Abstract
Cell adhesion and biofilm formation are critical processes in the pathogenicity of fungi and are mediated through a family of adhesin proteins conserved throughout yeasts and fungi. In Saccharomyces cerevisiae, Flo11 is the main adhesin involved in cell adhesion and biofilm formation, making the study of its function and regulation in this nonpathogenic budding yeast highly relevant. The S. cerevisiae FLO11 gene is driven by a TATA-box-containing promoter that is regulated through one of the longest regulatory upstream regions (3 kb) in yeast. We reported recently that two chromatin cofactor complexes, the Rpd3L deacetylase and the Swi/Snf chromatin-remodeling complexes, contribute significantly to the regulation of FLO11. Here, we analyze directly how these complexes impact on FLO11 promoter chromatin structure and dissect further the interplay between histone deacetylases, chromatin remodeling, and the transcriptional repressor Sfl1. We show that the regulation of chromatin structure represents an important layer of control in the highly complex regulation of the FLO11 promoter.
Keywords: FLO11, Swi/Snf, Rpd3, histone deacetylase (HDAC), chromatin
THE initiation of transcription is one of the main targets of gene expression regulation and is determined by regulator binding sequences both within the promoter region and in more distant regions. In yeasts, where the true promoter and associated regulatory sequences are usually close together, the combined regions are often referred to as “the promoter,” and for simplicity we will follow this convention in this work. Even though promoter regulation depends mainly on the identity of bound regulators, either activators or repressors, and on the position and affinities of their binding sites, it has become clear that the accessibility of the promoter region is an equally important level of control (Li et al. 2007). In eukaryotes, DNA accessibility is governed through chromatin structure, whereby DNA is packaged into nucleosomes. Nucleosomes are assemblies of basic histone proteins on a stretch of 147 bp of DNA. They can adopt different positions along the DNA and may contain different histone variants or post-translationally modified histones (Kamakaka and Biggins 2005; Kouzarides 2007; Li et al. 2007). Histone modifications or changes in nucleosome positions are strategies to alter DNA accessibility and therefore gene expression. Accordingly, the analysis of chromatin structure and dynamics has become a key area in the study of promoter regulation. Indeed, many multi-protein complexes that affect chromatin have been described, and these are often recruited to promoters by transcriptional regulators or through DNA-binding subunits of their own.
Histone deacetylase (HDAC) complexes, for example, may either activate or repress gene transcription and are involved not only in transcription initiation but also in elongation (Kurdistani and Grunstein 2003a; Li et al. 2007). There are >10 HDAC enzymes known in Saccharomyces cerevisiae, grouped into three classes: I, II, and III. The class I member Rpd3 is the most studied and contributes to either repression or activation of many genes (Vidal and Gaber 1991; Kadosh and Struhl 1997, 1998; De Nadal et al. 2004; Puig et al. 2004). Rpd3 is the HDAC subunit of the Rpd3L and Rpd3S complexes, together with associated proteins such as Sap30, Pho23, Sin3, Cti6, Sds3, or Rxt2. Sds3 and Pho23 are exclusive subunits of Rpd3L, while Rco1 is an exclusive subunit of the Rpd3S complex (Carrozza et al. 2005).
Another example of promoter regulation through chromatin is the repositioning, remodeling, or disassembly of nucleosomes mediated by a family of ATP-dependent Snf2-type helicases that are often part of large multisubunit complexes (Clapier and Cairns 2009). The founding member of this family is the Swi/Snf complex, which contains the Swi2/Snf2 ATPase subunit and 11 more subunits. Several of these subunits are involved in the activation of different genes. Moreover, the Swi/Snf complex may interact with HDACs in gene regulation (Deckert and Struhl 2002; Sertil et al. 2007).
We are particularly interested in the regulation of FLO11 expression. FLO11 encodes the main cell-surface protein involved in cell–cell and cell-surface adhesion in S. cerevisiae (Lo and Dranginis 1998; Reynolds and Fink 2001). Adhesion is critical for infection in pathogenic fungi, such as Candida albicans (Verstrepen et al. 2004). In the nonpathogenic yeast S. cerevisiae, adhesion is also essential for various properties, including flocculation, biofilm formation, or the dimorphic switch, which are responses to environmental stress (Gimeno et al. 1992; Guo et al. 2000; Verstrepen and Klis 2006). As S. cerevisiae is a highly tractable organism with many genetic and molecular tools, it represents a very powerful model for studying adhesion in fungi and dissecting the regulation of FLO11 gene expression (Reynolds and Fink 2001; Verstrepen et al. 2004; Verstrepen and Klis 2006). The FLO11 promoter, ∼3 kb in length, is the largest described in S. cerevisiae. Its regulation is very complex, with many different inputs and pathways affecting FLO11 expression (Madhani and Fink 1997; Rupp et al. 1999; Pan and Heitman 2000, 2002; Kohler et al. 2002; Kuchin et al. 2002; Braus et al. 2003; Zeitlinger et al. 2003; Halme et al. 2004; van Dyk et al. 2005; Bumgarner et al. 2009; Octavio et al. 2009). We have previously described that a high level of FLO11 expression is required for the formation of a liquid surface biofilm (Fidalgo et al. 2006). Moreover, we showed that only the promoter carried by the “flor” Saccharomyces yeast strain (used to produce sherry wine) is able to reach this level of expression. The promoter of this FLO11 allele bears several point mutations and a deletion of 111 bp that confers a higher level of expression than common laboratory alleles (Fidalgo et al. 2006).
Nonetheless, it remains an open question through which mechanism these point mutations and the deletion lead to higher expression levels. To gain further mechanistic insights, we previously performed a genetic screen for factors activating FLO11 expression. In this way, we identified subunits of the Swi/Snf nucleosome-remodeling complex and the HDAC complexes as important FLO11 activators (Barrales et al. 2008), suggesting that the FLO11 promoter may also be strongly regulated at the chromatin level.
Here, we extend our investigation into the role of these two chromatin-related complexes, the HDAC and Swi/Snf, in FLO11 regulation. We analyzed if these complexes impact on the chromatin structure of the FLO11 promoter and show that it is substantially altered by the removal of HDAC or Swi/Snf subunits. As no subunit with deacetylase activity was identified in our previous screen, we tested several candidate enzymes for their effect on FLO11 expression, finding that the Hos2 and Rpd3 deacetylases activate FLO11 in a partially redundant manner. Surprisingly, the role of these HDAC subunits was not mirrored by the loss of Pho23, a subunit of the Rpd3L deaceylation complex. Our data confirm an additional layer of FLO11 regulation at the level of chromatin through an intricate interplay between histone modifications and nucleosome remodeling.
Materials and Methods
Strains, media, and genetic methods
The S. cerevisiae strains used in this study are listed in Table 1. PCR-mediated disruption (Lorenz et al. 1995) was used for gene deletions. Double deletions were carried out as described (Guldener et al. 1996). Plasmids and disruption cassettes were introduced using the LiAc/SSDNA/PEG procedure (Gietz et al. 1995). Standard yeast extract peptone dextrose (YEPD) contained 2 or 0.5% of glucose. Synthetic complete dextrose (SCD) medium without the appropriate amino acids was used for plasmid selection. When necessary, YEPD medium was supplemented with 200 mg/liter geneticin for selection of geneticin-resistant transformants.
