Abstract
Genetic studies in mouse models have played an integral role in the discovery of the mechanisms underlying many human diseases. The primary mode of discovery has been the application of linkage analysis to mouse crosses. This approach results in high power to identify regions that affect traits, but in low resolution, making it difficult to identify the precise genomic location harboring the causal variant. Recently, a panel of mice referred to as the hybrid mouse diversity panel (HMDP) has been developed to overcome this problem. However, power in this panel is limited by the availability of inbred strains. Previous studies have suggested combining results across multiple panels as a means to increase power, but the methods employed may not be well suited to structured populations, such as the HMDP. In this article, we introduce a meta-analysis-based method that may be used to combine HMDP studies with F2 cross studies to gain power, while increasing resolution. Due to the drastically different genetic structure of F2s and the HMDP, the best way to combine two studies for a given SNP depends on the strain distribution pattern in each study. We show that combining results, while accounting for these patterns, leads to increased power and resolution. Using our method to map bone mineral density, we find that two previously implicated loci are replicated with increased significance and that the size of the associated is decreased. We also map HDL cholesterol and show a dramatic increase in the significance of a previously identified result.
Keywords: model organism, mouse genetics, meta-analysis, genome-wide association, population structure, statistics
MODEL organisms continue to play a pivotal role in the research of human diseases. The use of mouse models in particular has been extremely effective for the identification of genes underlying Mendelian disorders. The traditional mode of discovery used to identify loci underlying such disorders has been the F2 cross. In an F2 cross, two inbred mice are used to produce F1 progeny and then these progeny are crossed to obtain F2 mice, each of which have a genetic structure that is a mix of the two original inbred strains. By applying linkage analysis to F2 populations, regions harboring causal variants are identified with high statistical power. Unfortunately, these approaches have had limited success in identifying genetic variations underlying complex, polygenic traits due to the low resolution of the studies (Flint and Mott 2001; Bennett et al. 2010), meaning that the regions found to harbor causal variants are very large.
As an alternative to F2 mapping, a number of groups have proposed the use of genome-wide association study (GWAS) methodologies to map traits in inbred populations (Pletcher et al. 2004; Payseur et al. 2007). Such approaches result in increased resolution, as inbred strains have a more diverse genetic structure, in which only small portions of the genome are shared between any two strains. The initial results were promising, but it was later found that the significant population structure within inbred strains causes a large number of spurious associations and inflates the significance of true associations. Upon correction for population structure, most of the associations identified as significant were found to be spurious (Kang et al. 2008; Manenti et al. 2009). Also, when corrected for population structure, existing panels of classical inbred strains were underpowered to detect genetic variants explaining <10% of the phenotypic variation. To address these issues, Bennett et al. (2010) utilized a panel of mice called the hybrid mouse diversity panel (HMDP), which combines inbred strains with recombinant inbred (RI) strains, which resemble an inbred version of an F2 cross (Bennett et al. 2010). The idea is that inbred strains provide high resolution, while RI strains provide increased power. They showed that when performing association mapping within this panel they achieved higher resolution than when performing mapping only using RI strains and showed that they achieved higher power than when performing mapping with only inbred strains. However, the power to detect small effects remains quite low, a problem that is due to an inherent limitation in the design of the HMDP: the limit on the availability of inbred strains.
Limited power and resolution are noted problems in many mapping panels and to combat these issues, a number of groups have suggested methods with which to combine the results from multiple studies (Hitzemann et al. 2000, 2002; Li et al. 2005; Peirce et al. 2007). The core concepts behind these methods, all of which are formed on linkage-based methodologies, may be adapted to work in association analysis. However, such approaches may not be well suited for studies in structured populations. For example, a shared feature of these linkage-based methods is the attribution of equal informativeness to each study. Such an assumption may not hold in studies with population structure, as the informativeness of a given panel will be locus dependent. In this case, methods attributing equal weight to each population may result in suboptimal power.
In this article, we propose a method with which to combine studies in a locus-specific manner, weighting each study relative to its level of informativeness, and show that our method achieves optimal power within the proposed framework. Our method is based on the concept of meta-analysis. In a meta-analysis, the statistics obtained for each SNP in two separate studies are used to obtain a meta-statistic, which combines information across studies. The most common methods for performing meta-analysis are based on the fixed effect weighted sum of Z-scores (WSoZ) (de Bakker et al. 2008), in which Z-scores from each study are combined using a predefined weighting scheme. Typically, weights are set as proportional to the number of individuals in the study. Using this basic idea, we propose a meta-analysis method for combining the results obtained from mapping in the HMDP with those obtained from mapping within an F2 population. Since the best way to combine results from these two populations at a given SNP is dependent on the strain distribution pattern in each population at that SNP, current meta-analysis methodologies are not well suited. We introduce a method that accounts for the genetic structure within each population when combining results. Using a mixed-model-based approach to correct for population structure, we derive a meta-statistic based on the WSoZ. By applying an optimal weighting scheme, our method achieves both higher power and increased resolution over mapping performed only within one population. We note that the HMDP is only one of several recently proposed strategies for increasing the resolution of mouse genetic studies over traditional crosses. Other strategies include the collaborative cross (Aylor et al. 2011) and the use of heterogeneous stocks (Huang et al. 2009). The meta-analysis method we introduce is flexible and may be used to combine studies conducted within these panels as well.
