Abstract
Although the use of hydrogen exchange (HX) mass spectrometry (MS) to study proteins and protein conformation is now over 20 years old, the perception lingers that it still has “issues”. Is this method, in fact, still in the quicksand with many remaining obstacles to overcome? We do not think so. This critical insight addresses the “issues” and explores several broad questions including: have the limitations of HX MS been surmounted and has HX MS achieved “indispensable” status in the pantheon of protein structural analysis tools.
Introduction
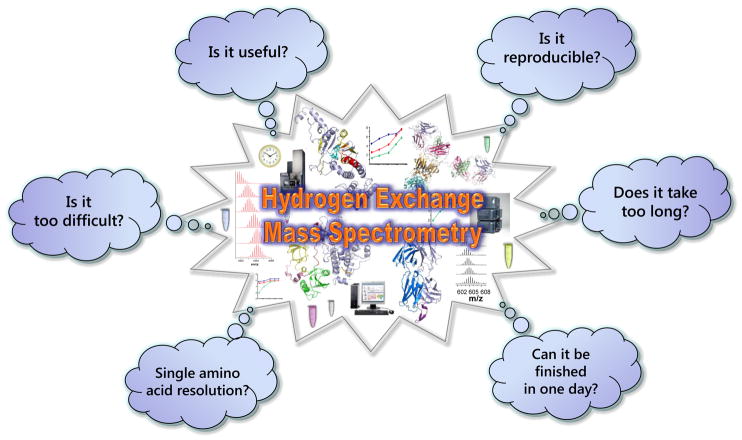
Twenty years ago, Katta and Chait [1] showed that mass spectrometry (MS) could be used to monitor hydrogen exchange (HX) – and therefore protein conformation. Since then, the field has been rapidly growing due to remarkable advances in protein availability, instrumentation and software. However, despite all the successes that hydrogen exchange mass spectrometry has witnessed over the years, there are still some who doubt that this method can provide anything useful and that it can be routinely applied to the study of protein conformation and dynamics. Is HX MS, in fact, still in the quicksand with many remaining obstacles to overcome? This critical insight is not intended to be a “classic” review but rather it is the authors’ collection of opinions which answer the basic questions (Figure 1): Have the limitations of HX MS been surmounted and has HX MS achieved “indispensable” status in the pantheon of protein structural analysis tools?
Figure 1.
Questions to ponder about modern hydrogen exchange mass spectrometry.
Is HX MS useful for protein structural analysis?
Since the early reports of Anfinsen [2,3], it is clear that protein structure is tied to protein function and vice versa; therefore understanding the structure of proteins provides tremendous insight into how proteins function and what their roles are in living organisms. The well-established structural methods (X-ray crystallography, NMR, and cryoEM) can routinely provide highly detailed structural data for many proteins. For these methods, structural analysis may require large quantities of soluble protein, often at a high concentration. However, some proteins cannot be produced in the quantities that might be required, some proteins may aggregate or precipitate at higher concentrations, or some are, for a variety of reasons, just simply not amenable to crystallization. HX MS is especially suited for analysis of these “difficult and otherwise incompatible” proteins that cannot be studied with methods like crystallography, NMR and cryoEM. With only a nanomole of relatively dilute protein, details of conformation and conformational changes can be obtained with HX MS [4–10]. A high-resolution structure cannot be obtained from HX MS and atomic coordinates cannot be generated, but, following the old adage, something is better than nothing for these proteins that cannot be analyzed by other methods.
For proteins that already have high-resolution structures, these structures become even more valuable with the addition of HX information. HX MS does not supersede X-ray crystallography or NMR structural analysis. On the contrary, it is highly complementary to these methods, and HX MS data becomes more powerful when interpreted in the light of three dimensional structural information, providing substantial evidence about flexibility and solution dynamics, changes in conformation during function, and interaction surfaces (e.g., [11–17]). Protein function is influenced by both protein structure and protein dynamics/flexibility. Monitoring hydrogen exchange (by MS or other means) provides information on protein motion/flexibility [11,12,18–23] and the impact of ligand binding on protein dynamics (e.g., [24–29]). For example Kong et al. [27] studied the binding reaction of the HIV-1 gp120 envelope glycoprotein to the CD4 receptor by HX MS and they noticed that neither unliganded gp120 nor free CD4 were substantially unstructured, suggesting that most of the diverse conformations that make up the gp120 unliganded state are reasonably ordered. In a study of nucleoside reverse transcriptase inhibitors against HIV [29], HX MS showed revealed allosteric changes in particular domains of the protein. Tiyanont et al. [30] recently described analysis of the Notch activation switch, revealing that in the autoinhibited conformation, an important site (S2) is protected against exchange but that conversion to the active state leads to accelerated deuteration in and around the S2 site. HX MS analysis of a Notch transcription complex described the details of conformational changes during protein-protein interactions, flexibility changes during complex formation, and the relative contributions of each member of the complex [31]. Details of the interactions between the molecular chaperone Hsp90 and its client transfer protein Sti1 [32] recently showed which regions of Hsp90 interact with Sti1 and new details about Hsp90 molecular mechanisms. And the list goes on…. There are many, many more examples of the positive and lasting impact of HX MS measurements on the field of structural biology. Overall, we think HX MS is very useful as a structural analysis tool, especially when combined with other structural techniques.