Table 1 . Yeast strains used in this study.
| Strains | Genotype | Source |
|---|---|---|
| 133d | MATa ura3-52 | Fidalgo et al. (2006) |
| 133d pho23Δ | MATa ura3-52 pho23Δ::KanMX4 | Barrales et al. (2008) |
| 133d hos1Δ | MATa ura3-52 hos1Δ::KanMX6 | This study |
| 133d hos2Δ | MATa ura3-52 hos2Δ::KanMX6 | This study |
| 133d hos3Δ | MATa ura3-52 hos3Δ::KanMX6 | This study |
| 133d rpd3Δ | MATa ura3-52 rpd3Δ::KanMX6 | This study |
| 133d hda1Δ | MATa ura3-52 hda1Δ::KanMX6 | This study |
| 133d hos2Δ rpd3Δ | MATa ura3-52 rpd3Δ hos2Δ::KanMX6 | This study |
| 133d sfl1Δ | MATa ura3-52 sfl1Δ::KanMX6 | This study |
| 133d flo8Δ | MATa ura3-52 flo8Δ::KanMX4 | Barrales et al. (2008) |
| 133d flo8Δ sfl1Δ | MATa ura3-52 sfl1Δ flo8Δ::KanMX4 | This study |
| 133d snf5Δ | MATa ura3-52 snf5Δ::KanMX4 | Barrales et al. (2008) |
| 133d snf5Δ sfl1Δ | MATa ura3-52 sfl1Δ snf5Δ::KanMX6 | This study |
| 133d pho23Δ sfl1Δ | MATa ura3-52 sfl1Δ pho23Δ::KanMX6 | This study |
| L5684 | MATa ura3-52 leu2Δ | G. R. Fink |
| L5684 pho23Δ | MATa ura3-52 leu2Δ pho23Δ::KanMX6 | Barrales et al. (2008) |
| L5684 hos1Δ | MATa ura3-52 leu2Δ hos1Δ::KanMX6 | This study |
| L5684 hos2Δ | MATa ura3-52 leu2Δ hos2Δ::KanMX6 | This study |
| L5684 hos3Δ | MATa ura3-52 leu2Δ hos3Δ::KanMX6 | This study |
| L5684 rpd3Δ | MATa ura3-52 leu2Δ rpd3Δ::KanMX6 | This study |
| L5684 hda1Δ | MATa ura3-52 leu2Δ hda1Δ::KanMX6 | This study |
| L5684 hos2Δ rpd3Δ | MATa ura3-52 leu2Δ rpd3Δ hos2Δ::KanMX6 | This study |
| L5684 sfl1Δ | MATa ura3-52 leu2Δ sfl1Δ::KanMX6 | This study |
| L5684 flo8Δ | MATa ura3-52 leu2Δ flo8Δ::KanMX4 | Barrales et al. (2008) |
| L5684 flo8Δ sfl1Δ | MATa ura3-52 leu2Δ sfl1Δ flo8Δ::KanMX4 | This study |
| L5684 snf5Δ | MATa ura3-52 leu2Δ snf5Δ::KanMX6 | Barrales et al. (2008) |
| L5684 snf5Δ sfl1Δ | MATa ura3-52 leu2Δ sfl1Δ snf5Δ::KanMX6 | This study |
| L5684 pho23Δ sfl1Δ | MATa ura3-52 leu2Δ sfl1Δ pho23Δ::KanMX6 | This study |
| L5684 flo8Δ hda1Δ | MATa ura3-52 leu2Δ hda1Δ::KanMX6 flo8Δ::hphMX4 | This study |
| L5684 snf5Δ hda1Δ | MATa ura3-52 leu2Δ hda1Δ::KanMX6 snf5Δ::hphMX4 | This study |
| L5684 pho23Δ hda1Δ | MATa ura3-52 leu2Δ hda1Δ::KanMX6 pho23Δ::hphMX4 | This study |
| L5684 pho23:Myc | MATa ura3-52 leu2Δ PHO23-13Myc::KanMX4 | This study |
| L5684 rpd3:Myc | MATa ura3-52 leu2Δ RPD3-13Myc::KanMX4 | This study |
Expression analysis
For FLO11, ICR1, and SFL1 gene expression analysis, cells were incubated overnight in YEPD liquid medium at 30°, transferred to fresh YEPD medium, and incubated to an optical density at 600 nm (OD600) of ∼0.8. Cells were washed with ice-cold water and total RNA was isolated (QIAGEN RNeasy Mini Kit). For the Northern blot analysis, total RNA was separated by denaturing formaldehyde agarose gel electrophoresis and transferred overnight by capillary action to nylon membranes. The 400-bp regions at the 5′ end of the FLO11 or SCR1 genes were used as probes (labeled using the AMERSHAM Megaprime DNA Labeling System Kit), and radioactive bands were visualized using a Molecular Dynamics PhosphorImager. The complete images obtained were manipulated using Adobe Photoshop CS2 to convert them into grayscale format and adjust levels. For expression analysis by quantitative RT-PCR (qRT-PCR) cDNA was synthesized using the Transcriptor First Strand cDNA Synthesis Kit (Roche), and cDNA was quantitatively measured in triplicate with the ABI Prism 7000 sequence detection system. SCR1 cDNA was used for normalization. Primers used are listed in the Supporting Information, Table S1.
Flow cytometry
To quantify GFP levels, cells were grown in SCD medium without uracil overnight at 30°, washed and resuspended in fresh medium, and incubated until they reached an OD600 of 0.8. Cells were pelleted, washed, and resuspended in 50 mM sodium citrate. The fluorescence of 20,000 cells was measured using a FACSCalibur flow cytometer (Becton Dickinson) with a 530/30 band-pass filter.
β-Galactosidase Assay
The β-galactosidase assays were performed essentially as described previously (Rose and Botstein 1983). Cells were grown in SCD medium without uracil overnight at 30°, washed and resuspended in fresh medium, and incubated until they reached an OD600 of 0.8. Specific β-galactosidase activity was then measured and normalized to the total protein content in each extract.
Chromatin immunoprecipitation analysis
The cells were incubated in YEPD liquid medium overnight at 30°, transferred to fresh YEPD medium, and incubated until they reached an OD600 of ∼1. Cells for L5684 pho23:Myc and L5684 rpd3:Myc strains were incubated 4 hr in 0.5% glucose medium . Cells were cross-linked with 1% formaldehyde for 20 min at room temperature. In the case of Rpd3 and Pho23 chromatin immunoprecipitation (ChIP), cells were pretreated with 10 mM dimethyl adipimidate dihydrochloride for 45 min as described previously (Kurdistani and Grunstein 2003b). Cross-linking was quenched by adding glycine to a final concentration of 125 mM. The cells were washed twice with ice-cold 0.9% NaCl, resuspended in HEG150 buffer [150 mM NaCl, 50 mM HEPES (pH 7.6), 10% glycerol, 1% Triton X-100, 1 mM EDTA, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride] and lysed with a FastPrep FP120 (two times for 15 sec at power 4.5 with a 60-sec pause on ice). Chromatin was sheared to an average size of 500-bp fragments by sonication using a Bioruptor (Diagenode, three times for 30 sec with a 60-sec pause, position on high, ice-water bath). Chromatin immunoprecipitation was performed as described (Strahl-Bolsinger et al. 1997). The antibodies used were anti-histone H3 antibody from Abcam (ab1791), anti-acetyl-histone H4 from Millipore (06-866), and anti-cMyc from Santa Cruz Biotechnology (9E10). Immunoprecipitated and input DNA was quantitatively measured in triplicates with the ABI Prism 7000 sequence detection system. Primers used are listed in Table S1.
Chromatin analysis
The preparation of yeast nuclei (Almer et al. 1986) and chromatin analysis of nuclei by restriction nucleases and MNase digestion with indirect end-labeling was performed as described (Svaren and Horz 1995; Gregory and Horz 1999). Secondary cleavage was with XcmI (+584), to monitor the whole promoter, or with XbaI (−461 in strain 133d, −460 in L5684), to analyze in more detail the promoter 5′ region. The probes are PCR products corresponding to bases +87 to +553 from the ATG of the FLO11 ORF of strain 133d for the XcmI secondary cleavage and to bases −975 to −589 for the XbaI secondary cleavage. Probes generated from strain 133d genomic DNA worked equally as well as probes generated from strain L5684 DNA (data not shown). The complete images obtained were manipulated using Adobe Photoshop CS2 to convert them into grayscale format and adjust levels. Sometimes parts of the image were reorganized to avoid space between lines.
