Abstract
Immunosenescence, the age-related decline in immune system function, is a general hallmark of aging. While much is known about the cellular and physiological changes that accompany immunosenescence, we know little about the genetic influences on this phenomenon. In this study we combined age-specific measurements of bacterial clearance ability following infection with whole-genome measurements of the transcriptional response to infection and wounding to identify genes that contribute to the natural variation in immunosenescence, using Drosophila melanogaster as a model system. Twenty inbred lines derived from nature were measured for their ability to clear an Escherichia coli infection at 1 and 4 weeks of age. We used microarrays to simultaneously determine genome-wide expression profiles in infected and wounded flies at each age for 12 of these lines. Lines exhibited significant genetically based variation in bacterial clearance at both ages; however, the genetic basis of this variation changed dramatically with age. Variation in gene expression was significantly correlated with bacterial clearance ability only in the older age group. At 4 weeks of age variation in the expression of 247 genes following infection was associated with genetic variation in bacterial clearance. Functional annotation analyses implicate genes involved in energy metabolism including those in the insulin signaling/TOR pathway as having significant associations with bacterial clearance in older individuals. Given the evolutionary conservation of the genes involved in energy metabolism, our results could have important implications for understanding immunosenescence in other organisms, including humans.
Keywords: genetic variation, bacterial clearance, wound response, aging, immunosenescence
UNDERSTANDING the genetic architecture of senescence, the physiological decline that occurs with increasing age, is a long-standing problem in biology. Most studies in this area have focused on elucidating the genetic basis of heritable variation in age-specific mortality and longevity (Dudycha and Tessier 1999; Guarente and Kenyon 2000; Wilson et al. 2007, 2008; Blomquist 2010; Fontana et al. 2010) and several candidate genes contributing to this variation have been identified (De Luca et al. 2003; Burger and Promislow 2006; Paaby and Schmidt 2008; Jazwinski et al. 2010; Soerensen et al. 2010). In contrast, we know little about the genes that influence age-related functional declines in life history or physiological traits that may in turn affect age-specific mortality and limit life span (Burger and Promislow 2006; Charmantier et al. 2006; Leips et al. 2006; Lesser et al. 2006; Nussey et al. 2008a). Functional decline in such traits with age can reduce fitness in natural environments (Dudycha and Tessier 1999; Wilson et al. 2007; Bowler and Terblanche 2008; Nussey et al. 2008b; Wilson et al. 2008; Blomquist 2010; Ceballos and Kiørboe 2011) and for humans influences the quality of life in older individuals. As such, knowledge of the genes that influence age-related deterioration in functional traits is important for understanding the genetic basis of evolutionary fitness as well as general aspects of health in the aging organism. In addition, this information would provide insight into the genetic interrelationships between these traits and longevity and increase our understanding of correlated evolutionary responses among them (Burger and Promislow 2006).
Immunosenescence, the age-related decline in immune function, is a general consequence of aging (Kaaya and Darji 1988; Kogut et al. 2002; Laws et al. 2004; Hillyer et al. 2005; Zerofsky et al. 2005; Pawelec 2006; Kovacs et al. 2009; Leips 2009; Scholz et al. 2009). Early studies of immunosenescence primarily focused on deterioration of the adaptive immune response with age, that component of the immune response that provides immunological memory and is uniquely found in vertebrates. However, an increasing number of studies indicate that the innate immune response, the nonspecific response to infection, also shows signs of deterioration with age (Butcher et al. 2001; Grewe 2001; Fulop et al. 2004; Mocchegiani and Malavolta 2004; Plackett et al. 2004; Ramsden et al. 2008; Panda et al. 2009; Agarwal and Busse 2010; Shaw et al. 2010). In humans, age-related declines in innate immune components include reduced efficacy of phagocytosis (Agaisse 2008), impaired killing efficiency of neutrophils (Knight et al. 2001; Fulop et al. 2004), compromised macrophage function (Sambhara et al. 2004), and impaired production of cytokines and chemokines by natural killer cells (Kovacs et al. 2004; Mocchegiani and Malavolta 2004) and dendritic cells (Agrawal et al. 2007, 2011; Ramsden et al. 2008). Given that the innate immune response also regulates aspects of adaptive immunity, a better understanding of how age affects the innate immune response could be useful for understanding immunosenescence in adaptive immunity.
While immunosenescence is ubiquitous, studies in mice, flies, and humans have found that the effect of age on the immune response varies among individuals and this variation has a genetic component (Pinti et al. 2002; Ponnappan et al. 1992; Hsu et al. 2003; Jackson et al. 2003; Cipriano et al. 2005; Lesser et al. 2006). One of the striking findings of these studies is that genetic influences on immune responses have age-specific effects; that is, alleles that produce variation in the immune response in younger animals appear to have different or no detectable effects on immunocompetence at later ages. Thus, the genetic architecture of immune function appears to change with age, possibly involving different physiological systems and regulatory genetic networks. The idea that alleles with age-specific effects contribute to the natural variation and evolution of senescence is a fundamental assumption of life-history theory and evolutionary theories of aging. Nevertheless, the age-specific effects of natural alleles on phenotypes are rarely studied (Hughes 2010; but see Cho et al. 2010). As a result, we know little about how common these effects are, how they are regulated at the genetic level, or what genes or pathways produce age-related phenotypic changes.
Here we characterize the genetic basis of natural variation in age-related changes in the immune response to infection, combining measures of the age-specific ability to clear bacterial infection with genome-wide transcriptional responses to both wounding and infection, using Drosophila melanogaster as a model system. In this study we address three general questions. First, what is the pattern of genetic variation in clearance of infection across ages and does the effect of age on this trait vary among genotypes? Variation among lines in age-specific immune response would imply that different genotypes vary in the rate or degree of immunosenescence. Second, to what extent does genetic variation in age-specific clearance ability arise from differences among lines in their transcriptional response to infection as they age? And third, if variation in expression is associated with variation in bacterial clearance, what are the genes/pathways involved and do the same genes influence clearance ability at different ages?
To address these questions we measured the ability of a set of inbred lines of flies derived from a natural population to clear Escherichia coli infection at two ages, 1 and 4 weeks of age, using a standard bacterial injection assay (McKean and Nunney 2001; Lesser et al. 2006). The advantage of using natural strains as opposed to using inbred laboratory or mutant strains is that our results will capture the effects of alleles that actually exist in nature. Thus our results are relevant for understanding the genetic basis of natural variation in age-specific bacterial clearance and the evolutionary potential of this trait. We simultaneously tested for an association between variation in the ability to clear infection and variation in gene expression across the genome to identify candidate genes affecting clearance ability at each age. We found significant genetic variation in age-specific bacterial clearance ability at each age and variation in gene expression that was associated with this trait. Notably variation in expression was significantly associated with clearance ability only at the older age. Our study suggests that variation in the expression of genes involved in energy metabolism is a significant contributor to natural variation in the immune response to wounding and infection at a later age. We discuss the likely role of these genes in immune function, including their potential to influence variation in tolerance to infection (Schneider and Ayres 2008). We also highlight candidate genes that may serve as potential targets for maintaining the functional integrity of the immune response in the aging organism.
Materials and Methods
Drosophila lines
Twenty inbred lines derived from a natural population in Raleigh, North Carolina (kindly provided by the laboratory of Trudy F. C. Mackay, North Carolina State University) were used in the bacterial clearance assay. Individuals from 12 of these lines were used in the microarray study. In all experiments, virgin females were collected from each line and maintained in vials containing cornmeal–molasses–yeast–agar medium (without live yeast) under standard conditions (25° and 12-hr light:12-hr dark cycles) until assayed.
Immune response assay
For this assay, we adapted the procedure from McKean and Nunney (2001; Lesser et al. 2006) to measure the ability of different lines to clear a bacterial infection when flies were 1 and 4 weeks old (1 and 4 weeks posteclosion). The experiment was carried out in five temporal blocks. To reduce the potential for bias in infection load within each block (e.g., due to differences in needle bore size and variation in infection load injected due to needle wear over time) we used a randomized design to determine the order in which individuals from each line were injected. With this design, any variation in initial bacterial load injected into each fly should reduce our ability to detect differences in clearance ability among genotypes and reduce our ability to detect correlations between infection level and transcript abundance. Prior to injection, the bacterial culture was grown in LB broth plus streptomycin (100 µg/ml) from a single colony of a streptomycin-resistant strain of E. coli until OD600 reached 1.0 ± 0.1 [Beckman (Fullerton, CA) DU-65 Spectrophotometer], corresponding to a final concentration of ∼5.5 × 108 colony-forming units/ml. The bacterial solution was then loaded into pulled-glass capillary needles. Bacteria-filled capillary needles were connected to an Eppendorf (Westbury, NY) Femtojet microinjector and flies were injected with the same volume of E. coli in the abdomen (microinjector settings: injection pressure = 0.46, injection time = 0.2 sec, and compensation pressure = 0.20). Twenty-four hours postinjection, all living flies were homogenized in 200 µl of Ringer’s solution. Fifty microliters of the homogenate was plated on LB/agar plates containing streptomycin (100 µg/ml). The resulting colonies were counted and used as a measure of bacterial clearance ability. In this assay flies with high colony counts have poor bacterial clearance ability whereas flies with a lower colony count have better clearance ability.
Flies from each line were also sham injected with sterile Ringer’s solution. The sham-injected control served three purposes: The first was to ensure that these lines were not carrying a naturally occurring streptomycin-resistant bacterial strain. Second, it served as a control for the transcriptional response to wounding for the microarray component of this experiment (we could separate the transcriptional response to wounding from that of infection with E. coli). Third, this allowed us to explore how the transcriptional response to wounding alone contributed to the ability to clear infection.