Is HX MS reproducible?
Reproducibility is an important question to address when thinking about HX MS. If an experiment is run today and a replicate experiment in a month or a year, how reproducible are the measurements? What are the error bars one can expect and can reliable, reproducible results be obtained? Given the large number of variables in an HX MS experiment, many things must be controlled to make the overall experiment a success.
Global reproducibility in HX MS is defined by the combined reproducibility of all the variables within the entire HX MS experiment, beginning to end. The major variables are: the protein preparation itself (equilibrium population, concentration, protein pI, minor components in the purified preparation); the composition of the buffers used to do the labeling; the pH and temperature of labeling and quenching; the digestion conditions (time, pH, temperature, and concentration of protein and enzyme); the LC/MS analysis (time, pH and temperature); and the mass accuracy of the mass spectrometer. In order to obtain the most reproducible results, each of these variables must be understood, controlled and made as nearly identical as possible.
Some variables (e.g., pH) have large effects on the overall experiment and others (e.g., mass spectrometer accuracy) have smaller effects. The mass accuracy of the mass spectrometer makes only a minor contribution to the reproducibility of measuring deuterium in a peptide or protein. The majority of modern instruments can easily measure the mass of a peptide with tolerance of ±0.01 Da or less. What contributes more to variability in the mass measurement is the fact that what we want to experimentally determine in HX MS is isotopic enrichment, which by its very nature changes the mass. So the variability associated with mass measurements does not come primarily from the mass spectrometer accuracy, but from the isotope pattern that is measured. The isotope pattern that is measured is a result of the way in which the isotope was incorporated. Because we always label an equilibrium population of protein molecules that are not all in the identical conformation state (the native state ensemble [33–35]), there will be variability in the HX that occurs into each of the different co-existing conformational forms. We always, therefore, measure the average of this ensemble by finding the center of the isotopic distribution [4,36]. Perturbing the native state ensemble (with buffer conditions, concentration, pH, and temperature) will change the average deuterium incorporation and therefore the mass that is measured. Just a slight variation in labeling pH (even 0.2 units or less) will change the deuterium incorporation rate [37–39] because exchange is a function of both acid and base concentration, and varies about 10-fold per pH unit with a dependence on primary structure [40,41]. Variability in the quench and LC analysis pH (as well as temperature and time) can alter both the deuterium back-exchange rate and the digestion pattern of the protein analyzed [1,4]. The better the pH is controlled – with buffers, reproducible pipetting and transfer time – the more reproducible each of the steps affected by pH will be, and therefore the higher the global HX MS reproducibility will be.
With the variables just described in mind, how are we doing? Is it possible to simultaneously control all these seemingly complex variables and be reproducible? Anecdotal evidence and experience have indicated that, yes, the measurements can be done with a fair degree of reproducibility. The anecdotal evidence has recently been supported by detailed measurements from several laboratories which have measured the error associated with HX MS experiments [42–44]. In the majority of the published cases, the inclusion of robotics (e.g., Ref. [45]) was essential to obtaining a large enough dataset to quantify the reproducibility and to making the measurements as reproducible as possible. Chalmers et al. [45] found that following triplicate experiments, the average standard deviation of the mass measurement across 52 different deuterated peptides was 0.08 Da, with a mean standard deviation of the percent deuterium incorporated of 1.4%. Chalmers et al. more recently [44] validated the use of HX MS for ligand screening and obtained data for 127 replicate measurements for a large number of deuterated peptides over an eight month period. Based on their measurements 96% of all measured percent deuterium values (4039 measurements in all) were within 10% of the mean value obtained during that period and 3452 values (or 82% of all measurements) were within 5% of the mean. Burkitt et al. [42] tested the reproducibility of an automated system over a two month period, and they found the intraday repeatability in measured exchange values of deuterated peptides to be between 0.2–0.9% deuterium [coefficients of variation (CVs) for intraday repeatability ranged from 0.3–1.5%]. For interday reproducibility, the variation was between 1.0–2.2% deuterium [CVs of 1.9–3.8%]. Houde et al. [43] found the global error of each mass measurement of deuterated peptides to be about ±0.14 Da, and this deviation was shown to be independent of peptide size, deuteration time, or the magnitude of the mass difference between undeuterated and deuterated. They calculated with 98% confidence that differences of ±0.5 Da between relative measurements were significant differences and well outside the error range of the experiment. Consistent with the Burkitt et al. and Chalmers et al. reports [42,44,45], Houde et al. described comparability studies and also concluded that interday analyses were significantly more variable than intraday analyses. This implies that, as discussed above, the experimental conditions all must be precisely controlled before interday comparisons can be conducted with high reliability. The studies that were just illustrated included robotics which were extremely valuable for reducing variability to the lowest levels. Robots can be helpful in all sample handling steps, including labeling and subsequent LC/MS analysis.