Results
FLO11 transcriptional activation exhibits an allele-specific requirement for Rpd3 and Hos2 deacetylases
We showed previously that many subunits of the Rpd3L complex (Pho23, Sds3, Rxt2, Sap30, and Ash1) are involved in the activation of FLO11 expression; this occurred even when using glucose-containing media as the sole carbon source, where FLO11 expression should be repressed. These subunits activate the expression of both the “flor” and the laboratory alleles of FLO11 (Barrales et al. 2008). The Rpd3L complex is recruited to specific promoters through the DNA-binding proteins Ash1 and Ume6 (Carrozza et al. 2005). Once recruited, the HDAC complex subunit Rpd3 is responsible for repressing or activating target gene transcription via its deacetylase activity (Kadosh and Struhl 1997; Kurdistani et al. 2002; De Nadal et al. 2004). However, in our screen we did not identify any catalytic HDAC subunits as a FLO11 activator. This could be because (i) the screen did not reach saturation, (ii) the activation by the Rpd3L complex does not require HDAC activity, or (iii) other histone deacetylases present in S. cerevisiae act redundantly with the components of the Rpd3L complex in FLO11 activation.
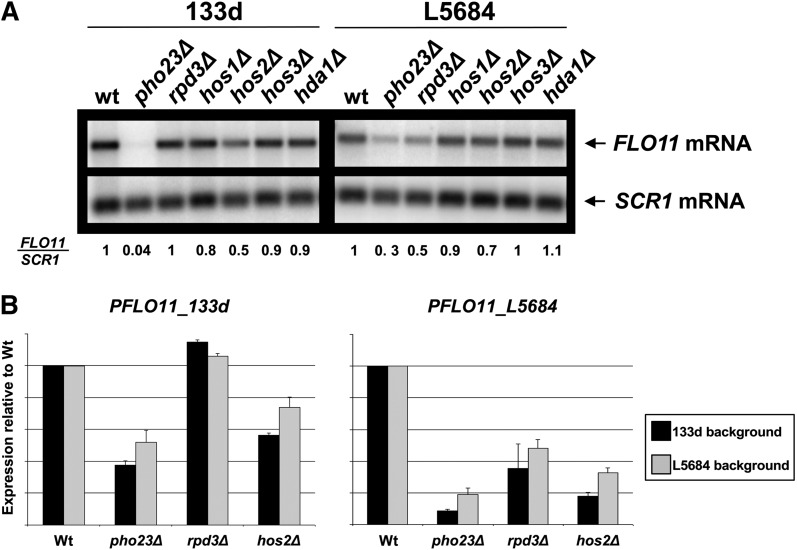
To directly address these possibilities, we first examined the effect of deleting RPD3 on FLO11 expression in both flor (133d) (Fidalgo et al. 2006) and laboratory (L5684) strains. Given possibility (iii), we also examined the effect of deleting other candidate deacetylases (HOS1, HOS2, HOS3, and HDA1). In the laboratory L5684 strain, FLO11 expression was reduced mainly in the rpd3Δ mutant (Figure 1A), which supports a role for Rpd3-mediated deacetylase activity in FLO11 regulation. Interestingly, a smaller but significant reduction in FLO11 expression was also observed in the hos2Δ mutant (Figure 1A). However, in the flor 133d strain, the deletion of rpd3Δ did not affect FLO11 expression, but was reduced only by the loss of HOS2 (Figure 1A). The Rpd3L complex subunit Pho23, previously identified as a FLO11 activator (Barrales et al. 2008), was included as a positive control. Interestingly, the decrease in FLO11 expression in both the 133d strain/hos2Δ and the L5684 strain/rpd3Δ mutants was smaller than the corresponding decrease in either pho23Δ mutant (Figure 1A).
Figure 1 .
Strain-dependent activation of FLO11 transcription by the Hos2 or Rpd3 deacetylases. (A) Northern blot analysis of FLO11 mRNA levels in the indicated mutants of 133d (flor) or the L5684 (laboratory) yeast strains. SCR1 mRNA was probed as loading control. Numbers below the blot indicate FLO11 expression levels normalized to SCR1 and the respective quotient of the wild type. (B) The differential roles of deacetylases are specific to the promoter alleles. Expression levels were measured by flow cytometry or β-galactosidase assays for strains transformed with a plasmid containing the GFP ORF under the control of the 133d FLO11 promoter (PFLO11_133d) or the lacZ ORF under the control of the laboratory one (PFLO11_L5684), respectively, and normalized to wild type. Error bars represent the standard deviation of three biologically independent measurements.
The strain-specific requirements for different HDACs in FLO11 activation could be due directly to the differences in the FLO11 promoter or indirectly to other differences in their genetic background. To distinguish between these possibilities, we transformed the L5684 and 133d strains with a plasmid harboring the GFP gene under the control of the FLO11 promoter from the 133d strain (pFLT133dGFP) or the lacZ ORF controlled by the FLO11 promoter from the L5684 strain (B3782) (Rupp et al. 1999). This allowed us to use flow cytometry and β-galactosidase assays to measure the effect of PHO23, HOS2, and RPD3 deletions on the activity of both FLO11 promoters in the two genetic backgrounds. As expected, FLO11 expression directed by the flor FLO11 promoter was reduced in the flor strain background by the deletion of PHO23 and HOS2, but not of RPD3 (Figure 1B). On the other hand, a reduction in expression was observed in the three mutants when the laboratory FLO11 promoter was analyzed in the laboratory background (Figure 1B). The reduction in expression observed in the hos2Δ mutant using this assay was stronger than the one observed by Northern blot. However, a difference in the medium used in these two assays could explain this difference. These data suggest that our assay faithfully recapitulates FLO11 promoter regulation. As before, the effect of the PHO23 deletion was somewhat stronger than that of the RPD3 or HOS2 deletions (Figure 1B). Significantly, the expression levels observed for the flor and laboratory promoters were unaffected by the genetic background of the strain used (Figure 1B). This confirms the results shown in Figure 1A and argues that the different deacetylase requirements are due to differences in the promoter sequence between the two FLO11 alleles.
Rpd3 and Hos2 deacetylases redundantly activate FLO11 expression
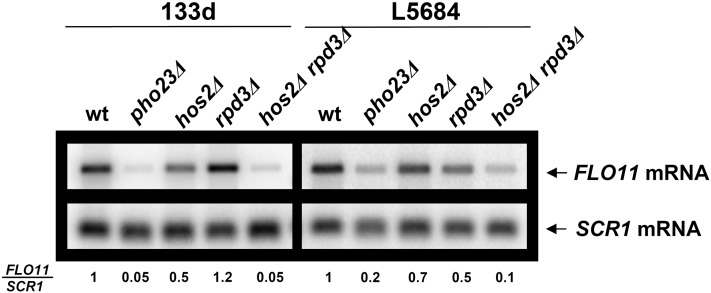
As Rpd3 and Hos2 were differentially involved in activating different FLO11 promoter alleles, we considered the possibility that these two deacetylases might be acting partially redundantly. To test this, we generated hos2Δ rpd3Δ double mutants in both the 133d and L5684 strain backgrounds and found that the double-mutant combination resulted in a significantly stronger reduction in FLO11 expression in both cases (Figure 2). Thus the activity of Hos2 and Rpd3 on FLO11 expression appears to be partially redundant in both strains, with each deacetylase probably having additional allele-specific functions. As the levels of FLO11 expression in the double mutants were approximately the same as in the corresponding pho23Δ mutants (Figure 2), it seems likely that the redundant activities of both deacetylases are mediated through the Rpd3L complex.
Figure 2 .
Hos2 and Rpd3 synergistically activate FLO11 expression. Northern blot analysis as in Figure 1A, but including the hos2Δ rpd3Δ double mutant in 133d and L5684 strains.
