits the Ccr4/Pop2 deadenylase (Rendl et al. 2008). Similarly, the Cth1 and Cth2 proteins are zinc-finger RNA-binding proteins that regulate the deadenylation of a subset of mRNAs, perhaps through interactions with Dhh1 that interacts with Pop2 (Puig et al. 2005; Pedro-Segura et al. 2008). One anticipates that a growing set of mRNA-specific binding proteins will regulate deadenylation either by direct recruitment of the deadenylase complexes or by inhibiting translation initiation and thereby indirectly promoting deadenylation.
Environmental control of deadenylation:
Deadenylation is also regulated on a global scale in response to environmental cues. For example, a variety of different stresses lead to a general inhibition of both Ccr4 and Pan2 deadenylation (Hilgers et al. 2006). Similar results occur in mammalian cells, suggesting that inhibition of deadenylation is a conserved aspect of the stress response (Gowrishankar et al. 2005, 2006). Inhibition of deadenylation during stress does not seem to require mRNAs to assemble in stress granules or P-bodies (see below), since deadenylation is still inhibited by stress in the presence of cyclohexmide (Hilgers et al. 2006), which prevents the formation of stress granules and P-bodies (Sheth and Parker 2003; Buchan et al. 2008). Deadenylation is also inhibited during stress. Since the stress response often leads to a global decrease in translation initiation, a general inhibition of deadenylation might be required to maintain a stable population of mRNAs. Deadenylation can also be reduced for some mRNAs when Hsp70 function is altered, which might mimic a stress response, although the basis or generality of this effect has not been determined (Duttagupta et al. 2003).
Normal rates of deadenylation also appear to be dependent on the activity of the Pkh1 and Pkh2 kinases, which are activated by sphingolipids (Luo et al. 2011). This suggests that aspects of mRNA metabolism are modulated in response to lipid signaling. This interpretation is also supported by the observation that, during heat stress, the formation of P-bodies, which are cytoplasmic mRNP aggregates of untranslating mRNAs, in conjunction with the mRNA decapping machinery (see below), requires sphingolipid synthesis, and exogenous phytosphingosine can stimulate P-body formation (Cowart et al. 2010). Interestingly, the effect of Pkh1 and Pkh2 on deadenylation rates is observed only in synthetic media, suggesting that this regulation is an integrative readout of both lipid and nutrient availability (Luo et al. 2011). An important area of future work will be to understand how deadenylation is regulated both globally and on specific mRNAs in response to environmental cues.
mRNA decapping
Nucleases of decapping and 5′ to 3′ degradation:
mRNA decapping is carried out by a complex of the Dcp1 and Dcp2 proteins and is influenced by several other factors (Table 2). Dcp2 is the catalytic subunit and is a member of the Nudix family of pyrophosphatases (Van Dijk et al. 2002; Steiger et al. 2003). Dcp2 cleaves the cap structure to release m7GDP and a 5′ monophosphate mRNA (She et al. 2008). Dcp1 is an EVH family protein (She et al. 2004) that interacts with Dcp2 to promote its catalytic activity (Deshmukh et al. 2008; She et al. 2008). The first 300 amino acids of Dcp2 are sufficient to promote decapping (Dunckley and Parker 1999) and fold into a two-domain structure wherein the N-terminal domain interacts with Dcp1 and the Nudix domain is present in the C-terminal domain (residues 100–245) (She et al. 2006, 2008). Dcp2 has a conserved region between 245 and 286, which contains binding sites for the Edc3 protein and possibly other decapping activators (Harigaya et al. 2010). Yeast Dcp2 has an extended C-terminal region that is not required for general mRNA decapping. Since Dcp2 can shuttle into the nucleus (Grousl et al. 2009) and this region has sites that can enhance transcription (Gaudon et al. 1999), one possibility is that this region plays some role in controlling transcription (Shalem et al. 2011).
Table 2 . Decapping and 5′ → 3′ exonuclease factors.
| Factor | Function | References |
|---|---|---|
| Dcp1/Dcp2 | mRNA decapping enzymeDcp2: catalytic subunit: Nudix family memberReleases m7GDP and 5′p-RNADcp1: stimulatory subunit, Evh1/WH1 family memberBlocked by eIF4E bound to cap | Schwartz and Parker (2000); She et al. (2004, 2008); Deshmukh et al. (2008) |
| Xrn1 | Major cytoplasmic 5′ to 3′ exonucleaseProcessive and requires 5′ monophosphateStimulated by Dcs1/Dcs2 | Kenna et al. (1993); Poole and Stevens (1995); Van Dijk et al. (2003); Malys et al. (2004); |
| Dcs1 (DcpS)/Dcs2 | mRNA decapping enzymes with preference for short RNAs | Liu and Kiledjian (2005); Jinek et al. (2011) |
| Releases m7Gp and ppN− | ||
| Cleaves m7GDP produced by Dcp1/Dcp2 to m7GMP and P | ||
| Can affect stress responses | ||
| Rat1 | Major nuclear 5′ to 3′ nuclease | Johnson (1997); Xiang et al. (2009) |
| Paralog of Xrn1 | ||
| Functions in nuclear RNA processing and decay | ||
| Rai1 | Interacts with and stimulates Rat1Contains mRNA cleavage siteReleases m7GpppN− and N−May function in cap quality control | Xue et al. (2000); Xiang et al. (2009); Jiao et al. (2010) |
| Pat1 | Activates general mRNA decappingServes as scaffolding protein for decapping complexesBoth represses translation initiation and stimulates Dcp2Interacts with Lsm1–7 complex and prefers to bind 3′ end of oligoadenylated mRNAPromotes P-body assemblyAfter deadenylation stabilizes 3′ ends to 3′ trimmingTarget of PKA kinase | Bouveret et al. (2000); Tharun et al. (2000); Chowdhury et al. (2007); Pilkington and Parker (2008); Nissan et al. (2010); Ramachandran et al. (2011) |
| Lsm1–7 complex | Required for efficient decappingForms heptometric ring complex and binds oligo- or deadenylated mRNAsMay promote Pat1 conformational change to activate Dcp2After deadenylation stabilizes 3′ ends to 3′ trimming | Boeck et al. (1998); Bouveret et al. (2000); Tharun et al. (2000); Chowdhury et al. (2007) |
| Dhh1 | Required for efficient decapping of translating mRNAsMember of ATP-dependent DExD/H box RNA helicase familyInhibits translation initiation in vitro upstream of 48S complex formationAccumulates in both stress granules and P-bodiesInteracts with Dcp2, Pat1, Scd6, Edc3 | Coller et al. (2001); Fischer and Weiss (2002); Coller and Parker (2005); Swisher and Parker (2009); Nissan et al. (2010) |
| Edc3 | RNA-binding proteinBinds and directly stimulates Dcp2Plays major role in aggregation of P-bodies and serves as scaffold for decapping factorsNot generally required for mRNA decapping unless Dcp1/Dcp2 is limited | Badis et al. (2004); Kshirsagar and Parker (2004); Decker et al. (2007); Dong et al. (2007); Harigaya et al. (2010) |
| Scd6 | RNA-binding protein related to Edc3Genetic interaction with Edc3 and synthetic decapping defect in edc3Δ scd6Δ Represses translation by binding eIF4GInteracts with Dhh1, Dcp2, Pat1May be mRNA-specific decapping/translation regulatory factor | Decourty et al. (2008); Nissan et al. (2010); Rajyaguru et al. (2011) |
| Edc1/Edc2 | Two small RNA-binding proteinsDirectly bind and stimulate Dcp1/Dcp2 | Dunckley et al. 2001; Schwartz et al. (2003); Neef and Thiele (2009); Borja et al. (2011) |
| Stm1 | Ribosome-binding protein | Balagopal and Parker (2009, 2011) |
| Can stimulate Dhh1-dependent decapping | ||
| Typically required only for subset of mRNAs decapping | ||
| Stalls 80S complex after translation initiation | ||
| Sbp1 | Abundant RNA-binding protein | Segal et al. (2006); Rajyaguru et al. (2011) |
| Overexpression suppresses pat1Δ defects by enhancing Dhh1 function | ||
| Binds eIF4G to repress translation initiation | ||
| Tif51A | Translation initiation factor eIF5A | Zuk and Jacobson (1998) |
| Specific mutations inhibit decapping | ||
| Mechanism is not known | ||
| Mrt4, Grc5, Sla2, Ths1 | Additional proteins affecting mRNA turnover by unknown mechanism | Zuk et al. (1999) |
Dcp2’s catalytic mechanism is a typical Nudix family reaction wherein Mg++ ions coordinated by a set of glutamic acid residues promote catalysis (Dunckley and Parker 1999; Steiger et al. 2003; She et al. 2006). Dcp2 catalysis is promoted by the closing of the bi-lobed Dcp2 structure to create a more active enzyme and more stable substrate binding (Deshmukh et al. 2008; She et al. 2008; Floor et al. 2010). Dcp1 is thought to enhance decapping by promoting the formation of this closed and more active structure (Deshmukh et al. 2008; She et al. 2008). The Dcp1/Dcp2 holoenzyme or Dcp2 alone prefer longer mRNA substrates in vitro, which is consistent with Dcp2 containing an extended RNA-binding site and having a reaction mechanism that consists of an initial binding to the substrate followed by sliding to the cap structure (Steiger et al. 2003). However, the presence of structures near the 5′ end is unlikely to inhibit decapping in vivo since even mRNAs with poly(G) tracts very near their 5′ end undergo rapid decapping in vivo (Muhlrad et al. 1994, 1995), presumably because Dcp2 catalysis is not generally rate limiting for decapping in vivo (see below).
Yeast cells contain additional decapping enzymes. The Dcs1 and Dcs2 proteins are members of the HIT family of pyrophosphatases and in vitro appear to cleave short RNA substrates (Liu et al. 2002). One function for Dcs1 in yeast is to cleave the m7GDP produced by decapping to m7GMP (Van Dijk et al. 2003), although how the m7GMP is further recycled is not known. Dcs2 can form heterodimers with Dcs1 and inhibit its activity, which occurs as cells enter diauxie (Malys and McCarthy 2006), although the significance of this effect is not clear. The nuclear Rai1 protein is known to function as a endonuclease that can cleave near the 5′ ends of mRNAs, and this has been suggested to function as a quality control mechanism for mRNA capping (Jiao et al. 2010).
Following decapping, mRNAs are degraded in a 5′ to 3′ direction by the Xrn1 nuclease (Hsu and Stevens 1993; Muhlrad et al. 1994), which prefers mRNA substrates with a 5′ monophosphate (Stevens 2001). Xrn1 has two highly conserved domains that fold into the active region of the enzyme, which is then stabilized by interactions with additional domains (Chang et al. 2011; Jinek et al. 2011). The active site of Xrn1 couples unwinding of duplexes to the processivity of the enzyme, which explains how it can degrade through structures without a helicase (Jinek et al. 2011). A paralog of Xrn1 is Rat1, which is typically localized to the nucleus and functions in nuclear RNA processing and/or degradation pathways (see below). However, Rat1 can substitute for Xrn1 when it is localized to the cytoplasm due to mutation, indicating that no Xrn1-specific protein–protein interactions are required for mRNA degradation (Johnson 1997). Xrn1 is inhibited by the adenosine 3′, 5′ biphosphate (pAp), which is produced by sulfate assimilation (Dichtl et al. 1997), and cells can utilize this circuit to limit Xrn1 activity during various responses (Benard 2004; Todeschini et al. 2006).
Model of mRNA decapping:
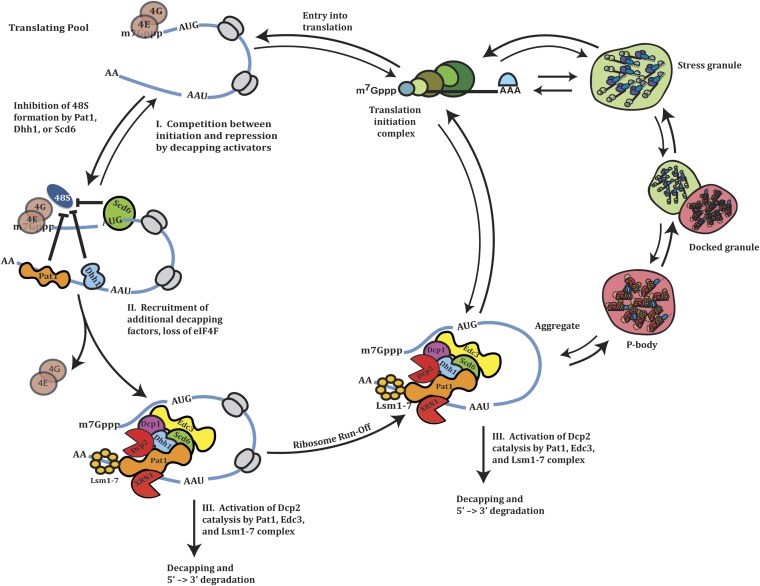
A working model for mRNA decapping has three critical steps (Figure 2). First, the 5′ cap structure must be exposed, and therefore the cytoplasmic cap-binding complex consisting of eIF4E and eIF4G needs to be lost from the mRNA. Second, the decapping enzyme must be recruited to the mRNA, which appears to be coordinated with the formation of a larger decapping complex, including the decapping enzyme, Xrn1, and several decapping activators (see below). Third, catalysis by Dcp2 occurs, leading to rapid 5′ to 3′ degradation of the mRNA. Consistent with this model, proteins enhancing decapping can function by interfering with translation initiation factors, by binding RNA and forming scaffolds for assembly of the decapping machinery, or by promoting Dcp2 catalysis. Untranslating mRNPs complexed with the decapping machinery can also aggregate into cytoplasmic RNP granules referred to as P-bodies although the specific role of these macromolecular complexes is not yet clear (see below).
Figure 2 .
Model for mRNA decapping.
Decapping and translation initiation:
Several observations argue that decapping is in competition with translation initiation and that decapping requires the loss of the cap-binding complex. This was first suggested since the cap structure, which is recognized for decapping, also functions in promoting translation initiation by recruiting the eIF4E/eIF4G translation initiation complex. Moreover, when translation initiation is decreased by mutations in translation initiation factors, a poor AUG context, or 5′ UTR structures, there is a concomitant increase in decapping rates (Muhlrad et al. 1995; Lagrandeur and Parker 1999; Schwartz and Parker 1999). In addition, the eIF4E cap-binding protein can directly inhibit decapping in vitro (Schwartz and Parker 2000). These observations argue that decapping requires the mRNP to exchange the eIF4E/eIF4G cap-binding complex for the decapping enzyme.
Stimulation of decapping by general activators:
Several protein factors, referred to as either decapping enhancers or activators, are known to function to stimulate the rate of decapping in vivo (Table 2). The core set of proteins affecting decapping includes Dhh1, a DEAD-box helicase, Pat1, Edc1, Edc2, Edc3, Scd6, and the Lsm1–7 complex. Some of these decapping activators promote decapping by inhibiting translation initiation. For example, Dhh1, a member of the DEAD family of ATPases, represses translation in vitro, and its overexpression in cells inhibits translation and leads to the accumulation of cytoplasmic mRNP granules (Coller and Parker 2005; Swisher and Parker 2010; Carroll et al. 2011). Similarly, Pat1, Scd6, and Stm1, which affect the decapping of some mRNAs (Balagopal and Parker 2009), repress translation both in vivo and in vitro (Pilkington and Parker 2008; Nissan et al. 2010; Balagopal and Parker 2011; Rajyaguru et al. 2012).
Decapping activators can inhibit translation at different steps. For example, the Pat1, Dhh1, and Scd6 proteins all appear to block translation before the formation of a 48S pre-initiation complex (Coller and Parker 2005; Nissan et al. 2010). For Scd6, this translation repression appears to occur by direct binding to eIF4G and inhibition of the joining of the 43S complex (Rajyaguru et al. 2012). In contrast, the Stm1 protein, which promotes decapping of a subset of yeast mRNAs (Balagopal and Parker 2009), inhibits translation after formation of an 80S complex, likely through direct interactions with the ribosome (Balagopal and Parker 2011). An unresolved issue is how inhibition of translation initiation by these factors leads to decapping. One possibility is that, by stalling initiation, it gives more time for dissociation of the translation initiation factors to allow for decapping complexes to associate with the mRNA. Alternatively, such a transition may involve an ordered exchange of factors on the mRNA, which is suggested by decapping activators, such as Pat1 and Scd6, that can directly interact with translation factors and the decapping enzyme (Nissan et al. 2010; Rajyaguru et al. 2012). An important area for future research is determining how mRNPs are remodeled to allow decapping complexes to form and degrade the mRNA.
A second role of decapping activators is to promote the assembly of a larger decapping complex. The core set of decapping components shows an extensive network of direct interactions as determined by protein-binding experiments with recombinant proteins and supported by co-ip and two-hybrid analyses (Decker et al. 2007; Nissan et al. 2010). On the basis of coimmunoprecipitation (co-ip) experiments and the dependence of interactions on RNA, there appear to be two complexes that assemble on mRNAs targeted for decapping. One complex consists of the Pat1 protein, the Lsm1–7 complex, and Xrn1 (Bouveret et al. 2000; Tharun et al. 2000; Tharun and Parker 2001). This complex is thought to assemble on the 3′ end of deadenylated mRNAs on the basis of its binding specificity in vitro (Chowdhury et al. 2007) and the exonuclease trimming of the 3′ end of deadenylated mRNAs in pat1Δ or lsm1Δ strains (Boeck et al. 1998; Tharun et al. 2000; He and Parker 2001). A second set of interacting proteins consists of the Dcp1, Dcp2, Edc3, or Scd6 and Dhh1, although whether all these factors can associate at the same time remains to be determined. Within and between these complexes, Pat1 and Edc3 appear to play important scaffolding roles and interact with many components of the decapping machinery (Decker et al. 2007; Nissan et al. 2010).
A third role of decapping activators is to directly stimulate decapping by Dcp2. For example, the Edc3 and Pat1 proteins directly bind Dcp2 and enhance its activity in purified systems (Harigaya et al. 2010; Nissan et al. 2010). Similarly, the paralogs Edc1 and Edc2, which are high-copy suppressors of temperature-sensitive alleles in Dcp1 or Dcp2 (Dunckley et al. 2001), bind RNA and stimulate Dcp2 either in extracts or in reconstituted systems (Schwartz et al. 2003; Steiger et al. 2003). Edc1, and presumably Edc2 as well, directly bind Dcp1 to stimulate the decapping enzyme by enhancing both Km and kcat of Dcp2 (Borja et al. 2011).
Several observations suggest that Dcp2 catalysis is not generally rate limiting for decapping in vivo. First, conditional mutations in Dcp1 or Dcp2 that compromise decapping activity in vitro do not significantly affect mRNA decapping rates in vivo (Tharun and Parker 1999; Dunckley et al. 2001; Steiger et al. 2003). Second, strains lacking Edc1, Edc2, and/or Edc3 do not show defects in mRNA decay rates in vivo unless the decapping ability of Dcp2 is reduced by mutation (Dunckley et al. 2001; Kshirsagar and Parker 2004). Third, deletion of the C-terminal domain of Pat1, which stimulated Dcp2 in vitro, has only a marginal effect on mRNA decapping rates in vivo (Pilkington and Parker 2008; Nissan et al. 2010). Taken together, these observations suggest that, at least in mid-log growth, the rate-limiting step in decapping is the translation repression and assembly of a decapping complex on the mRNP.