Another way to reduce variability is to make relative measurements. Relative measurements are found throughout analytical chemistry, for example in double-beam UV-Vis spectrometers where the solvent is automatically compensated for with a cuvette in a reference beam, or in the thermal conductivity detector in gas chromatography where the reference flow gas is compared to the column flow via a Wheatstone bridge [46]. We have long been advocates of relative measurements in HX MS [9] wherein a reference protein condition is compared to a test condition. That is, HX is performed on two states of the protein in question (bound vs. free, wild-type vs. mutant, buffer A vs. buffer B, etc.) using the most similar conditions possible – same protein stock solution, same buffers, same instrumental setup, analyses done very close in time, etc. The difference between HX into the two states is monitored with the goal of first locating where HX differences exist, and second what the magnitude of the differences is. In relative measurements, many variables then cancel out, for example minor changes in temperature, pH, time of digestion, amounts of back-exchange, etc. apply equally to both the reference state and to the test state. It is still very important to know the error of each data point even in a relative experiment (see Ref. [43]).
We think reproducibility is presently not a limitation to the technique of HX MS.
Is HX MS just too difficult to do and does it take too long?
For historical reasons, HX MS is often perceived as a method that “…just takes way too long and is just way too hard” (anonymous, personal communication). If these perceptions are true, one might see how it would be difficult to justify adopting HX MS, especially in industrial settings where there is often not the luxury of doing an experiment in which it may take weeks or even months to develop the methods and interpret the results. These perceptions about HX MS might have been true in the early days, but they are certainly not true in 2012. The following section explores some of the issues and where we stand currently.
Are there too many variables to control?
As described above in the section on reproducibility, there are a lot of variables one must keep under control to be successful. This can be a daunting task to the uninitiated, but it just requires careful planning to surmount. In the words of Norman Vincent Peale, “Plan your work–work your plan” [47]. One way to ease the control over all the variables is to automate. A number of recent articles have described robotics that can significantly reduce variability [42,43,45,48–50]. One caveat is that the initial setup of the sample handling system takes more time than simply pipetting deuterium into an eppendorf tube. However, once set up, fully operational and well understood, robotic systems can help control the many variables and make HX MS experiments significantly easier to perform reproducibly.
Are the quench conditions too restrictive?
Another complaint is that due to the restrictive conditions one must adopt in order to preserve the deuterium label, namely short analysis times and chromatography at zero degrees, it is too difficult to do the experiment and maintain the deuterium label (i.e., too much back-exchange). For those used to long separation times and for not being fastidious with the details of the separation, adapting to the fast separation times required can be a challenge. It is not just doing a separation fast; it is also doing a high-quality, efficient separation fast and at 0 °C (see next section). One solution that may seem to simplify the problem is to use MALDI instead of LC electrospray MS, therefore eliminating the entire LC step. While MALDI can be used successfully in HX MS (e.g., [51–55]), the disadvantage is that there is more back-exchange than well controlled LC electrospray MS experiments and that all peptides one wishes to study are in a single spectrum. Lack of chromatography limits the protein size that can be analyzed owing to peak capacity problems and ionization efficiencies. Another reported solution to get around back-exchange was to use super-critical fluid chromatography (SFC) [56] but this solution is not practical as it requires a new separation column for each injection. We think the restrictive quench conditions actually sound much harder than they are.
Is separation too poor at quench conditions?
Related to the previous section, chromatographic performance at low temperature is not stellar. One way to fix this is to use higher performance separations systems such as UPLC/UHPLC [57,58]. The most important factor is to do the chromatography with sub-2-micron particles where the separation efficiency at 0 °C is vastly superior to that of more conventional HPLC particles of 3.0–3.5 microns in diameter. With sub-2-micron particles, the deuterium recovery is generally higher since the gradient time can be made shorter while still maintaining separation quality. The Waters Corporation commercialized (both pumps and columns) the use of 1.7 micron particles in 2004 [59] and there are now many other instrument vendors that also provide commercial systems compatible with small diameter separation particles. We adopted UPLC separations for HX MS [60,61] and now use UPLC exclusively; sub-2-micron particles are now also used by other groups practicing HX MS (e.g., [62–64]. Peptides from the digestion of protein systems in excess of 200 kDa of unique sequence can be efficiently separated using UPLC at HX MS quench conditions [64]. Other alternatives to the traditional 3.5 micron reversed-phase HPLC particles could also be used to significantly improve the separation efficiency. We have recently explored whether additional resolving power could be incorporated into the overall HX MS experiment after the chromatography step by using ion mobility spectrometry (IMS) [65]. Including IMS to increase the resolving power is not a new concept (e.g., [66–69]). The deconvolution of overlapping peptides unresolved in the chromatographic separation or isotope distributions that become unresolved with addition of deuterium can be separated in many cases with IMS. More work in this area will hopefully see IMS routinely incorporated to increase resolving power in the overall HX MS experiment, especially now that commercially available MS IMS instruments have appeared.