Pho23, but not HDACs Rpd3 and Hos2, has a clear role in generating a nucleosome-depleted FLO11 promoter region
Rpd3 mainly deacetylates histones H4 and H3 (Kurdistani and Grunstein 2003a), acting redundantly with Hos2 in the case of histone H4 (Sharma et al. 2007). If Hos2 and Rpd3 are the catalytic subunits of the complexes involved in FLO11 activation, we would expect to see changes in histone acetylation at the FLO11 promoter in single or double mutants for these catalytic subunits, as well as in pho23Δ mutants. To check this prediction, we used ChIP to measure levels of acetylated H4 at specific points along the FLO11 promoter in the 133d and L5684 strains and compared the effect of single pho23Δ, hos2Δ, and rpd3Δ mutants as well the double hos2Δ rpd3Δ mutant on H4 deacetylase activity (Figure 3, A–E). Surprisingly, the levels of H4 acetylation at the promoter were somewhat decreased by the loss of PHO23 and HOS2 in both backgrounds (Figure 3, A–C). H4 acetylation levels were almost unchanged in hos2Δ rpd3Δ double mutants and increased only in the 133d strain/rpd3Δ mutant, even though we had not observed an effect on FLO11 expression in this strain (Figure 3, A, D, and E; also see Figures 1 and 2). Thus, we did not observe a clear-cut relationship between the effects on expression levels and histone H4 acetylation.
Figure 3 .
Mutant phenotypes of histone depletion and acetylation at the FLO11 promoter. The level of acetylated histone H4 (A–E) and total histone H3 (F–J) occupancy over the FLO11 promoter and coding region as well as over the INO1 promoter was monitored by ChIP assay in the indicated wild type and mutants for both the 133d and the L5684 strain. Acetylated histone H4 levels are shown relative to histone H3 levels, and histone H3 levels were normalized to an amplicon at the telomere. Error bars represent the standard deviation of three biologically independent measurements. Diagrams above the panels show amplicon positions. The gray box in the FLO11 promoter corresponds to the 111-bp region that is deleted in the flor promoter allele. As a positive control, we used an amplicon at the INO1 promoter to test H3 levels and observed a strong decrease in nucleosome occupancy in the rpd3Δ and hos2Δ rpd3Δ mutants (F, I, and J) in agreement with increased INO1 transcription in these mutants (Rundlett et al. 1998).
In this experiment, the amount of histone H3 was used as a control to determine total nucleosome occupancy. However, when we directly compared levels of histone H3 between wild-type and mutant strains, we obtained some very interesting results. In wild-type L5684 and 133d strains, the FLO11 promoter showed strikingly low levels of nucleosome occupancy, especially over the central region (Figure 3F). Given the differential FLO11 expression in these two strains, it suggests that this promoter nucleosome depletion is not a consequence of high transcription levels. In contrast, nucleosome occupancy in the FLO11-coding region was low only in the 133d background, presumably reflecting the high expression levels of this strain (Figure 3F).
Interestingly, neither the promoter in either strain, nor the coding region in the 133d strain, was nucleosome depleted in the pho23Δ mutant (Figure 3, F and G). This is consistent with the greatly FLO11-reduced expression of this mutant (Figures 1 and 2) and suggests that Pho23 is somehow involved in maintaining an open chromatin structure at the FLO11 promoter. A similar but somewhat less pronounced increase in nucleosome occupancy was observed for the hos2Δ mutant in the 133d background (Figure 3, F and H), which also correlates with the lower expression levels in this strain (Figures 1 and 2). Conversely, the rpd3Δ single mutant showed increased nucleosome occupancy in the FLO11 promoter region mainly in the upstream region in the L5684 but not in the 133d strain (Figure 3, F and I, amplicon 2). Moreover, there was a general tendency toward higher nucleosome occupancy levels at the FLO11 promoter in the hos2Δ rpd3Δ double mutant. Thus, there was a correlation between an increase in promoter nucleosome occupancy and a decrease of FLO11 expression. Nonetheless, it remains to be determined if the increase in nucleosome occupancy is a cause or a consequence of decreased expression. Interestingly, the difference in nucleosome occupancy between pho23Δ and rpd3Δ hos2Δ mutants (Figure 3, G and J), even with the same level of FLO11 expression, suggests that Pho23 is actively involved in maintaining a nucleosome-depleted FLO11 promoter and that this effect is at least partially independent of Rpd3 and Hos2.
Deacetylase activity of Rpd3 is required for FLO11 expression
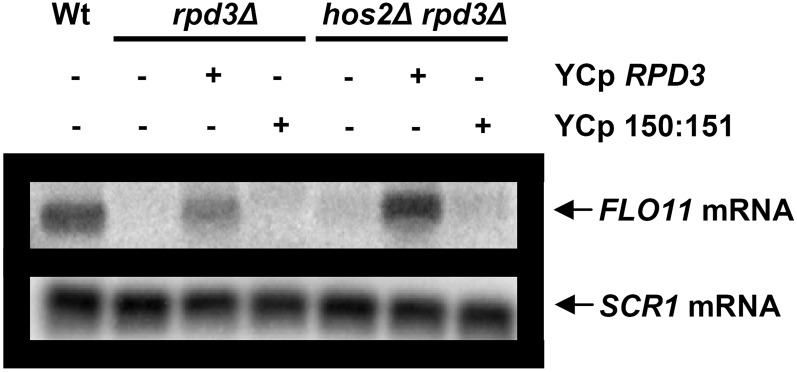
To study if the reduction in FLO11 expression obtained in the laboratory strain was due to the loss of Rpd3 deacetylase activity, we transformed L5684 strain yeasts harboring RPD3 and HOS2RPD3 deletions with a plasmid containing either the wild-type RPD3 allele or a point mutant allele where His150 and His151 were replaced with alanines resulting in defective deacetylase activity (De Nadal et al. 2004). The level of FLO11 expression was restored only if transformed with the wild-type but not with the mutant allele (Figure 4), showing that the enzymatic activity of Rpd3 is essential for complete FLO11 activation in the laboratory L5684 background.
Figure 4 .
The enzymatic activity of Rpd3 is required for FLO11 activation in the laboratory strain. Northern blot analysis (as in Figure 1A) of FLO11 mRNA from wild type (L5684) and rpd3Δ or hos2Δ rpd3Δ mutant transformed (+) or not (−) with the plasmid containing either the RPD3 wild-type gene (YCp RPD3) or the RPD3 allele defective for deacetylase activity (YCp 150:151).
Rpd3-dependent activation of the laboratory FLO11 allele may be partly due to reduced Sfl1 repression
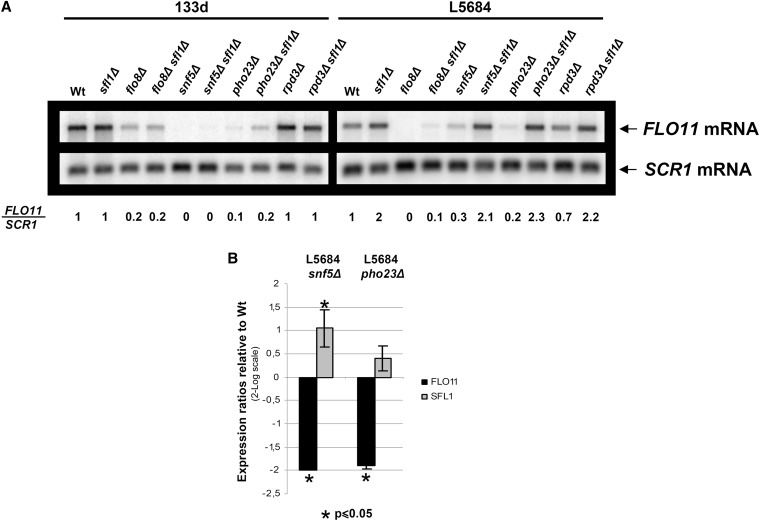
Rpd3 has been described to activate the expression of some genes by counteracting the effect of gene repressors (Sertil et al. 2003, 2007). Sfl1 and the Rpd3L complex have recently been described to mediate antagonistic regulatory effects on the FLO11 promoter regulation (Bumgarner et al. 2009). Sfl1 is one of the main repressors of FLO11 expression, and a region for Sfl1 binding has been identified in the FLO11 promoter (Pan and Heitman 2002). This region includes the 111-bp sequence that is absent in the 133d strain. Thus, an explanation of why Rpd3 deacetylase has an effect in the laboratory allele but not in the flor one could be that Sfl1 is counteracted by Rpd3 in yeast carrying the laboratory FLO11 allele but not the flor one. To check this hypothesis, we deleted SFL1 from both laboratory (L5684) and flor (133d) backgrounds. Northern blot analysis showed that, as predicted, Sfl1 had a repressor effect on the L5684 strain but not on the 133d one (Figure 5A). To check a possible relation between this repressor and Rpd3, we generated a rpd3Δ sfl1Δ double mutant. As controls, we also generated double mutants with PHO23, as a member of the complex, and with FLO8 and SNF5, as other FLO11 activators, to discard a general effect. In the L5684 strains, FLO11 expression was completely restored by the deletion of SFL1 in all the activator mutants except flo8Δ, where we observed only a partial restoration (Figure 5A). As expected, the reduced expression of FLO11 in the 133d strain produced by deletion of the described activators was not restored in sfl1Δ double mutants (Figure 5A). These data suggest that Rpd3 deacetylase may be required to counteract FLO11 repression by Sfl1 and explain why in the 133d strain—where this repression does not exist—we do not observe an activator effect of this deacetylase. The question of why the activation of FLO11 by the Swi/Snf and Rpd3L complexes is Sfl1-independent and much stronger in the 133d background than in the laboratory one is an open question that remains to be studied.