Several observations suggest that Pat1 and the Lsm1–7 complex function in an mRNP rearrangement that enhances decapping activity. First, lsm1Δ strains accumulate mRNA and the decapping machinery in P-bodies, arguing that the Lsm1–7 complex functions at a late stage in decapping after the mRNA has exited translation (Sheth and Parker 2003; Teixeira and Parker 2007). Second, the middle and carboxy-terminal domains of Pat1 interact with themselves, and, while the C-terminal domain is sufficient to bind Dcp2 and activate decapping in vitro, its interaction with Dcp2 is blocked by the middle domain of Pat1 (Nissan et al. 2010). This raises the possibility that, for Pat1 to interact with Dcp2 and activate decapping catalysis, a conformational change in Pat1 between the middle and C-terminal domains is required. Strikingly, the Lsm1–7 complex appears to interact with both the middle and the C-terminal domain of Pat1 (Pilkington and Parker 2008; Nissan et al. 2010). Thus, a working model is that the binding of Pat1 to the mRNA with the Lsm1–7 complex allows for the formation of a binding site for Dcp2 in Pat1’s C-terminal domain that is sufficient to activate catalysis.
Several other factors have been identified as promoting decapping (Zuk et al. 1999; Table 2). Most notably, mutations in one of the genes encoding eIF5a lead to slower decapping of mRNAs, perhaps because of alteration to translation initiation and/or translation elongation (Zuk and Jacobson 1998; Saini et al. 2009). The Stm1 and Sbp1 proteins interact with the ribosome and eIF4G, respectively, and thereby appear to promote the ability of Dhh1 to promote decapping (Segal et al. 2006; Balagopal and Parker 2009, 2011; Rajyaguru et al. 2012), although the mechanism by which these proteins stimulate Dhh1 function is unknown.
Control of decapping:
Poly(A) tails as inhibitors of decapping:
Several observations argue that the ability of the poly(A) tail to inhibit decapping is partially mediated through the poly(A)-binding protein (Pab1). First, it has been demonstrated in yeast that decapping occurs when the poly(A) tail length has been shortened to an oligo(A) length of ∼12 residues (Decker and Parker 1993). This is approximately the minimum length required for Pab1 binding (Sachs et al. 1987). Second, in pab1 mutant strains, decapping is uncoupled from deadenylation (Caponigro and Parker 1995; Morrissey et al. 1999). In this case, intermediates in mRNA decay, trapped by inhibiting 5′ to 3′ degradation in cis with strong secondary structures, are produced as decapped mRNA fragments with long poly(A) species (Caponigro and Parker 1995). This indicates that, in the absence of Pab1, the absolute requirement for prior deadenylation before decapping is not necessary.
A second reason for decapping occurring after deadenylation is that the Pat1/Lsm1–7 complex prefers to bind oligoadenylated mRNAs in vitro (Chowdhury et al. 2007) and associates with mRNAs after deadenylation in vivo (Tharun and Parker 2001). Thus, a working model is that polyadenylated mRNAs are protected by Pab1 binding and by promoting translation initiation, and deadenylated mRNAs are enhanced for decapping by the Pat1/Lsm1–7 complex binding and promoting translation repression and decapping (Tharun 2009).
Control decapping on specific mRNAs:
Differences in mRNA decapping rates appear to arise due to two features of individual mRNAs. First, since decapping is inversely related to translation initiation, features that inherently decrease translation initiation are expected to increase decapping rates. For example, the faster decapping rate of the MFA2 mRNA as compared to the PGK1 mRNA is due to a poor AUG context on the MFA2 mRNA (Lagrandeur and Parker 1999). In addition, mRNAs contain sites for sequence-specific mRNA-binding proteins that enhance decapping. For example, the binding of Puf3 to the Cox17 mRNA enhances decapping of this mRNA following deadenylation (Olivas and Parker 2000).
An important correlation is that many features that increase the decapping rate—including 3′ UTR or coding region elements, AUG context, or 5′ stem-loop structures—also increase deadenylation rate (Muhlrad and Parker 1992; Caponigro et al. 1993; Muhlrad et al. 1995; Caponigro and Parker 1996; Lagrandeur and Parker 1999; Olivas and Parker 2000). This suggests that a critical aspect of controlling cytoplasmic mRNA function is an exchange of mRNAs between mRNPs complexed with translation factors and protected from decay and an mRNP associated with decay factors and having reduced translation rate and enhanced deadenylation and decapping. A key issue for future work is determining the specifics of these mRNPs and how transitions between both states occur.
Decapping of specific mRNAs can also be triggered independently of deadenylation. For example, the Edc1 mRNA is decapped without prior deadenylation, and this is due to a poly(U) tract in the 3′ UTR that sequesters the poly(A) tail and renders it nonfunctional (Muhlrad and Parker 2005). Alternatively, in an autoregulatory loop, the Rps28b mRNA assembles a decapping complex through Rps28 binding to a specific stem loop in its 3′ UTR that allows decapping to proceed independently of deadenylation (Badis et al. 2004). An unresolved issue is how many different modes of decapping exist in yeast cells and therefore how the diversity of mRNA decay rates is achieved.
Decapping can also be negatively regulated on specific mRNAs. For example, the Khd1 protein binds to and limits the decapping on the Mtl1 mRNA (Mauchi et al. 2010). Since Khd1 can bind eIF4G and inhibit translation (Paquin et al. 2007), this may allow for the formation of a mRNP complex that is limited for translation, but protected from decapping. Since Khd1 can affect mRNA localization (Hasegawa et al. 2008), such a mechanism may be important in keeping mRNAs in a stable, but untranslating, state while the mRNAs localize to specific regions of the cell. This may be a common mechanism of mRNA control since multiple translation repressors that can affect decapping also bind directly to eIF4G and can repress translation (Rajyaguru et al. 2012). Similarly, the Pub1 protein binds to, and stabilizes, a significant subset of yeast mRNAs, presumably by inhibiting decapping, although this has not been directly demonstrated (Ruiz-Echevarria and Peltz 2000; Duttagupta et al. 2005).
mRNA decapping and P-bodies:
The mRNA degradation machinery can also be concentrated in specific cytoplasmic mRNP aggregates referred to as P-bodies (Sheth and Parker 2003; see Figure 2). P-bodies are aggregates of untranslating mRNAs that are associated with the mRNA decapping machinery, and to a lesser extent, with the deadenylases (Parker and Sheth 2007). P-bodies are proportional to the pool of untranslating mRNA associated with the decapping machinery (Teixeira et al. 2005). Consistent with this view, if mRNAs are trapped in polysomes, P-bodies decrease, whereas decreases in translation initiation increase the pool of mRNAs in P-bodies (Teixeira et al. 2005). Moreover, when mRNA decay is limited at the actual step of decapping or 5′ to 3′ degradation, P-bodies increase (Sheth and Parker 2003; Teixeira and Parker 2007). mRNAs within P-bodies can return to translation (Brengues et al. 2005), and this may occur through the transition of mRNAs from P-bodies to stress granules, which are aggregates of untranslating mRNAs assocatied with translation initation factors and RNA-binding proteins (reviewed in Buchan and Parker 2009). Note that the putative transfer of an mRNA from a P-body to a stress granule would correspond to an exchange of the mRNA decapping machinery for translation initiation factors (Buchan et al. 2008). This constitutes an “mRNA cycle” wherein mRNAs can exit translation either for degradation or to eventually return (Figure 2), which may play a role in the regulation of both translation and mRNA degradation (Balagopal and Parker 2009). Interestingly, recent genomic analyses have identified at least 400 different mRNAs, compromising a large percentage of the transcriptome, that can recycle from repression to translation upon stress relief (Arribere et al. 2011).
An unresolved issue is the role of P-body formation per se in the decapping of mRNAs. The aggregation of individual mRNPs into larger P-bodies is largely dependent on the YjeF domain of the Edc3 protein (Decker et al. 2007), although P-bodies can still form to some extent on the basis of a “prion” domain on Lsm4 (Decker et al. 2007; Reijns et al. 2008) and some Pat1-dependent aggregation (Buchan et al. 2008). How these aggregation motifs affect mRNA decay rates is unclear. In one report, strains lacking Edc3 and/or the aggregation domain of Lsm4 did not show any changes in the decay rates of the MFA2 mRNA (Decker et al. 2007), although in another report a strain lacking the Lsm4 aggregation motif did show a modest change in MFA2 mRNA decay rates (Reijns et al. 2008). The current simplest interpretation is that the formation of large P-body aggregates is not required for mRNA decapping but might affect the rate of decapping either in certain conditions or for subsets of mRNAs.
Relationship of decapping to ongoing translation elongation:
An unresolved issue is how ongoing translation elongation affects the decapping of yeast mRNAs. Some evidence argues that decapping can be inhibited by elongating ribosomes. First, inhibition of translation elongation by chemicals such as cycloheximide or sodarin leads to decreases in the rate of decapping (Beelman and Parker 1994; Cereghino et al. 1995; S. Jain and R. Parker, unpublished observation). However, this could also be due to indirect effects since cycloheximide can stabilize mRNAs that are never translated as well (Beelman and Parker 1994). Second, the rate of decapping of NMD substrates is proportional to the length of the ORF (Cao and Parker 2003). Moreover, in some cases, shortening the length of the ORF can lead to faster decay of specific mRNAs, although this could be due to loss of specific stabilizing sequences (Heaton et al. 1992). These results raise the possibility that mRNAs harboring elongating ribosomes have slower rates of decapping than ribosome-free mRNAs.
In contrast, several observations argue that decapping can occur while ribosomes are still associated with mRNAs. First, in the presence of cycloheximide mRNAs are seen to be shortened from their 5′ ends and degraded in a 5′ to 3′ manner to internal sites that are thought to be stalled ribosomes, although this rate is slower than normal rates of decapping (Beelman and Parker 1994; Cereghino et al. 1995). Similarly, mRNAs with strong stalls in translation elongation have been argued to generate mRNA fragments by decapping and 5′ to 3′ degradation to the stalled ribosomes (Hu et al. 2009), but more recent studies argue that these mRNA fragments may be produced by an endonuclease cleavage triggered by stalled ribosomes, referred to as no-go decay (NGD; see below) (D. Muhlrad and R. Parker, unpublished data). Additional evidence for decapping occurring on mRNAs engaged in elongation is that decapped mRNAs, either in wild-type cells or in xrn1Δ strains, appear to associate with polysomes, suggesting they are decapped while bound to elongating ribosomes (Hu et al. 2009). Taken together, the best current interpretation is that decapping can occur while mRNAs are associated with ribosomes, but that ribosomes may also play a role in limiting the rates of decapping.
Regulation by signal transduction paths:
Multiple signal transduction pathways impinge on mRNA decapping. First, Ste20 has been observed to phosphorylate Dcp2 and affect its assembly into P-bodies. Ste20 phosphorylation of Dcp2 also affects the degradation of certain mRNAs (Yoon et al. 2010). Second, Pat1 is a target of PKA, and phosphorylation of Pat1 limits its ability to assemble into P-bodies (Ramachandran et al. 2011), although how this affects specific protein interactions or mRNA decay is not clear. Third, when the growth regulatory Tor kinase is inhibited, the activation of the Rim15 kinase leads to phosphorylation of the paralogous Igo1 and Igo2 proteins, which then interact with Dhh1 and stabilize mRNAs required for entry into G0 (Talarek et al. 2010). One anticipates that other signal transduction pathways will regulate the mRNA turnover machinery. On the basis of genome-wide studies Dhh1, Edc1, Edc3, Pop2, Ccr4, Xrn1, Dcp2, and Dcp1 are also known to be phosphoproteins and could be targets of such signal transduction pathways.
3′ to 5′ mRNA degradation
The second pathway of mRNA decay following deadenylation is 3′ to 5′ degradation, which is catalyzed by the exosome and various cofactors (Table 3) (Anderson and Parker 1998). The exosome is a multiprotein complex consisting of 10 main proteins, including six members of the RNase PH protein family and three small RNA-binding proteins (Allmang et al. 1999), which together form a ring structure analogous to bacterial PNPase (Liu et al. 2006), and the Rrp44/Dis3 protein, which has both an exonuclease and endonuclease domain (Lebreton et al. 2008; Schaeffer et al. 2009). In addition to its roles in the cytoplasm, the exosome is involved in numerous nuclear RNA processing and degradation processes (see below and reviewed in Lykke-Andersen et al. 2009). In the nucleus, the exosome is also associated with Rrp6, another 3′ to 5′ exonuclease, Rrp47, and Mpp6 (Mitchell et al. 2003; Milligan et al. 2008; Synowsky et al. 2009), which have roles in nuclear function of the exosome (see below).
Table 3 . Exosome and associated proteins involved in 3′ to 5′ degradation of RNAs.
| Component | Features | Reference |
|---|---|---|
| Core exosome | Six RNasePH domain proteins (no active sites) Rrp41, Rrp42, Rrp43, Rrp45, Rrp46, Mtr3 | Lykke-Andersen et al. (2011) |
| 3 RNA-binding subunits (Rrp4, Rrp40, Csl4) | ||
| One catalytic subunit, Rrp44, with both endo and exo active sites | ||
| Functions in both RNA processing and degradation in cytoplasm and nucleus | ||
| Cytoplasmic cofactors | ||
| Ski7 | Binds Csl4 subunit of coreRequired for 3′ to 5′ decay of mRNAsGTPase domain required for non-stop decayInteracts with Ski2/Ski3/Ski8 complex | Van Hoof et al. (2000b, 2002); Wang et al. (2005) |
| Ski2/Ski3/Ski8 complex | Required for 3′ to 5′ mRNA decaySki2 is member of ATPase RNA helicase familySki8 is WD40 proteinSki3 may function as scaffold | Anderson and Parker (1998); Brown et al. (2000); Araki et al. (2001); Wang et al. (2005) |
| Nuclear cofactors | ||
| Rrp6 | 3′ to 5′ exonuclease of RNAseD family | Butler and Mitchell (2011) |
| Associated with nuclear exosome | ||
| Required for RNA processing and decay of RNAs in the nucleus | ||
| Functions in retention of aberrant mRNAs at sites of transcription | ||
| Rrp47 (Lrp1) | RNA-binding protein | Butler and Mitchell (2011) |
| Required for RNA processing and nuclear RNA decay | ||
| Associated with nuclear exosome | ||
| Mpp6 | RNA-binding protein | Milligan et al. (2008) |
| Associated with nuclear exosome | ||
| Required for RNA processing and nuclear RNA decay | ||
| Tramp complexes (Trf4 or Trf5) | Consist of Mtr4 with 1 noncanonical poly(A) polymerase (Trf4 or Trf5) and 1 RNA-binding protein (Air1 or Air2)Required for several RNA-processing and nuclear RNA decay pathwaysCan promote processing/degradation in poly(A)-dependent and -independent manners by recruiting exosome to substrates | Houseley and Tollervey (2006); San Paolo et al. (2009); Butler and Mitchell (2011) |
Despite the similarity of the core ring domain to active exonucleases, the only active nuclease sites in the exosome appear to be present in the Rrp44/Dis3 protein (Liu et al. 2006; Dziembowski et al. 2007). The exosome is then thought to function by the core ring structure serving as a binding platform for proteins targeting the exosome to various substrates and to channel the RNA to the active sites of Rrp44/Dis3 (Bonneau et al. 2009).
For the degradation of cytoplasmic mRNA, the exosome requires the Ski proteins (Anderson and Parker 1998; Van Hoof et al. 2000b; Araki et al. 2001). The Ski7 protein appears stably bound to the cytoplasmic exosome through the Ski4 subunit (Van Hoof et al. 2002). The Ski2, Ski3, and Ski8 proteins form a separate protein complex (Brown et al. 2000; Wang et al. 2005). The Ski2/3/8 complex interacts with Ski7 (Araki et al. 2001; Wang et al. 2005), and this interaction, which appears to occur between Ski7 and the Ski3 and Ski8 proteins, is required for 3′ to 5′ degradation of mRNAs. The Ski2 protein is an ATPase of the RNA helicase family and presumably utilizes the energy of ATP hydrolysis to unwind substrates and/or dissociate bound proteins to deliver the RNA to the exosome. Interestingly, RNA processing and degradation by the nuclear exosome require the related ATPase Mtr4, suggesting that this is a general feature of exosome function (reviewed in Lykke-Andersen et al. 2009).
There are several unresolved issues with regards to 3′ to 5′ degradation of mRNAs. For example, the role of the Ski complex is not well understood. Moreover, although mRNAs can have differences in their rates of 3′ to 5′ degradation (Cao and Parker 2001), the features of mRNAs that dictate different rates of 3′ to 5′ degradation are not understood. In mammalian cells, sequence-specific RNA-binding proteins can recruit the exosome directly to mRNAs, and similar events might exist in yeast cells (e.g., Chen et al. 2001). Finally, whether some mRNAs are normally degraded by the exosome or whether 3′ to 5′ degradation is the major pathway of cytoplasmic mRNA under some growth conditions is not resolved.
Other mRNA decay pathways
Some observations imply that there will be two additional mechanisms by which yeast mRNAs are degraded. First, the vacuolar nuclease Rny1 can degrade tRNAs, rRNAs, and even small nuclear RNA (snRNAs) (Thompson and Parker 2009; N. Luhtala and R. Parker, unpublished observations). Given this, one anticipates some mRNAs will also be degraded by Rny1 either during the process of autophagy or specific targeting of some mRNAs to the vacuole or because Rny1 can enter the cytosol and degrade mRNAs under some conditions (Thompson and Parker 2009). A second nuclease that probably targets mRNAs during stress is Nuc1. Nuc1 is a general nuclease that is localized to the mitochondria, but during stress or in high cell densities is released to the cytosol through the mitochondrial porins and then is transported to the nucleus where it can play a role in apoptosis (Buttner et al. 2007). Interestingly, Nuc1 is also known to target the mRNAs produced by the double-stranded RNA killer virus, suggesting that it could also target some, or all, cytoplasmic mRNAs under these conditions (Liu and Dieckmann 1989). A potential role of Nuc1 in mRNA decay is suggested by its negative genetic interactions with Ski2, -3, -7, -8, and Xrn1 (Costanzo et al. 2010).
mRNA Quality Control Pathways
Cytoplasmic quality control
Several cytoplasmic quality control mechanisms degrade eukaryotic mRNAs that are defective in translation (Doma and Parker 2007). An emerging principle is that aberrant mRNAs are distinguished from the normal mRNAs by adaptor proteins that interact with the translation machinery and direct the aberrant mRNA into a degradation pathway. Key issues for each quality control pathway are the biological role, the specificity of distinguishing normal from aberrant mRNAs, and the mechanism by which mRNAs are recognized and degraded.
Nonsense-mediated decay:
NMD is an mRNA quality control system that degrades mRNAs with aberrant translation termination. NMD was first described as a system that degrades mutant mRNAs with premature translation termination codons (Losson and Lacroute 1979). However, NMD degrades a wide variety of mRNAs that have aberrant translation termination events. Such substrates include mRNAs with long 3′ UTRs that alter the relationship of the poly(A) tail to the stop codon (Muhlrad and Parker 1999a; Kebaara and Atkin 2009; Deliz-Aguirre et al. 2011), mRNAs with alternative translation initiation sites that that are out of frame with the main ORF and lead to premature termination (Welch and Jacobson 1999), mRNAs with upstream ORFs (Gaba et al. 2005; Guan et al. 2006), pre-mRNAs that contain introns with stop codons (He et al. 1993; Sayani et al. 2008), and mRNAs with frameshifts, where a proportion of the ribosomes are shifted into alternative reading frames containing premature termination codons (Belew et al. 2011). In addition, one anticipates that errors in transcription or mis-splicing that introduce premature stop codons will generate substrates for NMD at a low level across many different genes.
Consistent with this wide range of substrates, several genomic analyses have revealed that NMD targets a wide range of different mRNAs. As such, NMD is not just a quality control system but is also utilized by cells to degrade a subset of “normal” mRNAs, particularly those involved in cell-surface dynamics and chromosome structure (Lelivelt and Culbertson 1999; He et al. 2003; Guan et al. 2006). For many of these mRNAs it is not clear why they are substrates of NMD. One possibility is that they are lacking features that specify proper translation termination and as such are targeted by NMD (see below).