Quantitative genetic analyses
We performed two analyses to examine the genetic basis of variation in age-specific bacterial clearance. First, a mixed-model analysis of variance was used to test the model, y = c + age + line + age × line + block +ε, where y is colony count 24 hr after infection, c is constant, the fixed-effect age tested for overall differences in clearance ability between the two ages, the line effect (random) tested for genetically based differences among lines in clearance ability, and the age × line interaction effect (random) tested for differences among lines in the effect of age on clearance ability. The second analysis tested for genetic differences in clearance ability among lines at each age separately. In these analyses we used a random-effects model, y = c + line + block + ε, where y is the colony count for each individual 24 hr postinfection, c is a constant, and line is the random effect of line. Colony counts were transformed to the natural log to satisfy assumptions of analysis of variance (ANOVA) (Zar 1999). The line effects and residual variance components from each model were used to calculate heritability (broad sense), genetic correlations, and the coefficients of genetic and residual variation (below). Unless otherwise stated, all statistical procedures were performed in SAS version 9.1.
Broad sense heritability (H2) was calculated at each age [σ2L/(σ2L + σ2E)], using the variance components from the above random-effects model, where σ2L is the variance due to the line effect and σ2E is the residual variance. We also calculated the coefficient of genetic variation (CVG) for young and old flies as 100(√σ2L)/mean (Houle 1992), where σ2L, the among-line variance component, was standardized by the average colony count for all lines at each age. H2 and CVG were used to estimate the proportion of phenotypic variation explained by genetic differences among the lines. The coefficient of residual variation (CVE) for young and old flies was calculated for each age as 100(√σ2E)/mean, where σ2E, the residual variance, was standardized by the average colony count for all lines at each age. CVE estimated the proportion of phenotypic variation that could not be explained by genetic differences among lines (Hill and Mulder 2010).
The genetic correlation (rGA) was calculated as σ2L1,4/√(σ2L1σ2L4) (Robertson 1959), where σ2L1,4 is the among-line variance component when data are combined across ages, σ2L1 is the among-line variance component at 1 week, and σ2L4 is the among-line variance component at 4 weeks.
Microarray sample preparation
During the immune response assay, a subset of the bacterially infected and sham-injected flies from each of the lines was frozen at −80° 6 hr postinjection for use in the microarray study. From this set of 20 lines we chose 12 for our microarray study after we had obtained data on the age-specific clearance ability. Our clearance assay indicated a difference among lines in the way that age influenced bacterial clearance and so we selected lines for the microarray study on the basis of an altered bacterial clearance response at the older age. In this case all of the expression data came from RNA extracted from individuals from each line (we pooled the RNA from 8–15 individuals per line) at each age from each treatment (sham injected vs. infected). Because this experimental design precludes our ability to assess expression variation within lines (we had only one sample of RNA for each of the 12 lines), we could not analyze genetic variation in gene expression among lines. Instead our level of replication comes from the variation in expression of the same gene among the lines. We chose this approach to maximize our sample of genotypes (and to potentially maximize transcriptional variation) to increase our ability to identify genes whose variation in expression was associated with variation in bacterial clearance ability.
Flies were frozen 6 hr postinjection because known genes involved in the immune response (phagocytosis and antimicrobial peptides) are typically expressed at high levels by this time (De Gregorio et al. 2001; Irving et al. 2001; Vodovar et al. 2005). This experimental design then asks, To what extent does variation in gene expression at 6 hr following wounding or infection predict the bacterial clearance 24 hr after infection? Total RNA was extracted using the TRIzol extraction method, followed by an RNeasy cleanup. Both steps were implemented according to the manufacturer’s instructions (Invitrogen/QIAGEN). RNA was labeled using a Message-Amp II kit (Ambion). Labeled RNA was hybridized to Drosophila Genome 2.0 arrays (Affymetrix). In the end, there were a total of 46 chips in this study (12 lines × 2 treatments × 2 ages for a total of 48 chips, minus 2 chips that were excluded due to poor hybridization).
Microarray data analyses
Raw expression values were log2 transformed and normalized using the GCRMA statistical package available through Bioconductor (Irizarry et al. 2003; Wu and Irizarry 2004). To identify transcripts associated with differences across age and infection status, we used a gene-specific linear model with expression level as the dependent variable and age, infection status, and age-by-infection as fixed independent variables. Gene-specific variance component estimates were adjusted using an empirical Bayes approach to improve variance estimates (Feng et al. 2006). We calculated the false discovery rate (FDR) using the Benjamini and Hochberg method (Benjamini and Hochberg 1995). All candidate genes were annotated using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) (Huang et al. 2009a,b); Flymine (Lyne et al. 2007); and FlyBase (Drysdale and Crosby 2005).
Spearman’s rank tests were performed for all transcripts (genes) represented on the microarray to identify genes whose variation in expression was associated with genetic variation in clearance ability at each age. The correlation was between the average colony count of each line at each age and the expression level of each gene by each line at each age. We carried out this analysis for both sham-injected (wounded) and E. coli-infected flies. We used this conservative nonparametric test because it makes no assumptions about normality of the data, which is difficult to assess with this small number of lines. FDR was implemented to correct for multiple testing at a significance level of 0.05. For candidate genes identified in this study (effect of age, wounding, and infection), the molecular function of Gene Ontology (GO) information was used to group candidate genes into functional categories. When GO analysis (by DAVID or Flymine) identified no functional categories, we assigned categories manually when five or more genes had similar annotation terms according to FlyBase (Tweedie et al. 2009).
Results
The effect of age on bacterial clearance ability differs among lines
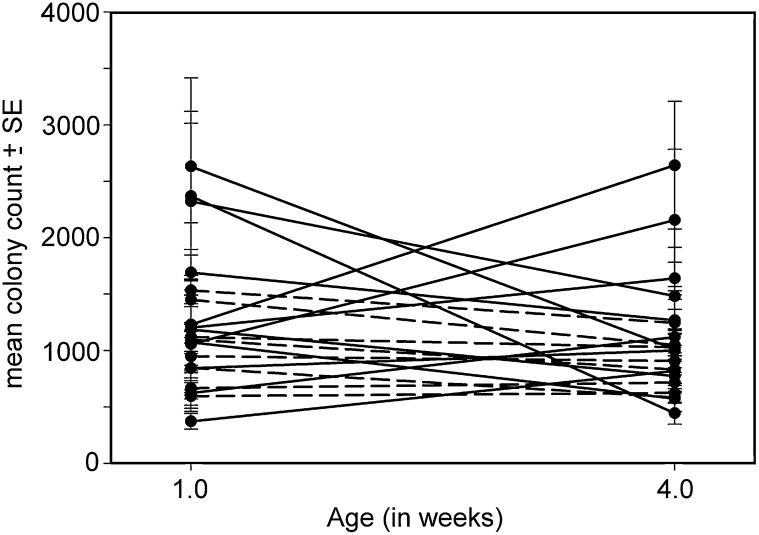
We first tested for the effects of age, line, and line × age interaction on clearance ability, using a mixed-model ANOVA. Neither the main effects of age (F1,19 = 3.84, P = 0.06) nor those of line were significant (χ21 = 2.2, P = 0.14); however, there was a significant line × age interaction (χ21 = 6.9, P < 0.01), indicating that the effect of age on bacterial clearance ability differed among genotypes. This was also evident in the patterns of age-specific clearance ability among lines, with some lines declining with age, others showing no effect of age, and some lines improving with age (Figure 1). The genetic correlation in clearance ability across ages was moderate (rGA = 0.47, Table 1).
Figure 1 .
Mean colony counts (±SE) for the 20 lines screened for ability to clear E. coli infection at 1 and 4 weeks of age. The solid lines indicate the lines used in the microarray study. Note that a high colony count represents poor bacterial clearance ability whereas a lower colony counts represents better clearance ability.
Table 1 . Statistical genetic analyses for clearance ability at each age for the 20 wild-derived inbred lines.
| Age | Mean colony count ± SE (CFU) | H2 (%) | CVG | CVE | rGA |
|---|---|---|---|---|---|
| 1 wk | 1189 ± 85 | 10 | 39.7 | 130 | 0.47 |
| 4 wk | 1077 ± 65 | 16 | 43.4 | 101 |
H2, broad-sense heritability; CVG, coefficient of genetic variation; CVE, coefficient of residual variation; rGA, genetic correlation between clearance ability at 1 week and 4 weeks of age.
Genetic variation in clearance ability increases from 1 to 4 weeks of age
Because of the significant line × age interaction, we tested for genetic differences in clearance ability among lines within each age in separate analyses, using a random-effects model. We found significant variation in clearance ability among lines at both ages (week 1, χ21 = 19, P < 0.0001; week 4, χ21 = 27.9, P < 0.0001). We estimated broad-sense heritability, H2, and the coefficient of genetic variation, CVG, of bacterial clearance ability to explore how genetic differences among lines contributed to the phenotypic variation in this trait at each age. Both of these estimates increased with age (Table 1), indicating that genetic differences among the lines explained a larger proportion of the phenotypic variation in bacterial clearance in older flies (Table 1). This result is consistent with our earlier study (Lesser et al. 2006).