Figure 5 .
Sfl1-mediated repression is important for FLO11 regulation but is absent in the 133d strain. (A) Northern blot analysis of FLO11 mRNA, as described in Figure 1A, for the indicated wild-type and mutant strains. (B) qRT-PCR analysis of FLO11 and SFL1 expression in the laboratory strain. Expression was normalized to SCR1 expression and to wild type. Error bars represent the standard deviation of three biologically independent measurements.
To know if the activation of FLO11 mediated by both complexes in the laboratory background is a consequence of SFL1 expression regulation, we performed qRT-PCR to measure the level of SFL1 expression in the snf5Δ and pho23Δ mutants. As can be observed in Figure 5B, SFL1 expression was increased in the snf5Δ mutant but not in the pho23Δ one, indicating that at least part of the activation effect observed for the Swi/Snf complex could be mediated through SFL1 repression. The fact that SFL1 transcription is unaffected in the pho23Δ mutant, and that in the 133d strain FLO11 activation by the Swi/Snf and Rpd3L complexes was Sfl1-independent, suggests a direct effect of these complexes on the FLO11 promoter.
Sfl1 has been proposed to recruit the Hda1 repressor to the FLO11 promoter, contributing to an epigenetic regulation (Halme et al. 2004; Octavio et al. 2009). To check if deletion of HDA1 restores FLO11 expression in a similar way to sfl1Δ mutants, we deleted HDA1 in flo8Δ, snf5Δ, and pho23Δ single mutants. However, as is shown in Figure 6A, the drop in FLO11 expression is not restored in any of the double mutants.
Figure 6 .
Rpd3 counteracts the action of Sfl1 on the laboratory FLO11 allele by regulating ICR1 ncRNA. (A) Northern blot analysis of FLO11 mRNA, as described in Figure 1A, for the indicated wild-type and mutant strains. (B) qRT-PCR analysis of ICR1 ncRNA expression in both laboratory and 133d strains. Expression was normalized to SCR1 expression and to wild type. Error bars represent the standard deviation of three biologically independent measurements.
On the other hand, Sfl1 has been described to repress FLO11 expression by increasing the amount of ICR1 noncoding RNA (ncRNA), which is transcribed in the FLO11 promoter region. Sfl1 activates ICR1 ncRNA transcription by repressing the transcription of PWR1 ncRNA that partially interferes with the ICR1 one. In this interesting regulatory mechanism, the Rpd3L complex has been described to have the opposite role (Bumgarner et al. 2009, 2012). We carried out qRT-PCR to study the level of ICR1 ncRNA in mutants from both backgrounds (Figure 6B) and to study the effect of histone deacetylases and Sfl1 over this regulation. As it has been previously described, ICR1 ncRNA amounts increased significantly in the pho23Δ and rpd3Δ mutants in the L5684 laboratory background (Figure 6B). Interestingly, this increase was not observed in the 133d flor background (Figure 6B). Moreover, the increase in the ICR1 ncRNA in an rpd3Δ mutant was abolished in a double mutant with SFL1 (Figure 6B). These observations support the idea that the Rpd3L complex counteracts the repression of FLO11 expression exerted by Sfl1 through the regulation of ICR1. Deletion of HOS2 had no significant effect over ICR1 ncRNA expression (Figure 6B), indicating that the redundant role of Rpd3 and Hos2 in controlling FLO11 expression is not mediated by regulating this ncRNA. This result and the fact that the ncRNA system seems to be unregulated in the 133d strain support the idea that the opposing actions of Rpd3 and Sfl1 on FLO11 expression via ncRNA is the additional mechanism by which Rpd3 affects laboratory FLO11 allele expression independently of its redundant activity with Hos2.
Pho23 is important for the maintenance of proper nucleosome organization at the FLO11 promoter
Our observations described above and the strong effect on overall promoter nucleosome occupancy in the pho23Δ mutant (Figure 3, F and G) led us to look for direct effects of the Rpd3L complex on FLO11 promoter chromatin. To this end, we assessed if there were changes in chromatin structure at the level of nucleosome positioning. First, we characterized FLO11 promoter chromatin structure in both the 133d and L5684 wild-type strains by MNase indirect end-labeling analysis. We used YEPD medium, where the deletion of PHO23 leads to an obvious drop in FLO11 expression. We observed a characteristic pattern for each strain, which differed from that of free DNA (Figure 7, A and B). A small deletion in the ORF of the 133d flor FLO11 allele caused a shift of the banding patterns relative to the L5684 laboratory strain if XcmI was used for secondary cleavage (Figure 7A). Taking this shift into account, both patterns were quite similar to each other. However, closer examination suggested subtle differences in the region close to the 111-bp flor-specific promoter deletion. Downstream of the deletion, two distinct hypersensitive bands flanking a protected region were observed in the 133d strain (Figure 7A). In contrast, the laboratory strain showed more uniformly smeared accessibility in the same region. Using XbaI for secondary cleavage, which cuts closer to this region and allows us to zoom in on it (Figure 7B), we identified a protected area in the L5684 strain just at the position where the 111 bp are deleted in the flor strain; the flor strain showed a more pronounced hypersensitive band just downstream of this region. Even though subtle, these differences may be highly relevant because the 133d specific 111-bp deletion is the main sequence difference between the two promoter alleles. Moreover, targeted deletion of this region in the laboratory allele leads to increased FLO11 expression (Fidalgo et al. 2006).
Figure 7 .
