Abstract
Identifying transcription factor (TF) binding sites is essential for understanding regulatory networks. The specificity of most TFs is currently modeled using position weight matrices (PWMs) that assume the positions within a binding site contribute independently to binding affinity for any site. Extensive, high-throughput quantitative binding assays let us examine, for the first time, the independence assumption for many TFs. We find that the specificity of most TFs is well fit with the simple PWM model, but in some cases more complex models are required. We introduce a binding energy model (BEM) that can include energy parameters for nonindependent contributions to binding affinity. We show that in most cases where a PWM is not sufficient, a BEM that includes energy parameters for adjacent dinucleotide contributions models the specificity very well. Having more accurate models of specificity greatly improves the interpretation of in vivo TF localization data, such as from chromatin immunoprecipitation followed by sequencing (ChIP-seq) experiments.
TRANSCRIPTION factor proteins (TFs) function by binding to specific sequences in the genome and activating or repressing the expression of their target genes. Identifying the sequences that each TF binds to can help map out transcriptional regulatory networks as well as predict how genetic variation may disrupt normal gene expression, which is often associated with disease. In vivo TF binding locations can be determined experimentally using techniques such as chromatin immunoprecipitation (ChIP) followed by microarray hybridization (ChIP-chip) (Ren et al. 2000) or chromatin immunoprecipitation followed by sequencing (ChIP-seq) (Johnson et al. 2007). In addition to its intrinsic specificity for DNA, in vivo TF binding preferences are influenced by other cellular factors such as the presence of cooperating or competing TFs as well as the local chromatin state. It is possible to gain mechanistic insight by comparing the intrinsic TF specificity measured in vitro, which reflects only the bimolecular interaction between the TF and DNA, with in vivo binding locations, which are influenced by many other cellular factors. For example, in vivo binding to a genomic region without strong binding sites indicates that the TF is either binding indirectly (the TF itself is not bound to DNA, but is in complex with some other factor that is bound to DNA) or bound to a weak site that is stabilized by a cooperative interaction with another factor (Gordan et al. 2009).
Currently, the most widely used mathematical representation of TF specificity is the position weight matrix (PWM) model (Stormo 2000). This model assumes the positions within the binding site are independent, and the contribution at one position of the binding site to the overall affinity does not depend on the identity of nucleotides in other positions of the site. Despite the restrictions imposed by this strong independence assumption, the PWM model has been successfully used to identify TF binding sites (TFBS) in sets of coexpressed genes (Stormo and Hartzell 1989; Roth et al. 1998; Tavazoie et al. 1999; Bussemaker et al. 2001) as well as model TF binding site evolution (Doniger and Fay 2007; Mustonen et al. 2008; Bradley et al. 2010). Quantitative analysis of high-throughput binding data has also shown that PWMs are a good quantitative model for most TFs (Zhao et al. 2009; Zhao and Stormo 2011).
Despite the success of PWM-based methods, it has long been recognized that TF–DNA interaction is highly complex structurally. Many cases of a single amino acid interacting with multiple bases simultaneously have been observed in crystal structures of TF–DNA complexes (Luscombe et al. 2001), the overwhelming majority of which are neighboring bases. In addition to direct contact with bases, TFs can recognize DNA sequence indirectly through sequence-specific DNA conformations, distortions, or water-mediated contacts (Sarai and Kono 2005; Rohs et al. 2009, 2010). For example, drastic DNA deformations have been observed in some TF–DNA complexes, including catabolite gene activator protein (CAP or CRP) and TATA binding protein (TBP) (Schultz et al. 1991; Kim et al. 1993). The identity of bases in neighboring positions is particularly important for DNA deformation energy through their stacking interactions.
In addition to structural analysis, detailed biochemical studies of specific proteins have also shown dependencies between adjacent positions (Man and Stormo 2001; Bulyk et al. 2002; Berger et al. 2006). Interactions between nonadjacent bases are possible (Jacobson et al. 1997), but they appear to be much less common than interactions between adjacent positions (Luscombe et al. 2001).
Statistical analyses of collections of known binding sites have also offered evidence of interactions between positions within binding sites for some TFs. Several groups (Barash et al. 2003; Zhou and Liu 2004; Tomovic and Oakeley 2007) have analyzed collections of TF binding sites in the TRANSFAC and JASPAR databases (Matys et al. 2006; Portales-Casamar et al. 2010) and found statistically significant correlated positions. Although these studies successfully identified the existence of correlated positions within TFBS, the type of binding data then available imposed severe limits on their analyses. The most serious problem was the small number of known binding sites available. For example, on average, only 30 binding sites per TF were used in the Tomovic and Oakeley (2007) study. Another problem is that TFBS collected in databases such as TRANSFAC (Matys et al. 2006) and JASPAR (Portales-Casamar et al. 2010) generally do not include affinity data; a sequence is simply labeled as a binding site. This binarization results in a loss of information and is especially problematic in the setting of small sample sizes. Finally, because of the nonlinearity between binding probability and binding affinity, the nonindependence in base frequencies in different positions may be observed even when the binding energy contributions are independent (Djordjevic et al. 2003; Homsi et al. 2009; Zhao et al. 2009). Therefore analyses that specifically model the binding energy contributions of each base at each position can better address the issue of whether they act independently.