We evaluate our method through simulation and by applying it to real phenotype data for which previous discoveries have been made. First, we evaluate both power and resolution through a simulation framework. We find under many different settings that the meta-analysis approach results in higher power when compared to either single panel. We also find that when applying the meta-analysis approach, resolution is increased 1.5-fold with respect to the HMDP and 3.5-fold with respect to an F2 panel. Next, we apply the meta-analysis approach to map bone mineral density (BMD), which was measured from the femurs of 865 HMDP mice and 161 F2 mice, a cross between C57BL/6 and C3H (Farber et al. 2009, 2011). In our results, two previously implicated loci are recovered with increased significance. We also find that our method results in increased resolution over results obtained through linkage analysis. Finally, we apply our method to map HDL cholesterol in 687 HMDP mice and 164 F2 mice (van Nas et al. 2009) and find that a gene (Apoa2; Warden et al. 1993) known to be associated is identified with increased significance.
Materials and Methods
Association studies
Let us assume that we have measured a phenotype within a population i that contains ni individuals. We denote the ni × 1 column vector of phenotype measurements as . To test the association between the phenotype and a given SNP r, we test the hypothesis β = 0 under the model in Equation 1, where μ is the global phenotype mean and xi is a vector of minor allele counts of SNP r for individuals in population i:
| (1) |
A test statistic for testing β = 0 is derived by noting the distribution of the estimate of β under the assumption of normality. We denote the estimate of β in population i as , where and denotes the squared standard error of the estimate in population i. The z-score statistic for SNP r in population i, Zi, is given in Equation 2 and may be used to test the hypothesis β = 0 or may be used to derive other statistics, such as a chi-square or F-statistic:
| (2) |
Traditional Meta-analysis
Most traditional methods for meta-analysis employ the WSoZ approach (Willer et al. 2008; Zeggini et al. 2008; Soranzo et al. 2009). In this method, a meta-statistic for each SNP is calculated using Equation 3, where wi denotes a weight given to each Z-score for a population i. We note that our meta-statistic formulation in Equation 3 uses a different notation, with respect to standard meta-analysis literature, which represents the meta-statistic as a sum over effect sizes. However, both formulations are equivalent:
| (3) |
The weights, wi, are often a function of the sample size of their respective population, so that larger population samples obtain a higher weight (de Bakker et al. 2008). This weighting scheme make sense intuitively as we may want to attribute greater confidence to studies with more individuals. Alternatively, weights are set as the inverse of the standard error of the estimate of the β-coefficient, so that wi = 1/si. The resulting meta-statistic is the so-called pooled inverse variance-weighted β-coefficient (de Bakker et al. 2008). As has been done for case-control studies (Zaitlen and Eskin 2010), it is possible to show that this particular weighting scheme is optimal in the sense that these weights maximize the power of detecting an effect of size β.