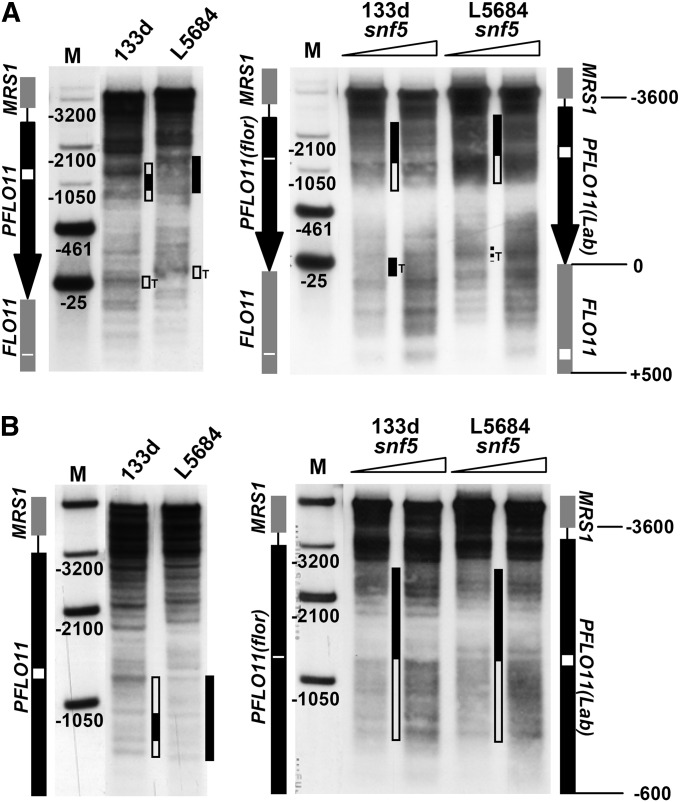
Chromatin structure at the FLO11 promoter is generally very similar between flor and laboratory alleles but is distinct in the vicinity of the 111-bp deletion. MNase indirect end-labeling analysis of chromatin structure at the FLO11 promoter in 133d and L5684 strains. Ramps above the lanes denote increasing MNase concentrations. MNase patterns of free DNA are shown for comparison. Secondary cleavage with XcmI is shown in A and with XbaI in B. The five marker bands of lanes M in A were generated by double digestion with XcmI and either StuI, XbaI, HpaI, XmnI, or AflI (from bottom to top, corresponding to positions −25, −461, −1050, −2100, and −3200 bp from FLO11 ATG). The three marker bands in lanes M of B were generated by double digestion with XbaI and HpaI, XmnI, or AflI (from bottom to top). Diagrams outlining the FLO11 promoter (black arrow in A and black line in B) and flanking ORFs (gray lines) are shown on either side of the blots. The regions where the flor allele has deletions in the promoter and ORF relative to the laboratory allele are marked by a white box for the laboratory and by a white line for the flor allele. Hypersensitive regions of interest are highlighted by open bars in between lanes, and more protected regions by solid bars. Note the protected region flanked by two bands next to the 111-bp deletion region (white box) in the L5684 allele in B. “T” denotes the TATA box region.
Next we asked whether or not Pho23 was involved in generating this chromatin structure at the FLO11 promoter. Indeed, MNase mapping of the FLO11 promoter chromatin structure in pho23Δ mutants revealed differences from the wild-type pattern in both flor and laboratory backgrounds (Figure 8). We observed changes in the banding pattern of the region around the 111-bp deletion and the appearance of additional very strong hypersensitive sites downstream of the TATA box, as well as in a somewhat more protected TATA box region (Figure 8A). A closer look at the upstream FLO11 promoter region using secondary XbaI cleavage showed that the pho23Δ mutant patterns in the vicinity of the −1050 position were similar to the wild-type pattern in terms of the position of bands, but the relative band intensities were affected, especially in the laboratory background (Figure 8B). Importantly, these differences in intensities were not due to differences in the degree of digestion as the band intensities in the vicinity of the −2100 position were all very similar. These observations, combined with the histone H3 ChIP analysis (Figure 3B), suggest not only that the average occupancy levels of the FLO11 promoter nucleosomes are increased in the pho23Δ mutant, but also that the nucleosomes are more strongly associated with the DNA.
Figure 8 .
Pho23 is essential for the correct chromatin structure at the FLO11 promoter. MNase indirect end-labeling analysis of the FLO11 promoter, as in Figure 7 but including the pho23Δ mutants in the 133d and L5684 backgrounds. Secondary cleavage with XcmI is shown in A and with XbaI in B. Wild-type patterns taken from Figure 7 are shown on the left for comparison.
The additional hypersensitive bands downstream of the TATA box were not present in the free DNA pattern (Figure 7A) and flank a protected region that may correspond to a strongly positioned nucleosome. Furthermore, the wild-type pattern showed a weak but substantial band at the −25 position, which is close to the TATA box itself. This band was also present in the free DNA pattern (Figure 7A), but became protected in the pho23Δ mutant patterns (Figure 8A). This may argue for a less accessible TATA box and a strongly positioned nucleosome downstream of the TATA box in this mutant. In summary, our findings suggest that Pho23 is important for correct FLO11 promoter chromatin structure especially in the region of the 111-bp flor-specific deletion and around the TATA box.
Swi/Snf complex is involved in organizing FLO11 promoter chromatin structure
Contrasting types of interactions between Rpd3 and Swi/Snf complexes have been described at different target promoters. So, Rpd3 inhibits the recruitment of the Swi/Snf complex at several promoters that are repressed by the Rpd3 complex (Deckert and Struhl 2002). Conversely, it has been proposed that Rpd3 complex-mediated deacetylation helps the Swi/Snf complex to bind promoters where Rpd3 activates gene expression (Sertil et al. 2007). Because we have demonstrated that subunits of both complexes are strong activators of FLO11 (Barrales et al. 2008) and because we saw substantial effects on FLO11 promoter chromatin structure in the pho23Δ mutant (Figure 8), we repeated the FLO11 promoter MNase indirect end-labeling analysis for the snf5Δ mutants in both backgrounds (Figure 9, A and B).
Figure 9 .
Snf5 is involved in nucleosome positioning at the FLO11 promoter. MNase indirect end-labeling analysis as in Figure 7, but including the snf5Δ mutants in flor and laboratory backgrounds. Secondary cleavage with XcmI is shown in A and with XbaI in B. Wild-type patterns taken from Figure 7 are shown on the left for comparison.
As before, we found substantial differences in the chromatin structure between the mutant and the wild-type in both backgrounds. Downstream of the 111-bp deletion there was more pronounced hypersensitivity, while upstream, close to the −2100 position, the region was somewhat more protected (Figure 9A). XbaI secondary cleavage revealed that the deletion is located just at the intersection between the more protected and more hypersensitive regions (Figure 9B). Moreover, the TATA box region (−25 position) in the 133d snf5Δ mutant appeared more protected than in the wild type, similar to that seen for the pho23Δ mutant (Figure 8A), but this difference was not as clear in the laboratory background. Thus, the Swi/Snf complex also has a role in shaping the FLO11 promoter chromatin structure. Even though the snf5Δ mutant patterns were different from the pho23Δ mutant patterns, the same promoter subregions in the vicinity of the 111-bp deletion and the TATA box were affected.
Discussion
FLO11 promoter regulation at the chromatin level
Regulation of FLO11 is exceedingly complex because of an unusually long promoter region and the numerous regulators that act on it. Recently, we added new elements to this complex system by showing that the Rpd3L and Swi/Snf complexes are major activators for FLO11 expression (Barrales et al. 2008). These two complexes were previously described to control gene expression by alterations in chromatin structure through histone modification or chromatin remodeling, respectively. Here we show that both complexes clearly affect chromatin structure at the FLO11 promoter. Deletion of PHO23 led to increased nucleosome occupancy over the whole FLO11 promoter (Figure 3, F and G), to the protection of a hypersensitive site in the TATA box area, and, mostly for the laboratory strain, to the appearance of a more strongly positioned nucleosome in the region where the 111-bp flor-specific deletion is located (Figure 8, A and B). Similarly, deletion of SNF5 resulted in protection of the TATA box region as well as in accessibility changes in the vicinity of the deletion, which is different from that observed in the pho23Δ mutant (Figure 9, A and B).
Our repeated observations of altered chromatin structure close to the 111-bp region led us to postulate a key role for this region in the regulation of FLO11 expression. This may explain why a deletion of this region is sufficient to give rise to a new capability for S. cerevisiae, i.e., overexpression of FLO11 and the consequent formation of a biofilm on top of a liquid surface. Alteration of the sequence and nucleosome structure in this region could affect the accessibility for previously described regulators, such as the activator Flo8 and the wide-ranging repressor Sfl1. The Swi/Snf complex is thought to be required as a general activator for genes under the control of global repressors (Sertil et al. 2007). We demonstrated that the activation effect exerted by Swi/Snf in the laboratory strain may work in part by counteracting the repression by Sfl1. Swi/Snf is also described to enhance binding of Flo8 to the STA1 promoter (Kim et al. 2004), a promoter very similar to the FLO11 promoter. Interestingly, Flo8 binds to the FLO11 promoter closely upstream of the 111-bp deletion region where the Swi/Snf complex maintains an accessible chromatin structure (Figure 9). Moreover, downstream of this region, Swi/Snf generates a protected area that includes a binding site for the Sfl1 repressor. Thus, the Swi/Snf complex could maintain a chromatin structure in this area that prevents Sfl1 binding and enhances binding of Flo8, resulting in FLO11 activation. Interestingly, the repression exerted by Sfl1 is absent from the 133d flor strain, perhaps as a result of the changes to the FLO11 genomic region in this strain. Nevertheless, a role for changes in the genetic background cannot be ruled out, and more studies must be done to better understand the important role of this repression and the basis for the differences between the two strains.