Substrates for NMD are identified by the action of the interacting Upf1, Upf2, and Upf3 proteins (reviewed in Baker and Parker 2004). The recognition of an mRNA by the NMD pathway has several effects on the metabolism of the mRNA. Specifically, the mRNA is targeted for enhanced deadenylation (Muhlrad and Parker 1994; Cao and Parker 2003; Mitchell and Tollervey 2003), rapid deadenylation-independent decapping (Muhlrad and Parker 1994), slightly increased rates of 3′ to 5′ degradation after deadenylation (Cao and Parker 2003; Mitchell and Tollervey 2003), and translation repression (Muhlrad and Parker 1999b). Consistent with NMD targeting mRNAs for translation repression, when decapping or 5′ to 3′ degradation is blocked, NMD substrates accumulate as repressed mRNAs in P-bodies in an Upf1-dependent manner (Sheth and Parker 2006).
NMD has been suggested to be coupled to degradation of the nascent peptide in an Upf1-dependent manner (Kuroha et al. 2009). In this manner, not only would the mRNA be degraded, but any potential dominant-negative peptides produced would also be rapidly destroyed. Interestingly, Upf1 has been suggested to have ubiquitin ligase activity, and mutations that affect this activity alter the process of NMD for RNA degradation (Takahashi et al. 2008), although the sites of these mutations would also be predicted to disrupt Upf1 interaction with Upf2, which is known to be required for NMD (He et al. 1996, 1997; Clerici et al. 2009). However, how general NMD stimulated protein decay remains to be established since a peptide from a different NMD mRNA substrate shows the same decay rates in wild-type and upf1Δ cells (Muhlrad and Parker 1999b).
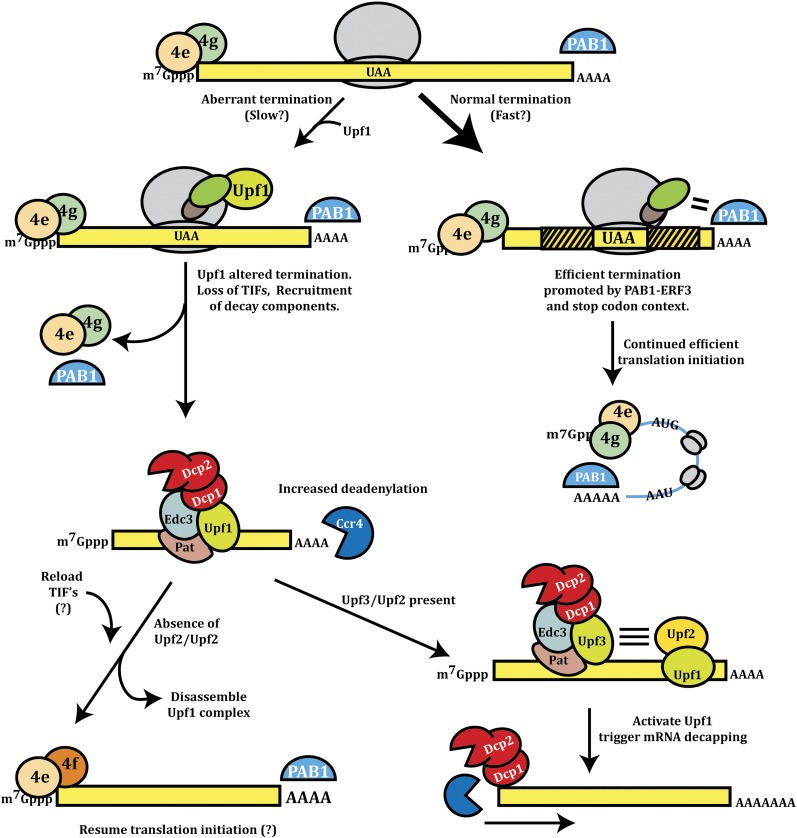
Transcripts appear to be targeted for the diverse effects of NMD in two steps (Figure 3). Several observations suggest that, in an initial step, Upf1, which is a member of the SF1 protein superfamily of nucleic acid helicases (Fairman-Williams et al. 2010), interacts with the translation termination complex and alters the nature of translation termination (reviewed in Baker and Parker 2004). The most direct evidence is that a ribosome toeprint generated at a normal translation termination codon is distinct from the toeprint of a ribosome terminating at a premature termination codon in a manner dependent on Upf1p (Amrani et al. 2004). In addition, the Upf1, -2, and -3 proteins co-immunoprecipitate with the eukaryotic translation termination factors eRF1 and/or eRF3 (Czaplinski et al. 1998; Wang et al. 2001). Although upf1Δ mutants can show increased rates of stop-codon readthrough, this effect appears to be due to stabilization and increased expression in the upf1Δ strain of a magnesium transporter, which increases intracellular Mg++ and leads to increased miscoding of stop codons (Johansson and Jacobson 2010). Interestingly, a defect in upf2Δ and upf3Δ strains in stop codon readthrough can be suppressed by overexpression of Upf1, suggesting that Upf1 directly affects translation, independently of Upf2 and Upf3, of mRNAs with some stop codons, perhaps by inhibiting translation initation (Muhlrad and Parker 1999b; Maderazo et al. 2000). Consistent with this view, Upf1 can associate with polysomes independently of Upf2 and Upf3 (Atkin et al. 1997). The upf2Δ and upf3Δ strains may have decreased Upf1 function since in these strains Upf1 accumulates in P-bodies, and therefore the majority of Upf1 may not be available to affect translation termination (Sheth and Parker 2006). Moreover, because NMD substrates accumulate in P-bodies independently of Upf2 and Upf3, the simplest model is that Upf1 is sufficient to repress translation of the mRNA (Sheth and Parker 2006).
Figure 3 .
Model for the nonsense-mediated decay.
A second step in NMD appears to be the interaction of Upf2 and Upf3 with Upf1, and this triggers the degradation of the mRNA. This conclusion is supported by the Upf2 and Upf3 independent effects of Upf1 on translation readthrough and targeting of NMD substrates to P-bodies (Maderazo et al. 2000; Sheth and Parker 2006). Upf3 is an RNA-binding protein that is proposed to load on mRNAs in the nucleus (Shirley et al. 2002). Upf2 interacts with both Upf3 and Upf1, and its binding to Upf1 reduces the Upf1–RNA interaction and enhances the helicase activity of Upf1 (Chakrabarti et al. 2011). This suggests that, following translation termination altered by Upf1, interaction of Upf2 with Upf1 would enhance Upf1 catalytic properties and lead to mRNP rearrangements that trigger mRNA degradation, possibly by rearrangements of the mRNP or by altering the fate of the terminating ribosome (Ghosh et al. 2010). Consistent with that model, mutations inactivating the ATPase activity of Upf1 also accumulate mRNAs in P-bodies (Sheth and Parker 2006).
A key issue is how the specificity of NMD is determined. In principle, an mRNA will be targeted for NMD on the basis of the nature of translation termination (is it “aberrant” or not?) and whether Upf2/Upf3 can influence Upf1 after altered termination. Thus, the specificity of NMD is determined by factors that influence translation termination and whether Upf2/Upf3 is associated with the mRNA after termination. One factor that contributes to proper translation termination is Pab1, which is known to interact with the translation termination factors, and, when tethered to the mRNA near a premature stop codon, can prevent NMD on that mRNA (Amrani et al. 2004). However, strains lacking Pab1, as well as poly(A)-mRNAs, still show Upf1-dependent degradation of mRNAs with premature stop codons, indicating that additional factors also contribute to NMD targeting, although whether those factors influence Upf1 effects on termination or effect a downstream step in NMD is not resolved (Caponigro and Parker 1995; Meaux et al. 2008). Thus, a key issue is determining what other features of an mRNA influence the nature of translation termination and Upf2/Upf3 interaction with the mRNA. Although currently controversial, one potential contribution is elements in coding regions, sometimes referred to as downstream sequence elements (reviewed in Gonzalez et al. 2001), that might recruit Upf3 and Upf2 to mRNAs, and, if Upf2 and Upf3 are not removed by elongating ribosomes, might lead to triggering NMD after upstream termination.
The multistep process of NMD in yeast is also revealed by the observation that 5′ proximal stop codons trigger faster mRNA degradation than stop codons farther into the ORF (Losson and Lacroute 1979; Peltz et al. 1993; Cao and Parker 2003). Strikingly, as judged by Upf1-promoted deadenylation, all premature stop codons are recognized as aberrant, but the position of the stop codon simply dictates differences in the actual rates of Upf1-promoted decapping (Cao and Parker 2003). Although the molecular mechanism that leads to distal stop codons leading to slower rates of decapping is not known, it does demonstrate that there are multiple steps in the targeting of an mRNA for NMD.
Additional factors can also influence the process of NMD in yeast. Both Upf1 and Upf2 are phosphorylated (De Pinto et al. 2004; Wang et al. 2006), and phosphorylation of Upf2 may affect NMD (Wang et al. 2006). In addition, strains lacking the Ebs1 protein, which is homologous to the metazoan NMD factor Smg7 and contains a 14-3-3 domain for binding phosphoproteins, show partial defects in NMD (Luke et al. 2007). Interestingly, Ebs1 is also regulated by the NMD pathway, which might provide a feedback regulatory loop for maintaining active NMD under some conditions (Ford et al. 2006).
No-go decay:
A second quality control system for mRNA translation, NGD, leads to endonucleolytic cleavage of mRNAs with strong stalls in translation elongation (Doma and Parker 2006; reviewed in detail in Harigaya et al. 2010). After such cleavage, the 3′ mRNA fragment is degraded by Xrn1, and the 5′ fragment is degraded primarily by the cytoplasmic exosome (Doma and Parker 2006). No-go decay can occur at a wide range of translation elongation stalls, including strong stem loops, rare codons, polyLys or polyArg runs, sites of depurination, and possibly at frameshift sites (Doma and Parker 2006; Gandhi et al. 2008; Chen et al. 2010; Kuroha et al. 2010; Letzring et al. 2010; Belew et al. 2011). To date, no specific mRNAs that are predominantly degraded by NGD have been identified, and the suggestion is that NGD primarily functions to degrade aberrant or damaged mRNAs, which could be produced by chemicals or ultraviolet light exposure (Y. Harigaya and R. Parker, unpublished data). The endonuclease(s) that cleaves the mRNA during NGD has not been identified.
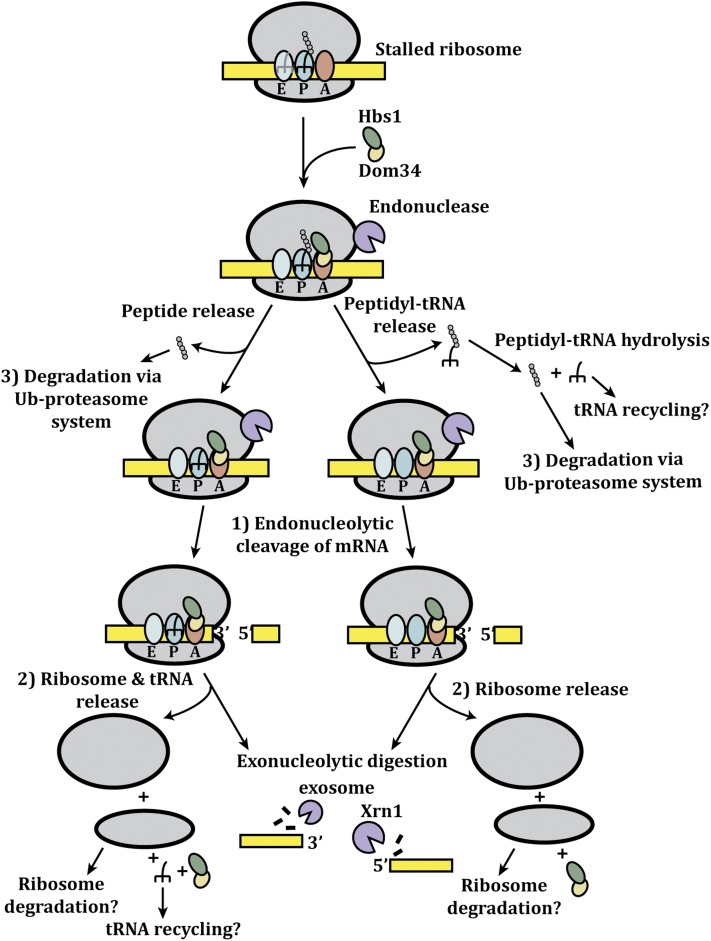
At some translation stalls, NGD is promoted by the Dom34 and Hbs1 proteins, which are paralogs of the translation termination factors eRF1 and eRF3 (Doma and Parker 2006). Structural analyses indicate that Dom34 and Hbs1 fold and interact similarly to eRF1 and eRF3, respectively (Lee et al. 2007; Graille et al. 2008; Chen et al. 2010; Van Den Elzen et al. 2010). Moreover, Dom34 and Hbs1 bind the ribosome in the empty A site (Becker et al. 2011). This similarity to translation termination complexes suggested that Dom34 and Hbs1 function at elongation stalls to terminate translation, which has been demonstrated using reconstituted systems from both yeast and mammals (Shoemaker et al. 2010; Pisareva et al. 2011). However, under some conditions, or at very strong translation pauses, Hbs1 and Dom34 are not required for NGD (Passos et al. 2009; Chen et al. 2010; Kuroha et al. 2010).
On the basis of these observations, a working model for NGD can be proposed (Figure 4). During translation elongation, the ribosome can be paused for a variety of reasons. If the A site is empty during a prolonged elongation stall, it allows for a Dom34/Hbs1/GTP complex, rather than cognate aminoacyl-tRNA, to interact with the A site in the stalled ribosome, leading to dissociation of the peptide and tRNA or a peptide-tRNA conjugate, while maintaining the ribosome on the mRNA. At this stage, three events can occur. First, the mRNA can be cleaved at the vicinity of the “terminated” ribosome. Although the identity of the nuclease is unknown, it is likely to be physically associated with the stalled ribosome. Note that the mRNA cleavage could possibly occur before the release of the peptide-tRNA conjugate. Second, the ribosomes can be released by an unknown mechanism, which may or may not be similar to ribosome recycling at a regular termination codon. Release of the ribosomes is predicted to limit cleavage of the mRNA by preventing the recruitment of the nuclease. Third, the released nascent peptide or peptide tRNA-conjugate would be subject to ubiquitin-proteasome-mediated degradation, possibly in conjunction with peptide-tRNA hydrolysis.
Figure 4 .
Model for no-go decay.
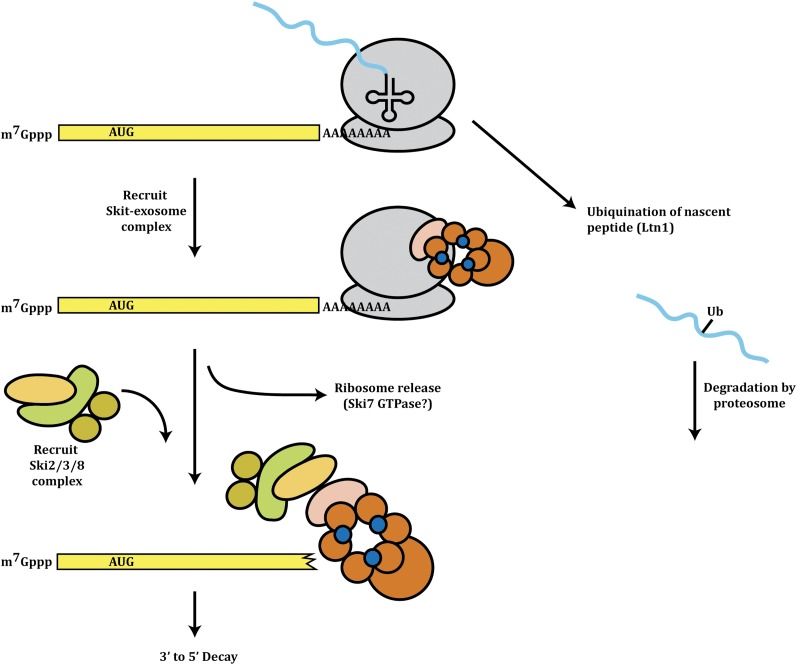
Non-stop decay:
Another mechanism of mRNA quality control is the rapid degradation of mRNAs that do not contain translation termination codons, which is referred to as non-stop decay (NSD) (Frischmeyer et al. 2002; Van Hoof et al. 2002) (Figure 5). Such mRNAs naturally occur due to use of polyadenylation sites within open reading frames, which are estimated to occur at ∼10% of the transcriptional events (Van Hoof et al. 2002). In addition, such non-stop mRNAs could also arise due to mutations, readthrough of stop codons due to PSI+ that limits the translation termination factor function (Wilson et al. 2005), or endonuclease cleavage sites with the open reading frame.
Figure 5 .
Model for non-stop decay.
The process of NSD appears triggered by a ribosome reaching the 3′ end of the mRNA and being unable to terminate translation. In this situation, the mRNA is rapidly degraded in a process that requires the exosome and the Ski7, Ski2, Ski3, and Ski8 proteins. NSD is distinguished from 3′ to 5′ decay of mRNAs by requiring the GTPase domain of Ski7, which is similar to Ef1a and is thought to interact with the ribosome (Van Hoof et al. 2002). NSD is also mechanistically different from normal 3′ to 5′ decay of mRNAs in that it can utilize either endonuclease or exonuclease activity of Rrp44, whereas 3′ to 5′ decay of normal mRNAs appears to require the exonuclease activity of Rrp44 (Schaeffer and Van Hoof 2011). Given this, the prevailing model of NSD is that a stalled ribosome at the 3′ end of the mRNA is recognized by the Ski7 protein, which then recruits the Ski2/Ski3/Ski8 complex and the exosome to degrade the mRNA in a 3′ to 5′ direction. In the absence of Ski7 or the exosome, nonstop mRNAs are subject to accelerated decapping and 5′ to 3′ degradation (Inada and Aiba 2005), perhaps because of the failure to recycle terminating ribosomes for continued efficient translation initiation. An unresolved issue is how the ribosome is removed from the mRNA and if Ski7 hydrolyzes GTP to promote its dissociation.
The peptides produced by non-stop mRNAs are also subject to rapid degradation in a proteasome-mediated manner. In this case, two different ubiquitin ligases have been proposed to function. Two reports have suggested that the Ltn1 protein, which is an E3 ubiquitin ligase that interacts with ribosomes, is required for rapid decay of the nascent peptide (Wilson et al. 2007; Bengtson and Joazeiro 2010). In a second study, it has been suggested that Not4 promotes the ubiquitination and degradation of such nascent peptides (Dimitrova et al. 2009), although whether this is due to differences in the reporter constructs used is yet to be resolved. In either case, the rapid degradation of the nascent and aberrant polypeptide would ensure that only proteins of the proper length are produced.
The specific features of the mRNAs may affect how they become substrates for NSD or other mRNA quality control pathways. For example, while NSD mRNAs generated by poly(A) addition within the coding region require the GTPase domain of Ski7 for their degradation, non-stop mRNAs generated by a ribozyme within the coding region do not (Meaux and Van Hoof 2006). One possibility is that this difference is due to the specific loading of proteins during nuclear polyadenylation that affect NSD in the cytosol. Alternatively, it could be that mRNAs with translated poly(A) tails are subject to a hybrid type of mRNA decay that involves aspects of both NGD and NSD. This possibility is suggested by the fact that poly(A) tracts, which encode for lysine, can stall elongating ribosomes and trigger NGD (Ito-Harashima et al. 2007; Kuroha et al. 2010). An interesting area for future research will be to determine how specific types of mRNAs are recognized and targeted for these quality control systems.