Given the distribution of , we have that when β ≠ 0, Zmeta ∼ (λ,1), where λ is a noncentrality parameter with . λ is maximized, when wi = 1/si; thus the meta-statistic has optimal power to detect an effect of size of β. The optimality of the weight (wi = 1/si) is shown by using the Cauchy–Schwarz inequality
Under the assumption that β is the same across all populations, equality holds when wi = 1/si
Association studies in structured populations
Although the traditional approach to association mapping is often used, a number of issues arise when performing this basic analysis. One problem is that of population structure or cryptic relatedness (Devlin et al. 2001; Voight and Pritchard 2005), in which genetic similarities between individuals both inhibit the ability to identify true associations as well as cause the appearance of a large number of false or spurious associations. Mixed effects models are often used to correct this problem (Yu et al. 2006; Kang et al. 2008, 2010). Methods employing a mixed effects correction account for the genetic similarity between individuals with the introduction of a random variable into the traditional model from Equation 1:
| (4) |
In the model in Equation 4, the random variable ui represents the vector of genetic contributions to the phenotype for individuals in population i. This random variable is assumed to follow a normal distribution with , where Ki is the ni × ni kinship coefficient matrix for population i. With this assumption, the total variance of yi is given by . A z-score statistic is derived for the test β = 0 by noting the distribution of the estimate of . To avoid complicated notation, we introduce a more basic matrix form of the model in Equation 4:
| (5) |
In Equation 5, Xi is a ni × 2 matrix encoding the global mean and SNP vectors and Γ is a 2 × 1 coefficient vector. We note that this form also easily extends to models with multiple covariates. The maximum-likelihood estimate for Γ in population i is given by , which follows a normal distribution with a mean equal to the true Γ and variance . The z-score statistic for testing β = 0 is then given in Equation 7, where R = [0 1] is a vector used to select the appropriate entry in the vector :
| (6) |
| (7) |
| (8) |
Meta-analysis in structured populations
To perform meta-analysis using multiple structured populations, we adopt the weighted sum of z-scores approach shown in Equation 3, where the z-score for population i is given in Equation 7. When β ≠ 0, Zmeta will have a normal distribution with variance 1 and mean . Again we employ the use of the Cauchy–Schwarz inequality, shown in Equation 9, to show that the optimal weights are given by . We may also arrive at this result by noting that from Equation 7 is the mixed-model equivalent to s from section (traditional meta-analysis). However, this result is more general, allowing for a more flexible hypothesis-testing framework in which any linear combinations of the elements of Γ may be evaluated:
| (9) |
By substituting the optimal weights we arrive at the final meta-statistic given in Equation 10 with its distribution under the alternative hypothesis given in Equation 11:
| (10) |
| (11) |
It should be noted that when Σi is unknown, it must be estimated from the data. In this case, Zmeta may not follow a standard normal distribution under the null, due to the unaccounted uncertainty in the estimation of Σi. However, we are able to side step this issue by using a global search technique (Kang et al. 2008, 2010) to find an optimal estimate of Σi for each population.
Simulations
Simulations were performed using a previously designed framework (Kirby et al. 2010; Bennett et al. 2010). For both power and resolution, phenotypes were generated by sampling a phenotype for each strain while assuming the model from Equation 4. The genetic variance was determined for a given genetic background (g2) by using Equation 12, where S = In − 1/nJn (Jn is an n × n matrix of ones):
| (12) |
The power and resolution for each effect size (β) was determined by first applying the association mapping procedures to each simulated phenotype. Power was calculated as the percentage of associations for the known causal SNP that reached significance. For resolution, association was applied to each SNP on the same chromosome as the causal SNP. The distance between the true causal SNP and the peak associations were recorded. If the peak association is >15 Mb away from the causal SNP, then the value is recorded as 15 Mb. This procedure helps to reduce the mean shift that occurs because of low power within a region.
Significance threshold estimation
Significance thresholds were estimated for each method using a technique utilized previously (Bennett et al. 2010; Kirby et al. 2010). Ten thousand null phenotypes were generated and association statistics were calculated for each phenotype over all SNPs. We selected the minimum P-value for each phenotype, resulting in a set of 10,000 minimum null P-values. The threshold was chosen by selecting the P-value for which only 5% of the minimum P-values were smaller. This P-value then represents our threshold controlling for 5% FDR. Thresholds for the HMDP, F2, and meta-analysis approach were found as follows: 3.715 × 10−6, 2.4637 × 10−4, and 2.7 × 10−6.
Mouse association data
Genotypes for the F2 cross were obtained from a previous study (Estrada-Smith et al. 2004; Wang et al. 2006; van Nas et al. 2009; Farber et al. 2011). The original cross contained 311 mice, but we randomly sampled only 300 for our simulation studies. Each mouse was genotyped at about 1200 markers spread across the genome and it was this set of markers that was used previously to perform linkage analysis. To apply the meta-analysis approach outlined in this article, we require that the F2 mice be typed at the same markers as the HMDP. Fortunately, since the parental strains for the F2s are part of the HMDP, genotyping is not necessary. Instead we perform imputation to determine the state of each marker, which is typed in the HMDP but is not part of the markers typed in the F2 cross. By applying the imputation algorithm described below, we obtained a set of 113,650 SNPs that were polymorphic in both the HMDP and the F2 cross. This is compared to the total set of markers available for the HMDP, which is of size 132,285.
We utilize a straightforward approach to imputation by noting the simple structure of the F2 genomes. For any two adjacent markers in a given F2 mouse, the state of the intervening markers will be determined by the state of the two adjacent markers. Let two adjacent markers be xi and xi+k, where k is the number of intervening markers. If both xi and xi+k are in the same state as parent 1, then the markers from xi+1 to xi+k−1 will be set to be the same as the corresponding markers in parent 1. Likewise, if both xi and xi+k share the same state as parent 2, the intervening markers will be set to those from parent 2. If there is a switch in state between the two adjacent markers, this indicates a recombination. In this case, we are not able to determine the state of the intervening markers and these will be labeled as unknown. This process assumes that the probability of a double recombination occurring between genotyped markers is close to zero.