Regarding the Rpd3L complex, the effect of the pho23Δ mutation on chromatin structure in the 111-bp deletion region of the 133d strain was smaller than for the snf5Δ mutation, although a clear effect on total nucleosome promoter occupancy was observed. These results support the idea that this complex has a general function in FLO11 activation, such as in maintaining the whole promoter in an open state.
All these data strongly support our hypothesis that these complexes directly affect chromatin structure at the FLO11 promoter. This is further supported by the direct demonstration of Swi3 binding, a subunit of the Swi/Snf complex, to the FLO11 promoter and coding region (Venters and Pugh 2009) and Rpd3 binding to the FLO11 promoter (Bumgarner et al. 2009). We also observed a slightly increased occupancy of Pho23 and Rpd3 in the central region of the FLO11 promoter (Figure S1). In addition to this direct effect, our evidence also suggests that the Swi/Snf complex could be indirectly activating FLO11 by repressing SFL1 expression.
Activation of FLO11 by both Rpd3 and Hos2
Rpd3 and Hos2 have been described to have generally overlapping specificities in gene expression regulation, but with different functions—Rpd3 as a repressor and Hos2 as an activator (Wang et al. 2002). However, both deacetylases have been shown to act redundantly to positively regulate the expression of DNA damage-inducible genes (Sharma et al. 2007). Here we have observed a strong reduction of FLO11 expression in the 133d strain when both RPD3 and HOS2 were deleted. However, there was no change in FLO11 expression when only RPD3 was deleted, suggesting that Rpd3 and Hos2 also act redundantly in activating this promoter. Rpd3 and Hos2 differentially contribute to FLO11 activation, depending on the allele. Both Rpd3 and Hos2 activate the laboratory FLO11 allele. While Rpd3 exerts a stronger effect on the laboratory allele, only Hos2 has a clear effect on the 133d flor allele. So we postulate a model in which a basal level of FLO11 expression in both alleles is redundantly governed by Rpd3 and Hos2, for example, by promoting polymerase initiation complex assembly as described for DAN1 regulation (Sharma et al. 2007). The Pho23-dependent accessibility at the TATA box region could correspond to this effect. Nonetheless, it could also be a consequence of reduced FLO11 transcription levels. Recently, an expanded Rpd3 complex (Rpd3LE) has been described (Shevchenko et al. 2008). The Rpd3LE complex contains five additional members from the Set3 complex, including Hos2. Therefore, it is possible that both deacetylases are recruited to the FLO11 promoter through this complex and take part in FLO11 activation. Additionally, Rpd3 and Hos2 might have an extra role in this process. However, the extra effect of Rpd3 would not matter for regulation of the flor promoter allele.
In this work, we have shown that Sfl1 repression is a major regulator of FLO11 expression in the laboratory strain but has no effect on its expression in the flor strain. Interestingly, we have found that reduced FLO11 expression in the laboratory strain caused by a rpd3Δ single mutant is restored when SFL1 is deleted, suggesting that at least part of the activation effect of Rpd3 could be due to its counteracting Sfl1 repression. Accordingly, the lack of such a repression mechanism in the flor allele could be the reason why the deletion of RPD3 did not affect FLO11 expression in this case.
Rpd3 and Sfl1 have been described to have opposing roles in controlling FLO11 expression via the regulation of the transcription of the ICR1 ncRNA. Rpd3 activates FLO11 expression by repression of ICR1 ncRNA and Sfl1 has the opposite role (Bumgarner et al. 2009, 2012). We have observed that this effect of Rpd3 is not shared by Hos2, suggesting that this effect may be the additional mechanism independent of Hos2 observed for Rpd3. Moreover, we show that the ICR1 regulation seems to be lost in the 133d strain, further supporting this hypothesis.
To analyze the mechanism by which the histone deacetylase complex regulates FLO11 transcription, we studied the levels of nucleosome occupancy as well as the degree of histone H4 acetylation along the promoter. Surprisingly, we did not observe a clear correlation between FLO11 expression and nucleosome occupancy or H4 acetylation for the different mutants studied. This would suggest that the interplay of factors regulating FLO11 expression leads to a more complex readout of promoter chromatin states than expected. Nonetheless, at this stage it is already very interesting that the deletion of PHO23 had a stronger effect on nucleosome occupancy than was observed for the rpd3Δ hos2Δ double mutant. This suggests a novel function for the Rpd3L complex, where it can act to maintain a generally open state of FLO11 promoter chromatin at least partly independently of Rpd3 and Hos2 histone deacetylases.
Supplementary Material
Acknowledgments
We thank Valle Rubio, Sandra Romero, and Victor Carranco for excellent technical assistance; and John R. Pearson for critical reading of the manuscript. We also thank Francesc Posas and Gerald R. Fink for contributing plasmids used in this work. This work was supported by Ministerio de Ciencia e Innovacion and Fondo Europeo de Desarrollo Regional grant BIO2010-16787. Centro Andaluz de Biología del Desarrollo is institutionally supported by Consejo Superior de Investigaciones Científicas, Universidad Pablo de Olavide, and Junta de Andalucía.