Quality control of nuclear mRNA processing
Numerous quality control systems target mRNA defective in pre-mRNA splicing, polyadenylation, or mRNA export. These nuclear quality control systems prevent the function of the aberrant mRNA by triggering nuclear degradation or by nuclear export leading to cytoplasmic degradation. In addition, aberrant or unprocessed nuclear mRNAs can also be retained within the nucleus. Nuclear retention may be important both to give time for RNA processing to be completed and to allow for a kinetically disfavored nuclear degradation pathway to degrade the RNA (see below). Examples of nuclear retention of aberrant mRNAs include the retention of mRNAs with defects in polyadenylation (Hilleren et al. 2001; Jensen et al. 2001). Interestingly, these aberrant RNAs are retained in the vicinity of the gene (Thomsen et al. 2003), which has the potential to have feedback effects on transcription.
Quality control of pre-mRNA splicing:
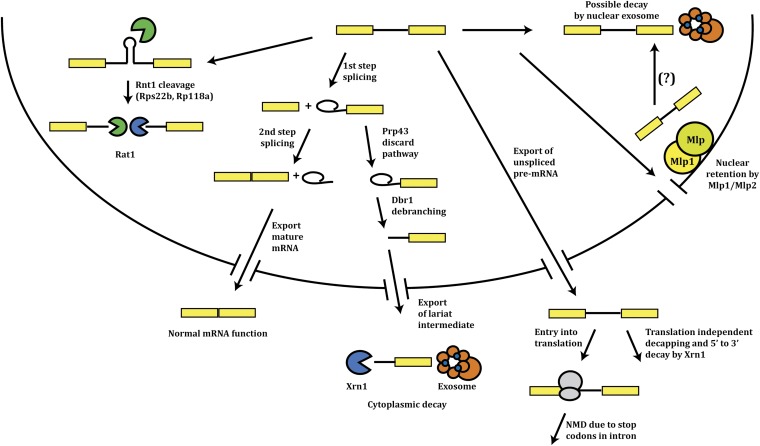
Several RNA degradation systems contribute to degrading unspliced or mis-spliced pre-mRNAs (Figure 6). For the Rps22b and Rpl18a pre-mRNAs, the intron contains a cleavage site for the yeast RNAseIII enzyme, Rnt1, and endonuclease cleavage within the intron reduces the pool of both pre-mRNA and the excised intron (Danin-Kreiselman et al. 2003). This reaction appears to occur in the nucleus due to Rnt1 being concentrated in the nucleus, and the observation that the pre-mRNA cleavage products are degraded by the nuclear Rrp6 and Rat1 nucleases (Danin-Kreiselman et al. 2003). Nuclear pre-mRNA degradation has also been proposed for pre-mRNAs that fail to enter splicing or are trapped as lariat intermediates (Bousquet-Antonelli et al. 2000).
Figure 6 .
Mechanisms of degradation for unspliced pre-mRNAs.
In contrast to nuclear degradation, multiple experiments suggest that unspliced pre-mRNA are exported to the cytoplasm and degraded. This was first suggested by the observation that the CYH2 pre-mRNA was exported to the cytoplasm and degraded by NMD (He et al. 1993). Genome-wide analysis has shown that a number of pre-mRNAs with weak splicing signals are exported to the cytosol and then degraded by NMD (Sayani et al. 2008). In addition, in strains with defective splicing machinery, NMD is seen to degrade pre-mRNAs from even more genes (Kawashima et al. 2009). Bioinformatics analysis suggests that this is an effective way to monitor introns since intron sequences contain an overrepresentation of translation termination signals and would tend to channel pre-mRNAs that enter translation into NMD (reviewed in Egecioglu and Chanfreau 2011). NMD would also be expected to degrade errors in splice site choice that lead to frameshifting during translation and thereby to premature translation termination.
Unspliced pre-mRNAs can also be degraded in the cytoplasm independently of NMD (Hilleren and Parker 2003), perhaps because the retained intron represses translation initiation and untranslated mRNAs in yeast tend to be rapidly deadenylated and decapped (Muhlrad et al. 1995).
Pre-mRNAs can also be degraded after the formation of the lariat intermediate. In this case, when the lariat intermediate is stuck—either due to mutations in the intron (Hilleren and Parker 2003; Mayas et al. 2010) or to errors in 5′ splice site choice or stochastic events in endogenous genes (Y. Harigaya and R. Parker, unpublished data)—the 3′ intron–exon lariat is debranched, exported to the cytoplasm, and degraded by Xrn1 or the cytoplasmic exosome. Moreover, the Prp43 ATPase is required for this discard pathway, presumably to facilitate spliceosome disassembly, thereby allowing export (Mayas et al. 2010).
Unspliced pre-mRNAs from reporter mRNAs can also be retained at the nuclear pore by the Mlp1 and Mlp2 proteins (Galy et al. 2004), although this system appears to part of a more general quality control system for mRNA export that is not limited to pre-mRNAs (Vinciguerra et al. 2005).
Quality control of 3′ end generation:
Multiple types of mutations lead to defects in mRNA 3′ end generation and polyadenylation. For example, mutations in proteins required for recognition of the polyadenylation site or the poly(A) polymerase Pap1 can alter the site of 3′ end formation and, in the case of pap1 alleles, lead to the production of unadenylated mRNAs (Patel and Butler 1992; Mandart and Parker 1995). Similarly, defects in mRNA export factors or in the Tho complex, which couples transcription and mRNP assembly, lead to hyperadenylation of mRNAs, perhaps due to a failure to recycle mRNP proteins to the nucleus (Hilleren and Parker 2001; Jensen et al. 2001; Libri et al. 2002).
Several studies suggest that yeast mRNAs with aberrant 3′ ends are retained at the site of transcription (Hilleren et al. 2001; Jensen et al. 2001; Thomsen et al. 2003). Moreover, this retention appears to be due to the absence of a proper poly(A) tail since mRNAs where the 3′ end is generated by a ribozyme are often retained (Libri et al. 2002) unless a poly(A) tract is encoded 5′ of the site of ribozyme cleavage (Dower et al. 2004).
The retention of aberrant mRNAs with aberrant ends requires the 3′ to 5′ exonuclease activity of the nuclear exosome and is reduced in rrp6Δ strains, or by mutations in the exonuclease active sites of Rrp6 or Dis3/Rrp44 (Hilleren et al. 2001; Libri et al. 2002; Thomsen et al. 2003; Assenholt et al. 2008). The role of the exosome suggested that these aberrant mRNAs might be degraded in the nucleus by the nuclear exosome. However, in some cases the actual levels of mRNAs do not increase in the rrp6Δ strains, although the mRNAs are no longer nuclear retained, suggesting that the role of the exosome might be to allow retention, and not degrade, the mRNAs (Hilleren et al. 2001). In contrast, in other studies it has been suggested that the exosome degrades these mRNAs since, in strains defective in the THO complex, the 3′ end of some mRNAs is reduced compared to the 5′ end, implying that some mRNAs have been degraded in a 3′ to 5′ direction (Libri et al. 2002).
In principle, mRNAs generated in mutants with defects in 3′-end formation might be subject to three different fates and that population studies might give misleading results due to different populations behaving in different ways. For example, in export or THO mutant strains a subset of the transcripts is produced with normal 3′-end generation and poly(A) tails, and these appear to undergo normal export, deadenylation, and cytoplasmic decapping (Hilleren and Parker 2001; Libri et al. 2002). A second population of mRNAs are those that are produced with either hypo- or hyperadenylated 3′ ends. Strikingly, the observable population of both hyperadenylated and hypoadenylated mRNAs is quite stable, suggesting that these mRNAs are retained within the nucleus and are stable in that state (Mandart and Parker 1995; Hilleren and Parker 2001; Rougemaille et al. 2007). Finally, it may be that there is a pool of mRNAs that are recognized as aberrant and subjected to extremely rapid degradation (Rougemaille et al. 2007). Since these mRNAs are proposed to degrade very rapidly, it would not be possible to measure their decay rates from steady state as the steady-state pool will consist only of mRNAs that have escaped this rapid degradation. An important issue in future work is determining if such rapid degradation does occur and determining its mechanism.
Intergenic, Intragenic, Promoter-Associated, and Antisense RNAs
From genome-wide analysis it is now clear that yeast cells produce numerous untraditional transcripts. Such transcripts include intergenic and intragenic transcripts (referred to as cryptic unstable transcripts, stable unannotated transcripts, or Xrn1-sensitive unstable transcripts); antisense RNAs; transcripts associated with or overlapping promoter regions (referred to as promoter-associated RNAs); and RNAs from heterochromatin, including telomeric, centromere, and rRNA spacer regions (Olivas et al. 1997; Wyers et al. 2005; Houseley et al. 2007, 2008; Luke et al. 2008). These transcripts can be produced from bidirectional transcription from promoters of known genes (Neil et al. 2009; Xu et al. 2009) or from distinct promoters (Rhee and Pugh 2012) and have been suggested to have a variety of functions. For example, some antisense RNAs, or their act of transcription, can play a role in modulating the expression of the corresponding sense gene via histone modification (Camblong et al. 2007; Berretta et al. 2008; Houseley et al. 2008; Geisler et al. 2012). At least in the case of the GAL1 mRNA, the degradation of an antisense RNA appears to be required for efficient induction of the corresponding sense transcript (Geisler et al. 2012). This might be a general feature of genes induced by environmental stimuli since antisense RNAs stabilized in xrn1Δ and dcp2Δ strains are often antisense to induced genes (Van Djik et al. 2011; Geisler et al. 2012), An important and unresolved future issue will be determining why degradation of this antisense RNA is required for efficient gene induction.
In other cases, transcripts that overlap the promoter, or even the ORF, can alter the transcription of the associated mRNA (e.g., Martens et al. 2004; Thiebaut et al. 2008; Huang et al. 2010; Toesca et al. 2011). Finally, since some of these RNAs are associated with ribosomes and can contain short ORFs, it seems likely that some of these RNAs encode short polypeptides (Olivas et al. 1997; Thompson and Parker 2007; Wilson and Masel 2011). Because many of these RNAs are most easily detected in strains defective in RNA degradation pathways, it is thought that many such RNAs are highly unstable.
This set of RNAs is subject to a variety of different and overlapping RNA degradation pathways. For example, many intergenic, antisense, and promoter-associated transcripts increase in levels and/or show increased stability in xrn1Δ, dcp1Δ, or dcp2Δ strains, suggesting that they are subject to degradation by decapping and Xrn1 action (Thompson and Parker 2007; Berretta et al. 2008; Van Dijk et al. 2011; Geisler et al. 2012). A significant number of intergenic RNAs are also increased in strains defective in NMD, suggesting that these RNAs might enter translation and then be recognized as aberrant RNAs and degraded by NMD (Thompson and Parker 2007; Toesca et al. 2011). Some of these RNAs also appear to be degraded in a mechanism dependent on RNaseP (Marvin et al. 2011).
Numerous studies have also shown that intergenic, intragenic, promoter-associated, and antisense RNAs increase in levels in strains defective in the nuclear exosome, most commonly the Rrp6 subunit, leading to the suggestion that these RNAs are subject to nuclear 3′ to 5′ degradation (Wyers et al. 2005; Davis and Ares 2006; Thompson and Parker 2007; Milligan et al. 2008; Lardenois et al. 2011). However, examination of RNA decay rates has shown that while the steady-state levels of RNA actually increase for many RNAs in rrp6Δ strains, the actual decay rates increase and appear dependent on Xrn1 (Thompson and Parker 2007), although some reports indicate an increased half-life in rrp6Δ strains (Wyers et al. 2005). One possibility is that rrp6Δ strains show increased transcription, but this model is inconsistent with nuclear run-on and RNA polymerase II chromatin immunoprecipitation experiments (Wyers et al. 2005; Rougemaille et al. 2007). An alternative is that the nuclear degradation of such transcripts occurs extremely rapidly and cannot be measured at steady state, an issue that remains to be rigorously resolved in future work.
The reduction in cryptic unstable transcript and promoter-associated RNA levels by the nuclear exosome is facilitated by the TRAMP complexes (Lacava et al. 2005; Wyers et al. 2005; Egecioglu et al. 2006; Houseley et al. 2007; San Paolo et al. 2009; Callahan and Butler 2010). The TRAMP complexes consist of one of two related RNA-binding proteins, Air1 and Air2, the Mtr4 RNA helicase, and one of two noncanonical poly(A) polymerases, Trf4 or Trf5 (Lacava et al. 2005; Vanacova et al. 2005; Wyers et al. 2005). Analogous to the situation in bacteria, the TRAMP complex can promote 3′ to 5′ degradation of RNAs by the addition of poly(A) tails to their 3′ end, which is then thought to promote the targeting of those RNAs to the exosome (Kadaba et al. 2004; Lacava et al. 2005; Vanacova et al. 2005; Wyers et al. 2005). However, in some cases the TRAMP complex can also promote exosome-mediated RNA degradation independently of polyadenylation activity and may in that case serve as a scaffolding complex to recruit the exosome to the RNA (Houseley et al. 2007; Rougemaille et al. 2007; San Paolo et al. 2009). The TRAMP complexes also serve important roles in targeting the exosome for degradation or processing of small nucleolar RNAs (snoRNAs), snRNAs, tRNA, and other RNA species (see below).
The targeting of RNAs to the TRAMP and exosome complexes appears to be modulated in part by the Nrd1 and Nab3 RNA-binding proteins. The Nrd1 and Nab3 proteins bind to specific elements in nascent transcripts (Creamer et al. 2011; Jamonnak et al. 2011; Wlotzka et al. 2011) and can direct termination of transcription for snoRNAs, snRNAs, some mRNAs, and numerous intergenic transcripts (Arigo et al. 2006; Thiebaut et al. 2006; Carroll et al. 2007; Rondon et al. 2009; Kim and Levin 2011). Nrd1 and Nab3 associate with the carboxy-terminal doman of RNA polymerase II when phosphorylated at serine 5 and serine 7 (Gudipati et al. 2008; Vasiljeva et al. 2008; Kim et al. 2010) and may act preferentially on shorter transcripts as serine 5 and 7 phosphorylation can be lost with ongoing elongation (Komarnitsky et al. 2000; Schroeder et al. 2000). Nrd1 and Nab3 then appear to recruit the exosome to degrade intergenic transcripts or to promote processing of snoRNAs and snRNAs (Arigo et al. 2006; Thiebaut et al. 2006; Grzechnik and Kufel 2008; Vasiljeva et al. 2008). This effect is not limited to RNA polymerase II transcripts as Nab3 and Nrd1 can also target RNA polymerase III transcripts for polyadenylation and degradation (Wlotzka et al. 2011).
Decay of tRNA, rRNAs, snRNAs, and snoRNAs
Most functional noncoding RNAs such as rRNAs, tRNAs, snRNAs, and snoRNAs are quite stable, although they are probably degraded at some rate even under optimal growth conditions. However, these RNAs are clearly subject to increased turnover during nutrient limitations (see below) and are also subject to a variety of quality control systems to ensure their proper biogenesis and function.
tRNAs
Three mechanisms can target tRNAs for degradation. First, when tRNAs have defects in their maturation and/or modification, pre-tRNAs can be adenylated by TRAMP complexes for targeting to the nuclear exosome (Kadaba et al. 2004, 2006; Vanacova et al. 2005). Similarly, defects in 3′-end processing of pre-tRNAs can also lead to adenylation by TRAMP complexes (Copela et al. 2008; Ozanick et al. 2009). Adenylation of defective tRNAs is thought to provide a single-stranded extension of RNA that the nuclear exosome can then load on and then degrade through the structured tRNA.
A second mechanism of tRNA decay occurs when tRNAs are defective in their modifications and/or stability of the combined acceptor and T stems. For example, in trm4Δ trm8Δ strains, which produce undermodified tRNA[Val(AAC)], the tRNA[Val(AAC)] is degraded more rapidly (Alexandrov et al. 2006). This degradation utilizes the 5′ to 3′ exonucleases Rat1 and Xrn1 (Chernyakov et al. 2008). Moreover, mutations that weaken the stability of the combined acceptor and T stems of the tRNA increase degradation (Whipple et al. 2011). This suggests a model wherein the strength of the tRNA structure at the 5′ end dictates whether it can be subject to 5′ to 3′ degradation.
The stability of the 5′ stem loop in tRNAs can also affect whether tRNAs receive an additional CCA addition, which can promote their degradation (Wilusz et al. 2011). This occurs when tRNAs have a weak acceptor stem and the first two nucleotides of the tRNA are guanosines. In this case, the tRNA appears to adopt an alternative fold whereby the CCA addition enzyme can add a second CCA, which then appears to promote degradation of the tRNA, perhaps by providing a 3′ single-stranded region for the recruitment of the exosome (Wilusz et al. 2011). Strains defective in the CCA addition enzyme also produce tRNAs with shortened 3′ ends that are degraded more rapidly (Aebi et al. 1990), although the specific mechanism is not yet known. One anticipates that the stability of tRNAs will also be influenced by tRNA-binding proteins (e.g., eEF1 and synthetases) that will limit nuclease accessibility.
A final mechanism of tRNA cleavage and degradation involves the nuclease Rny1. Rny1 is an RNaseT2 family member that is secreted and targeted to membrane-bound compartments, most notably the vacuole (Macintosh et al. 2001; Thompson and Parker 2009). Rny1 has been shown to cleave tRNAs in the anticodon loop, at least during various stresses and high cell density (Thompson et al. 2008; Thompson and Parker 2009). Rny1 may be released from membrane compartments to degrade tRNAs during stress, although it remains possible that tRNAs are also imported into the vacuole, or other membrane-bound compartment, by some type of autophagy-related process for degradation by Rny1.
rRNA decay
To date, three types of RNA degradation have been described to occur to rRNAs. First, when rRNA processing or assembly into ribosomes is altered by a variety of mutations, the defective rRNA is targeted for degradation by polyadenylation by TRAMP complexes and recruitment of the nuclear exosome (e.g., Allmang et al. 2000; Kuai et al. 2004; Fang et al. 2005; Lacava et al. 2005; Dez et al. 2006; Kadaba et al. 2006). Polyadenylated rRNA species can also be detected in strains defective in nuclear exosome function, suggesting that a certain percentage of “normal” rRNA transcripts, either due to errors in transcription, processing, or assembly, are subject to this nuclear RNA degradation pathway.
Ribosomes that are accurately assembled but fail to function are exported to the cytoplasm and degraded by a process referred to as nonfunctional ribosomal decay. For 25S rRNAs that are defective in peptidyl transferase activity, the 60S subunit appears to be recognized as aberrant prior to assembly into 80S complexes and targeted for degradation by ubiquitination in a process dependent on the ubiquitin ligase Rtt101 and its associated protein Mms1 (Lariviere et al. 2006; Fujii et al. 2009). The exact nuclease(s) that degrades these defective 25S RNAs is not yet known, although the exosome may have some role (Cole et al. 2009).