General transcriptional changes with age
We first looked for general changes in transcript levels across the two ages, using the combined data from both wounded and infected flies. We found that 1166 transcripts exhibited significant changes in expression with age, 588 transcripts were upregulated, and 578 transcripts were downregulated with age (see supporting information, Table S1 for a complete list of genes). Genes upregulated with age were enriched for immunity-related gene ontology classes (Table 2) that included Relish, Thiolester containing protein (Tep) IV, and Eiger, as well as genes encoding antimicrobial peptides (AMPs) (e.g., Attacin-A, -B, and -D; Cecropin B and C; and Diptericin B) and peptidoglycan receptor proteins (e.g., PGRP-SD, PGRP-LF, and PGRP-SB1). Genes with peptidase activity were also overrepresented [e.g., Death associated molecule related to Mch2 (Damm), Matrix metalloproteinase1, Death executioner caspase related to Apopain/Yama (decay), and CG17571], as were solute transporter genes (e.g., Na[+]-dependent inorganic phosphate cotransporter, Sodium-dependent multivitamin transporter, CG7881, and CG31547) and transmembrane proteins (e.g., Epithelial membrane protein, Moody, Methuselah-like 8, Rhodopsin 4, and CG2930).
Table 2 . List of overrepresented functional groups (classified by gene ontology) within candidate genes that upregulated with age.
| GO term | No. genes | Benjamini P |
|---|---|---|
| Biological process | ||
| Immune response | 22 | 5.90E-04 |
| Response to bacterium | 15 | 4.00E-04 |
| Defense response | 21 | 1.20E-03 |
| Defense response to bacterium | 13 | 1.40E-03 |
| Proteolysis | 50 | 2.00E-03 |
| Innate immune response | 14 | 3.90E-03 |
| Molecular function | ||
| Peptidase activity | 54 | 2.10E-04 |
| Peptidase activity, acting on l-amino acid peptides | 49 | 1.30E-03 |
| Alkaline phosphatase activity | 7 | 2.20E-03 |
| Solute cation symporter activity | 14 | 1.80E-03 |
| Symporter activity | 14 | 2.50E-03 |
| Solute: sodium symporter activity | 12 | 2.30E-03 |
| Cellular component | ||
| Extracellular space | 18 | 2.20E-06 |
| Extracellular region | 47 | 4.20E-06 |
| Extracellular region part | 23 | 4.60E-06 |
| Integral to membrane | 87 | 1.80E-05 |
| Intrinsic to membrane | 87 | 3.10E-05 |
For a complete list of genes see Table S1.
Genes that were downregulated with age were enriched for roles in a wide range of biological processes such as the DNA damage response (e.g., CG3240, Spindle B, CG5524, CG6318, and Meiotic 41), the cell cycle (e.g., Spindle D, Cyclin C, Cyclin J, and Giant nuclei), and transcriptional regulation (Rigor mortis, Negative elongation factor E, TBP-related factor, Suppressor of hairless, and mediator complex subunit genes) (Table 3 and Table S1).
Table 3 . List of overrepresented functional groups (classified by gene ontology) within candidate genes that were downregulated with age.
| GO term | No. genes | Benjamini P |
|---|---|---|
| Biological process | ||
| Chromosome organization | 43 | 3.3E-10 |
| DNA metabolic process | 37 | 6.4E-10 |
| Cell cycle | 65 | 1.3E-09 |
| Cell-cycle phase | 55 | 1.1E-08 |
| Mitotic cell cycle | 45 | 1.4E-08 |
| Cell-cycle process | 57 | 3.4E-08 |
| Chromosome segregation | 24 | 6.2E-08 |
| M phase | 51 | 1.3E-07 |
| M phase of mitotic cell cycle | 26 | 2.3E-07 |
| Mitosis | 25 | 7.9E-07 |
| Nuclear division | 25 | 9.5E-07 |
| Organelle fission | 25 | 1.7E-06 |
| Chromatin modification | 21 | 2.8E-06 |
| Mitotic sister chromatid segregation | 13 | 2.3E-05 |
| Sister chromatid segregation | 13 | 2.8E-05 |
| Transcription | 44 | 2.9E-05 |
| Chromatin organization | 24 | 3.9E-05 |
| DNA-dependent transcription | 21 | 3.8E-05 |
| RNA biosynthetic process | 21 | 5.2E-05 |
| DNA repair | 18 | 5.7E-05 |
| Response to DNA damage | 19 | 6.7E-05 |
| Regulation of transcription | 62 | 9.0E-05 |
| Regulation of cell-cycle process | 16 | 8.9E-05 |
| Meiosis I | 12 | 8.7E-05 |
| Regulation of mitotic cell cycle | 17 | 1.3E-04 |
| Transcription from RNA polymerase II promoter | 17 | 1.4E-04 |
| Regulation of cell cycle | 22 | 2.3E-04 |
| Macromolecular complex subunit organization | 30 | 2.4E-04 |
| Chromosome condensation | 9 | 1.0E-03 |
| Mitotic chromosome condensation | 7 | 1.2E-03 |
| Molecular function | ||
| DNA binding | 77 | 1.2E-07 |
| DNA-dependent atpase activity | 14 | 4.3E-07 |
| Adenyl ribonucleotide binding | 62 | 2.3E-04 |
| Adenyl nucleotide binding | 65 | 2.7E-04 |
| ATP binding | 61 | 2.7E-04 |
| Purine nucleoside binding | 65 | 2.3E-04 |
| Nucleoside binding | 65 | 2.6E-04 |
| General RNA polymerase II transcription factor activity | 16 | 4.1E-04 |
| Purine ribonucleotide binding | 68 | 1.1E-03 |
| Ribonucleotide binding | 68 | 1.1E-03 |
| Purine nucleotide binding | 71 | 1.5E-03 |
| Cellular component | ||
| Chromosome | 49 | 5.8E-13 |
| Chromosomal part | 41 | 2.4E-11 |
| Intracellular non-membrane–bounded organelle | 83 | 9.9E-09 |
| Non-membrane–bounded organelle | 83 | 9.9E-09 |
| Nuclear lumen | 46 | 7.4E-08 |
| Nucleoplasm | 35 | 7.6E-07 |
| Organelle lumen | 54 | 1.7E-06 |
| Intracellular organelle lumen | 54 | 1.7E-06 |
| Membrane-enclosed lumen | 54 | 3.5E-06 |
| Nuclear chromosome | 16 | 3.8E-05 |
| Nucleoplasm part | 28 | 2.1E-04 |
| Chromatin | 17 | 9.9E-04 |
For a complete list of genes see Table S1.
Differences in the transcriptional response to E. coli infection vs. wounding
We next examined differences in the transcriptional response to wounding vs. E. coli infection. Only 80 transcripts were differentially expressed in infected vs. wounded flies. Genes upregulated (48/80) in infected flies were enriched for immune response-related GO-annotated categories (e.g., Diptericin B, Relish, Fondue, Eiger, and PGRP-LF). Genes that were downregulated (32/80) in infected flies were enriched for cellular carbohydrate catabolic processes (e.g., Glycerol 3 phosphate dehydrogenase, Aldolase, Hexokinase C, and CG6910) and basement membrane (e.g., Collagen type IV, Glutactin, and Papilin) (Table S2). Downregulation of genes involved in metabolism following infection in flies has been reported in several independent studies and so appears to be a general response to infection (De Gregorio et al. 2001; Dionne et al. 2006; Zheng et al. 2011).
When we tested the full model to look at the effect of age (results given above), treatment (infected vs. wounded), and their interaction, no transcripts exhibited significant age × treatment (wound vs. infection) interaction; however, we attribute this pattern to low statistical power to detect interaction effects (the probability of type II error, β = 0.93, based on effect sizes in the top quartile and standard errors in the bottom quartile of the data). To identify transcripts with different effects at young and old ages we conducted separate tests of the effect of infection at each age. We found 45 genes that were differentially expressed between wounded and infected flies at 1 week of age and 74 genes that were differentially expressed in 4-week-old flies. Twenty-seven of these genes were involved in the response to infection at both ages (Table S2) and most of these were known immune response genes. The remaining transcripts were unique to each age group. To assess the possibility that these tests detected different transcripts at each age due to a high rate of false negatives, we evaluated the overlap among the top-ranked genes (with respect to P-value) in each list. Among the 100 genes with the smallest P-values in the week-4 list, 47 were not among the top 400 genes in the week-1 list. Similarly, of the 100 genes with the smallest P-values in the week-1 list, 46 were not among the 400 genes in the week-4 list. We conclude that aging influences the differential response to wounding vs. infection.
Variation in gene expression is associated with age-specific bacterial clearance
We hypothesized that differences among lines in the age-specific transcriptional response to infection gave rise to age-specific variation in clearance ability. To identify candidate genes responsible for variation in age-specific immune function we performed a genome-wide test for association between the average bacterial clearance ability of each line and transcript abundance of each line, using a Spearman’s rank test (Zar 1999). Transcripts for which this statistic had FDR-adjusted P-values <0.05 were considered strong candidates.
For the wounded flies (sham injected) at 1 week of age we found 14 genes that were significantly associated with bacterial clearance ability (FDR < 0.05, Table S3). For 6 of these genes higher expression was associated with higher bacterial loads while for the remaining 8 higher expression was associated with better clearance. While GO analysis found no functional categories overrepresented in either group, one gene had previously been implicated in the immune response to infection (CG7532, l(2)34Fc). In contrast, at 4 weeks of age we found 114 genes that were significantly associated with bacterial clearance ability (Table S3). Higher expression levels were associated with poorer clearance ability (higher bacterial loads) for 43 of these genes. No functional categories were identified by GO analysis. For 67 of these genes, higher levels were associated with improved clearance ability. GO analysis found enrichment for genes involved in core RNA polymerase II-binding transcription factor activity (Cyclin C, Mediator complex subunit 20, Mediator complex subunit 22, TBP-associated factor 13, and Transcription factor IIF alpha) and zinc ion binding [CG11971, CG14962, CG17721, CG4854, lethal (2) k10201, and trade embargo].