The genotypes and phenotypes utilized in this article have been made available online at http://genetics.cs.ucla.edu/mousemeta/.
Results
Combining the HMDP with an F2 cross increases power
We show that by combining the mapping results obtained in the HMDP with those obtained in an F2 cross through meta-analysis, we achieve higher power than when mapping within only one panel. Simulations are performed with genotypes for 300 F2 mice, which were obtained from a previously generated cross (Estrada-Smith et al. 2004; Wang et al. 2006; van Nas et al. 2009). The F2s were genotyped at about 1200 markers and imputation was performed (see Materials and Methods) to obtain genotypes at all markers typed in the HMDP strains.
Power simulations were performed as described in previous studies (Bennett et al. 2010; Kirby et al. 2010). We randomly selected a set of 10,000 SNPs that are polymorphic in both the F2 cross and the HMDP. For each SNP we generated a phenotype with a 25% genetic background effect and a SNP effect of a given size. The genetic background effect can be thought of as the heritability of the trait. Association between each SNP and its corresponding set of generated phenotypes was tested using the efficient mixed-model association (EMMA) method (Kang et al. 2008) for the F2 and HMDP panels alone. Power for each SNP effect size was calculated as the percentage of tests that resulted in a significant P-value. Significance thresholds for each panel were obtained through a parametric bootstrap procedure (see Materials and Methods).
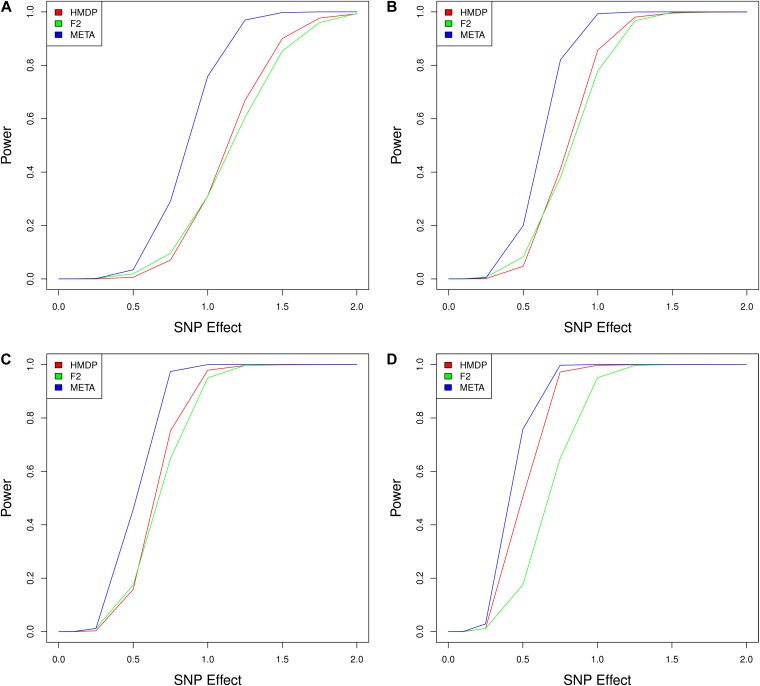
Figure 1 shows the comparison of power between the meta-analysis approach and mapping within the individual panels. In these simulations, we varied both the number of F2 mice and the number of HMDP replicates. Power is reported on the y-axis and the magnitude of the SNP effect is reported on the x-axis. The SNP effect is reported in terms of β from Equation 4 and the actual variance explained for a given value will depend on the SNP as well as the genetic background. Therefore, we determine the variance explained by a given effect size under a given genetic background by taking the average variance explained in the HMDP across all SNPs. The meta-analysis method has higher power than mapping within the single populations in all simulations. As power within each of the single populations increases, so does the power of the meta-analysis method. For a large number of F2 mice and HMDP mice, the power to detect small effects increases dramatically by applying meta-analysis. For example, for a SNP effect accounting for 5% of the phenotypic variance (β = 0.5), we find that mapping within only the HMDP with five replicates results in a power of 50%, while mapping within only the F2 cross results in a power of 17%. When combining the results through meta-analysis, the power increases to 75% (Figure 1D).
Figure 1 .
Mapping power is increased by combining populations using meta-analysis. (A) 100 HMDP, 100 F2s. (B) 200 HMDP, 200 F2s. (C) 300 HMDP, 300 F2s. (D) 500 HMDP, 300 F2s. We performed simulations assuming a background genetic effect of 25%. Power was calculated as the percentage of associations detected at a given level of significance for SNPs simulated to be causal. The meta-analysis method is shown to provide increased power at all effect sizes.