For the small ribosomal subunit, mutations in the decoding site of the 18S rRNA lead to 40S complexes that can assemble into 80S complexes on the translation start site, but appear to be blocked for subsequent elongation (Lariviere et al. 2006). In this case, the degradation of the defective 18S rRNAs utilizes the same Dom34/Hbs1 complex that is involved in the release of ribosomes stalled by defects in the mRNA (NGD; see above), which leads to degradation by the cytoplasmic exosome as well as Xrn1 (Cole et al. 2009).
rRNAs are also subject to degradation during stress or nutrient limitations. For example, in response to oxidative stress or entry into stationary phase, the rRNAs are fragmented to some extent (Mroczek and Kufel 2008; Thompson et al. 2008). This degradation is at least partially dependent on the Rny1 nuclease, which is concentrated in vacuoles (Thompson and Parker 2009). One possibility is that these stress conditions lead to release of Rny1 from the vacuole and cleavage of cytoplasmic rRNAs (Thompson and Parker 2009). Since the Nuc1 nuclease is also released from mitochondria under these conditions (Buttner et al. 2007), it might also play some role in rRNA degradation during stress. Another possibility is that rRNAs are targeted to vacuoles under these conditions by selective autophagy. Consistent with that model, based on following GFP-tagged ribosomal proteins, ribosomes have been inferred to be targeted to vacuoles during nitrogen starvation in a process referred to as ribophagy, which requires ubiquitination and de-ubiquitination (Kraft et al. 2008; Ossareh-Nazari et al. 2010). Ribosomes may also be targeted for degradation in the vacuole by a piecemeal microautophagy of the nucleus, wherein regions of the nucleus are directly targeted to the vacuole by invagination of the nuclear envelope into the vacuolar lumen (Roberts et al. 2003).
snRNAs/snoRNAs
Several observations suggest that defects in snRNP or snoRNP assembly lead to degradation of these RNA species. For example, mutations in the Naf1 protein, which is required for the assembly of H/ACA snoRNPs, lead to the loss of that class of snoRNAs (Fatica et al. 2002; Dez et al. 2002). Similarly, mutations in the SM-binding sites of yeast U1, U2, and U5 or the telomerase RNA and TLC1 lead to decreased levels of these RNAs (Jones and Guthrie 1990; Seipelt et al. 1999; Seto et al. 1999). Although effects on transcription have not been ruled out, the simplest model is that the failure to properly assemble the snoRNP/snRNPs leads to their accelerated degradation by a yet-to-be-determined degradation pathway.
Defects in U6 snRNA biogenesis also give increased decay of RNA. For example, an internal deletion within the U6 snRNA leads to unstable transcripts, which are adenylated and degraded by the nuclear exosome (Kadaba et al. 2006). Similarly, defects in the Lsm2–8 complex, which binds the 3′ end of the U6 snRNA, lead to decreased levels of the U6 snRNA (Pannone et al. 2001, 1998; Luhtala and Parker 2009), although the mechanism of U6 degradation in this case is not known. Taken together, these observations argue that systems exist to degrade snRNAs and snoRNAs that fail to be properly assembled into RNPs.
Degradation of RNAs in Mitochondria
tRNAs, rRNAs, and mRNAs are also transcribed within the mitochondria of yeast cells in 13 complex transcription units. These transcripts are subject to RNA processing, including splicing of group I and group II introns, endonuclease cleavage, and 5′ and 3′ trimming. The biogenesis of mitochondrial mRNAs typically involves a 5′ endonucleolytic cleavage, often followed by 5′ to 3′ trimming of the exposed 5′ end to specific sites and a 3′ endonuclease cleavage just downstream of a so-called dodecamer element (Dieckmann and Staples 1994). The dodecamer element is proposed to bind an unknown protein factor and may play a role in translation and/or stabilization of the mRNAs (reviewed in Gagliardi et al. 2004). In contrast to cytoplasmic mRNAs, mitochondrial mRNAs in yeast are not polyadenylated.
The major functional RNAse in the mitochondria mechanism is termed the mitochondrial degradasome or mtEXO and consists of a 1:1 ratio of Dss1, a member of the RNaseII family, and Suv3, a DEVH family member. Strains defective in Suv3 or Dss1 show accumulation of mitochondrial mRNAs, indicating a defect in their turnover (Dziembowski et al. 2003; Malecki et al. 2008). Suv3 and Dss1 mutants also accumulate excised introns, indicating that mtEXO is required for their degradation (Szczesny et al. 2011). Interestingly, such strains also accumulate incompletely processed mitochondrial rRNAs, which has led to the suggestion that the mtEXO also functions in a quality control system for the degradation of rRNAs or mRNAs that are not completely processed. The activity of the mtEXO on mitochondrial mRNAs may be limited by the 3′ features of mRNAs, possibly including a factor binding the dodecamer element (Min and Zassenhaus 1993).
The activity of mtEXO is due to the concerted action of the Suv3 helicase activity and the Dss1 nuclease in response to a RNA substrate. For example, Suv3 has intrinsic ATPase activity, but, in a complex with Dss1, the ATPase actitivy is enhanced by a single-stranded nucleic acid substrate (Malecki et al. 2007). Moreover, the helicase activity of Suv3 is dependent on Dss1 (Malecki et al. 2007). Similarly, the exonuclease activity of Dss1 is increased by Suv3 and made dependent on ATP (Malecki et al. 2007). These observations suggest a working model whereby Suv3 acts as an ATP-dependent motor to feed RNA into the active site of Dss1 (Szczesny et al. 2011).
There is also a 5′ to 3′ exonuclease activity in mitochondria that is dependent on the Pet127 protein (Wiesenberger and Fox 1997; Fekete et al. 2008), although whether Pet127 is actually a nuclease remains to be determined. The Pet127-dependent 5′ to 3′ exonuclease may also be able to stimulate and/or degrade mitochondrial RNAs since dominant mutations in, or overexpression of, Pet127 partially suppresses the defects seen in suv3 or dss1 mutants (Wegierski et al. 1998; Chen et al. 1999). Strains lacking Pet127 also show misregulation of some mitochondrial mRNAs, although whether this is due to misprocessing of their 5′ ends or due to disruption of normal mRNA degradation pathways is not yet clear (Wiesenberger and Fox 1997; Fekete et al. 2008).
Two additional nucleases have been localized to mitochondria but do not appear to affect RNA degradation. These include Nuc1, which is the major DNAse/RNAse in mitochondria by enzymatic activity (Dake et al. 1988; Vincent et al. 1988), and Rex2, which is a 3′ to 5′ exonuclease that is found both in the mitochondria (Hanekamp and Thorsness 1999) and in the nucleus, where it plays a role in the trimming of 5S and 5.8S rRNAs and U4 and U5 snRNA (Van Hoof et al. 2000a). Although no clear role for these nucleases has been identified in yeast mitochondrial RNA degradation, it is notable that nuc1Δ and rex2Δ strains show a negative genetic interaction for growth, suggesting some degree of functional overlap (Costanzo et al. 2010).
Future Perspectives
There are several areas of importance in the future study of RNA turnover in yeast cells. For example, although many RNA degradation pathways, enzymes, and cofactors have been identified, a precise understanding of their biochemical function and mechanisms of action will need further experimentation, including the development of robust in vitro systems for mechanistic studies. Moreover, one anticipates that there are yet-to-be-identified nucleases that play roles in RNA degradation.
It will also be important to understand how RNA degradation systems interface with other cellular processes. Although this is a general issue, three examples stand out. First, the tight inverse coupling between translation and mRNA degradation highlights that an understanding of the regulation of mRNA function will require mechanistic insight into how mRNAs transition between mRNP states capable of translation or deadenylation and mRNA decapping. A second, and related issue, is that an understanding of mRNA degradation will require a broader understanding of mRNP biogenesis, dynamics, and how the specific proteins bound to individual mRNAs act, either in isolation or in combination, to modulate translation and mRNA degradation.
A third area of interest will be in determining the relationship between mRNA degradation and transcription. This issue is of interest since several observations suggest that transcription and mRNA degradation are coupled. For example, dcp1Δ strains were observed to have prolonged decay rates of reporter mRNAs without a corresponding increase in steady-state levels, suggesting alterations in transcription (Muhlrad and Parker 1999b). In addition, strains lacking the decapping activator Edc1 were unable to induce new transcription during a shift in carbon source (Schwartz et al. 2003). Moreover, the Rpb4 and Rpb7 subunits of RNA polymerase II were shown to shuttle to the cytoplasm and to affect the decay of mRNAs (Lotan et al. 2005, 2007; Selitrennik et al. 2006) in a manner coupled to their recruitment to RNA polymerase II (Goler-Baron et al. 2008). This has led to the suggestion that transcription of new mRNAs is dependent on decay of pre-existing mRNAs in the cytosol, which would be an elegant feedback mechanism to couple mRNA biogenesis and degradation (Dori and Choder 2007). More recently, it has been suggested that the rate of mRNA degradation can be determined by the promoter (Bregman et al. 2011; Trcek et al. 2011), possibly through the loading of proteins on the nascent transcript in a promoter-dependent manner. Given these new-found connections between mRNA biogenesis and degradation, it will be important to determine how the mechanisms by which these two processes are coupled, whether this is general to most mRNAs and even other RNA classes, and the how such coupling is used by the cell to regulate gene expression in response to environmental cues.
Footnotes
Communicating editor: A. Hopper
Literature Cited
- Aebi M., Kirchner G., Chen J. Y., Vijayraghavan U., Jacobson A., et al. , 1990. Isolation of a temperature-sensitive mutant with an altered tRNA nucleotidyltransferase and cloning of the gene encoding tRNA nucleotidyltransferase in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 265: 16216–16220 [PubMed] [Google Scholar]
- Alexandrov A., Chernyakov I., Gu W., Hiley S. L., Hughes T. R., et al. , 2006. Rapid tRNA decay can result from lack of nonessential modifications. Mol. Cell 21: 87–96 [DOI] [PubMed] [Google Scholar]
- Allmang C., Petfalski E., Podtelejnikov A., Mann M., Tollervey D., et al. , 1999. The yeast exosome and human PM-Scl are related complexes of 3′ → 5′ exonucleases. Genes Dev. 13: 2148–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang C., Mitchell P., Petfalski E., Tollervey D., 2000. Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res. 28: 1684–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrani N., Ganesan R., Kervestin S., Mangus D. A., Ghosh S., et al. , 2004. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432: 112–118 [DOI] [PubMed] [Google Scholar]
- Anderson J. S., Parker R. P., 1998. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 17: 1497–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki Y., Takahashi S., Kobayashi T., Kajiho H., Hoshino S., et al. , 2001. Ski7p G protein interacts with the exosome and the Ski complex for 3′-to-5′ mRNA decay in yeast. EMBO J. 20: 4684–4693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arigo J. T., Eyler D. E., Carroll K. L., Corden J. L., 2006. Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol. Cell 23: 841–851 [DOI] [PubMed] [Google Scholar]
- Arribere J. A., Doudna J. A., Gilbert W. V., 2011. Reconsidering movement of eukaryotic mRNAs between polysomes and P bodies. Mol. Cell 44: 745–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assenholt J., Mouaikel J., Andersen K. R., Brodersen D. E., Libri D., et al. , 2008. Exonucleolysis is required for nuclear mRNA quality control in yeast THO mutants. RNA 14: 2305–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkin A. L., Schenkman L. R., Eastham M., Dahlseid J. N., Lelivelt M. J., et al. , 1997. Relationship between yeast polyribosomes and Upf proteins required for nonsense mRNA decay. J. Biol. Chem. 272: 22163–22172 [DOI] [PubMed] [Google Scholar]
- Aviv T., Lin Z., Ben-Ari G., Smibert C. A., Sicheri F., 2006. Sequence-specific recognition of RNA hairpins by the SAM domain of Vts1p. Nat. Struct. Mol. Biol. 13: 168–176 [DOI] [PubMed] [Google Scholar]
- Badis G., Saveanu C., Fromont-Racine M., Jacquier A., 2004. Targeted mRNA degradation by deadenylation-independent decapping. Mol. Cell 15: 5–15 [DOI] [PubMed] [Google Scholar]
- Baker K. E., Parker R., 2004. Nonsense-mediated mRNA decay: terminating erroneous gene expression. Curr. Opin. Cell Biol. 16: 293–299 [DOI] [PubMed] [Google Scholar]
- Balagopal V., Parker R., 2009. Stm1 modulates mRNA decay and Dhh1 function in Saccharomyces cerevisiae. Genetics 181: 93–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagopal V., Parker R., 2011. Stm1 modulates translation after 80S formation in Saccharomyces cerevisiae. RNA 17: 835–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T., Armache J. P., Jarasch A., Anger A. M., Villa E., et al. , 2011. Structure of the no-go mRNA decay complex Dom34-Hbs1 bound to a stalled 80S ribosome. Nat. Struct. Mol. Biol. 18: 715–720 [DOI] [PubMed] [Google Scholar]
- Beelman C. A., Parker R., 1994. Differential effects of translational inhibition in cis and in trans on the decay of the unstable yeast MFA2 mRNA. J. Biol. Chem. 269: 9687–9692 [PubMed] [Google Scholar]
- Beelman C. A., Stevens A., Caponigro G., Lagrandeur T. E., Hatfield L., et al. , 1996. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature 382: 642–646 [DOI] [PubMed] [Google Scholar]
- Beilharz T. H., Preiss T., 2007. Widespread use of poly(A) tail length control to accentuate expression of the yeast transcriptome. RNA 13: 982–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belew A. T., Advani V. M., Dinman J. D., 2011. Endogenous ribosomal frameshift signals operate as mRNA destabilizing elements through at least two molecular pathways in yeast. Nucleic Acids Res. 39: 2799–2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benard L., 2004. Inhibition of 5′ to 3′ mRNA degradation under stress conditions in Saccharomyces cerevisiae: from GCN4 to MET16. RNA 10: 458–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtson M. H., Joazeiro C. A., 2010. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature 467: 470–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berretta J., Pinskaya M., Morillon A., 2008. A cryptic unstable transcript mediates transcriptional trans-silencing of the Ty1 retrotransposon in S. cerevisiae. Genes Dev. 22: 615–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeck R., Tarun S., Jr, Rieger M., Deardorff J. A., Muller-Auer S., et al. , 1996. The yeast Pan2 protein is required for poly(A)-binding protein-stimulated poly(A)-nuclease activity. J. Biol. Chem. 271: 432–438 [DOI] [PubMed] [Google Scholar]
- Boeck R., Lapeyre B., Brown C. E., Sachs A. B., 1998. Capped mRNA degradation intermediates accumulate in the yeast spb8–2 mutant. Mol. Cell. Biol. 18: 5062–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneau F., Basquin J., Ebert J., Lorentzen E., Conti E., 2009. The yeast exosome functions as a macromolecular cage to channel RNA substrates for degradation. Cell 139: 547–559 [DOI] [PubMed] [Google Scholar]
- Borja M. S., Piotukh K., Freund C., Gross J. D., 2011. Dcp1 links coactivators of mRNA decapping to Dcp2 by proline recognition. RNA 17: 278–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousquet-Antonelli C., Presutti C., Tollervey D., 2000. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell 102: 765–775 [DOI] [PubMed] [Google Scholar]
- Bouveret E., Rigaut G., Shevchenko A., Wilm M., Seraphin B., 2000. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 19: 1661–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bregman A., Avraham-Kelbert M., Barkai O., Duek L., Guterman A., et al. , 2011. Promoter elements regulate cytoplasmic mRNA decay. Cell 147: 1473–1483 [DOI] [PubMed] [Google Scholar]
- Brengues M., Teixeira D., Parker R., 2005. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 310: 486–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C. E., Sachs A. B., 1998. Poly(A) tail length control in Saccharomyces cerevisiae occurs by message-specific deadenylation. Mol. Cell. Biol. 18: 6548–6559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C. E., Tarun S. Z., Jr, Boeck R., Sachs A. B., 1996. PAN3 encodes a subunit of the Pab1p-dependent poly(A) nuclease in Saccharomyces cerevisiae. Mol. Cell. Biol. 16: 5744–5753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. T., Bai X., Johnson A. W., 2000. The yeast antiviral proteins Ski2p, Ski3p, and Ski8p exist as a complex in vivo. RNA 6: 449–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan J. R., Parker R., 2009. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell 36: 932–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan J. R., Muhlrad D., Parker R., 2008. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J. Cell Biol. 183: 441–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler J. S., Mitchell P., 2011. Rrp6, rrp47 and cofactors of the nuclear exosome. Adv. Exp. Med. Biol. 702: 91–104 [DOI] [PubMed] [Google Scholar]
- Buttner S., Eisenberg T., Carmona-Gutierrez D., Ruli D., Knauer H., et al. , 2007. Endonuclease G regulates budding yeast life and death. Mol. Cell 25: 233–246 [DOI] [PubMed] [Google Scholar]
- Callahan K. P., Butler J. S., 2010. TRAMP complex enhances RNA degradation by the nuclear exosome component Rrp6. J. Biol. Chem. 285: 3540–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camblong J., Iglesias N., Fickentscher C., Dieppois G., Stutz F., 2007. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell 131: 706–717 [DOI] [PubMed] [Google Scholar]
- Cao D., Parker R., 2001. Computational modeling of eukaryotic mRNA turnover. RNA 7: 1192–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D., Parker R., 2003. Computational modeling and experimental analysis of nonsense-mediated decay in yeast. Cell 113: 533–545 [DOI] [PubMed] [Google Scholar]
- Caponigro G., Parker R., 1995. Multiple functions for the poly(A)-binding protein in mRNA decapping and deadenylation in yeast. Genes Dev. 9: 2421–2432 [DOI] [PubMed] [Google Scholar]
- Caponigro G., Parker R., 1996. Mechanisms and control of mRNA turnover in Saccharomyces cerevisiae. Microbiol. Rev. 60: 233–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caponigro G., Muhlrad D., Parker R., 1993. A small segment of the MAT alpha 1 transcript promotes mRNA decay in Saccharomyces cerevisiae: a stimulatory role for rare codons. Mol. Cell. Biol. 13: 5141–5148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll J. S., Munchel S. E., Weis K., 2011. The DExD/H box ATPase Dhh1 functions in translational repression, mRNA decay, and processing body dynamics. J. Cell Biol. 194: 527–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll K. L., Ghirlando R., Ames J. M., Corden J. L., 2007. Interaction of yeast RNA-binding proteins Nrd1 and Nab3 with RNA polymerase II terminator elements. RNA 13: 361–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cereghino G. P., Atencio D. P., Saghbini M., Beiner J., Scheffler I. E., 1995. Glucose-dependent turnover of the mRNAs encoding succinate dehydrogenase peptides in Saccharomyces cerevisiae: sequence elements in the 5′ untranslated region of the Ip mRNA play a dominant role. Mol. Biol. Cell 6: 1125–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S., Jayachandran U., Bonneau F., Fiorini F., Basquin C., et al. , 2011. Molecular mechanisms for the RNA-dependent ATPase activity of Upf1 and its regulation by Upf2. Mol. Cell 41: 693–703 [DOI] [PubMed] [Google Scholar]
- Chang J. H., Xiang S., Xiang K., Manley J. L., Tong L., 2011. Structural and biochemical studies of the 5′→3′ exoribonuclease Xrn1. Nat. Struct. Mol. Biol. 18: 270–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. Y., Gherzi R., Ong S. E., Chan E. L., Raijmakers R., et al. , 2001. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107: 451–464 [DOI] [PubMed] [Google Scholar]
- Chen J., Chiang Y. C., Denis C. L., 2002. CCR4, a 3′-5′ poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO J. 21: 1414–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Muhlrad D., Hauryliuk V., Cheng Z., Lim M. K., et al. , 2010. Structure of the Dom34-Hbs1 complex and implications for no-go decay. Nat. Struct. Mol. Biol. 17: 1233–1240 [DOI] [PubMed] [Google Scholar]