Recently, experimental techniques for high-throughput, quantitative measurements of TF binding specificity have been developed (Stormo and Zhao 2010). The wealth of data generated by these high-throughput experiments provides us with an opportunity to answer questions left open by previous analyses. One such unanswered question is the average effect size of interactions between positions. Even though statistically significant interactions may exist between positions within the binding site of a TF, if their effect size is small, then the PWM model may still provide a good approximation of the true TF specificity (Benos et al. 2002). However, if the effect size is large, then more complex statistical models, requiring additional parameters, must be used to adequately model the specificity of the TF. This has important practical implications as the vast majority of existing bioinformatics software use the PWM model. Models designed to accommodate complex interactions within a binding site have been developed (Stormo et al. 1986; Zhang and Marr 1993; Barash et al. 2003; King and Roth 2003; Zhou and Liu 2004; Sharon et al. 2008; Stormo 2011) but have not seen wide adoption.
In this article, we report the results of a quantitative analysis of >400 TF specificity data obtained using the universal protein-binding microarray (PBM) technology (Berger et al. 2006; Berger and Bulyk 2009) and available in the UniPROBE database (Robasky and Bulyk 2011). We use the binding energy estimate by maximum likelihood for PBM (BEEML-PBM) (Zhao and Stormo 2011) program to parameterize specificity models of varying complexity and find that improvements from incorporating interactions between positions are usually small, although there are some significant exceptions. Moreover, we find that interactions between neighboring bases are stronger than interactions between nonneighboring bases.
Materials and Methods
Model of TF specificity
We use an equilibrium model of binding (Djordjevic et al. 2003; Zhao et al. 2009) where the probability that DNA sequence Si is bound by the TF is given by
| (1) |
where E(Si) is the free energy of the TF binding to Si and µ is the chemical potential that is related to the TF concentration.
We introduce the binding energy model (BEM) as a vector of energy contributions, . For any sequence, Si, the binding energy predicted by the model is , where is the vector encoding of the sequence Si that can include whatever features of the sequence are relevant to its binding energy (Schneider et al. 1984; Stormo et al. 1986; Sharon et al. 2008; Stormo 2011). If the only relevant features are which bases occur at each position within the binding site, then will be a PWM with the characteristic that each element is an energy contribution (Djordjevic et al. 2003; Zhao et al. 2009). PWMs have been developed using a variety of techniques. The first use was as a discriminant function learned to separate sequences into two classes (Stormo et al. 1982). Staden introduced a probabilistic version of the PWM (Staden 1984) and a variety of other, mostly ad hoc, methods have also been used. Under certain conditions the probabilistic model is equivalent to an energy model (Berg and von Hippel 1987; Heumann et al. 1994; Stormo and Fields 1998; Lassig 2007), but those conditions are violated at high concentrations of the TF (Djordjevic et al. 2003; Zhao et al. 2009). By using an energy model directly and including the TF concentration as a separate parameter, µ, one can model the nonlinear relationship between the logarithm of binding probability and the binding free energy described in Equation 1.
When the energy contributions of each position are independent, is explicitly
where L is the length of the binding site, ε(b, m) are the energy contributions of base b at position m, and Si(b, m) is an indicator variable with Si(b, m) = 1 if base b occurs at position m of sequence Si and Si(b, m) = 0 otherwise (Stormo et al. 1982; Stormo 2000). If we find that the positions are not independent, we can include pairwise interactions between adjacent positions by adding interaction terms to the energy function such that is
where ε(b, m, c, n) is the energy contribution of having base b at position m and base c at position n. Since the single-base contributions are included explicitly, in the first set of sums, the dinucleotide contributions explicitly represent the deviations from additivity of the individual bases, the energy residuals not captured by the single-base contributions. Higher-order models can be constructed similarly and in each case can be represented as , where both vectors include the relevant elements that contribute to the binding energy.
Encoding of DNA sequence
In learning the energy parameters we use the WYK encoding scheme (Stormo 2011), which uses the minimum number of parameters required of the model and enforces the clear separation of the contributions of the individual bases from the contributions of the interactions between the bases. This encoding has the further advantage, especially for some machine-learning and optimization methods (Stormo 2011), that all sequence vectors have the same Euclidian length so the energy of a sequence is proportional to the cosine of the angle between it and the energy vector where θ is the angle between the vectors. For convenience of display and interpretation the models are converted to standard ACGT encoding as described above.
Scoring sequences with binding energy models
Given a binding energy model , the program BEMSER scans a sequence and determines the predicted binding energy at every location within the sequence (including both orientations if desired). If contains only independent base contributions (it is a PWM), then BEMSER returns the same scores as would be obtained using PatSer (Hertz and Stormo 1999), a commonly used program for scanning sequences with a PWM. But if also contains energy contributions from combinations of bases, BEMSER uses those contributions as well to score each potential site (subsequence) in the sequence. BEMSER as well as all of the BEMs are available for download from http://stormo.wustl.edu/TF-BEMs.
Parameterization of TF specificity model
The models are all determined from PBM data available from the UniPROBE database (Robasky and Bulyk 2011). All models were parameterized using the BEEML-PBM algorithm, which has been described in detail previously (Zhao and Stormo 2011). Briefly, BEEML-PBM employs nonlinear regression to obtain the parameters of the model, and μ, that maximize the fit to the data.
Evaluation of model performance on PBM data
Given a binding energy model the fluorescence of each probe is predicted by summing the predicted binding probabilities for each position in the probe sequence in both orientations. Because the individual probe measurements are somewhat noisy, we determine the median fluorescence intensity for all 8mers, each of which occurs 32 times on the array (counting both orientations). We assess the performance of different models by determining the square of the correlation coefficient (r2) between the predicted and measured 8mer intensities. When only one array exists for a specific TF, this is useful in determining how much better fit is obtained (increase in r2) for models that include dinucleotide contributions than for those that assume independent base contributions. Because of the large number of 8mers, even very small changes in r2 are statistically significant (P < 0.05), but may make almost no difference in prediction accuracy.