We next used Spearman’s rank test to identify genes whose transcriptional variation was significantly associated with variation in bacterial clearance ability, using flies that were infected by E. coli. Surprisingly, at 1 week of age, there were no significant associations between gene expression and clearance ability with FDR-adjusted P-values <0.05, despite substantial variation in gene expression among lines. In contrast, at 4 weeks of age we identified 247 transcripts with variation in expression that was significantly correlated with bacterial clearance ability (Figure 2, Table 5, and Table S4).
Figure 2 .
Examples of candidate genes with significant correlations between variation in transcript level and bacterial-clearance ability at 4 weeks of age. Peptidoglycan receptor protein-LC (A) and 18 wheeler (B) are immune response candidate genes; Fatty acid synthase (C) and Gonadotropin-releasing hormone receptor (D) are lipid metabolism candidate genes; and RAC serine/threonine-protein kinase or Akt (E) and mos/CG8767 (F) are also shown. Note that variation in the expression of gonadotropin-releasing hormone receptor and mos was negatively correlated with colony counts and expression levels of the remaining candidates were positively correlated with colony counts.
Table 5 . Twenty of the candidate genes with significant correlations between variation in transcript level and clearance ability at 4 weeks of age (in flies that were infected with E. coli).
| FlyBase ID | Gene | BP | MF | Correlation | FDR P | Raw P |
|---|---|---|---|---|---|---|
| FBgn0032433 | Organic anion transporting polypeptide 33Ea | Organic anion transport | Organic anion transmembrane transporter activity | 0.94 | 0.001 | 0.00E+00 |
| FBgn0027571 | Fatty acid synthase | Metabolism | Fatty acid synthase activity | 0.90 | 0.01 | 1.00E-05 |
| FBgn0030114 | CG17754 | Phagocytosis, engulfment | — | 0.87 | 0.02 | 7.00E-05 |
| FBgn0035976 | Peptidoglycan recognition protein LC | Immune response | Peptidoglycan binding | 0.84 | 0.02 | 2.60E-04 |
| FBgn0010379 | RAC serine/threonine-protein kinase (Akt1) | Anti-apoptosis, insulin receptor signaling | Protein serine/threonine kinase activity | 0.84 | 0.02 | 2.60E-04 |
| FBgn0014020 | Ras-like GTP-binding protein Rho1 | Actin cytoskeleton organization, wound healing, etc | Protein/kinase binding | 0.82 | 0.03 | 5.50E-04 |
| FBgn0025595 | Gonadotropin-releasing hormone receptor | Carbohydrate/lipid homeostasis | Peptide receptor activity | 0.81 | 0.04 | 7.00E-04 |
| FBgn0024326 | MAP kinase kinase 4 (Mkk4) | Activation of JUN kinase | JUN kinase kinase activity | 0.81 | 0.04 | 7.00E-04 |
| FBgn0004364 | 18 wheeler | Immune response | Transmembrane receptor activity | 0.80 | 0.04 | 8.60E-04 |
| FBgn0261444 | Tweety-2; CG3638 | Phagocytosis, engulfment | — | 0.80 | 0.04 | 8.60E-04 |
| FBgn0002652 | Squash | Gene silencing by RNA, oogenesis | — | −0.92 | 0.003 | 0.00E+00 |
| FBgn0031739 | CG14005 | — | — | −0.86 | 0.02 | 1.00E-04 |
| FBgn0004106 | Cell division control protein 2 homolog | Response to DNA damage stimulus, asymmetric neuroblast division, etc. | Cyclin-dependent protein kinase activity, protein binding | −0.86 | 0.02 | 1.00E-04 |
| FBgn0017418 | Ariadne | Negative regulation of transcription | Specific transcriptional repressor activity, zinc ion binding | −0.85 | 0.02 | 1.90E-04 |
| FBgn0013531 | Mediator complex subunit 20 | Regulation of transcription from RNA polymerase II promoter | Transcription coactivator activity | −0.82 | 0.03 | 5.50E-04 |
| FBgn0040286 | SC35; CG5442 | Regulation of alternative nuclear mrna splicing, via spliceosome | Protein binding | −0.82 | 0.03 | 5.50E-04 |
| FBgn0000579 | Enolase | Glycolysis | Magnesium ion binding | −0.81 | 0.04 | 7.00E-04 |
| FBgn0027903 | DNA polymerase delta small subunit, CG12018 | Response to DNA damage stimulus | DNA binding | −0.80 | 0.04 | 8.60E-04 |
| FBgn0033773 | mos; CG8767 | Positive regulation of TOR signaling cascade | Protein kinase activity | −0.80 | 0.045 | 1.07E-03 |
| FBgn0029712 | CG15912 | Metabolism | Phosphoglycolate phosphatase activity | −0.80 | 0.05 | 1.13E-03 |
Positively correlated genes are at the top of the table and negatively correlated genes are at the bottom of the table. Positive correlations indicate that a higher expression level is associated with higher infection levels (poorer clearance) and negative correlations indicate that a higher expression level is associated with a lower infection level (better clearance). The complete list of candidate genes can be found in Table S3 (for the flies that were wounded only) and Table S4 (for the flies that were infected with E. coli).
Spearman’s rank test is a conservative statistical test. This, combined with the fact that there was greater residual variation in the clearance assay in younger flies (Table 1), prompted us to qualitatively assess whether the same transcripts were associated with variation in clearance ability at both ages. To do this we used the same approach that we took to evaluate the effect of age on the transcriptional response to wounding vs. infection across the two ages. In this case, we evaluated the overlap among the top 100 ranking transcripts that had the strongest association with clearance ability at both ages. In the sham-injected flies, only three genes overlapped between the 1- and 4-week-old flies, fne (found in neurons), Pino (Pinocchio), and Frq1 (Frequenin 1). Interestingly, two of these genes (fne and Frq1) are involved in neuronal regulation. In the flies infected with E. coli, only one transcript, lola (longitudinals lacking), was found within the top ranking genes at both ages. Notably, lola was also significantly associated with clearance ability in the 4-week-old sham-injected flies. The striking picture that emerges from these results is that the genetic basis of variation in bacterial clearance following infection changes dramatically with age.
As an additional test of the idea that the genetic basis of variation in clearance changes with age, we reanalyzed the correlation between gene expression and bacterial clearance, using four lines whose clearance changed the least between 1 and 4 weeks of age. For this analysis we used Pearson’s correlation coefficient instead of Spearman’s rank test. We did this, not to test significance, but instead to allow us to rank the top 100 genes associated with clearance across ages. With only four replicates, there are few possible values for Spearman’s rho (the test statistic) so it is not possible to find the top 100 ranked genes using this statistic. Comparing the top 100 genes from the sham-infected flies, only 1 gene overlapped both lists, the gene IntS1 (Integrator 1). For the infected flies no genes were found in common across ages in the top 100 genes that were correlated with clearance ability. Taking these results together with the larger analyses of all 12 lines, we conclude that the transcriptional response to infection changes dramatically with age.
Focusing on the genes in the infected flies that were significantly associated with bacterial clearance ability at 4 weeks of age (Table S4), 141 exhibited positive correlations between transcript abundance and bacterial load, meaning that higher expression levels of these particular candidates were associated with poor clearance ability (for examples see Figure 2). For 106 candidate genes, expression levels and bacterial loads were negatively correlated, meaning that higher transcript levels were associated with improved clearance ability.
GO analysis of genes whose higher expression was associated with poorer clearance (higher bacterial loads) implicated those involved in cell signaling (Fmr1, cac, unc-13, Caps, alpha-Adaptin, endoA, unc-104, Amph, Nrx-IV, and sif) (see Table 4). For genes whose higher expression levels were associated with improved clearance (reduced bacterial load) GO analysis indicated enrichment of genes involved in transition metal ion binding (CG1134, CG12219, CG13126, CG15514, CG17186, CG31053, CG7357, CG8159, Non-structural maintenance of chromosomes element 1 homolog, and Zinc finger protein 330 homolog) (see Table 4). We also categorized functional groups on the basis of the annotation information available in FlyBase (Figure 3). On the basis of this information 66 genes encode enzymes such as kinases [e.g., MAP kinase kinase 4 (Mkk4), RAC serine/threonine-protein kinase (Akt1), and Atypical protein kinase C] and peptidases [e.g., Death executioner caspase related to Apopain/Yama (decay), CG14642, and Methionine aminopeptidase] and other enzymes with roles in metabolism (e.g., Fatty acid synthase, CG15912, and Adenylyl cyclase 35C). A number of candidates encode transcription factors and genes involved in transcriptional regulation (e.g., Prospero, Bric-a-brac 2, Ariadne, MED19, and MED20), cytoskeletal binding proteins (e.g., Spire and Rho1), and transport proteins (CG7777, Surfeit 6, and AP-2 complex subunit). A large number of genes have unknown functions (84 genes). One of the more surprising findings was that of the 247 candidates whose expression levels were correlated with late age clearance ability, only 7 had been previously implicated to be involved in Drosophila immunity (18w, PGRP-LC, CG17754, lola, GATAe, prospero, and tweety-2).