- Chen W., Islas-Osuna M. A., Dieckmann C. L., 1999. Suppressor analysis of mutations in the 5′-untranslated region of COB mRNA identifies components of general pathways for mitochondrial mRNA processing and decay in Saccharomyces cerevisiae. Genetics 151: 1315–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernyakov I., Whipple J. M., Kotelawala L., Grayhack E. J., Phizicky E. M., 2008. Degradation of several hypomodified mature tRNA species in Saccharomyces cerevisiae is mediated by Met22 and the 5′-3′ exonucleases Rat1 and Xrn1. Genes Dev. 22: 1369–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury A., Mukhopadhyay J., Tharun S., 2007. The decapping activator Lsm1p-7p-Pat1p complex has the intrinsic ability to distinguish between oligoadenylated and polyadenylated RNAs. RNA 13: 998–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chritton J. J., Wickens M., 2011. A role for the poly(A)-binding protein Pab1p in PUF protein-mediated repression. J. Biol. Chem. 286: 33268–33278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark L. B., Viswanathan P., Quigley G., Chiang Y. C., McMahon J. S., et al. , 2004. Systematic mutagenesis of the leucine-rich repeat (LRR) domain of CCR4 reveals specific sites for binding to CAF1 and a separate critical role for the LRR in CCR4 deadenylase activity. J. Biol. Chem. 279: 13616–13623 [DOI] [PubMed] [Google Scholar]
- Clerici M., Mourao A., Gutsche I., Gehring N. H., Hentze M. W., et al. , 2009. Unusual bipartite mode of interaction between the nonsense-mediated decay factors, UPF1 and UPF2. EMBO J. 28: 2293–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole S. E., Lariviere F. J., Merrikh C. N., Moore M. J., 2009. A convergence of rRNA and mRNA quality control pathways revealed by mechanistic analysis of nonfunctional rRNA decay. Mol. Cell 34: 440–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J. M., Tucker M., Sheth U., Valencia-Sanchez M. A., Parker R., 2001. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylation complexes. RNA 7: 1717–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J., Parker R., 2005. General translational repression by activators of mRNA decapping. Cell 122: 875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copela L. A., Fernandez C. F., Sherrer R. L., Wolin S. L., 2008. Competition between the Rex1 exonuclease and the La protein affects both Trf4p-mediated RNA quality control and pre-tRNA maturation. RNA 14: 1214–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson B., Couturier A., Chabelskaya S., Kiktev D., Inge-Vechtomov S., et al. , 2002. Poly(A)-binding protein acts in translation termination via eukaryotic release factor 3 interaction and does not influence [PSI(+)] propagation. Mol. Cell. Biol. 22: 3301–3315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E. D., et al. , 2010. The genetic landscape of a cell. Science 327: 425–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowart L. A., Gandy J. L., Tholanikunnel B., Hannun Y. A., 2010. Sphingolipids mediate formation of mRNA processing bodies during the heat-stress response of Saccharomyces cerevisiae. Biochem. J. 431: 31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creamer T. J., Darby M. M., Jamonnak N., Schaughency P., Hao H., et al. , 2011. Transcriptome-wide binding sites for components of the Saccharomyces cerevisiae non-poly(A) termination pathway: Nrd1, Nab3, and Sen1. PLoS Genet. 7: e1002329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czaplinski K., Ruiz-Echevarria M. J., Paushkin S. V., Han X., Weng Y., et al. , 1998. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev. 12: 1665–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dake E., Hofmann T. J., McIntire S., Hudson A., Zassenhaus H. P., 1988. Purification and properties of the major nuclease from mitochondria of Saccharomyces cerevisiae. J. Biol. Chem. 263: 7691–7702 [PubMed] [Google Scholar]
- Danin-Kreiselman M., Lee C. Y., Chanfreau G., 2003. RNAse III-mediated degradation of unspliced pre-mRNAs and lariat introns. Mol. Cell 11: 1279–1289 [DOI] [PubMed] [Google Scholar]
- Daugeron M. C., Mauxion F., Seraphin B., 2001. The yeast POP2 gene encodes a nuclease involved in mRNA deadenylation. Nucleic Acids Res. 29: 2448–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis C. A., Ares M., Jr, 2006. Accumulation of unstable promoter-associated transcripts upon loss of the nuclear exosome subunit Rrp6p in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 103: 3262–3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker C. J., Parker R., 1993. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 7: 1632–1643 [DOI] [PubMed] [Google Scholar]
- Decker C. J., Teixeira D., Parker R., 2007. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J. Cell Biol. 179: 437–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decourty L., Saveanu C., Zemam K., Hantraye F., Frachon E., et al. , 2008. Linking functionally related genes by sensitive and quantitative characterization of genetic interaction profiles. Proc. Natl. Acad. Sci. USA 105: 5821–5826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deliz-Aguirre R., Atkin A. L., Kebaara B. W., 2011. Copper tolerance of Saccharomyces cerevisiae nonsense-mediated mRNA decay mutants. Curr. Genet. 57: 421–430 [DOI] [PubMed] [Google Scholar]
- Deluen C., James N., Maillet L., Molinete M., Theiler G., et al. , 2002. The Ccr4-not complex and yTAF1 (yTaf(II)130p/yTaf(II)145p) show physical and functional interactions. Mol. Cell. Biol. 22: 6735–6749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis C. L., Chen J., 2003. The CCR4-NOT complex plays diverse roles in mRNA metabolism. Prog. Nucleic Acid Res. Mol. Biol. 73: 221–250 [DOI] [PubMed] [Google Scholar]
- De Pinto B., Lippolis R., Castaldo R., Altamura N., 2004. Overexpression of Upf1p compensates for mitochondrial splicing deficiency independently of its role in mRNA surveillance. Mol. Microbiol. 51: 1129–1142 [DOI] [PubMed] [Google Scholar]
- Deshmukh M. V., Jones B. N., Quang-Dang D. U., Flinders J., Floor S. N., et al. , 2008. mRNA decapping is promoted by an RNA-binding channel in Dcp2. Mol. Cell 29: 324–336 [DOI] [PubMed] [Google Scholar]
- Dez C., Noaillac-Depeyre J., Caizergues-Ferrer M., Henry Y., 2002. Naf1p, an essential nucleoplasmic factor specifically required for accumulation of box H/ACA small nucleolar RNPs. Mol. Cell. Biol. 22: 7053–7065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dez C., Houseley J., Tollervey D., 2006. Surveillance of nuclear-restricted pre-ribosomes within a subnucleolar region of Saccharomyces cerevisiae. EMBO J. 25: 1534–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichtl B., Stevens A., Tollervey D., 1997. Lithium toxicity in yeast is due to the inhibition of RNA processing enzymes. EMBO J. 16: 7184–7195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieckmann C. L., Staples R. R., 1994. Regulation of mitochondrial gene expression in Saccharomyces cerevisiae. Int. Rev. Cytol. 152: 145–181 [DOI] [PubMed] [Google Scholar]
- Dimitrova L. N., Kuroha K., Tatematsu T., Inada T., 2009. Nascent peptide-dependent translation arrest leads to Not4p-mediated protein degradation by the proteasome. J. Biol. Chem. 284: 10343–10352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doma M. K., Parker R., 2006. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature 440: 561–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doma M. K., Parker R., 2007. RNA quality control in eukaryotes. Cell 131: 660–668 [DOI] [PubMed] [Google Scholar]
- Dong S., Li C., Zenklusen D., Singer R. H., Jacobson A., et al. , 2007. YRA1 autoregulation requires nuclear export and cytoplasmic Edc3p-mediated degradation of its pre-mRNA. Mol. Cell 25: 559–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dori D., Choder M., 2007. Conceptual modeling in systems biology fosters empirical findings: the mRNA lifecycle. PLoS ONE 2: e872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dower K., Kuperwasser N., Merrikh H., Rosbash M., 2004. A synthetic A tail rescues yeast nuclear accumulation of a ribozyme-terminated transcript. RNA 10: 1888–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunckley T., Parker R., 1999. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 18: 5411–5422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunckley T., Tucker M., Parker R., 2001. Two related proteins, Edc1p and Edc2p, stimulate mRNA decapping in Saccharomyces cerevisiae. Genetics 157: 27–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duttagupta R., Vasudevan S., Wilusz C. J., Peltz S. W., 2003. A yeast homologue of Hsp70, Ssa1p, regulates turnover of the MFA2 transcript through its AU-rich 3′ untranslated region. Mol. Cell. Biol. 23: 2623–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duttagupta R., Tian B., Wilusz C. J., Khounh D. T., Soteropoulos P., et al. , 2005. Global analysis of Pub1p targets reveals a coordinate control of gene expression through modulation of binding and stability. Mol. Cell. Biol. 25: 5499–5513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziembowski A., Piwowarski J., Hoser R., Minczuk M., Dmochowska A., et al. , 2003. The yeast mitochondrial degradosome. Its composition, interplay between RNA helicase and RNase activities and the role in mitochondrial RNA metabolism. J. Biol. Chem. 278: 1603–1611 [DOI] [PubMed] [Google Scholar]
- Dziembowski A., Lorentzen E., Conti E., Seraphin B., 2007. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat. Struct. Mol. Biol. 14: 15–22 [DOI] [PubMed] [Google Scholar]
- Egecioglu D. E., Chanfreau G., 2011. Proofreading and spellchecking: a two-tier strategy for pre-mRNA splicing quality control. RNA 17: 383–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egecioglu D. E., Henras A. K., Chanfreau G. F., 2006. Contributions of Trf4p- and Trf5p-dependent polyadenylation to the processing and degradative functions of the yeast nuclear exosome. RNA 12: 26–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairman-Williams M. E., Guenther U. P., Jankowsky E., 2010. SF1 and SF2 helicases: family matters. Curr. Opin. Struct. Biol. 20: 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang F., Phillips S., Butler J. S., 2005. Rat1p and Rai1p function with the nuclear exosome in the processing and degradation of rRNA precursors. RNA 11: 1571–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A., Dlakic M., Tollervey D., 2002. Naf1 p is a box H/ACA snoRNP assembly factor. RNA 8: 1502–1514 [PMC free article] [PubMed] [Google Scholar]
- Fekete Z., Ellis T. P., Schonauer M. S., Dieckmann C. L., 2008. Pet127 governs a 5′ → 3′-exonuclease important in maturation of apocytochrome b mRNA in Saccharomyces cerevisiae. J. Biol. Chem. 283: 3767–3772 [DOI] [PubMed] [Google Scholar]
- Fischer N., Weis K., 2002. The DEAD box protein Dhh1 stimulates the decapping enzyme Dcp1. EMBO J. 21: 2788–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floor S. N., Jones B. N., Hernandez G. A., Gross J. D., 2010. A split active site couples cap recognition by Dcp2 to activation. Nat. Struct. Mol. Biol. 17: 1096–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford A. S., Guan Q., Neeno-Eckwall E., Culbertson M. R., 2006. Ebs1p, a negative regulator of gene expression controlled by the Upf proteins in the yeast Saccharomyces cerevisiae. Eukaryot. Cell 5: 301–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischmeyer P. A., Van Hoof A., O’Donnell K., Guerrerio A. L., Parker R., et al. , 2002. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science 295: 2258–2261 [DOI] [PubMed] [Google Scholar]
- Fujii K., Kitabatake M., Sakata T., Miyata A., Ohno M., 2009. A role for ubiquitin in the clearance of nonfunctional rRNAs. Genes Dev. 23: 963–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funakoshi Y., Doi Y., Hosoda N., Uchida N., Osawa M., et al. , 2007. Mechanism of mRNA deadenylation: evidence for a molecular interplay between translation termination factor eRF3 and mRNA deadenylases. Genes Dev. 21: 3135–3148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaba A., Jacobson A., Sachs M. S., 2005. Ribosome occupancy of the yeast CPA1 upstream open reading frame termination codon modulates nonsense-mediated mRNA decay. Mol. Cell 20: 449–460 [DOI] [PubMed] [Google Scholar]
- Gagliardi D., Stepien P. P., Temperley R. J., Lightowlers R. N., Chrzanowska-Lightowlers Z. M., 2004. Messenger RNA stability in mitochondria: different means to an end. Trends Genet. 20: 260–267 [DOI] [PubMed] [Google Scholar]
- Galy V., Gadal O., Fromont-Racine M., Romano A., Jacquier A., et al. , 2004. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell 116: 63–73 [DOI] [PubMed] [Google Scholar]
- Gandhi R., Manzoor M., Hudak K. A., 2008. Depurination of Brome mosaic virus RNA3 in vivo results in translation-dependent accelerated degradation of the viral RNA. J. Biol. Chem. 283: 32218–32228 [DOI] [PubMed] [Google Scholar]
- Gaudon C., Chambon P., Losson R., 1999. Role of the essential yeast protein PSU1 in p6anscriptional enhancement by the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 18: 2229–2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S., Lojek L., Khalil A. M., Baker K. E., Coller J., 2012. Decapping of long noncoding RNAs regulates inducible genes. Mol. Cell. 45: 279–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber A. P., Herschlag D., Brown P. O., 2004. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2: E79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S., Ganesan R., Amrani N., Jacobson A., 2010. Translational competence of ribosomes released from a premature termination codon is modulated by NMD factors. RNA 16: 1832–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G., Chu A. M., Ni L., Connelly C., Riles L., et al. , 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391 [DOI] [PubMed] [Google Scholar]
- Goldstrohm A. C., Wickens M., 2008. Multifunctional deadenylase complexes diversify mRNA control. Nat. Rev. Mol. Cell Biol. 9: 337–344 [DOI] [PubMed] [Google Scholar]
- Goldstrohm A. C., Hook B. A., Seay D. J., Wickens M., 2006. PUF proteins bind Pop2p to regulate messenger RNAs. Nat. Struct. Mol. Biol. 13: 533–539 [DOI] [PubMed] [Google Scholar]
- Goldstrohm A. C., Seay D. J., Hook B. A., Wickens M., 2007. PUF protein-mediated deadenylation is catalyzed by Ccr4p. J. Biol. Chem. 282: 109–114 [DOI] [PubMed] [Google Scholar]
- Goler-Baron V., Selitrennik M., Barkai O., Haimovich G., Lotan R., et al. , 2008. Transcription in the nucleus and mRNA decay in the cytoplasm are coupled processes. Genes Dev. 22: 2022–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez C. I., Bhattacharya A., Wang W., Peltz S. W., 2001. Nonsense-mediated mRNA decay in Saccharomyces cerevisiae. Gene 274: 15–25 [DOI] [PubMed] [Google Scholar]
- Gowrishankar G., Winzen R., Bollig F., Ghebremedhin B., Redich N., et al. , 2005. Inhibition of mRNA deadenylation and degradation by ultraviolet light. Biol. Chem. 386: 1287–1293 [DOI] [PubMed] [Google Scholar]
- Gowrishankar G., Winzen R., Dittrich-Breiholz O., Redich N., Kracht M., et al. , 2006. Inhibition of mRNA deadenylation and degradation by different types of cell stress. Biol. Chem. 387: 323–327 [DOI] [PubMed] [Google Scholar]
- Graille M., Chaillet M., Van Tilbeurgh H., 2008. Structure of yeast Dom34: a protein related to translation termination factor Erf1 and involved in No-Go decay. J. Biol. Chem. 283: 7145–7154 [DOI] [PubMed] [Google Scholar]
- Grigull J., Mnaimneh S., Pootoolal J., Robinson M. D., Hughes T. R., 2004. Genome-wide analysis of mRNA stability using transcription inhibitors and microarrays reveals posttranscriptional control of ribosome biogenesis factors. Mol. Cell. Biol. 24: 5534–5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grousl T., Ivanov P., Frydlova I., Vasicova P., Janda F., et al. , 2009. Robust heat shock induces eIF2alpha-phosphorylation-independent assembly of stress granules containing eIF3 and 40S ribosomal subunits in budding yeast, Saccharomyces cerevisiae. J. Cell Sci. 122: 2078–2088 [DOI] [PubMed] [Google Scholar]
- Grzechnik P., Kufel J., 2008. Polyadenylation linked to transcription termination directs the processing of snoRNA precursors in yeast. Mol. Cell 32: 247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Q., Zheng W., Tang S., Liu X., Zinkel R. A., et al. , 2006. Impact of nonsense-mediated mRNA decay on the global expression profile of budding yeast. PLoS Genet. 2: e203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudipati R. K., Villa T., Boulay J., Libri D., 2008. Phosphorylation of the RNA polymerase II C-terminal domain dictates transcription termination choice. Nat. Struct. Mol. Biol. 15: 786–794 [DOI] [PubMed] [Google Scholar]
- Hanekamp T., Thorsness P. E., 1999. YNT20, a bypass suppressor of yme1 yme2, encodes a putative 3′-5′ exonuclease localized in mitochondria of Saccharomyces cerevisiae. Curr. Genet. 34: 438–448 [DOI] [PubMed] [Google Scholar]
- Harigaya Y., Jones B. N., Muhlrad D., Gross J. D., Parker R., 2010. Identification and analysis of the interaction between Edc3 and Dcp2 in Saccharomyces cerevisiae. Mol. Cell. Biol. 30: 1446–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa Y., Irie K., Gerber A. P., 2008. Distinct roles for Khd1p in the localization and expression of bud-localized mRNAs in yeast. RNA 14: 2333–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F., Peltz S. W., Donahue J. L., Rosbash M., Jacobson A., 1993. Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1- mutant. Proc. Natl. Acad. Sci. USA 90: 7034–7038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F., Brown A. H., Jacobson A., 1996. Interaction between Nmd2p and Upf1p is required for activity but not for dominant-negative inhibition of the nonsense-mediated mRNA decay pathway in yeast. RNA 2: 153–170 [PMC free article] [PubMed] [Google Scholar]
- He F., Brown A. H., Jacobson A., 1997. Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol. Cell. Biol. 17: 1580–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F., Li X., Spatrick P., Casillo R., Dong S., et al. , 2003. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol. Cell 12: 1439–1452 [DOI] [PubMed] [Google Scholar]
- He W., Parker R., 2001. The yeast cytoplasmic LsmI/Pat1p complex protects mRNA 3′ termini from partial degradation. Genetics 158: 1445–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton B., Decker C., Muhlrad D., Donahue J., Jacobson A., et al. , 1992. Analysis of chimeric mRNAs derived from the STE3 mRNA identifies multiple regions within yeast mRNAs that modulate mRNA decay. Nucleic Acids Res. 20: 5365–5373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henri J., Rispal D., Bayart E., Van Tilbeurgh H., Seraphin B., et al. , 2010. Structural and functional insights into Saccharomyces cerevisiae Tpa1, a putative prolylhydroxylase influencing translation termination and transcription. J. Biol. Chem. 285: 30767–30778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrick D., Parker R., Jacobson A., 1990. Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 10: 2269–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgers V., Teixeira D., Parker R., 2006. Translation-independent inhibition of mRNA deadenylation during stress in Saccharomyces cerevisiae. RNA 12: 1835–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren P., Parker R., 2001. Defects in the mRNA export factors Rat7p, Gle1p, Mex67p, and Rat8p cause hyperadenylation during 3′-end formation of nascent transcripts. RNA 7: 753–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren P. J., Parker R., 2003. Cytoplasmic degradation of splice-defective pre-mRNAs and intermediates. Mol. Cell 12: 1453–1465 [DOI] [PubMed] [Google Scholar]
- Hilleren P., McCarthy T., Rosbash M., Parker R., Jensen T. H., 2001. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature 413: 538–542 [DOI] [PubMed] [Google Scholar]
- Hook B. A., Goldstrohm A. C., Seay D. J., Wickens M., 2007. Two yeast PUF proteins negatively regulate a single mRNA. J. Biol. Chem. 282: 15430–15438 [DOI] [PubMed] [Google Scholar]
- Hosoda N., Kobayashi T., Uchida N., Funakoshi Y., Kikuchi Y., et al. , 2003. Translation termination factor eRF3 mediates mRNA decay through the regulation of deadenylation. J. Biol. Chem. 278: 38287–38291 [DOI] [PubMed] [Google Scholar]
- Houseley J., Tollervey D., 2006. Yeast Trf5p is a nuclear poly(A) polymerase. EMBO Rep. 7: 205–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J., Lacava J., Tollervey D., 2006. RNA-quality control by the exosome. Nat. Rev. Mol. Cell Biol. 7: 529–539 [DOI] [PubMed] [Google Scholar]