A subset of 147 TFs, from the complete set of 401 available in UniPROBE, was assayed on two independent arrays with different probe sequences, but each containing all possible 10mers (Berger and Bulyk 2009). In those cases we can assess how well a model obtained from one array predicts the 8mer intensities of the other array for the same TF; we use the average of the two cross-predictions as the performance of the model. The squared correlation coefficient between the observed 8mer intensities for the two arrays is referred to as the replicate reproducibility. In previous work we considered models whose performance was at least 90% of the replicate reproducibility to provide very good fits to the data (Zhao and Stormo 2011). But it should be noted that it is possible, in fact common, for the model performance to exceed the replicate reproducibility as we showed in the supplemental materials of that article. This can occur when one of the two datasets is considerably noisier than the other. The replicate reproducibility can then be fairly low and the predictions using the model from the good array have low r2 on the noisier array. But we showed that the noisy array is still capable of generating fairly good models with much higher r2 on the good array. The average of the two model r2 values can then exceed the replicate reproducibility.
Analysis of ChIP-seq data
The DNA-binding domain of Hepatocyte nuclear factor 4, alpha (Hnf4a) is completely conserved between human and mouse. Since Badis et al. (2009) used only the DNA-binding domain of mouse Hnf4a to determine its in vitro binding specificity, we used this information to analyze Verzi et al.’s (2010) ChIP-seq data even though it was carried out in human cell lines. Hnf4a ChIP-seq data were downloaded from Gene Expression Omnibus (accession no. GSM575227) from the study conducted by Verzi et al. (2010) and we used the peak location and summit information as provided. Binding site location analysis was conducted by aligning all peaks by their annotated summit and scoring genomic sequences within 200 bp on each side of the summit, using each of the different models for predicting the binding sites. For each model, the predicted lowest energy site within that window was considered the binding site for that peak. In the vast majority of cases the predicted best binding site was the same for all models within that 400-bp window. We then determined the number of reads that overlap that predicted binding site after extending each read by 100 bp on each side. While the choices of 400- and 200-bp windows are somewhat arbitrary, they are consistent with typical fragmentation sizes for ChIP-seq experiments. Most importantly, since most of the predicted sites are the same for all of the models, we can directly compare their predicted relative affinities for each identified peak without the confounding effect of different models predicting different binding sites. For each model the predicted relative binding affinities for the predicted binding site within each peak were calculated as , where Econ is the energy of the lowest possible energy site (the consensus sequence for the model) and Esite is the energy of the site being considered.
Primary and secondary PWMs for Hnf4a obtained by the seed-and-wobble method (Berger et al. 2006) were downloaded from the UniPROBE database (Robasky and Bulyk 2011) and converted to energy PWMs. Positions with low information content were trimmed off so both PWMs were 8-long, the same size as the BEEML models. The energy of an 8mer was calculated as the minimum of the energies predicted by primary and secondary PWMs. If longer models were used, the predicted relative affinities would either stay the same (in cases where the predicted site had the consensus base in the additional position) or decrease (if it had a nonconsensus base). So by trimming the UniPROBE models to be 8-long, the differences in the predicted relative affinities between different models are minimized.
Results
PBM technology uses double-stranded DNA microarrays to measure the binding of TFs to many sequences in a highly parallel fashion (Bulyk et al. 2001; Mukherjee et al. 2004; Berger et al. 2006). In the current design (Berger et al. 2006; Berger and Bulyk 2009) all possible 10-nucleotide-long binding sites (10mers) are contained in the sequences of microarray probes. In a recent PBM study, Badis et al. (2009) measured the binding specificity of 104 mouse TFs and found many cases where the best PWM model they obtained seemed inadequate to represent TF specificity. In fact, the authors invoked a model where TFs could use alternative modes of binding, each represented by independent PWMs, to explain the PBM data. We developed a nonlinear regression method, BEEML-PBM, to estimate PWM parameters from PBM data (Zhao and Stormo 2011). Using BEEML-PBM, we showed that, contrary to the conclusions of Badis et al. (2009), the PWM model provides a good quantitative model of specificity for most of the TFs in their study.