Table 4 . List of overrepresented functional groups (classified by gene ontology) within candidate genes that were upregulated (top) and downregulated (bottom) in infected flies.
| GO term | No. genes | Benjamini P |
|---|---|---|
| Upregulated genes | ||
| Biological process | ||
| Defense response | 18 | 9.1E-19 |
| Immune response | 17 | 8.6E-18 |
| Response to bacterium | 15 | 6.9E-19 |
| Innate immune response | 14 | 1.7E-16 |
| Antimicrobial humoral response | 10 | 6.6E-15 |
| Defense response to Gram-negative bacterium | 8 | 2.8E-10 |
| Defense response to Gram-positive bacterium | 7 | 4.8E-09 |
| Carbohydrate catabolic process | 5 | 2.8E-03 |
| Peptidoglycan metabolic process | 5 | 2.1E-06 |
| Polysaccharide catabolic process | 5 | 8.5E-05 |
| Glycosaminoglycan metabolic process | 5 | 5.0E-05 |
| Response to fungus | 4 | 1.7E-03 |
| Molecular function | ||
| Peptidoglycan binding | 5 | 9.1E-07 |
| N-acetylmuramoyl-l-alanine amidase activity | 5 | 1.3E-06 |
| Glycosaminoglycan binding | 5 | 3.3E-06 |
| Hydrolase activity, acting on carbon–nitrogen (but not peptide) bonds, in linear amides | 5 | 3.1E-04 |
| Cellular component | ||
| Extracellular region | 18 | 1.2E-09 |
| Extracellular space | 8 | 1.8E-06 |
| Downregulated genes | ||
| Biological process | ||
| Cellular carbohydrate catabolic process | 4 | 4.6E-02 |
| Cellular component | ||
| Basement membrane | 3 | 9.2E-03 |
| Lipid particle | 6 | 1.0E-02 |
For a complete list of genes see Table S2.
Figure 3 .
Functional categorization of candidate genes with significant correlations between variation in transcript level and bacterial-clearance ability at 4 weeks of age. Functional groups shown contain ≥5 genes with similar molecular function gene ontology annotation terms according to FlyBase. The 260 candidate genes were categorized into 11 groups that included enzymes, transcription factors, transcriptional regulatory proteins, transport proteins, cytoskeletal-binding proteins, metal ion-binding proteins, nucleic acid-binding proteins, protein-binding proteins, unknown function, miscellaneous, and immune response. The miscellaneous group contains annotated genes that had <5 members and unknown genes had no recorded molecular function in FlyBase.
Discussion
Our data provide additional support for previous studies that identified natural genetic variation in the ability to clear infection (Jackson et al. 2003; Lazzaro et al. 2004; Lesser et al. 2006). While we found no “general” effect of age on bacterial clearance ability, there was a significant genotype × age interaction. This result confirms our earlier finding that used an independent set of lines derived from the same natural population (Lesser et al. 2006). These results are contradictory to those found in another study that reported that age did not influence the ability of Drosophila to clear a bacterial infection (Ramsden et al. 2008). A possible explanation for the discrepancy in these two studies is the small number of different genotypes examined by Ramsden et al. (two strains). Indeed many of the lines tested in this study as well as in our earlier study (Lesser et al. 2006) exhibited no obvious effect of age on clearance ability.
One of the more important results of this study was our finding that genetic architecture underlying the variation in clearance ability differed markedly at different ages. This was evident in the data obtained from all 12 lines as well as the data from the four lines that exhibited the least amount of variation in clearance with age. Not only did the majority of the genes that were associated with clearance ability differ across ages (in both the wounded and the infected treatments) but also in both the 12- and the 4-line analysis the relative influence of variation in gene expression on clearance appeared to change with age. This finding was evident in the comparisons of the age-specific transcriptional responses to wounding vs. infection and also in the correlation tests between bacterial clearance ability and transcript level in both the wounded and the infected flies.
One potential explanation for an increase in the influence of transcriptional variation on immune clearance at an older age is that the regulatory control over gene expression declines with age and the degree to which this regulation is influenced by age varies among lines. Age-related decline in the control of gene regulation has been suggested as a potential cause of immunosenescence in a previous study (Zerofsky et al. 2005) and this hypothesis may be supported by our data. We are not aware of additional studies that have tested this idea. Another, and not mutually exclusive, explanation is that genotypes vary in the rate of decline in the efficiency of phagocytosis with age. The two major components of the adult innate immune response following infection in Drosophila are phagocytosis, engulfment, and killing of bacteria by blood cells and the humoral response involving the production of AMPs, primarily by cells of the fat body. Of these two components, recent evidence suggests that the process of phagocytosis acts immediately upon infection and clears the majority of pathogens (Haine et al. 2008) while AMP production plays a relatively minor role in bacterial clearance. Age-related declines in the efficiency of phagocytosis have been reported in other organisms (Fearon and Locksley 1996; Butcher et al. 2000, 2001; Ginaldi et al. 2001; DeVeale et al. 2004; Terrón et al. 2004; Panda et al. 2009) and also in Drosophila (Mackenzie et al. 2011). Recent evidence also suggests that the number of blood cells (phagocytes) declines with age in Drosophila (Mackenzie et al. 2011). Age-related decline in the number of phagocytes combined with a decline in the efficiency of phagocytosis would lead to greater reliance on AMP production to clear infection, a process that involves extensive cell-signaling events leading to the production, transport, and secretion of antimicrobial peptides into the hemolymph (Ferrandon et al. 2007). While it is not clear that the production of AMPs is more energetically demanding than phagocytosis, reduced efficiency of phagocytosis would delay the response to infection, leaving higher bacterial loads for the humoral component to clear. Greater reliance on the humoral component to clear infection could produce changes in expression of genes other than those directly involved in antimicrobial peptide production, such as those involved in energy metabolism. Variation in the expression of such genes among lines then may contribute to (or be the product of) variation in infection levels observed 24 hr after infection.
This hypothesis is supported by our identification of many candidate genes with metabolic functions associated with bacterial clearance in older flies. For example, variation in expression of genes involved in lipid metabolism [e.g., fatty acid synthase (FAS) and gonadotropin-releasing hormone receptor (GRHR) (an adipokinetic hormone-binding gene)] was associated with variation in bacterial clearance. FAS is the enzyme responsible for the de novo synthesis of fatty acids (Furuta et al. 2008) and overexpression of GRHR in the fat body has been shown to block lipid storage (Grönke et al. 2007). Additionally, alterations in the expression of Akt and mos were also associated with variation in bacterial clearance at an older age. Akt is a serine/threonine protein kinase that interacts with other proteins to modulate the insulin-signaling and TOR pathways (Wu and Brown 2006). Akt also inhibits the activation of FOXO, a transcription factor that is activated by stress (reviewed in Salih and Brunet 2008) and that has been recently implicated in regulating the production of antimicrobial peptides (Becker et al. 2010). Furthermore, in Drosophila, PI3K/Akt signaling is known to be influenced by infection (DiAngelo et al. 2009) with functional consequences for lipid metabolism (Dionne et al. 2006). Variation among our lines in the extent to which infection alters insulin signaling may have contributed directly to variation in clearance. mos was identified in a mutant screen as a kinase that promotes TOR signaling to S6K (Findlay et al. 2007). Activation of the TOR pathway results in the phosphorylation of S6K, which mediates protein synthesis, cell growth, and other cellular processes (Miron and Sonenberg 2001; Ma and Blenis 2009; Katewa and Kapahi 2011). Earlier work in our lab found that a hypomorphic mutation in S6k, a gene acting downstream of TOR, resulted in decreased bacterial ability to clear infection by E. coli (Cho et al. 2010). Therefore, it is possible that bacterial clearance is mediated by activation of TOR signaling by higher mos levels. Of course, this experimental design does not allow us to separate cause from effect. Because most genes that we identified have no known immune function, it is impossible to determine whether the altered gene expression is a cause or a result of high bacterial loads. Further work is necessary to understand how alterations in gene expression either influence or are influenced by wounding and infection and how changes in gene expression influence functional aspects of immune responses (such as the efficiency of phagocytosis or production of AMPs).
An alternative perspective from which our results could be viewed is that at least a subset of the genes we found to be associated with variation in infection level do not directly affect bacterial clearance (a measure of resistance to infection), but instead contribute to variation in tolerance to infection. Tolerance is a measure of the ability of an individual to minimize the detrimental effects of infection (Painter 1958; Schneider and Ayres 2008). And so high expression of genes in flies with high infection load (such as those involved in lipid metabolism discussed above) may influence tolerance, offsetting the potential detrimental effects of high infection levels on fitness. In the current context it is not possible to ascribe the genes we identified as genes conferring variation in tolerance. E. coli is not a natural pathogen of Drosophila and does not influence survival (an indicator of tolerance) at the concentrations we used, although it can affect survival under extremely high concentrations (Ramsden et al. 2008). We also did not measure any other fitness-related trait in these flies (concurrently with the infection) and so we do not know whether there is variation among lines in their ability to tolerate infection. Future studies are necessary to evaluate this possibility.