- Houseley J., Kotovic K., El Hage A., Tollervey D., 2007. Trf4 targets ncRNAs from telomeric and rDNA spacer regions and functions in rDNA copy number control. EMBO J. 26: 4996–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J., Rubbi L., Grunstein M., Tollervey D., Vogelauer M., 2008. A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol. Cell 32: 685–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu C. L., Stevens A., 1993. Yeast cells lacking 5′→3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol. Cell. Biol. 13: 4826–4835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Sweet T. J., Chamnongpol S., Baker K. E., Coller J., 2009. Co-translational mRNA decay in Saccharomyces cerevisiae. Nature 461: 225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. C., Chen H. T., Teng S. C., 2010. Intragenic transcription of a noncoding RNA modulates expression of ASP3 in budding yeast. RNA 16: 2085–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada T., Aiba H., 2005. Translation of aberrant mRNAs lacking a termination codon or with a shortened 3′-UTR is repressed after initiation in yeast. EMBO J. 24: 1584–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito-Harashima S., Kuroha K., Tatematsu T., Inada T., 2007. Translation of the poly(A) tail plays crucial roles in nonstop mRNA surveillance via translation repression and protein destabilization by proteasome in yeast. Genes Dev. 21: 519–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamonnak N., Creamer T. J., Darby M. M., Schaughency P., Wheelan S. J., et al. , 2011. Yeast Nrd1, Nab3, and Sen1 transcriptome-wide binding maps suggest multiple roles in post-transcriptional RNA processing. RNA 17: 2011–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen T. H., Patricio K., McCarthy T., Rosbash M., 2001. A block to mRNA nuclear export in S. cerevisiae leads to hyperadenylation of transcripts that accumulate at the site of transcription. Mol. Cell 7: 887–898 [DOI] [PubMed] [Google Scholar]
- Jiao X., Xiang S., Oh C., Martin C. E., Tong L., et al. , 2010. Identification of a quality-control mechanism for mRNA 5′-end capping. Nature 467: 608–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Coyle S. M., Doudna J. A., 2011. Coupled 5′ nucleotide recognition and processivity in Xrn1-mediated mRNA decay. Mol. Cell 41: 600–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M. J., Jacobson A., 2010. Nonsense-mediated mRNA decay maintains translational fidelity by limiting magnesium uptake. Genes Dev. 24: 1491–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. W., 1997. Rat1p and Xrn1p are functionally interchangeable exoribonucleases that are restricted to and required in the nucleus and cytoplasm, respectively. Mol. Cell. Biol. 17: 6122–6130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M. H., Guthrie C., 1990. Unexpected flexibility in an evolutionarily conserved protein-RNA interaction: genetic analysis of the Sm binding site. EMBO J. 9: 2555–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba S., Krueger A., Trice T., Krecic A. M., Hinnebusch A. G., et al. , 2004. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 18: 1227–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba S., Wang X., Anderson J. T., 2006. Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA. RNA 12: 508–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima T., Pellegrini M., Chanfreau G. F., 2009. Nonsense-mediated mRNA decay mutes the splicing defects of spliceosome component mutations. RNA 15: 2236–2247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebaara B. W., Atkin A. L., 2009. Long 3′-UTRs target wild-type mRNAs for nonsense-mediated mRNA decay in Saccharomyces cerevisiae. Nucleic Acids Res. 37: 2771–2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling K. M., Salas-Marco J., Osherovich L. Z., Bedwell D. M., 2006. Tpa1p is part of an mRNP complex that influences translation termination, mRNA deadenylation, and mRNA turnover in Saccharomyces cerevisiae. Mol. Cell. Biol. 26: 5237–5248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenna M., Stevens A., McCammon M., Douglas M. G., 1993. An essential yeast gene with homology to the exonuclease-encoding XRN1/KEM1 gene also encodes a protein with exoribonuclease activity. Mol. Cell. Biol. 13: 341–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. S., Kim H. L., Kim K. H., Kim Do J., Lee S. J., et al. , 2010. Crystal structure of Tpa1 from Saccharomyces cerevisiae, a component of the messenger ribonucleoprotein complex. Nucleic Acids Res. 38: 2099–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. Y., Levin D. E., 2011. Mpk1 MAPK association with the Paf1 complex blocks Sen1-mediated premature transcription termination. Cell 144: 745–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Funakoshi Y., Hoshino S., Katada T., 2004. The GTP-binding release factor eRF3 as a key mediator coupling translation termination to mRNA decay. J. Biol. Chem. 279: 45693–45700 [DOI] [PubMed] [Google Scholar]
- Komarnitsky P., Cho E. J., Buratowski S., 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14: 2452–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft C., Deplazes A., Sohrmann M., Peter M., 2008. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat. Cell Biol. 10: 602–610 [DOI] [PubMed] [Google Scholar]
- Kruk J. A., Dutta A., Fu J., Gilmour D. S., Reese J. C., 2011. The multifunctional Ccr4-Not complex directly promotes transcription elongation. Genes Dev. 25: 581–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kshirsagar M., Parker R., 2004. Identification of Edc3p as an enhancer of mRNA decapping in Saccharomyces cerevisiae. Genetics 166: 729–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuai L., Fang F., Butler J. S., Sherman F., 2004. Polyadenylation of rRNA in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 101: 8581–8586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroha K., Tatematsu T., Inada T., 2009. Upf1 stimulates degradation of the product derived from aberrant messenger RNA containing a specific nonsense mutation by the proteasome. EMBO Rep. 10: 1265–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroha K., Akamatsu M., Dimitrova L., Ito T., Kato Y., et al. , 2010. Receptor for activated C kinase 1 stimulates nascent polypeptide-dependent translation arrest. EMBO Rep. 11: 956–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacava J., Houseley J., Saveanu C., Petfalski E., Thompson E., et al. , 2005. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121: 713–724 [DOI] [PubMed] [Google Scholar]
- Lagrandeur T., Parker R., 1999. The cis acting sequences responsible for the differential decay of the unstable MFA2 and stable PGK1 transcripts in yeast include the context of the translational start codon. RNA 5: 420–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lardenois A., Liu Y., Walther T., Chalmel F., Evrard B., et al. , 2011. Execution of the meiotic noncoding RNA expression program and the onset of gametogenesis in yeast require the conserved exosome subunit Rrp6. Proc. Natl. Acad. Sci. USA 108: 1058–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lariviere F. J., Cole S. E., Ferullo D. J., Moore M. J., 2006. A late-acting quality control process for mature eukaryotic rRNAs. Mol. Cell 24: 619–626 [DOI] [PubMed] [Google Scholar]
- Lebreton A., Tomecki R., Dziembowski A., Seraphin B., 2008. Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature 456: 993–996 [DOI] [PubMed] [Google Scholar]
- Lee D., Ohn T., Chiang Y. C., Quigley G., Yao G., et al. , 2010. PUF3 acceleration of deadenylation in vivo can operate independently of CCR4 activity, possibly involving effects on the PAB1-mRNP structure. J. Mol. Biol. 399: 562–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. H., Kim Y. S., Kim K. H., Heo I., Kim S. K., et al. , 2007. Structural and functional insights into Dom34, a key component of no-go mRNA decay. Mol. Cell 27: 938–950 [DOI] [PubMed] [Google Scholar]
- Lelivelt M. J., Culbertson M. R., 1999. Yeast Upf proteins required for RNA surveillance affect global expression of the yeast transcriptome. Mol. Cell. Biol. 19: 6710–6719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letzring D. P., Dean K. M., Grayhack E. J., 2010. Control of translation efficiency in yeast by codon-anticodon interactions. RNA 16: 2516–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libri D., Dower K., Boulay J., Thomsen R., Rosbash M., et al. , 2002. Interactions between mRNA export commitment, 3′-end quality control, and nuclear degradation. Mol. Cell. Biol. 22: 8254–8266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Kiledjian M., 2005. Scavenger decapping activity facilitates 5′ to 3′ mRNA decay. Mol. Cell. Biol. 25: 9764–9772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Rodgers N. D., Jiao X., Kiledjian M., 2002. The scavenger mRNA decapping enzyme DcpS is a member of the HIT family of pyrophosphatases. EMBO J. 21: 4699–4708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Greimann J. C., Lima C. D., 2006. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell 127: 1223–1237 [DOI] [PubMed] [Google Scholar]
- Liu Y. X., Dieckmann C. L., 1989. Overproduction of yeast viruslike particles by strains deficient in a mitochondrial nuclease. Mol. Cell. Biol. 9: 3323–3331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losson R., Lacroute F., 1979. Interference of nonsense mutations with eukaryotic messenger RNA stability. Proc. Natl. Acad. Sci. USA 76: 5134–5137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan R., Bar-On V. G., Harel-Sharvit L., Duek L., Melamed D., et al. , 2005. The RNA polymerase II subunit Rpb4p mediates decay of a specific class of mRNAs. Genes Dev. 19: 3004–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan R., Goler-Baron V., Duek L., Haimovich G., Choder M., 2007. The Rpb7p subunit of yeast RNA polymerase II plays roles in the two major cytoplasmic mRNA decay mechanisms. J. Cell Biol. 178: 1133–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luhtala N., Parker R., 2009. LSM1 over-expression in Saccharomyces cerevisiae depletes U6 snRNA levels. Nucleic Acids Res. 37: 5529–5536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke B., Azzalin C. M., Hug N., Deplazes A., Peter M., et al. , 2007. Saccharomyces cerevisiae Ebs1p is a putative ortholog of human Smg7 and promotes nonsense-mediated mRNA decay. Nucleic Acids Res. 35: 7688–7697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke B., Panza A., Redon S., Iglesias N., Li Z., et al. , 2008. The Rat1p 5′ to 3′ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol. Cell 32: 465–477 [DOI] [PubMed] [Google Scholar]
- Luo G., Costanzo M., Boone C., Dickson R. C., 2011. Nutrients and the Pkh1/2 and Pkc1 protein kinases control mRNA decay and P-body assembly in yeast. J. Biol. Chem. 286: 8759–8770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen S., Brodersen D. E., Jensen T. H., 2009. Origins and activities of the eukaryotic exosome. J. Cell Sci. 122: 1487–1494 [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen S., Tomecki R., Jensen T. H., Dziembowski A., 2011. The eukaryotic RNA exosome: same scaffold but variable catalytic subunits. RNA Biol. 8: 61–66 [DOI] [PubMed] [Google Scholar]
- Macintosh G. C., Bariola P. A., Newbigin E., Green P. J., 2001. Characterization of Rny1, the Saccharomyces cerevisiae member of the T2 RNase family of RNases: Unexpected functions for ancient enzymes? Proc. Natl. Acad. Sci. USA 98: 1018–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maderazo A. B., He F., Mangus D. A., Jacobson A., 2000. Upf1p control of nonsense mRNA translation is regulated by Nmd2p and Upf3p. Mol. Cell. Biol. 20: 4591–4603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malecki M., Jedrzejczak R., Stepien P. P., Golik P., 2007. In vitro reconstitution and characterization of the yeast mitochondrial degradosome complex unravels tight functional interdependence. J. Mol. Biol. 372: 23–36 [DOI] [PubMed] [Google Scholar]
- Malecki M., Jedrzejczak R., Puchta O., Stepien P. P., Golik P., 2008. In vivo and in vitro approaches for studying the yeast mitochondrial RNA degradosome complex. Methods Enzymol. 447: 463–488 [DOI] [PubMed] [Google Scholar]
- Malys N., McCarthy J. E., 2006. Dcs2, a novel stress-induced modulator of m7GpppX pyrophosphatase activity that locates to P bodies. J. Mol. Biol. 363: 370–382 [DOI] [PubMed] [Google Scholar]
- Malys N., Carroll K., Miyan J., Tollervey D., McCarthy J. E., 2004. The ‘scavenger’ m7GpppX pyrophosphatase activity of Dcs1 modulates nutrient-induced responses in yeast. Nucleic Acids Res. 32: 3590–3600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandart E., Parker R., 1995. Effects of mutations in the Saccharomyces cerevisiae RNA14, RNA15, and PAP1 genes on polyadenylation in vivo. Mol. Cell. Biol. 15: 6979–6986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens J. A., Laprade L., Winston F., 2004. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature 429: 571–574 [DOI] [PubMed] [Google Scholar]
- Marvin M. C., Clauder-Munster S., Walker S. C., Sarkeshik A., Yates J. R., III, et al. , 2011. Accumulation of noncoding RNA due to an RNase P defect in Saccharomyces cerevisiae. RNA 17: 1441–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauchi N., Ohtake Y., Irie K., 2010. Stability control of MTL1 mRNA by the RNA-binding protein Khd1p in yeast. Cell Struct. Funct. 35: 95–105 [DOI] [PubMed] [Google Scholar]
- Mayas R. M., Maita H., Semlow D. R., Staley J. P., 2010. Spliceosome discards intermediates via the DEAH box ATPase Prp43p. Proc. Natl. Acad. Sci. USA 107: 10020–10025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaux S., Van Hoof A., 2006. Yeast transcripts cleaved by an internal ribozyme provide new insight into the role of the cap and poly(A) tail in translation and mRNA decay. RNA 12: 1323–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaux S., Van Hoof A., Baker K. E., 2008. Nonsense-mediated mRNA decay in yeast does not require PAB1 or a poly(A) tail. Mol. Cell 29: 134–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan L., Decourty L., Saveanu C., Rappsilber J., Ceulemans H., et al. , 2008. A yeast exosome cofactor, Mpp6, functions in RNA surveillance and in the degradation of noncoding RNA transcripts. Mol. Cell. Biol. 28: 5446–5457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J., Zassenhaus H. P., 1993. Identification of a protein complex that binds to a dodecamer sequence found at the 3′ ends of yeast mitochondrial mRNAs. Mol. Cell. Biol. 13: 4167–4173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P., Tollervey D., 2003. An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3′→5′ degradation. Mol. Cell 11: 1405–1413 [DOI] [PubMed] [Google Scholar]
- Mitchell P., Petfalski E., Houalla R., Podtelejnikov A., Mann M., et al. , 2003. Rrp47p is an exosome-associated protein required for the 3′ processing of stable RNAs. Mol. Cell. Biol. 23: 6982–6992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey J. P., Deardorff J. A., Hebron C., Sachs A. B., 1999. Decapping of stabilized, polyadenylated mRNA in yeast pab1 mutants. Yeast 15: 687–702 [DOI] [PubMed] [Google Scholar]
- Mroczek S., Kufel J., 2008. Apoptotic signals induce specific degradation of ribosomal RNA in yeast. Nucleic Acids Res. 36: 2874–2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D., Parker R., 1992. Mutations affecting stability and deadenylation of the yeast MFA2 transcript. Genes Dev. 6: 2100–2111 [DOI] [PubMed] [Google Scholar]
- Muhlrad D., Parker R., 1994. Premature translational termination triggers mRNA decapping. Nature 370: 578–581 [DOI] [PubMed] [Google Scholar]
- Muhlrad D., Parker R., 1999a. Aberrant mRNAs with extended 3′ UTRs are substrates for rapid degradation by mRNA surveillance. RNA 5: 1299–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D., Parker R., 1999b. Recognition of yeast mRNAs as “nonsense containing” leads to both inhibition of mRNA translation and mRNA degradation: implications for the control of mRNA decapping. Mol. Biol. Cell 10: 3971–3978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D., Parker R., 2005. The yeast EDC1 mRNA undergoes deadenylation-independent decapping stimulated by Not2p, Not4p, and Not5p. EMBO J. 24: 1033–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D., Decker C. J., Parker R., 1994. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′→3′ digestion of the transcript. Genes Dev. 8: 855–866 [DOI] [PubMed] [Google Scholar]
- Muhlrad D., Decker C. J., Parker R., 1995. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol. Cell. Biol. 15: 2145–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munchel S. E., Shultzaberger R. K., Takizawa N., Weis K., 2011. Dynamic profiling of mRNA turnover reveals gene-specific and system-wide regulation of mRNA decay. Mol. Biol. Cell 22: 2787–2795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neef D. W., Thiele D. J., 2009. Enhancer of decapping proteins 1 and 2 are important for translation during heat stress in Saccharomyces cerevisiae. Mol. Microbiol. 73: 1032–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil H., Malabat C., D’Aubenton-Carafa Y., Xu Z., Steinmetz L. M., et al. , 2009. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature 457: 1038–1042 [DOI] [PubMed] [Google Scholar]
- Nissan T., Rajyaguru P., She M., Song H., Parker R., 2010. Decapping activators in Saccharomyces cerevisiae act by multiple mechanisms. Mol. Cell 39: 773–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivas W., Parker R., 2000. The Puf3 protein is a transcript-specific regulator of mRNA degradation in yeast. EMBO J. 19: 6602–6611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivas W. M., Muhlrad D., Parker R., 1997. Analysis of the yeast genome: identification of new non-coding and small ORF-containing RNAs. Nucleic Acids Res. 25: 4619–4625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossareh-Nazari B., Bonizec M., Cohen M., Dokudovskaya S., Delalande F., et al. , 2010. Cdc48 and Ufd3, new partners of the ubiquitin protease Ubp3, are required for ribophagy. EMBO Rep. 11: 548–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozanick S. G., Wang X., Costanzo M., Brost R. L., Boone C., et al. , 2009. Rex1p deficiency leads to accumulation of precursor initiator tRNAMet and polyadenylation of substrate RNAs in Saccharomyces cerevisiae. Nucleic Acids Res. 37: 298–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannone B. K., Xue D., Wolin S. L., 1998. A role for the yeast La protein in U6 snRNP assembly: evidence that the La protein is a molecular chaperone for RNA polymerase III transcripts. EMBO J. 17: 7442–7453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannone B. K., Kim S. D., Noe D. A., Wolin S. L., 2001. Multiple functional interactions between components of the Lsm2-Lsm8 complex, U6 snRNA, and the yeast La protein. Genetics 158: 187–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquin N., Menade M., Poirier G., Donato D., Drouet E., et al. , 2007. Local activation of yeast ASH1 mRNA translation through phosphorylation of Khd1p by the casein kinase Yck1p. Mol. Cell 26: 795–809 [DOI] [PubMed] [Google Scholar]
- Parker R., Sheth U., 2007. P bodies and the control of mRNA translation and degradation. Mol. Cell 25: 635–646 [DOI] [PubMed] [Google Scholar]
- Passos D. O., Parker R., 2008. Analysis of cytoplasmic mRNA decay in Saccharomyces cerevisiae. Methods Enzymol. 448: 409–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passos D. O., Doma M. K., Shoemaker C. J., Muhlrad D., Green R., et al. , 2009. Analysis of Dom34 and its function in no-go decay. Mol. Biol. Cell 20: 3025–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D., Butler J. S., 1992. Conditional defect in mRNA 3′ end processing caused by a mutation in the gene for poly(A) polymerase. Mol. Cell. Biol. 12: 3297–3304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedro-Segura E., Vergara S. V., Rodriguez-Navarro S., Parker R., Thiele D. J., et al. , 2008. The Cth2 ARE-binding protein recruits the Dhh1 helicase to promote the decay of succinate dehydrogenase SDH4 mRNA in response to iron deficiency. J. Biol. Chem. 283: 28527–28535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltz S. W., Brown A. H., Jacobson A., 1993. mRNA destabilization triggered by premature translational termination depends on at least three cis-acting sequence elements and one trans-acting factor. Genes Dev. 7: 1737–1754 [DOI] [PubMed] [Google Scholar]
- Pilkington G. R., Parker R., 2008. Pat1 contains distinct functional domains that promote P-body assembly and activation of decapping. Mol. Cell. Biol. 28: 1298–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisareva V. P., Skabkin M. A., Hellen C. U., Pestova T. V., Pisarev A. V., 2011. Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. EMBO J. 30: 1804–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole T. L., Stevens A., 1995. Comparison of features of the RNase activity of 5′-exonuclease-1 and 5′-exonuclease-2 of Saccharomyces cerevisiae. Nucleic Acids Symp. Ser., 79–81 [PubMed] [Google Scholar]