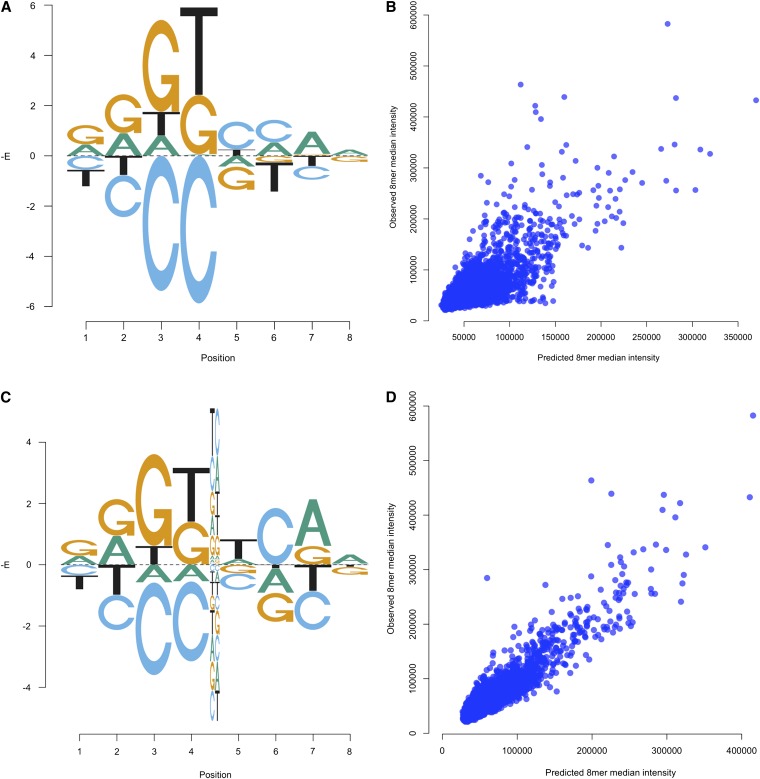
Despite the good performance of the PWM model in general, there were cases where the simple PWM performed poorly. For example, an 8-long BEEML-PBM PWM for Hnf4a trained on the data from one array is able to predict the median 8mer intensities of probes on the test array with only an r2 = 0.55 (Figure 1, A and B), much less than the experimental reproducibility between the training and testing data (r2 = 0.82). A model that includes interactions between all adjacent positions, which requires an additional 63 parameters over the PWM, results in significantly improved performance (r2 = 0.81). We also tested the performance of all 28 possible 8-long models that include a single pairwise interaction between two positions and found that a model with interaction terms only between positions 4 and 5 is able to achieve an r2 = 0.78 (Figure 1, C and D). This model appears to capture most of the relevant features of Hnf4a binding and includes only 9 more parameters than the PWM.
Figure 1 .
Binding energy model including interactions makes more accurate predictions of in vitro binding specificity than the PWM for Hnf4a. (A) Graphical representation of Hnf4a binding energies estimated from PBM data under the PWM model (Supporting Information, Figure S1). Negatives of binding energy (in units of RT) are plotted on the y-axis. Energies are normalized such that the average energy at each position is 0. This energy logo is equivalent to the “affinity logo” from Foat et al. (2006). (B) Performance of model shown in A on test PBM data. (C) Binding energy model estimated from the same training data but including interaction energies between positions 4 and 5 (Figure S2). (D) Performance of the energy model including interactions on test PBM data.
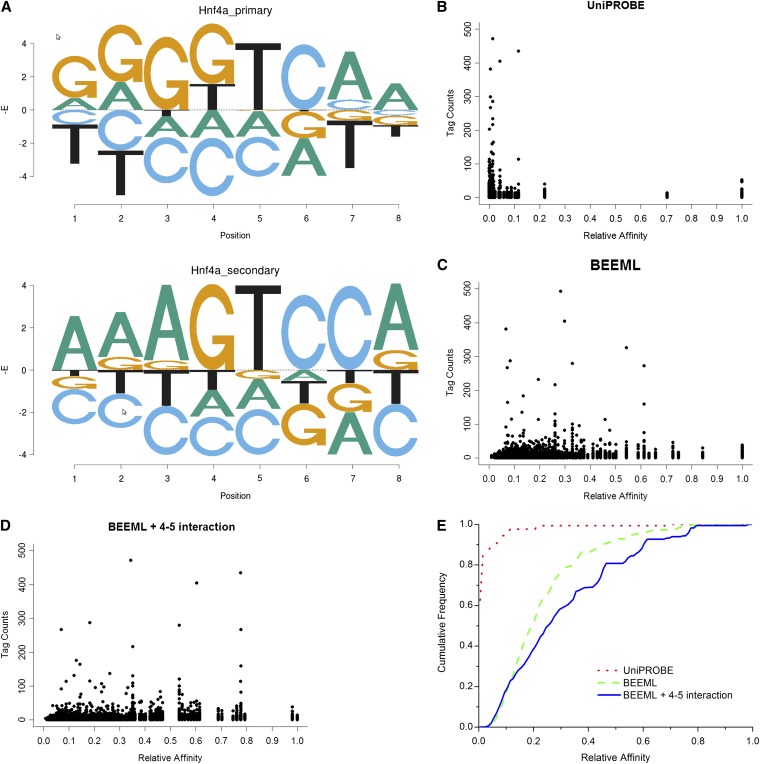
To determine the biological significance of the position dependence observed in vitro, we compared different specificity models of Hnf4a learned from PBM data with in vivo binding data from the ChIP-seq experiment conducted by Verzi et al. (2010). Figure 2A shows the primary and secondary PWMs obtained for Hnf4a from the UniProbe database (Robasky and Bulyk 2011). Figure 2B shows that the vast majority of best binding sites for the ChIP-seq peaks have very low predicted relative affinity using the UniProbe PWMs. Those PWMs are highly specific and the vast majority of peaks do not contain consensus sites for either the primary or the secondary motif or even for variations from the consensus that are predicted to be of high relative affinity. Figure 2C shows that more of the peaks have higher predicted relative affinity using the PWM obtained by BEEML-PBM on the same training data (Figure 1A), consistent with the previous results that those PWMs fit the in vitro PBM binding data better than the PWMs obtained using the seed-and-wobble algorithm (Zhao and Stormo 2011). Figure 2D shows that the energy model that includes dinucleotide energy contributions between positions 4 and 5 (Figure 1C) has even more predicted high-affinity binding sites. Figure 2E shows the cumulative frequency of the number of reads at increasing relative binding affinities for each of the three models.
Figure 2 .