Meta-analysis leads to an increase in resolution over HMDP and F2 Mapping
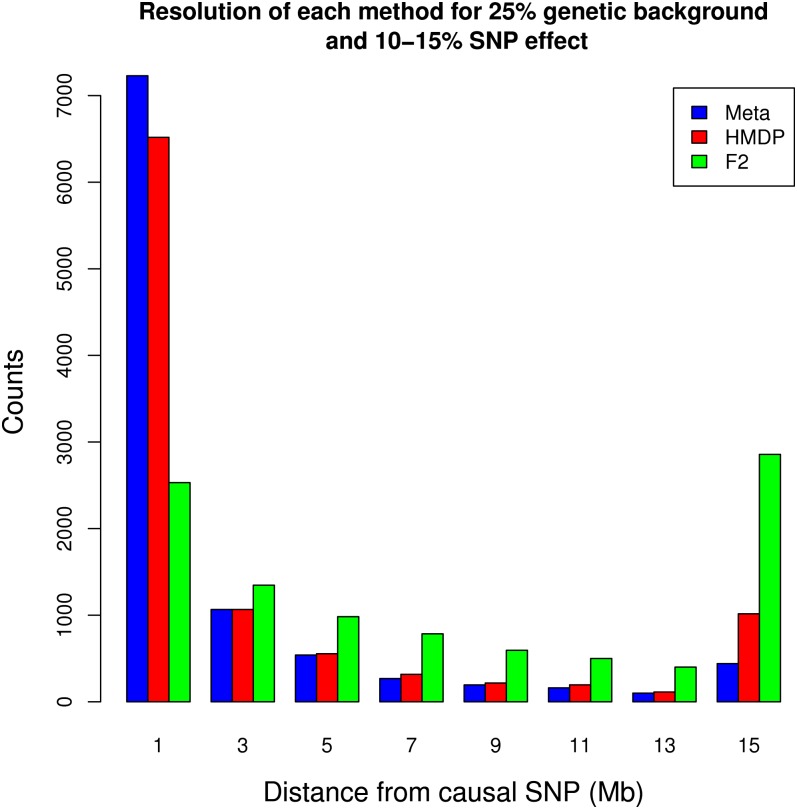
We evaluate the mapping resolution when using the HMDP, F2, and the meta-analysis approaches through simulation. Resolution was evaluated by calculating the genetic distance between a SNP simulated to be causal and the peak-associated SNP, while only considering the region within 15 Mb of the causal SNP. Figure 2 compares the distribution of these distances under each mapping method. Simulations were performed assuming a 25% genetic background effect and a SNP effect accounting for 10–15% of the phenotypic variance.
Figure 2 .
The combined mapping result has higher resolution than that of the HMDP or F2 mapping. The distribution of distances from the true causal SNP to the most significant association are shown in units of megabases. We considered peak associations that are within 15 Mb of the true causal SNP. As expected, the HMDP has much higher resolution than the F2 cross. The combined result achieves an even higher resolution.
Using one replicate for the HMDP, we find that the mean distance of the peak association to the true causal SNP is 3.17 Mb. This compares with a mean of 7.5 Mb obtained when mapping within the F2 panel. When combining results through the meta-analysis approach, the mean distance is decreased to 2.21 Mb. This is an almost 1.5-fold increase in resolution over the HMDP and an almost 3.5-fold increase in resolution over the F2 panel.
Application to bone mineral density
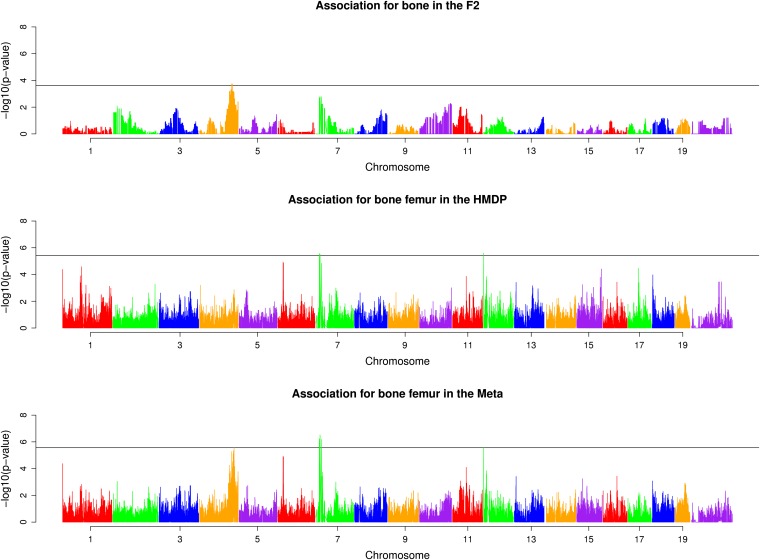
We obtained a set of BMD measurements from the femurs of 865 HMDP mice and 161 male F2 mice. We applied association mapping in each panel separately using EMMA (Kang et al. 2008) and applied the meta-analysis approach as well. Manhattan plots summarizing these results are shown in Figure 3. Two loci (chromosomes 4 and 7) showed an increase in significance relative to the associations in either the F2 or HMDP. The significance of the chromosome 7 meta-analysis peak was an order of magnitude more significant (3 × 10−7) than either the HMDP (3.1 × 10−6) or the F2 (1.6 × 10−3) peaks. The original QTL on chromosome 7 (Bmd41) had a 1.5 LOD support interval of 80 Mb (24.9–104.9 Mb) (Farber et al. 2009). We approximate the associated region obtained via meta-analysis by employing a simple approach. We define the associated region as that surrounding the peak SNP and containing SNPs with P-values ≤ 10−6. Thus defined, the chromosome 7 meta-analysis interval extending from 17.2 to 25.2 Mb is much smaller than the previously obtained support interval. This result indicates an increase in resolution for Bmd41.