While we know a fair amount about resistance mechanisms and genes involved in the immune response, we know less about physiological and genetic mechanisms involved in tolerance (Read et al. 2008; Schneider and Ayres 2008). Tolerance has long been recognized as a trait that could evolve in response to selection although conditions favoring the maintenance of variation in tolerance are often argued to be more stringent that those maintaining variation in resistance traits in natural populations (Roy and Kirchner 2000; Miller et al. 2005, 2006). Indeed variation in tolerance is not always found (Lefevre et al. 2011) but there is ample evidence of its existence in natural populations (Fineblum and Rausher 1995; Mauricio et al. 1997; Blanchet et al. 2010) and more empirical work is necessary to explain the maintenance of this variation (Read et al. 2008; Svensson and Raberg 2010). Mutant studies have identified a few genes/pathways that play a role in tolerance to infection (Ayres and Schneider 2008; Ayres et al. 2008; Shinzawa et al. 2009) but as far as we aware no genes have been identified that contribute to natural variation in tolerance. Until such studies are done we can only speculate that because many candidate genes from this study are involved in metabolic processes, they are candidates for contributing to variation in tolerance.
This idea is further supported by another surprising finding, the fact that natural variation in the transcript levels of known immunity genes did not explain much of the variation in clearance ability among lines (although when we compare the results for wounded vs. infected flies, it is clear that such genes responded to bacterial infection). This is consistent with results from a previous study that found few associations between expression of immune response genes and bacterial loads in Drosophila (Sackton et al. 2010). Genes involved in regulating the immune response to infection have been shown to be under strong directional and purifying selection in many organisms (Oliver et al. 2010; Alcaide and Edwards 2011; Tschirren et al. 2011; Voolstra et al. 2011), including Drosophila (Schlenke and Begun 2003; Lazzaro 2008; Morales-Hojas et al. 2009; Obbard et al. 2009). We suspect that expression of these genes would also be under strong selection and so limit genetically based variation in the transcriptional response to infection. As a result, variation in physiological traits that support the immune response (e.g., those controlling energy availability and so influence tolerance) could be expected to underlie variation in this trait.
We found higher expression levels of immune response genes at an older age. The upregulation of Drosophila immunity genes with age is often observed even in the absence of infection (Pletcher et al. 2002; Landis et al. 2004; Lai et al. 2007; Sarup et al. 2011) and age-related increases in immune response genes have also been noted in human fibroblast cells (Shelton et al. 1999) and mouse brain tissue (Lee et al. 2000). The factors responsible for the upregulation of Drosophila immune response genes are not well understood but it has been suggested to result from cumulative exposure to pathogens over time (Ren et al. 2007) or the loss in the transcriptional regulation of immune response genes (Zerofsky et al. 2005). Whatever the cause, our study suggests that these age-dependent changes in the expression of immune response genes themselves have little effect on producing natural variation in immune function.
Age-related changes in transcriptional profiles are common in aging organisms (Pletcher et al. 2002; de Magalhães et al. 2009; Lui et al. 2010) and studies of immunosenescence suggest that an age-related breakdown (Youngman et al. 2011) or misregulation of signaling pathways (Zerofsky et al. 2005) plays an important role in the declines in immune function. More work is needed to understand why expression changes with age and what genes contribute to interindividual variation in age-specific gene expression. One promising avenue of research is to use genome-wide mapping studies to identify the genetic basis of variation in functional traits across different ages and combine this information with age-specific transcriptional profiles. In this way, sequence polymorphisms that affect trait variation at a given age could be compared with expression data at that age. Combining this information would allow us to identify those polymorphisms that influence age-specific expression of transcripts that in turn affect the phenotype (Gilad et al. 2008; Majewski and Pastinen 2011).
Supplementary Material
Acknowledgments
We thank Maria De Luca, Michelle Starz-Gaiano, Maricel Kann, and Joseph Travis for insightful discussion of the results. Michelle Starz-Gaiano and Maria De Luca provided helpful comments on the manuscript. We thank Tim Ford for formatting the figures. T.M.F. was supported by U.S. Department of Education award P200A060197, National Institute of General Medical Sciences Initiative for Maximizing Student Development grant R25-GM55036, and Procter and Gamble. K.A.H. was supported by National Science Foundation grant DEB 0848337. This research was supported by National Institutes of Health grants 1R03 AG023339-01 and 5R01 DK084219-02 (to J.L.).
Footnotes
Communicating editor: L. Harshman
Literature Cited
- Agaisse H., 2008. Investigating the involvement of host factors involved in intracellular pathogen infection by RNAi in Drosophila cells. Methods Mol. Biol. 415: 395–402 [DOI] [PubMed] [Google Scholar]
- Agarwal S., Busse P. J.2010. Innate and adaptive immunosenescence. Ann. Allergy Asthma Immunol. 104:183–190; quiz 190–182, 210.
- Agrawal A., Agrawal S., Cao J. N., Su H., Osann K., et al. , 2007. Altered innate immune functioning of dendritic cells in elderly humans: a role of phosphoinositide 3-kinase-signaling pathway. J. Immunol. 178: 6912–6922 [DOI] [PubMed] [Google Scholar]
- Agrawal A., Sridharan A., Esposo M., Kaushal K., Tay J., et al. , 2011. Age-associated impaired plasmacytoid dendritic cell functions lead to decreased CD4 and CD8 T cell immunity. Age (Omaha) 33: 363–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcaide M., Edwards S. V., 2011. Molecular evolution of the toll-like receptor multigene family in birds. Mol. Biol. Evol. 28: 1703–1715 [DOI] [PubMed] [Google Scholar]
- Ayres J. S., Schneider D. S., 2008. A signaling protease required for melanization in Drosophila affects resistance and tolerance of infections. PLoS Biol. 6: 2764–2773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayres J. S., Freitag N., Schneider D. S., 2008. Identification of Drosophila mutants altering defense of and endurance to Listeria monocytogenes infection. Genetics 178: 1807–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T., Loch G., Beyer M., Zinke I., Aschenbrenner A. C., et al. , 2010. FOXO-dependent regulation of innate immune homeostasis. Nature 463: 369–373 [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y., 1995. Controlling the False Discovery Rate: a Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc., B 57: 289–300 [Google Scholar]
- Blanchet S., Rey O., Loot G., 2010. Evidence for host variation in parasite tolerance in a wild fish population. Evol. Ecol. 24: 1129–1139 [Google Scholar]
- Blomquist G., 2010. Heritability of individual fitness in female macaques. Evol. Ecol. 24: 657–669 [Google Scholar]
- Bowler K., Terblanche J. S., 2008. Insect thermal tolerance: what is the role of ontogeny, ageing and senescence? Biol. Rev. Camb. Philos. Soc. 83: 339–355 [DOI] [PubMed] [Google Scholar]
- Burger J. M., Promislow D. E., 2006. Are functional and demographic senescence genetically independent? Exp. Gerontol. 41: 1108–1116 [DOI] [PubMed] [Google Scholar]
- Butcher S., Chahel H., Lord J. M., 2000. Review article: ageing and the neutrophil: no appetite for killing? Immunology 100: 411–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher S. K., Chahal H., Nayak L., Sinclair A., Henriquez N. V., et al. , 2001. Senescence in innate immune responses: reduced neutrophil phagocytic capacity and CD16 expression in elderly humans. J. Leukoc. Biol. 70: 881–886 [PubMed] [Google Scholar]
- Ceballos S., Kiørboe T., 2011. Senescence and sexual selection in a pelagic copepod. PLoS ONE 6: e18870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charmantier A., Perrins C., McCleery R. H., Sheldon B. C., 2006. Quantitative genetics of age at reproduction in wild swans: support for antagonistic pleiotropy models of senescence. Proc. Natl. Acad. Sci. USA 103: 6587–6592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho I., Horn L., Felix T., Foster L., Gregory G., et al. , 2010. Age- and diet-specific effects of variation at S6 kinase on life history, metabolic, and immune response traits in Drosophila melanogaster. DNA Cell Biol. 29: 473–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipriano C., Caruso C., Lio D., Giacconi R., Malavolta M., et al. , 2005. The -308G/A polymorphism of TNF-alpha influences immunological parameters in old subjects affected by infectious diseases. Int. J. Immunogenet. 32: 13–18 [DOI] [PubMed] [Google Scholar]
- De Gregorio E., Spellman P. T., Rubin G. M., Lemaitre B., 2001. Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proc. Natl. Acad. Sci. USA 98: 12590–12595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca M., Roshina N. V., Geiger-Thornsberry G. L., Lyman R. F., Pasyukova E. G., et al. , 2003. Dopa decarboxylase (Ddc) affects variation in Drosophila longevity. Nat. Genet. 34: 429–433 [DOI] [PubMed] [Google Scholar]