Footnotes
Communicating editor: M. Hampsey
Literature Cited
- Almer A., Rudolph H., Hinnen A., Horz W., 1986. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 5: 2689–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrales R. R., Jimenez J., Ibeas J. I., 2008. Identification of novel activation mechanisms for FLO11 regulation in Saccharomyces cerevisiae. Genetics 178: 145–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braus G. H., Grundmann O., Bruckner S., Mosch H. U., 2003. Amino acid starvation and Gcn4p regulate adhesive growth and FLO11 gene expression in Saccharomyces cerevisiae. Mol. Biol. Cell 14: 4272–4284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bumgarner S. L., Dowell R. D., Grisafi P., Gifford D. K., Fink G. R., 2009. Toggle involving cis-interfering noncoding RNAs controls variegated gene expression in yeast. Proc. Natl. Acad. Sci. USA 106: 18321–18326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bumgarner S. L., Neuert G., Voight B. F., Symbor-Nagrabska A., Grisafi P., et al. , 2012. Single-cell analysis reveals that noncoding RNAs contribute to clonal heterogeneity by modulating transcription factor recruitment. Mol. Cell 45: 470–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza M. J., Florens L., Swanson S. K., Shia W. J., Anderson S., et al. , 2005. Stable incorporation of sequence specific repressors Ash1 and Ume6 into the Rpd3L complex. Biochim. Biophys. Acta 1731: 77–87, discussion 75–76 [DOI] [PubMed] [Google Scholar]
- Clapier C. R., Cairns B. R., 2009. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78: 273–304 [DOI] [PubMed] [Google Scholar]
- Deckert J., Struhl K., 2002. Targeted recruitment of Rpd3 histone deacetylase represses transcription by inhibiting recruitment of Swi/Snf, SAGA, and TATA binding protein. Mol. Cell. Biol. 22: 6458–6470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Nadal E., Zapater M., Alepuz P. M., Sumoy L., Mas G., et al. , 2004. The MAPK Hog1 recruits Rpd3 histone deacetylase to activate osmoresponsive genes. Nature 427: 370–374 [DOI] [PubMed] [Google Scholar]
- Fidalgo M., Barrales R. R., Ibeas J. I., Jimenez J., 2006. Adaptive evolution by mutations in the FLO11 gene. Proc. Natl. Acad. Sci. USA 103: 11228–11233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R. D., Schiestl R. H., Willems A. R., Woods R. A., 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11: 355–360 [DOI] [PubMed] [Google Scholar]
- Gimeno C. J., Ljungdahl P. O., Styles C. A., Fink G. R., 1992. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68: 1077–1090 [DOI] [PubMed] [Google Scholar]
- Gregory P. D., Horz W., 1999. Mapping chromatin structure in yeast. Methods Enzymol. 304: 365–376 [DOI] [PubMed] [Google Scholar]
- Guldener U., Heck S., Fielder T., Beinhauer J., Hegemann J. H., 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24: 2519–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B., Styles C. A., Feng Q., Fink G. R., 2000. A Saccharomyces gene family involved in invasive growth, cell-cell adhesion, and mating. Proc. Natl. Acad. Sci. USA 97: 12158–12163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halme A., Bumgarner S., Styles C., Fink G. R., 2004. Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell 116: 405–415 [DOI] [PubMed] [Google Scholar]
- Kadosh D., Struhl K., 1997. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell 89: 365–371 [DOI] [PubMed] [Google Scholar]
- Kadosh D., Struhl K., 1998. Histone deacetylase activity of Rpd3 is important for transcriptional repression in vivo. Genes Dev. 12: 797–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamakaka R. T., Biggins S., 2005. Histone variants: Deviants? Genes Dev. 19: 295–310 [DOI] [PubMed] [Google Scholar]
- Kim T. S., Kim H. Y., Yoon J. H., Kang H. S., 2004. Recruitment of the Swi/Snf complex by Ste12-Tec1 promotes Flo8-Mss11-mediated activation of STA1 expression. Mol. Cell. Biol. 24: 9542–9556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler T., Wesche S., Taheri N., Braus G. H., Mosch H. U., 2002. Dual role of the Saccharomyces cerevisiae TEA/ATTS family transcription factor Tec1p in regulation of gene expression and cellular development. Eukaryot. Cell 1: 673–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T., 2007. Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Kuchin S., Vyas V. K., Carlson M., 2002. Snf1 protein kinase and the repressors Nrg1 and Nrg2 regulate FLO11, haploid invasive growth, and diploid pseudohyphal differentiation. Mol. Cell. Biol. 22: 3994–4000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurdistani S. K., Grunstein M., 2003a. Histone acetylation and deacetylation in yeast. Nat. Rev. Mol. Cell Biol. 4: 276–284 [DOI] [PubMed] [Google Scholar]
- Kurdistani S. K., Grunstein M., 2003b. In vivo protein-protein and protein-DNA crosslinking for genomewide binding microarray. Methods 31: 90–95 [DOI] [PubMed] [Google Scholar]
- Kurdistani S. K., Robyr D., Tavazoie S., Grunstein M., 2002. Genome-wide binding map of the histone deacetylase Rpd3 in yeast. Nat. Genet. 31: 248–254 [DOI] [PubMed] [Google Scholar]
- Li B., Carey M., Workman J. L., 2007. The role of chromatin during transcription. Cell 128: 707–719 [DOI] [PubMed] [Google Scholar]
- Lo W. S., Dranginis A. M., 1998. The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by Saccharomyces cerevisiae. Mol. Biol. Cell 9: 161–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz M. C., Muir R. S., Lim E., McElver J., Weber S. C., et al. , 1995. Gene disruption with PCR products in Saccharomyces cerevisiae. Gene 158: 113–117 [DOI] [PubMed] [Google Scholar]
- Madhani H. D., Fink G. R., 1997. Combinatorial control required for the specificity of yeast MAPK signaling. Science 275: 1314–1317 [DOI] [PubMed] [Google Scholar]
- Octavio L. M., Gedeon K., Maheshri N., 2009. Epigenetic and conventional regulation is distributed among activators of FLO11 allowing tuning of population-level heterogeneity in its expression. PLoS Genet. 5: e1000673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Heitman J., 2000. Sok2 regulates yeast pseudohyphal differentiation via a transcription factor cascade that regulates cell-cell adhesion. Mol. Cell. Biol. 20: 8364–8372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Heitman J., 2002. Protein kinase A operates a molecular switch that governs yeast pseudohyphal differentiation. Mol. Cell. Biol. 22: 3981–3993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig S., Lau M., Thiele D. J., 2004. Cti6 is an Rpd3-Sin3 histone deacetylase-associated protein required for growth under iron-limiting conditions in Saccharomyces cerevisiae. J. Biol. Chem. 279: 30298–30306 [DOI] [PubMed] [Google Scholar]
- Reynolds T. B., Fink G. R., 2001. Bakers’ yeast, a model for fungal biofilm formation. Science 291: 878–881 [DOI] [PubMed] [Google Scholar]
- Rose M., Botstein D., 1983. Construction and use of gene fusions to lacZ (beta-galactosidase) that are expressed in yeast. Methods Enzymol. 101: 167–180 [DOI] [PubMed] [Google Scholar]
- Rundlett S. E., Carmen A. A., Suka N., Turner B. M., Grunstein M., 1998. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature 392: 831–835 [DOI] [PubMed] [Google Scholar]
- Rupp S., Summers E., Lo H. J., Madhani H., Fink G., 1999. MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. EMBO J. 18: 1257–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sertil O., Kapoor R., Cohen B. D., Abramova N., Lowry C. V., 2003. Synergistic repression of anaerobic genes by Mot3 and Rox1 in Saccharomyces cerevisiae. Nucleic Acids Res. 31: 5831–5837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sertil O., Vemula A., Salmon S. L., Morse R. H., Lowry C. V., 2007. Direct role for the Rpd3 complex in transcriptional induction of the anaerobic DAN/TIR genes in yeast. Mol. Cell. Biol. 27: 2037–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V. M., Tomar R. S., Dempsey A. E., Reese J. C., 2007. Histone deacetylases RPD3 and HOS2 regulate the transcriptional activation of DNA damage-inducible genes. Mol. Cell. Biol. 27: 3199–3210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A., Roguev A., Schaft D., Buchanan L., Habermann B., et al. , 2008. Chromatin Central: towards the comparative proteome by accurate mapping of the yeast proteomic environment. Genome Biol. 9: R167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl-Bolsinger S., Hecht A., Luo K., Grunstein M., 1997. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 11: 83–93 [DOI] [PubMed] [Google Scholar]
- Svaren J., Horz W., 1995. Interplay between nucleosomes and transcription factors at the yeast PHO5 promoter. Semin. Cell Biol. 6: 177–183 [DOI] [PubMed] [Google Scholar]
- van Dyk D., Pretorius I. S., Bauer F. F., 2005. Mss11p is a central element of the regulatory network that controls FLO11 expression and invasive growth in Saccharomyces cerevisiae. Genetics 169: 91–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venters B. J., Pugh B. F., 2009. A canonical promoter organization of the transcription machinery and its regulators in the Saccharomyces genome. Genome Res. 19: 360–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstrepen K. J., Klis F. M., 2006. Flocculation, adhesion and biofilm formation in yeasts. Mol. Microbiol. 60: 5–15 [DOI] [PubMed] [Google Scholar]
- Verstrepen K. J., Reynolds T. B., Fink G. R., 2004. Origins of variation in the fungal cell surface. Nat. Rev. Microbiol. 2: 533–540 [DOI] [PubMed] [Google Scholar]
- Vidal M., Gaber R. F., 1991. RPD3 encodes a second factor required to achieve maximum positive and negative transcriptional states in Saccharomyces cerevisiae. Mol. Cell. Biol. 11: 6317–6327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A., Kurdistani S. K., Grunstein M., 2002. Requirement of Hos2 histone deacetylase for gene activity in yeast. Science 298: 1412–1414 [DOI] [PubMed] [Google Scholar]
- Zeitlinger J., Simon I., Harbison C. T., Hannett N. M., Volkert T. L., et al. , 2003. Program-specific distribution of a transcription factor dependent on partner transcription factor and MAPK signaling. Cell 113: 395–404 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.