Figure 3 .
Meta-analysis results in increased significance and increased resolution for two loci known to be associated with BMD. Two loci, one on chromosome 4 and one on chromosome 7, were previously found to be associated with BMD (Bmd7 and Bmd41, respectively) (Farber et al. 2009). After applying meta-analysis, we found that the peak associated SNPs for both of these loci had increased significance with respect to the F2 and HMDP mapping panels. Thresholds for significance are indicated by the horizontal black bars in each plot.
The QTL on chromosome 4, previously referred to as Bmd7 (Farber et al. 2009), was the strongest locus affecting femoral BMD in the F2 (P = 7.8 × 10−4). The peak F2 SNP was moderately significant in the HMDP (1.3 × 10−3) and highly significant in the meta-analysis (2.8 × 10−6). Bmd7 was previously found to have a 1.5 LOD support interval of 11.0 Mb (126.2–137.2 Mb). In the meta-analysis SNPs with P-values of ≤ 10−6 spanned 10 Mb (from 129 to 139 Mb).
Application to HDL cholesterol
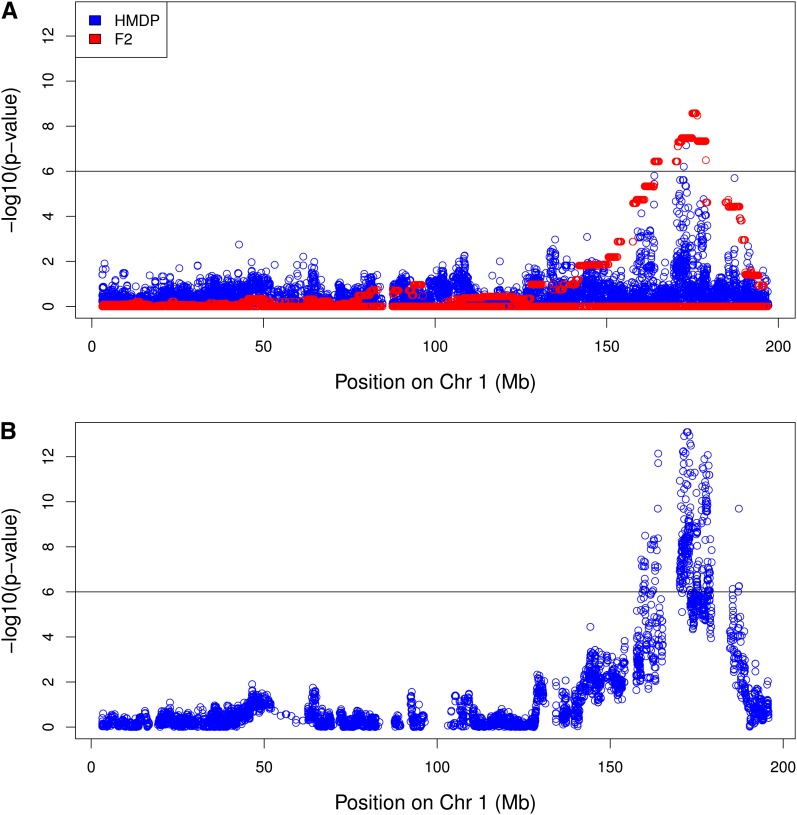
We obtained a set of HDL measurements for 687 male mice each a member of the HMDP and a set of 164 male F2s (van Nas et al. 2009). We applied association mapping in the HMDP and F2 panels separately using EMMA (Kang et al. 2008) and then applied our meta-analysis approach. Figure 4 shows the results of this experiment. As shown in the original article introducing the HMDP, the peak association for HDL is found on distal chromosome one, in which a well-known association with the Apoa2 (Doolittle et al. 1990) gene exists. The peak association is 25 kb upstream of the start site of the Apoa2 gene with a P-value of 7.06 × 10−8, which is significant at the 1 × 10−7 level estimated from a parametric bootstrap procedure. The mapping results obtained from the F2 panel (Figure 4A) resemble a linkage peak, due to the large amount of linkage disequilibrium within the F2 genomes. The peak association identified in the F2 population is over 2Mb downstream of the end site for the Apoa2 locus with a P-value of 2.67 × 10−09. Figure 4B shows the mapping result obtained with the meta-analysis procedure. Using the meta-analysis result, we again obtain the association, which is 25 kb from the start site of the gene; however, the P-value is greatly reduced to <1 × 10−15.