- de Magalhães J. P., Curado J., Church G. M., 2009. Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics 25: 875–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVeale B., Brummel T., Seroude L., 2004. Immunity and aging: the enemy within? Aging Cell 3: 195–208 [DOI] [PubMed] [Google Scholar]
- DiAngelo J., Bland M., Bambina S., Cherry S., Birnbaum M., 2009. The immune response attenuates growth and nutrient storage in Drosophila by reducing insulin signaling. Proc. Natl. Acad. Sci. USA 106: 20853–20858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionne M. S., Pham L. N., Shirasu-Hiza M., Schneider D. S., 2006. Akt and foxo dysregulation contribute to infection-induced wasting in Drosophila. Curr. Biol. 16: 1977–1985 [DOI] [PubMed] [Google Scholar]
- Drysdale R., Crosby M., 2005. FlyBase: genes and gene models. Nucleic Acids Res. 33: D390–D395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudycha J. L., Tessier A. J., 1999. Natural genetic variation of life span, reproduction and juvenile growth in Daphnia. Evolution 53: 1744–1756 [DOI] [PubMed] [Google Scholar]
- Fearon D., Locksley R., 1996. The instructive role of innate immunity in the acquired immune response. Science 272: 50–53 [DOI] [PubMed] [Google Scholar]
- Feng S., Wolfinger R. D., Chu T. M., Gibson G. C., McGraw L. A., 2006. Empirical Bayes analysis of variance component models for microarray data. J. Agric. Biol. Environ. Stat. 11: 197–209 [Google Scholar]
- Ferrandon D., Imler J., Hetru C., Hoffmann J., 2007. The Drosophila systemic immune response: sensing and signalling during bacterial and fungal infections. Nat. Rev. Immunol. 7: 862–874 [DOI] [PubMed] [Google Scholar]
- Findlay G. M., Yan L., Procter J., Mieulet V., Lamb R. F., 2007. A MAP4 kinase related to Ste20 is a nutrient-sensitive regulator of mTOR signalling. Biochem. J. 403: 13–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fineblum W. L., Rausher M. D., 1995. Tradeoff between resistance and tolerance to herbivore damage in a morning glory. Nature 377: 517–520 [Google Scholar]
- Fontana L., Partridge L., Longo V. D., 2010. Extending healthy life span–from yeast to humans. Science 328: 321–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulop T., Larbi A., Douziech N., Fortin C., Guerard K. P., et al. , 2004. Signal transduction and functional changes in neutrophils with aging. Aging Cell 3: 217–226 [DOI] [PubMed] [Google Scholar]
- Furuta E., Pai S., Zhan R., Bandyopadhyay S., Watabe M., et al. , 2008. Fatty acid synthase gene is up-regulated by hypoxia via activation of Akt and sterol regulatory element binding protein-1. Cancer Res. 68: 1003–1011 [DOI] [PubMed] [Google Scholar]
- Gilad Y., Rifkin S. A., Pritchard J. K., 2008. Revealing the architecture of gene regulation: the promise of eQTL studies. Trends Genet. 24: 408–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginaldi L., Loreto M., Corsi M., Modesti M., De Martinis M., 2001. Immunosenescence and infectious diseases. Microbes Infect. 3: 851–857 [DOI] [PubMed] [Google Scholar]
- Grewe M., 2001. Chronological ageing and photoageing of dendritic cells. Clin. Exp. Dermatol. 26: 608–612 [DOI] [PubMed] [Google Scholar]
- Grönke S., Müller G., Hirsch J., Fellert S., Andreou A., et al. , 2007. Dual lipolytic control of body fat storage and mobilization in Drosophila. PLoS Biol. 5: e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L., Kenyon C., 2000. Genetic pathways that regulate ageing in model organisms. Nature 408: 255–262 [DOI] [PubMed] [Google Scholar]
- Haine E. R., Moret Y., Siva-Jothy M. T., Rolff J., 2008. Antimicrobial Defense and Persistent Infection in Insects. Science 322: 1257–1259 [DOI] [PubMed] [Google Scholar]
- Hill W. G., Mulder H. A., 2010. Genetic analysis of environmental variation. Genet. Res. 92: 381–395 [DOI] [PubMed] [Google Scholar]
- Hillyer J., Schmidt S., Fuchs J., Boyle J., Christensen B., 2005. Age-associated mortality in immune challenged mosquitoes (Aedes aegypti) correlates with a decrease in haemocyte numbers. Cell. Microbiol. 7: 39–51 [DOI] [PubMed] [Google Scholar]
- Houle D., 1992. Comparing evolvability and variability of quantitative traits. Genetics 130: 195–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu H., Zhang H., Li L., Yi N., Yang P., et al. , 2003. Age-related thymic involution in C57BL/6J x DBA/2J recombinant-inbred mice maps to mouse chromosomes 9 and 10. Genes Immun. 4: 402–410 [DOI] [PubMed] [Google Scholar]
- Huang W., Sherman B. T., Lempicki R. A., 2009a. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Sherman B. T., Lempicki R. A., 2009b. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Hughes K. A., 2010. Mutation and the evolution of ageing: from biometrics to system genetics. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365: 1273–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry R. A., Hobbs B., Collin F., Beazer-Barclay Y. D., Antonellis K. J., et al. , 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Irving P., Troxler L., Heuer T. S., Belvin M., Kopczynski C., et al. , 2001. A genome-wide analysis of immune responses in Drosophila. Proc. Natl. Acad. Sci. USA 98: 15119–15124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A., Galecki A., Burke D., Miller R., 2003. Genetic polymorphisms in mouse genes regulating age-sensitive and age-stable T cell subsets. Genes Immun. 4: 30–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazwinski S. M., Kim S., Dai J., Li L., Bi X., et al. , 2010. HRAS1 and LASS1 with APOE are associated with human longevity and healthy aging. Aging Cell 9: 698–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaaya G. P., Darji N., 1988. The humoral defense system in tsetse: differences in response due to age, sex and antigen types. Dev. Comp. Immunol. 12: 255–268 [DOI] [PubMed] [Google Scholar]
- Katewa S. D., Kapahi P., 2011. Role of TOR signaling in aging and related biological processes in Drosophila melanogaster. Exp. Gerontol. 46: 382–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight C. G., Azevedo R. B., Leroi A. M., 2001. Testing life-history pleiotropy in Caenorhabditis elegans. Evolution 55: 1795–1804 [DOI] [PubMed] [Google Scholar]
- Kogut M., Rothwell L., Kaiser P., 2002. Differential effects of age on chicken heterophil functional activation by recombinant chicken interleukin-2. Dev. Comp. Immunol. 26: 817–830 [DOI] [PubMed] [Google Scholar]
- Kovacs E., Palmer J., Fortin C., Fülöp T. J., Goldstein D., et al. , 2009. Aging and innate immunity in the mouse: impact of intrinsic and extrinsic factors. Trends Immunol. 30: 319–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs E. J., Plackett T. P., Boehmer E. D., Faunce D. E., 2004. Aging and innate immune cells. J. Leukoc. Biol. 76: 291–299 [DOI] [PubMed] [Google Scholar]
- Lai C., Parnell L., Lyman R., Ordovas J., Mackay T., 2007. Candidate genes affecting Drosophila life span identified by integrating microarray gene expression analysis and QTL mapping. Mech. Ageing Dev. 128: 237–249 [DOI] [PubMed] [Google Scholar]
- Landis G. N., Abdueva D., Skvortsov D., Yang J. D., Rabin B. E., et al. , 2004. Similar gene expression patterns characterize aging and oxidative stress in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 101: 7663–7668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laws T. R., Harding S. V., Smith M. P., Atkins T. P., Titball R. W., 2004. Age influences resistance of Caenorhabditis elegans to killing by pathogenic bacteria. FEMS Microbiol. Lett. 234: 281–287 [DOI] [PubMed] [Google Scholar]
- Lazzaro B. P., 2008. Natural selection on the Drosophila antimicrobial immune system. Curr. Opin. Microbiol. 11: 284–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaro B. P., Sceurman B. K., Clark A. G., 2004. Genetic basis of natural variation in D-melanogaster antibacterial immunity. Science 303: 1873–1876 [DOI] [PubMed] [Google Scholar]
- Lee C. K., Weindruch R., Prolla T. A., 2000. Gene-expression profile of the ageing brain in mice. Nat. Genet. 25: 294–297 [DOI] [PubMed] [Google Scholar]
- Lefevre T., Williams A. J., de Roode J. C., 2011. Genetic variation in resistance, but not tolerance, to a protozoan parasite in the monarch butterfly. Proc. Biol. Sci. 278: 751–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leips J., 2009. Insect models of immunosenescence, pp. 87–106 Handbook on Immunosenescence: Basic Understanding and Clinical Applications, edited by Fulop T., Franceschi C., Hirokawa K., Pawelec G. Springer Netherlands, Dordrecht, The Netherlands [Google Scholar]
- Leips J., Gilligan P., Mackay T. F. C., 2006. Quantitative trait loci with age-specific effects on fecundity in Drosophila melanogaster. Genetics 172: 1595–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesser K. J., Paiusi I. C., Leips J., 2006. Naturally occurring genetic variation in the age-specific immune response of Drosophila melanogaster. Aging Cell 5: 293–295 [DOI] [PubMed] [Google Scholar]
- Lui J. C., Chen W., Barnes K. M., Baron J., 2010. Changes in gene expression associated with aging commonly originate during juvenile growth. Mech. Ageing Dev. 131: 641–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyne R., Smith R., Rutherford K., Wakeling M., Varley A., et al. , 2007. FlyMine: an integrated database for Drosophila and Anopheles genomics. Genome Biol. 8: R129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X. M., Blenis J., 2009. Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell Biol. 10: 307–318 [DOI] [PubMed] [Google Scholar]
- Mackenzie D. K., Bussiere L. F., Tinsley M. C., 2011. Senescence of the cellular immune response in Drosophila melanogaster. Exp. Gerontol. 46: 853–859 [DOI] [PubMed] [Google Scholar]
- Majewski J., Pastinen T., 2011. The study of eQTL variations by RNA-seq: from SNPs to phenotypes. Trends Genet. 27: 72–79 [DOI] [PubMed] [Google Scholar]