Hnf4a energy model including interactions makes more accurate predictions of in vivo ChIP-seq binding data. (A) Primary and secondary binding energy models for Hnf4a obtained by UniPROBE (Robasky and Bulyk 2011) (Figure S3). (B) Primary and secondary models predicted affinity of the best site under each peak vs. number of reads overlapping the best predicted sites. (C) BEEML-PBM PWM predicted affinities of best sites vs. number of reads. (D) Energy model including interaction between positions 4 and 5 predicted affinities of best sites vs. number of reads. (E) Cumulative frequency (fraction of total reads) for each model at increasing predicted relative affinities.
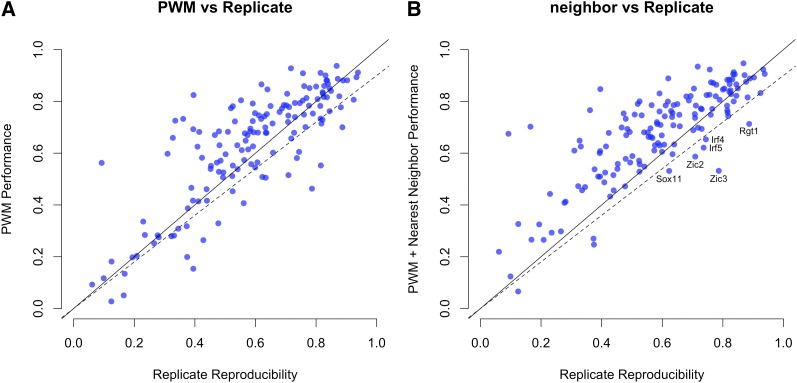
We next examined the 147 TFs in the UniPROBE database (Robasky and Bulyk 2011) for which replicate data are available. Figure 3A shows that the PWM model is unable to explain >90% of the reproducibility for 25 of these 147 TFs (17%). Note that the models obtained using BEEML-PBM are often better at predicting probe intensities than the reproducibility between arrays. This is because good models can be obtained even from noisy experimental data, as demonstrated in the Foxa2 example of Figure S4 of Zhao and Stormo (2011). For most of the TFs that are not well modeled by simple PWMs, predictive performance was substantially improved with the addition of interaction terms between adjacent positions (Figure 3B), indicating that the majority of interactions not captured by the PWM are between adjacent positions.
Figure 3 .
Comparisons of energy model predictions and experimental reproducibility. (A) The PWM energy model is unable to explain >90% of the reproducibility for 25 of these 147 TFs. (B) Predictive performance was substantially improved with the addition of interaction terms between adjacent positions, indicating that the majority of interactions not captured by the PWM are between adjacent positions.
For a more global perspective, we compared the fit of PWMs with those of nearest-neighbor and random interaction models for all 401 TFs in the UniPROBE database (Robasky and Bulyk 2011). Nearest-neighbor BEMs include the independent base contributions to binding (as in a PWM) as well as interaction energies between all adjacent positions. In this model, an L-long binding site requires 3L + 9(L − 1) independent parameters, approximately four times as many as a simple PWM model that requires 3L parameters. The random interaction model is defined as a PWM as well as interaction energies for the same number, L − 1, of randomly chosen nonadjacent position pairs in the binding site and its performance is calculated as the average of 10 random interaction assignments. The reason for comparing the nearest-neighbor to the random interaction model is twofold: first, it allows us to assess the importance of nearest-neighbor interactions; and second, since replicate data are not available for many of the TFs, the performance of the random interaction model, which has the same number of parameters as the nearest-neighbor model, gives an indication of the extent to which performance gain is simply due to these models having more parameters than the PWM.
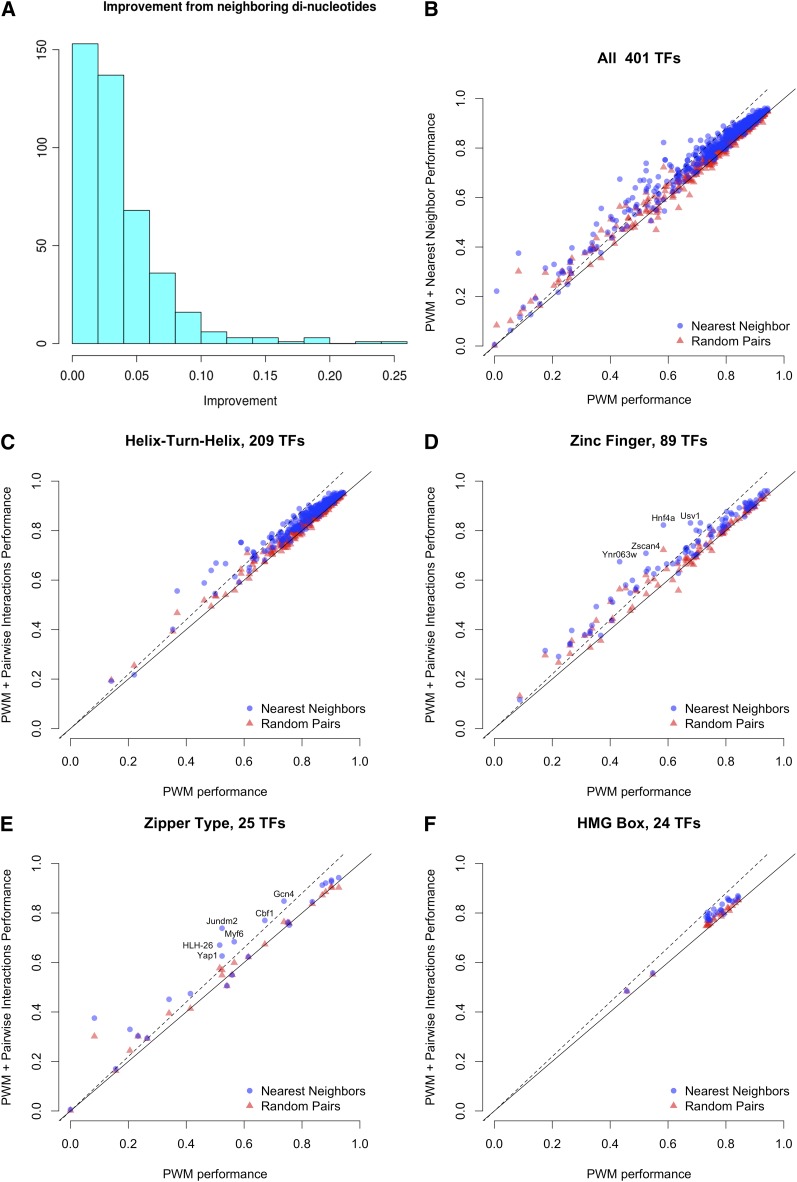
Figure 4A shows the distribution of increases in r2 for all of the nearest-neighbor BEMs compared to the PWMs. Figure 4B plots the PWM performance vs. the nearest-neighbor dinucleotide models for all 401 TFs. For most TFs, addition of interaction parameters did not substantially improve the fit; for <15% of TFs is the increase in r2 > 0.06. Furthermore, the nearest-neighbor model always outperformed the random interaction model, demonstrating the importance of nearest-neighbor interactions. Although nearest-neighbor and random interaction models have more parameters and therefore should always outperform the PWM model, the local optimization procedure used by BEEML sometimes fails to find optimal parameter values, resulting in few points falling below the main diagonal of Figure 4B.