- Puig S., Askeland E., Thiele D. J., 2005. Coordinated remodeling of cellular metabolism during iron deficiency through targeted mRNA degradation. Cell 120: 99–110 [DOI] [PubMed] [Google Scholar]
- Qiu H., Hu C., Yoon S., Natarajan K., Swanson M. J., et al. , 2004. An array of coactivators is required for optimal recruitment of TATA binding protein and RNA polymerase II by promoter-bound Gcn4p. Mol. Cell. Biol. 24: 4104–4117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajyaguru P., She M., Parker R., 2012. Scd6 targets eIF4G to repress translation: RGG motif proteins as a class of eIF4G-binding proteins. Mol. Cell 45: 244–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran V., Shah K. H., Herman P. K., 2011. The cAMP-dependent protein kinase signaling pathway is a key regulator of P body foci formation. Mol. Cell 43: 973–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijns M. A., Alexander R. D., Spiller M. P., Beggs J. D., 2008. A role for Q/N-rich aggregation-prone regions in P-body localization. J. Cell Sci. 121: 2463–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendl L. M., Bieman M. A., Smibert C. A., 2008. S. cerevisiae Vts1p induces deadenylation-dependent transcript degradation and interacts with the Ccr4p-Pop2p-Not deadenylase complex. RNA 14: 1328–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee H. S., Pugh B. F., 2012. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature 483: 295–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts P., Moshitch-Moshkovitz S., Kvam E., O’Toole E., Winey M., et al. , 2003. Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol. Biol. Cell 14: 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondon A. G., Mischo H. E., Kawauchi J., Proudfoot N. J., 2009. Fail-safe transcriptional termination for protein-coding genes in S. cerevisiae. Mol. Cell 36: 88–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougemaille M., Gudipati R. K., Olesen J. R., Thomsen R., Seraphin B., et al. , 2007. Dissecting mechanisms of nuclear mRNA surveillance in THO/sub2 complex mutants. EMBO J. 26: 2317–2326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Echevarria M. J., Peltz S. W., 2000. The RNA binding protein Pub1 modulates the stability of transcripts containing upstream open reading frames. Cell 101: 741–751 [DOI] [PubMed] [Google Scholar]
- Sachs A. B., Davis R. W., Kornberg R. D., 1987. A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability. Mol. Cell. Biol. 7: 3268–3276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini P., Eyler D. E., Green R., Dever T. E., 2009. Hypusine-containing protein eIF5A promotes translation elongation. Nature 459: 118–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Paolo S., Vanacova S., Schenk L., Scherrer T., Blank D., et al. , 2009. Distinct roles of non-canonical poly(A) polymerases in RNA metabolism. PLoS Genet. 5: e1000555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayani S., Janis M., Lee C. Y., Toesca I., Chanfreau G. F., 2008. Widespread impact of nonsense-mediated mRNA decay on the yeast intronome. Mol. Cell 31: 360–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer D., Van Hoof A., 2011. Different nuclease requirements for exosome-mediated degradation of normal and nonstop mRNAs. Proc. Natl. Acad. Sci. USA 108: 2366–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer D., Tsanova B., Barbas A., Reis F. P., Dastidar E. G., et al. , 2009. The exosome contains domains with specific endoribonuclease, exoribonuclease and cytoplasmic mRNA decay activities. Nat. Struct. Mol. Biol. 16: 56–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder S. C., Schwer B., Shuman S., Bentley D., 2000. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 14: 2435–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D. C., Parker R., 1999. Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 19: 5247–5256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D. C., Parker R., 2000. mRNA decapping in yeast requires dissociation of the cap binding protein, eukaryotic translation initiation factor 4E. Mol. Cell. Biol. 20: 7933–7942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D., Decker C. J., Parker R., 2003. The enhancer of decapping proteins, Edc1p and Edc2p, bind RNA and stimulate the activity of the decapping enzyme. RNA 9: 239–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal S. P., Dunckley T., Parker R., 2006. Sbp1p affects translational repression and decapping in Saccharomyces cerevisiae. Mol. Cell. Biol. 26: 5120–5130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seipelt R. L., Zheng B., Asuru A., Rymond B. C., 1999. U1 snRNA is cleaved by RNase III and processed through an Sm site-dependent pathway. Nucleic Acids Res. 27: 587–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selitrennik M., Duek L., Lotan R., Choder M., 2006. Nucleocytoplasmic shuttling of the Rpb4p and Rpb7p subunits of Saccharomyces cerevisiae RNA polymerase II by two pathways. Eukaryot. Cell 5: 2092–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto A. G., Zaug A. J., Sobel S. G., Wolin S. L., Cech T. R., 1999. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature 401: 177–180 [DOI] [PubMed] [Google Scholar]
- Shalem O., Groisman B., Choder M., Dahan O., Pilpel Y., 2011. Transcriptome kinetics is governed by a genome-wide coupling of mRNA production and degradation: a role for RNA Pol II. PLoS Genet. 7: e1002273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She M., Decker C. J., Sundramurthy K., Liu Y., Chen N., et al. , 2004. Crystal structure of Dcp1p and its functional implications in mRNA decapping. Nat. Struct. Mol. Biol. 11: 249–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She M., Decker C. J., Chen N., Tumati S., Parker R., et al. , 2006. Crystal structure and functional analysis of Dcp2p from Schizosaccharomyces pombe. Nat. Struct. Mol. Biol. 13: 63–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She M., Decker C. J., Svergun D. I., Round A., Chen N., et al. , 2008. Structural basis of dcp2 recognition and activation by dcp1. Mol. Cell 29: 337–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U., Parker R., 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300: 805–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U., Parker R., 2006. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell 125: 1095–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley R. L., Ford A. S., Richards M. R., Albertini M., Culbertson M. R., 2002. Nuclear import of Upf3p is mediated by importin-α/-β and export to the cytoplasm is required for a functional nonsense-mediated mRNA decay pathway in yeast. Genetics 161: 1465–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker C. J., Eyler D. E., Green R., 2010. Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science 330: 369–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon E., Seraphin B., 2007. A specific role for the C-terminal region of the poly(A)-binding protein in mRNA decay. Nucleic Acids Res. 35: 6017–6028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiger M., Carr-Schmid A., Schwartz D. C., Kiledjian M., Parker R., 2003. Analysis of recombinant yeast decapping enzyme. RNA 9: 231–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens A., 2001. 5′-Exoribonuclease 1: Xrn1. Methods Enzymol. 342: 251–259 [DOI] [PubMed] [Google Scholar]
- Struhl K., 2007. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat. Struct. Mol. Biol. 14: 103–105 [DOI] [PubMed] [Google Scholar]
- Swanson M. J., Qiu H., Sumibcay L., Krueger A., Kim S. J., et al. , 2003. A multiplicity of coactivators is required by Gcn4p at individual promoters in vivo. Mol. Cell. Biol. 23: 2800–2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swisher K. D., Parker R., 2009. Related mechanisms for mRNA and rRNA quality control. Mol. Cell 34: 401–402 [DOI] [PubMed] [Google Scholar]
- Swisher K. D., Parker R., 2010. Localization to, and effects of Pbp1, Pbp4, Lsm12, Dhh1, and Pab1 on stress granules in Saccharomyces cerevisiae. PLoS ONE 5: e10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Synowsky S. A., Van Wijk M., Raijmakers R., Heck A. J., 2009. Comparative multiplexed mass spectrometric analyses of endogenously expressed yeast nuclear and cytoplasmic exosomes. J. Mol. Biol. 385: 1300–1313 [DOI] [PubMed] [Google Scholar]
- Szczesny R. J., Borowski L. S., Malecki M., Wojcik M. A., Stepien P. P., et al. , 2011. RNA degradation in yeast and human mitochondria. Biochim. Biophys. Acta. DOI: 10.1016/j.bbagrm.2011.11.010 [DOI] [PubMed] [Google Scholar]
- Tadauchi T., Matsumoto K., Herskowitz I., Irie K., 2001. Post-transcriptional regulation through the HO 3′-UTR by Mpt5, a yeast homolog of Pumilio and FBF. EMBO J. 20: 552–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S., Araki Y., Ohya Y., Sakuno T., Hoshino S., et al. , 2008. Upf1 potentially serves as a RING-related E3 ubiquitin ligase via its association with Upf3 in yeast. RNA 14: 1950–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talarek N., Cameroni E., Jaquenoud M., Luo X., Bontron S., et al. , 2010. Initiation of the TORC1-regulated G0 program requires Igo1/2, which license specific mRNAs to evade degradation via the 5′-3′ mRNA decay pathway. Mol. Cell 38: 345–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D., Parker R., 2007. Analysis of P-body assembly in Saccharomyces cerevisiae. Mol. Biol. Cell 18: 2274–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D., Sheth U., Valencia-Sanchez M. A., Brengues M., Parker R., 2005. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 11: 371–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharun S., 2009. Lsm1–7-Pat1 complex: A link between 3′ and 5′-ends in mRNA decay? RNA Biol. 6: 228–232 [DOI] [PubMed] [Google Scholar]
- Tharun S., Parker R., 1999. Analysis of mutations in the yeast mRNA decapping enzyme. Genetics 151: 1273–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharun S., Parker R., 2001. Targeting an mRNA for decapping: displacement of translation factors and association of the Lsm1p-7p complex on deadenylated yeast mRNAs. Mol. Cell 8: 1075–1083 [DOI] [PubMed] [Google Scholar]
- Tharun S., He W., Mayes A. E., Lennertz P., Beggs J. D., et al. , 2000. Yeast Sm-like proteins function in mRNA decapping and decay. Nature 404: 515–518 [DOI] [PubMed] [Google Scholar]
- Thiebaut M., Kisseleva-Romanova E., Rougemaille M., Boulay J., Libri D., 2006. Transcription termination and nuclear degradation of cryptic unstable transcripts: a role for the nrd1-nab3 pathway in genome surveillance. Mol. Cell 23: 853–864 [DOI] [PubMed] [Google Scholar]
- Thiebaut M., Colin J., Neil H., Jacquier A., Seraphin B., et al. , 2008. Futile cycle of transcription initiation and termination modulates the response to nucleotide shortage in S. cerevisiae. Mol. Cell 31: 671–682 [DOI] [PubMed] [Google Scholar]
- Thompson D. M., Parker R., 2007. Cytoplasmic decay of intergenic transcripts in Saccharomyces cerevisiae. Mol. Cell. Biol. 27: 92–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson D. M., Parker R., 2009. Stressing out over tRNA cleavage. Cell 138: 215–219 [DOI] [PubMed] [Google Scholar]
- Thompson D. M., Lu C., Green P. J., Parker R., 2008. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA 14: 2095–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen R., Libri D., Boulay J., Rosbash M., Jensen T. H., 2003. Localization of nuclear retained mRNAs in Saccharomyces cerevisiae. RNA 9: 1049–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thore S., Mauxion F., Seraphin B., Suck D., 2003. X-ray structure and activity of the yeast Pop2 protein: a nuclease subunit of the mRNA deadenylase complex. EMBO Rep. 4: 1150–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todeschini A. L., Condon C., Benard L., 2006. Sodium-induced GCN4 expression controls the accumulation of the 5′ to 3′ RNA degradation inhibitor, 3′-phosphoadenosine 5′-phosphate. J. Biol. Chem. 281: 3276–3282 [DOI] [PubMed] [Google Scholar]
- Toesca I., Nery C. R., Fernandez C. F., Sayani S., Chanfreau G. F., 2011. Cryptic transcription mediates repression of subtelomeric metal homeostasis genes. PLoS Genet. 7: e1002163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trcek T., Larson D. R., Moldon A., Query C. C., Singer R. H., 2011. Single-molecule mRNA decay measurements reveal promoter-regulated mRNA stability in yeast. Cell 147: 1484–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker M., Valencia-Sanchez M. A., Staples R. R., Chen J., Denis C. L., et al. , 2001. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 104: 377–386 [DOI] [PubMed] [Google Scholar]
- Tucker M., Staples R. R., Valencia-Sanchez M. A., Muhlrad D., Parker R., 2002. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J. 21: 1427–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulbricht R. J., Olivas W. M., 2008. Puf1p acts in combination with other yeast Puf proteins to control mRNA stability. RNA 14: 246–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacova S., Wolf J., Martin G., Blank D., Dettwiler S., et al. , 2005. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 3: e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Den Elzen A. M., Henri J., Lazar N., Gas M. E., Durand D., et al. , 2010. Dissection of Dom34-Hbs1 reveals independent functions in two RNA quality control pathways. Nat. Struct. Mol. Biol. 17: 1446–1452 [DOI] [PubMed] [Google Scholar]
- Van Dijk E., Cougot N., Meyer S., Babajko S., Wahle E., et al. , 2002. Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 21: 6915–6924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijk E., Le Hir H., Seraphin B., 2003. DcpS can act in the 5′-3′ mRNA decay pathway in addition to the 3′-5′ pathway. Proc. Natl. Acad. Sci. USA 100: 12081–12086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijk E. L., Chen C. L., D’Aubenton-Carafa Y., Gourvennec S., Kwapisz M., et al. , 2011. XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature 475: 114–117 [DOI] [PubMed] [Google Scholar]
- Van Hoof A., Lennertz P., Parker R., 2000a. Three conserved members of the RNase D family have unique and overlapping functions in the processing of 5S, 5.8S, U4, U5, RNase MRP and RNase P RNAs in yeast. EMBO J. 19: 1357–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hoof A., Staples R. R., Baker R. E., Parker R., 2000b. Function of the ski4p (Csl4p) and Ski7p proteins in 3′-to-5′ degradation of mRNA. Mol. Cell. Biol. 20: 8230–8243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hoof A., Frischmeyer P. A., Dietz H. C., Parker R., 2002. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science 295: 2262–2264 [DOI] [PubMed] [Google Scholar]
- Vasiljeva L., Kim M., Mutschler H., Buratowski S., Meinhart A., 2008. The Nrd1-Nab3-Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain. Nat. Struct. Mol. Biol. 15: 795–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent R. D., Hofmann T. J., Zassenhaus H. P., 1988. Sequence and expression of NUC1, the gene encoding the mitochondrial nuclease in Saccharomyces cerevisiae. Nucleic Acids Res. 16: 3297–3312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinciguerra P., Iglesias N., Camblong J., Zenklusen D., Stutz F., 2005. Perinuclear Mlp proteins downregulate gene expression in response to a defect in mRNA export. EMBO J. 24: 813–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan P., Ohn T., Chiang Y. C., Chen J., Denis C. L., 2004. Mouse CAF1 can function as a processive deadenylase/3′-5′-exonuclease in vitro but in yeast the deadenylase function of CAF1 is not required for mRNA poly(A) removal. J. Biol. Chem. 279: 23988–23995 [DOI] [PubMed] [Google Scholar]
- Wang L., Lewis M. S., Johnson A. W., 2005. Domain interactions within the Ski2/3/8 complex and between the Ski complex and Ski7p. RNA 11: 1291–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Czaplinski K., Rao Y., Peltz S. W., 2001. The role of Upf proteins in modulating the translation read-through of nonsense-containing transcripts. EMBO J. 20: 880–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Cajigas I. J., Peltz S. W., Wilkinson M. F., Gonzalez C. I., 2006. Role for Upf2p phosphorylation in Saccharomyces cerevisiae nonsense-mediated mRNA decay. Mol. Cell. Biol. 26: 3390–3400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Liu C. L., Storey J. D., Tibshirani R. J., Herschlag D., et al. , 2002. Precision and functional specificity in mRNA decay. Proc. Natl. Acad. Sci. USA 99: 5860–5865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegierski T., Dmochowska A., Jablonowska A., Dziembowski A., Bartnik E., et al. , 1998. Yeast nuclear PET127 gene can suppress deletions of the SUV3 or DSS1 genes: an indication of a functional interaction between 3′ and 5′ ends of mitochondrial mRNAs. Acta Biochim. Pol. 45: 935–940 [PubMed] [Google Scholar]
- Welch E. M., Jacobson A., 1999. An internal open reading frame triggers nonsense-mediated decay of the yeast SPT10 mRNA. EMBO J. 18: 6134–6145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whipple J. M., Lane E. A., Chernyakov I., D’Silva S., Phizicky E. M., 2011. The yeast rapid tRNA decay pathway primarily monitors the structural integrity of the acceptor and T-stems of mature tRNA. Genes Dev. 25: 1173–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesenberger G., Fox T. D., 1997. Pet127p, a membrane-associated protein involved in stability and processing of Saccharomyces cerevisiae mitochondrial RNAs. Mol. Cell. Biol. 17: 2816–2824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson B. A., Masel J., 2011. Putatively noncoding transcripts show extensive association with ribosomes. Genome Biol. Evol. 3: 1245–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M. A., Meaux S., Parker R., Van Hoof A., 2005. Genetic interactions between [PSI+] and nonstop mRNA decay affect phenotypic variation. Proc. Natl. Acad. Sci. USA 102: 10244–10249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M. A., Meaux S., Van Hoof A., 2007. A genomic screen in yeast reveals novel aspects of nonstop mRNA metabolism. Genetics 177: 773–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz J. E., Whipple J. M., Phizicky E. M., Sharp P. A., 2011. tRNAs marked with CCACCA are targeted for degradation. Science 334: 817–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlotzka W., Kudla G., Granneman S., Tollervey D., 2011. The nuclear RNA polymerase II surveillance system targets polymerase III transcripts. EMBO J. 30: 1790–1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers F., Rougemaille M., Badis G., Rousselle J. C., Dufour M. E., et al. , 2005. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121: 725–737 [DOI] [PubMed] [Google Scholar]
- Xiang S., Cooper-Morgan A., Jiao X., Kiledjian M., Manley J. L., et al. , 2009. Structure and function of the 5'‐‐>3' exoribonuclease Rat1 and its activating partner Rai1. Nature 458: 784–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Wei W., Gagneur J., Perocchi F., Clauder-Munster S., et al. , 2009. Bidirectional promoters generate pervasive transcription in yeast. Nature 457: 1033–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Bai X., Lee I., Kallstrom G., Ho J., et al. , 2000. Saccharomyces cerevisiae RAI1 (YGL246c) is homologous to human DOM3Z and encodes a protein that binds the nuclear exoribonuclease Rat1p. Mol. Cell. Biol. 20: 4006–4015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao G., Chiang Y. C., Zhang C., Lee D. J., Laue T. M., et al. , 2007. PAB1 self-association precludes its binding to poly(A), thereby accelerating CCR4 deadenylation in vivo. Mol. Cell. Biol. 27: 6243–6253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J. H., Choi E. J., Parker R., 2010. Dcp2 phosphorylation by Ste20 modulates stress granule assembly and mRNA decay in Saccharomyces cerevisiae. J. Cell Biol. 189: 813–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosefzon Y., Koh Y. Y., Chritton J. J., Lande A., Leibovich L., et al. , 2011. Divergent RNA binding specificity of yeast Puf2p. RNA 17: 1479–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk D., Belk J. P., Jacobson A., 1999. Temperature-sensitive mutations in the Saccharomyces cerevisiae MRT4, GRC5, SLA2 and THS1 genes result in defects in mRNA turnover. Genetics 153: 35–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk D., Jacobson A., 1998. A single amino acid substitution in yeast eIF-5A results in mRNA stabilization. EMBO J. 17: 2914–2925 [DOI] [PMC free article] [PubMed] [Google Scholar]