- Mauricio R., Rausher M. D., Burdick D. S., 1997. Variation in the defense strategies of plants: Are resistance and tolerance mutually exclusive? Ecology 78: 1301–1311 [Google Scholar]
- McKean K., Nunney L., 2001. Increased sexual activity reduces male immune function in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 98: 7904–7909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. R., White A., Boots M., 2005. The evolution of host resistance: tolerance and control as distinct strategies. J. Theor. Biol. 236: 198–207 [DOI] [PubMed] [Google Scholar]
- Miller M. R., White A., Boots M., 2006. The evolution of parasites in response to tolerance in their hosts: the good, the bad, and apparent commensalism. Evolution 60: 945–956 [PubMed] [Google Scholar]
- Miron M., Sonenberg N., 2001. Regulation of translation via TOR signaling: insights from Drosophila melanogaster. J. Nutr. 131: 2988S–2993S [DOI] [PubMed] [Google Scholar]
- Mocchegiani E., Malavolta M., 2004. NK and NKT cell functions in immunosenescence. Aging Cell 3: 177–184 [DOI] [PubMed] [Google Scholar]
- Morales-Hojas R., Vieira C. P., Reis M., Vieira J., 2009. Comparative analysis of five immunity-related genes reveals different levels of adaptive evolution in the virilis and melanogaster groups of Drosophila. Heredity 102: 573–578 [DOI] [PubMed] [Google Scholar]
- Nussey D. H., Coulson T., Festa-Bianchet M., Gaillard J.-M., 2008a. Measuring senescence in wild animal populations: towards a longitudinal approach. Funct. Ecol. 22: 393–406 [Google Scholar]
- Nussey D. H., Wilson A. J., Morris A., Pemberton J., Clutton-Brock T., et al. , 2008b. Testing for genetic trade-offs between early- and late-life reproduction in a wild red deer population. Proc. Biol. Sci. 275: 745–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obbard D. J., Welch J. J., Kim K. W., Jiggins F. M., 2009. Quantifying adaptive evolution in the Drosophila immune system. PLoS Genet. 5: e1000698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver T. A., Garfield D. A., Manier M. K., Haygood R., Wray G. A., et al. , 2010. Whole-genome positive selection and habitat-driven evolution in a shallow and a deep-sea urchin. Genome Biol. Evol. 2: 800–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paaby A., Schmidt P., 2008. Functional significance of allelic variation at methuselah, an aging gene in Drosophila. PLoS One 3: e1987
- Painter R. H., 1958. Resistance of plants to insects. Annu. Rev. Entomol. 3: 276–290 [Google Scholar]
- Panda A., Arjona A., Sapey E., Bai F., Fikrig E., et al. , 2009. Human innate immunosenescence: causes and consequences for immunity in old age. Trends Immunol. 30: 325–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawelec G., 2006. Immunity and ageing in man. Exp. Gerontol. 41: 1239–1242 [DOI] [PubMed] [Google Scholar]
- Pinti M., Troiano L., Nasi M., Moretti L., Monterastelli E., et al. , 2002. Genetic polymorphisms of Fas (CD95) and FasL (CD178) in human longevity: studies on centenarians. Cell Death Differ. 9: 431–438 [DOI] [PubMed] [Google Scholar]
- Plackett T. P., Boehmer E. D., Faunce D. E., Kovacs E. J., 2004. Aging and innate immune cells. J. Leukoc. Biol. 76: 291–299 [DOI] [PubMed] [Google Scholar]
- Pletcher S. D., Macdonald S. J., Marguerie R., Certa U., Stearns S. C., et al. , 2002. Genome-wide transcript profiles in aging and calorically restricted Drosophila melanogaster. Curr. Biol. 12: 712–723 [DOI] [PubMed] [Google Scholar]
- Ponnappan U., Cinader B., Gerber V., Blaser K., 1992. Polymorphism of age-related changes in the antibody response to the hapten phosphorylcholine. Immunol. Invest. 21: 637–648 [DOI] [PubMed] [Google Scholar]
- Ramsden S., Cheung Y. Y., Seroude L., 2008. Functional analysis of the Drosophila immune response during aging. Aging Cell 7: 225–236 [DOI] [PubMed] [Google Scholar]
- Read A. F., Graham A. L., Raberg L., 2008. Animal defenses against infectious agents: is damage control more important than pathogen control. PLoS Biol. 6: e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren C., Webster P., Finkel S., Tower J., 2007. Increased internal and external bacterial load during Drosophila aging without life-span trade-off. Cell Metab. 6: 144–152 [DOI] [PubMed] [Google Scholar]
- Robertson A., 1959. The sampling variance of the genetic correlation coefficient. Biometrics 15: 469–485 [Google Scholar]
- Roy B. A., Kirchner J. W., 2000. Evolutionary dynamics of pathogen resistance and tolerance. Evolution 54: 51–63 [DOI] [PubMed] [Google Scholar]
- Sackton T. B., Lazzaro B. P., Clark A. G., 2010. Genotype and gene expression associations with immune function in Drosophila. PLoS Genet. 6: e1000797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salih D. A., Brunet A., 2008. FoxO transcription factors in the maintenance of cellular homeostasis during aging. Curr. Opin. Cell Biol. 20: 126–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambhara S., Plowden J., Renshaw-Hoelscher M., Engleman C., Katz J., 2004. Innate immunity in aging: impact on macrophage function. Aging Cell 3: 161–167 [DOI] [PubMed] [Google Scholar]
- Sarup P., Sorensen P., Loeschcke V., 2011. Flies selected for longevity retain a young gene expression profile. Age (Omaha) 33: 69–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenke T. A., Begun D. J., 2003. Natural selection drives Drosophila immune system evolution. Genetics 164: 1471–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider D. S., Ayres J. S., 2008. Two ways to survive infection: what resistance and tolerance can teach us about treating infectious diseases. Nat. Rev. Immunol. 8: 889–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz J. L., Quinn W. J., III, Cancro M. P., 2009. B-cell repertoire changes in model models of aging, pp. 393–414 Handbook on Immunosenescence: Basic Understanding and Clinical Applications, edited by Fulop T., Franceschi C., Hirokawa K., Pawelec G. Springer Netherlands, Dordrecht, The Netherlands [Google Scholar]
- Shaw A. C., Joshi S., Greenwood H., Panda A., Lord J. M., 2010. Aging of the innate immune system. Curr. Opin. Immunol. 22: 507–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton D. N., Chang E., Whittier P. S., Choi D., Funk W. D., 1999. Microarray analysis of replicative senescence. Curr. Biol. 9: 939–945 [DOI] [PubMed] [Google Scholar]
- Shinzawa N., Nelson B., Aonuma H., Okado K., Fukumoto S., et al. , 2009. p38 MAPK-dependent phagocytic encapsulation confers infection tolerance in Drosophila. Cell Host Microbe 6: 244–252 [DOI] [PubMed] [Google Scholar]
- Soerensen M., Dato S., Christensen K., McGue M., Stevnsner T., et al. , 2010. Replication of an association of variation in the FOXO3A gene with human longevity using both case-control and longitudinal data. Aging Cell 9: 1010–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson E. I., Raberg L., 2010. Resistance and tolerance in animal enemy-victim coevolution. Trends Ecol. Evol. 25: 267–274 [DOI] [PubMed] [Google Scholar]
- Terrón M. P., Paredes S. D., Barriga C., Ortega E., Rodríguez A. B., 2004. Comparative study of the heterophil phagocytic function in young and old ring doves (Streptopelia risoria) and its relationship with melatonin levels. J. Comp. Physiol. B 174: 421–427 [DOI] [PubMed] [Google Scholar]
- Tschirren B., Råberg L., Westerdahl H., 2011. Signatures of selection acting on the innate immunity gene Toll-like receptor 2 (TLR2) during the evolutionary history of rodents. J. Evol. Biol. 24: 1232–1240 [DOI] [PubMed] [Google Scholar]
- Tweedie S., Ashburner M., Falls K., Leyland P., McQuilton P., et al. , 2009. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 37: D555–D559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodovar N., Vinals M., Liehl P., Basset A., Degrouard J., et al. , 2005. Drosophila host defense after oral infection by an entomopathogenic Pseudomonas species. Proc. Natl. Acad. Sci. USA 102: 11414–11419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voolstra C. R., Sunagawa S., Matz M. V., Bayer T., Aranda M., et al. , 2011. Rapid evolution of coral proteins responsible for interaction with the environment. PLoS ONE 6: e20392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A., Charmantier A., Hadfield J., 2008. Evolutionary genetics of ageing in the wild: empirical patterns and future perspectives. Funct. Ecol. 22: 431–442 [Google Scholar]
- Wilson A. J., Nussey D. H., Pemberton J. M., Pilkington J. G., Morris A., et al. , 2007. Evidence for a genetic basis of aging in two wild vertebrate populations. Curr. Biol. 17: 2136–2142 [DOI] [PubMed] [Google Scholar]
- Wu Q., Brown M. R., 2006. Signaling and function of insulin-like peptides in insects. Annu. Rev. Entomol. 51: 1–24 [DOI] [PubMed] [Google Scholar]
- Wu Z., Irizarry R., 2004. Preprocessing of oligonucleotide array data. Nat. Biotechnol. 22: 656–658, author reply 658 [DOI] [PubMed] [Google Scholar]
- Youngman M. J., Rogers Z. N., Kim D. H., 2011. A decline in p38 MAPK signaling underlies immunosenescence in Caenorhabditis elegans. PLoS Genet. 7: e1002082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zar J. H., 1999. Biostatistical Analysis, Ed. 4 Prentice-Hall, Upper Saddle River, NJ [Google Scholar]
- Zerofsky M., Harel E., Silverman N., Tatar M., 2005. Aging of the innate immune response in Drosophila melanogaster. Aging Cell 4: 103–108 [DOI] [PubMed] [Google Scholar]
- Zheng Y., Wang J. L., Liu C., Wang C. P., Walker T., et al. , 2011. Differentially expressed profiles in the larval testes of Wolbachia infected and uninfected Drosophila. BMC Genomics 12: 595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.