Figure 4 .
Comparison of the fit of PWM, nearest-neighbor, and random interaction energy models. (A) Histogram of the improvement in r2 for the nearest-neighbor interaction models compared to the PWMs. (B–F) The performance of the PWM model is plotted on the x-axis, and performances of nearest-neighbor (circles) and random interaction (triangles) models are plotted on the y-axis. (B) Comparisons for all 401 TFs in the UniPROBE database. (C) Comparisons for the 209 helix-turn-helix TFs in the database. (D) Comparisons for the 89 zinc finger TFs, including C2H2, C4, C6, and GATA zinc finger domains. (E) Comparisons of 25 zipper class TFs, including the basic leucine zipper (bZIP) domain and the helix-loop-helix (bHLH) domain. (F) Comparisons of the 24 high-mobility group (HMG) TFs.
We examined the nearest-neighbor model performances for different TF structural classes (Figure 4, C–F). There are 209 helix-turn-helix TFs in the data set, including homeodomain and winged helix-turn-helix, such as ETS domain, TFs. Addition of interaction parameters typically resulted in relatively small gains in performance, but there are several cases where nearest-neighbor interactions are important (Figure 4C). This pattern holds true for the zinc finger class, which has 89 members including C2H2, C4, C6, and GATA zinc finger domains (Figure 4D). This includes the Hnf4a example analyzed in detail above. The 25 TFs of the zipper class, including the basic leucine zipper (bZIP) and the basic helix-loop-helix (bHLH) domains, appear to have benefited the most from the inclusion of nearest-neighbor interactions (Figure 4E). By contrast, none of the 24 high-mobility group (HMG) TFs benefitted substantially from the additional parameters (Figure 4F). While there are data showing nonindependence between positions for at least some HMG proteins (Jauch et al. 2011), those appear to be relatively minor contributions overall, as found previously for several zinc finger proteins (Benos et al. 2002; Bulyk et al. 2002).
Discussion
Our quantitative analysis of >400 in vitro quantitative TF specificities generated by PBM technology demonstrates that explicitly including interactions within the binding site generally results in relatively small improvements in performance. With a few exceptions, the PWM model provides a good approximation for TF specificity. Consistent with available structural information, when interactions between positions are important, most of them are found to occur between adjacent positions in the binding site. Some TF families are more likely to require interaction models than others. In particular the bZIP and bHLH families are commonly fitted much better by including adjacent dinucleotide energy contributions, consistent with previous information (Berger et al. 2006; Maerkl and Quake 2007; Stormo and Zhao 2007; Zhao et al. 2009; Nutiu et al. 2011).
Improved specificity models that are based on in vitro binding data can be very useful for assessing how consistent in vivo location data are with the expected binding sites. When predicted genomic binding sites are not observed in ChIP-seq data, one can usually assume that those locations are not accessible. But when binding is observed in locations without predicted binding sites, or with only very low predicted affinity sites, that implies either indirect or cooperative binding mediated through some other factor(s) that binds directly to the DNA (Gordan et al. 2009). Such indirect and cooperative binding events can lead to the discovery of interacting TFs that coordinately control gene expression. But to be confident about which ChIP-seq peaks are not due to direct binding one needs an accurate model for the specificity of the TF. As we show for Hnf4a, different models can lead to quite different conclusions about which peaks contain predicted high-affinity sites. For both the BEEML-PBM models, with and without dinucleotide contributions, a much greater fraction of the ChIP-seq reads can be explained by direct binding, with the dinucleotide model explaining the most.
In these analyses we have considered a model to be a good fit to the PBM data if it can capture >90% of the reproducible variance of the experiment. However, some of the data sets are fairly noisy and it is possible that cleaner data would show that even the interaction models do not capture the specificity well. For example, the Hnf4a PBM data have reproducibility between the two arrays of only r2 = 0.82. The BEM with interaction terms between only positions 4 and 5 predicts the test array data with r2 = 0.78, which is nearly all of the reproducible variance. But if additional data had higher consistency between experiments, it is possible that additional terms would be required to obtain an adequate model. We can only claim that, given the current experimental data sets, in most cases simple PWMs fit the data quite well and in most of the remaining cases an extended BEM, with energy terms for adjacent dinucleotides, captures most of the remaining variance.