Figure 4 .
Meta-analysis increases significance of known association. We compare the (A) association mapping results obtained from the HMDP, F2 cross and (B) meta-analysis for HDL cholesterol on chromosome 1. It was previously shown that the Apoa2 gene underlies the chromosome 1 HDL locus (Warden et al. 1993). The peak association in the HMDP result is 25 kb away from the start site of the gene with a P-value of 7.06 × 10−08, while the peak association in the F2 is 2 Mb downstream of the start site (P-value 2.67 × 10−09). After applying the meta-analysis method, we recover the same association identified in the HMDP with a P-value 2.36 × 10−15. The horizontal black bar is placed at −log10(p) = 6 for reference.
Discussion
In this article, we introduce a study design in which the HMDP inbred panel is combined with an F2 cross to perform association mapping. We show that by utilizing a meta-analysis approach that accounts for the genetic structure of the populations, both association power and resolution are increased when compared with mapping within either of the individual panels. The reason for increased power can be understood intuitively as, in general, increased sample sizes lead to increases in power. However, an increase in resolution when combining a high-resolution panel with a low-resolution panel is somewhat counterintuitive. One way to understand why we achieve higher resolution is by considering that by combining panels we are increasing the number of overall unique genomic break points.
Our results have focused on the case when the HMDP panel is combined with one F2 cross. However, by using the methodology we present, any number of panels can be combined. One obvious potential for this is that by adding additional F2 panels, we may increase power much further. A significant amount of cross data exist in publicly accessible databases such as MGI (Blake et al. 2011). By utilizing existing cross data, researchers will be able to use our technique to increase the power of their studies without spending money to generate F2s of their own.
Another advantage of our method is that it is general enough to be used to combine the HMDP with other types of study designs such as the Collaborative Cross (Aylor et al. 2011) and heterogeneous stock (Huang et al. 2009). However, one potential issue that may arise when combining the HMDP with such panels is that of heterogeneity of effect size. That is, the magnitude of main effects may vary between different mapping panels due to the difference in the overall genetic structure. In this case, our method may be easily extended to utilize approaches that account for such heterogeneity between effects (Han and Eskin 2011). Heterogeneity between effect sizes is also known to be a problem between sexes within the same population. Therefore, a similar approach may be utilized to combine results across sexes within the same mapping panel.
Footnotes
Communicating editor: D. W. Threadgill
Literature Cited
- Aylor D. L., Valdar W., Foulds-Mathes W., Buus R. J., Verdugo R. A., et al. , 2011. Genetic analysis of complex traits in the emerging collaborative cross. Genome Res. 21(8): 1213–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett B. J., Farber C. R., Orozco L., Kang H. M., Ghazalpour A., et al. , 2010. A high-resolution association mapping panel for the dissection of complex traits in mice. Genome Res. 20: 281–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake J., Bult C., Kadin J., Richardson J., Eppig J., 2011. The mouse genome database (mgd): premier model organism resource for mammalian genomics and genetics. Nucleic Acids Res. 39: D842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bakker P., Ferreira M., Jia X., Neale B., Raychaudhuri S., et al. , 2008. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum. Mol. Genet. 17: R122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin B., Roeder K., Bacanu S., 2001. Unbiased methods for population-based association studies. Genet. Epidemiol. 21: 273–284 [DOI] [PubMed] [Google Scholar]
- Doolittle M., LeBoeuf R., Warden C., Bee L., Lusis A., 1990. A polymorphism affecting apolipoprotein a-ii translational efficiency determines high density lipoprotein size and composition. J. Biol. Chem. 265: 16380. [PubMed] [Google Scholar]
- Estrada-Smith D., Castellani L., Wong H., Wen P., Chui A., et al. , 2004. Dissection of multigenic obesity traits in congenic mouse strains. Mamm. Genome 15: 14–22 [DOI] [PubMed] [Google Scholar]
- Farber C. R., van Nas A., Ghazalpour A., Aten J. E., Doss S., et al. , 2009. An integrative genetics approach to identify candidate genes regulating bmd: combining linkage, gene expression, and association. J. Bone Miner. Res. 24: 105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber C. R., Bennett B. J., Orozco L., Zou W., Lira A., et al. , 2011. Mouse genome-wide association and systems genetics identify asxl2 as a regulator of bone mineral density and osteoclastogenesis. PLoS Genet. 7: e1002038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J., Mott R., 2001. Finding the molecular basis of quantitative traits: successes and pitfalls. Nat. Rev. Genet. 2: 437–445 [DOI] [PubMed] [Google Scholar]