Supplementary Material
Acknowledgments
We thank all members of the Stormo laboratory for helpful comments on this work. Funding was provided by National Institutes of Health grant HG00249 (to G.D.S.).
Footnotes
Communicating editor: J. Boeke
Literature Cited
- Badis G., Berger M. F., Philippakis A. A., Talukder S., Gehrke A. R., et al. , 2009. Diversity and complexity in DNA recognition by transcription factors. Science 324: 1720–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barash Y., Elidan G., Kaplan T., Friedman N., 2003. Modeling dependencies in protein-DNA binding sites. Proceedings of the 7th Annual International Conference on Computational Molecular Biology (RECOMB), ACM, New York pp. 28–37 [DOI] [PubMed]
- Benos P. V., Bulyk M. L., Stormo G. D., 2002. Additivity in protein-DNA interactions: how good an approximation is it? Nucleic Acids Res. 30: 4442–4451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg O. G., von Hippel P. H., 1987. Selection of DNA binding sites by regulatory proteins. Statistical-mechanical theory and application to operators and promoters. J. Mol. Biol. 193: 723–750 [DOI] [PubMed] [Google Scholar]
- Berger M. F., Bulyk M. L., 2009. Universal protein-binding microarrays for the comprehensive characterization of the DNA-binding specificities of transcription factors. Nat. Protoc. 4: 393–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger M. F., Philippakis A. A., Qureshi A. M., He F. S., Estep P. W., 3rd, et al. , 2006. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat. Biotechnol. 24: 1429–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley R. K., Li X. Y., Trapnell C., Davidson S., Pachter L., et al. , 2010. Binding site turnover produces pervasive quantitative changes in transcription factor binding between closely related Drosophila species. PLoS Biol. 8: e1000343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulyk M. L., Huang X., Choo Y., Church G. M., 2001. Exploring the DNA-binding specificities of zinc fingers with DNA microarrays. Proc. Natl. Acad. Sci. USA 98: 7158–7163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulyk M. L., Johnson P. L., Church G. M., 2002. Nucleotides of transcription factor binding sites exert interdependent effects on the binding affinities of transcription factors. Nucleic Acids Res. 30: 1255–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussemaker H. J., Li H., Siggia E. D., 2001. Regulatory element detection using correlation with expression. Nat. Genet. 27: 167–171 [DOI] [PubMed] [Google Scholar]
- Djordjevic M., Sengupta A. M., Shraiman B. I., 2003. A biophysical approach to transcription factor binding site discovery. Genome Res. 13: 2381–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doniger S. W., Fay J. C., 2007. Frequent gain and loss of functional transcription factor binding sites. PLoS Comput. Biol. 3: e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foat B. C., Morozov A. V., Bussemaker H. J., 2006. Statistical mechanical modeling of genome-wide transcription factor occupancy data by MatrixREDUCE. Bioinformatics 22: e141–e149 [DOI] [PubMed] [Google Scholar]
- Gordan R., Hartemink A. J., Bulyk M. L., 2009. Distinguishing direct vs. indirect transcription factor-DNA interactions. Genome Res. 19: 2090–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz G. Z., Stormo G. D., 1999. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics 15: 563–577 [DOI] [PubMed] [Google Scholar]
- Heumann J. M., Lapedes A. S., Stormo G. D., 1994. Neural networks for determining protein specificity and multiple alignment of binding sites. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2: 188–194 [PubMed] [Google Scholar]
- Homsi D. S., Gupta V., Stormo G. D., 2009. Modeling the quantitative specificity of DNA-binding proteins from example binding sites. PLoS ONE 4: e6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson E. M., Li P., Leon-del-Rio A., Rosenfeld M. G., Aggarwal A. K., 1997. Structure of Pit-1 POU domain bound to DNA as a dimer: unexpected arrangement and flexibility. Genes Dev. 11: 198–212 [DOI] [PubMed] [Google Scholar]
- Jauch R., Ng C. K., Narasimhan K., Kolatkar P. R., 2012. The crystal structure of the Sox4 HMG domain-DNA complex suggests a mechanism for positional interdependence in DNA recognition. Biochem. J. 443: 39–47 [DOI] [PubMed] [Google Scholar]
- Johnson D. S., Mortazavi A., Myers R. M., Wold B., 2007. Genome-wide mapping of in vivo protein-DNA interactions. Science 316: 1497–1502 [DOI] [PubMed] [Google Scholar]
- Kim Y., Geiger J. H., Hahn S., Sigler P. B., 1993. Crystal structure of a yeast TBP/TATA-box complex. Nature 365: 512–520 [DOI] [PubMed] [Google Scholar]
- King O. D., Roth F. P., 2003. A non-parametric model for transcription factor binding sites. Nucleic Acids Res. 31: e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassig M., 2007. From biophysics to evolutionary genetics: statistical aspects of gene regulation. BMC Bioinformatics 8(Suppl. 6): S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscombe N. M., Laskowski R. A., Thornton J. M., 2001. Amino acid-base interactions: a three-dimensional analysis of protein-DNA interactions at an atomic level. Nucleic Acids Res. 29: 2860–2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maerkl S. J., Quake S. R., 2007. A systems approach to measuring the binding energy landscapes of transcription factors. Science 315: 233–237 [DOI] [PubMed] [Google Scholar]