- Han B., Eskin E., 2011. Random-effects model aimed at discovering associations in meta-analysis of genome-wide association studies. Am. J. Hum. Genet. 88: 586–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitzemann R., Demarest K., Koyner J., Cipp L., Patel N., et al. , 2000. Effect of genetic cross on the detection of quantitative trait loci and a novel approach to mapping qtls. Pharmacol. Biochem. Behav. 67: 767–772 [DOI] [PubMed] [Google Scholar]
- Hitzemann R., Malmanger B., Cooper S., Coulombe S., Reed C., et al. , 2002. Multiple cross mapping (mcm) markedly improves the localization of a qtl for ethanol-induced activation. Genes Brain Behav. 1: 214–222 [DOI] [PubMed] [Google Scholar]
- Huang G.-J. J., Shifman S., Valdar W., Johannesson M., Yalcin B., et al. , 2009. High resolution mapping of expression QTLs in heterogeneous stock mice in multiple tissues. Genome Res. 19: 1133–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H., Zaitlen N., Wade C., Kirby A., Heckerman D., et al. , 2008. Efficient control of population structure in model organism association mapping. Genetics 178: 1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H., Sul J., Service S., Zaitlen N., Kong S., et al. , 2010. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 42(4): 348–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby A., Kang H. M., Wade C. M., Cotsapas C. J., Kostem E., et al. , 2010. Fine mapping in 94 inbred mouse strains using a high-density haplotype resource. Genetics 185: 1081–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Lyons M. A., Wittenburg H., Paigen B., Churchill G. A., 2005. Combining data from multiple inbred line crosses improves the power and resolution of quantitative trait loci mapping. Genetics 169: 1699–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manenti G., Galvan A., Pettinicchio A., Trincucci G., Spada E., et al. , 2009. Mouse genome-wide association mapping needs linkage analysis to avoid false-positive loci. PLoS Genet. 5: e1000331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payseur B., Place Michael, 2007. Prospects for association mapping in classical inbred mouse strains. Genetics 175: 1999–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirce J., Broman K., Lu L., Williams R., 2007. A simple method for combining genetic mapping data from multiple crosses and experimental designs. PLoS ONE 2: e1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletcher M. T., McClurg P., Batalov S., Su A. I., Barnes S. W., et al. , 2004. Use of a dense single nucleotide polymorphism map for in silico mapping in the mouse. PLoS Biol. 2: e393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soranzo N., Rivadeneira F., Chinappen-Horsley U., Malkina I., Richards J., et al. , 2009. Meta-analysis of genome-wide scans for human adult stature identifies novel loci and associations with measures of skeletal frame size. PLoS Genet. 5: e1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nas A., Guhathakurta D., Wang S. S., Yehya N., Horvath S., et al. , 2009. Elucidating the role of gonadal hormones in sexually dimorphic gene coexpression networks. Endocrinology 150: 1235–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight B., Pritchard J., 2005. Confounding from cryptic relatedness in case-control association studies. PLoS Genet. 1: e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Yehya N., Schadt E., Wang H., Drake T., et al. , 2006. Genetic and genomic analysis of a fat mass trait with complex inheritance reveals marked sex specificity. PLoS Genet. 2: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warden C., Hedrick C., Qiao J., Castellani L., Lusis A., 1993. Atherosclerosis in transgenic mice overexpressing apolipoprotein A-II. Science 262(5131): 164. [DOI] [PubMed] [Google Scholar]
- Willer C., Speliotes E., Loos R., Li S., Lindgren C., et al. , 2008. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat. Genet. 41: 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Pressoir G., Briggs W., Vroh Bi I., Yamasaki M., et al. , 2006. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 38: 203–208 [DOI] [PubMed] [Google Scholar]
- Zaitlen N., Eskin E., 2010. Imputation aware meta-analysis of genome-wide association studies. Genet. Epidemiol. 34(6): 537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeggini E., Scott L., Saxena R., Voight B., Marchini J., et al. , 2008. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat. Genet. 40: 638–645 [DOI] [PMC free article] [PubMed] [Google Scholar]