- Man T. K., Stormo G. D., 2001. Non-independence of Mnt repressor-operator interaction determined by a new quantitative multiple fluorescence relative affinity (QuMFRA) assay. Nucleic Acids Res. 29: 2471–2478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V., Kel-Margoulis O. V., Fricke E., Liebich I., Land S., et al. , 2006. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 34: D108–D110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Berger M. F., Jona G., Wang X. S., Muzzey D., et al. , 2004. Rapid analysis of the DNA-binding specificities of transcription factors with DNA microarrays. Nat. Genet. 36: 1331–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustonen V., Kinney J., Callan C. G., Jr, Lassig M., 2008. Energy-dependent fitness: a quantitative model for the evolution of yeast transcription factor binding sites. Proc. Natl. Acad. Sci. USA 105: 12376–12381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutiu R., Friedman R. C., Luo S., Khrebtukova I., Silva D., et al. , 2011. Direct measurement of DNA affinity landscapes on a high-throughput sequencing instrument. Nat. Biotechnol. 29: 659–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portales-Casamar E., Thongjuea S., Kwon A. T., Arenillas D., Zhao X., et al. , 2010. JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res. 38: D105–D110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B., Robert F., Wyrick J. J., Aparicio O., Jennings E. G., et al. , 2000. Genome-wide location and function of DNA binding proteins. Science 290: 2306–2309 [DOI] [PubMed] [Google Scholar]
- Robasky K., Bulyk M. L., 2011. UniPROBE, update 2011: expanded content and search tools in the online database of protein-binding microarray data on protein-DNA interactions. Nucleic Acids Res. 39: D124–D128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohs R., West S. M., Sosinsky A., Liu P., Mann R. S., et al. , 2009. The role of DNA shape in protein-DNA recognition. Nature 461: 1248–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohs R., Jin X., West S. M., Joshi R., Honig B., et al. , 2010. Origins of specificity in protein-DNA recognition. Annu. Rev. Biochem. 79: 233–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth F. P., Hughes J. D., Estep P. W., Church G. M., 1998. Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole-genome mRNA quantitation. Nat. Biotechnol. 16: 939–945 [DOI] [PubMed] [Google Scholar]
- Sarai A., Kono H., 2005. Protein-DNA recognition patterns and predictions. Annu. Rev. Biophys. Biomol. Struct. 34: 379–398 [DOI] [PubMed] [Google Scholar]
- Schneider T. D., Stormo G. D., Yarus M. A., Gold L., 1984. Delila system tools. Nucleic Acids Res. 12: 129–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz S. C., Shields G. C., Steitz T. A., 1991. Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees. Science 253: 1001–1007 [DOI] [PubMed] [Google Scholar]
- Sharon E., Lubliner S., Segal E., 2008. A feature-based approach to modeling protein-DNA interactions. PLoS Comput. Biol. 4: e1000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R., 1984. Computer methods to locate signals in nucleic acid sequences. Nucleic Acids Res. 12: 505–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo G. D., 2000. DNA binding sites: representation and discovery. Bioinformatics 16: 16–23 [DOI] [PubMed] [Google Scholar]
- Stormo G. D., 2011. Maximally efficient modeling of DNA sequence motifs at all levels of complexity. Genetics 187: 1219–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo G. D., Fields D. S., 1998. Specificity, free energy and information content in protein-DNA interactions. Trends Biochem. Sci. 23: 109–113 [DOI] [PubMed] [Google Scholar]
- Stormo G. D., Hartzell G. W., III, 1989. Identifying protein-binding sites from unaligned DNA fragments. Proc. Natl. Acad. Sci. USA 86: 1183–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo G. D., Zhao Y., 2007. Putting numbers on the network connections. BioEssays 29: 717–721 [DOI] [PubMed] [Google Scholar]
- Stormo G. D., Zhao Y., 2010. Determining the specificity of protein-DNA interactions. Nat. Rev. Genet. 11: 751–760 [DOI] [PubMed] [Google Scholar]
- Stormo G. D., Schneider T. D., Gold L., Ehrenfeucht A., 1982. Use of the ’Perceptron’ algorithm to distinguish translational initiation sites in E. coli. Nucleic Acids Res. 10: 2997–3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo G. D., Schneider T. D., Gold L., 1986. Quantitative analysis of the relationship between nucleotide sequence and functional activity. Nucleic Acids Res. 14: 6661–6679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazoie S., Hughes J. D., Campbell M. J., Cho R. J., Church G. M., 1999. Systematic determination of genetic network architecture. Nat. Genet. 22: 281–285 [DOI] [PubMed] [Google Scholar]
- Tomovic A., Oakeley E. J., 2007. Position dependencies in transcription factor binding sites. Bioinformatics 23: 933–941 [DOI] [PubMed] [Google Scholar]
- Verzi M. P., Shin H., He H. H., Sulahian R., Meyer C. A., et al. , 2010. Differentiation-specific histone modifications reveal dynamic chromatin interactions and partners for the intestinal transcription factor CDX2. Dev. Cell 19: 713–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M. Q., Marr T. G., 1993. A weight array method for splicing signal analysis. Comput. Appl. Biosci. 9: 499–509 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Stormo G. D., 2011. Quantitative analysis demonstrates most transcription factors require only simple models of specificity. Nat. Biotechnol. 29: 480–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Granas D., Stormo G. D., 2009. Inferring binding energies from selected binding sites. PLoS Comput. Biol. 5: e1000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Liu J. S., 2004. Modeling within-motif dependence for transcription factor binding site predictions. Bioinformatics 20: 909–916 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.