Are both LC and MS skills required?
In the early days of HX MS, now some 15 years ago or so, combining LC with ESI was not as routine as it is now in 2012. Because both an LC system and mass spectrometer must be working well at the same time, and interfaced together, there were some difficulties for those that were not skilled in making this entire two-instrument setup function all at the same time. Those problems are mostly overcome at this point and many operators are very skilled in the combination of LC with MS.
Is it too hard to identify pepsin fragments?
Because the digestion step must be done with an acid protease active in quench conditions [4,70,71], pepsin is often used (see also below). Pepsin digestion is not predictable like digestion with trypsin or other enzymes (e.g., Asp-N, Glu-C, etc.) and likely has much to do with the acid-induced molten globule conformation at low pH. Therefore, all the peptide fragments must be identified. Modern MS/MS techniques, many of them automated, have made this task straightforward and much less of an impediment than it once was. Even as recently as 10 years ago, the rapid identification of pepsin fragments was often challenging.
Is digestion efficiency a problem?
To retain as much deuterium as possible in labeled samples, the time for analysis must be short – this includes digestion time. There is not the luxury of digesting overnight at 37 °C. To improve digestion efficiency, many have adopted online pepsin digestion (first described in 1999 [72] and expanded upon later [73]) because the enzyme:protein ratio can be made much higher than in-solution digestions. Alternatives to pepsin have also impacted efficiency [74–78]. Another modification that improves the digestion efficiency is including denaturants and reducing agents in the digestion step – first described by Woods et al. [79] and used by many since (e.g., [80,81]). With these modern digestion techniques, it is possible to digest very resistant proteins completely within 20–30 seconds at zero degrees, and produce a number of overlapping peptides (see also below).
Are the mass spectrometers good enough?
Historically, instrument sensitivity was a problem for proteins that were only available in small quantities. Huge improvements in instrument sensitivity and duty cycle in recent years have almost made this a non-issue. Analysis can be done currently with as little as 1–5 pmoles of protein per injection, which is perhaps an improvement of 50–100 fold over that of a decade or more ago [4,82–86]. We find that response is still variable depending on the protein, so the values listed here are not a hard rule. Given the high-quality and sensitive mass spectrometers available today, instrument sensitivity is a very minor issue for HX MS.
Does it take too long?
An HX MS experiment can be broken into three main components/stages: deuterium labeling, mass analysis and data processing. The maximum amount of time required for the deuterium labeling portion of the experiment is a function of the longest time point of deuterium exposure, typically 4–8 hours or more. The amount of time required to acquire the MS data is a function of the digestion, chromatography and system recovery time. The time required for both of these first two components of the total HX MS experiment has stayed relatively the same since the first experiments were reported, although these two parts can be done in parallel (i.e., LC/MS analysis of shorter labeling times while the longer labeling times are incubating in D2O, etc.) to save some time. Better technology, including faster computers, better operating systems and instrument control software, and user-friendly graphic interfaces have all contributed to making the analysis easier and faster. There have been huge reductions in the time required for the final component, the data analysis step. Data processing and analysis software has significantly reduced the amount of time required for interpreting the results. Before covering the software advancements in detail, we will take a slight detour to discuss protein size limits as that is related to the overall issue of analysis time.
What about protein size?
Given that manual data processing and interpretation can be an extremely long and laborious process in HX MS (taking weeks or months perhaps), there was an artificial limit placed on the maximum size of protein that people wanted to work with – big: it will take “forever” to process the data so it is not a practical/manageable experiment; small: data processing was manageable. Until several years ago, before the advent of software to aid in processing, the “too big” limit was something around 50 kDa whereas today with progress in software, the limit is closer to 300 kDa. Therefore a benefit of developing software for data processing is that it raises the size limit imposed by the processing burden. A goal of ours is for the maximum size/complexity that can be analyzed with HX MS to be limited by separation power rather than by processing capability. We are approaching that point now, hence the need to further improve separation capacity as described above.
Is there software for HX MS data analysis?
Without data processing software, routine HX MS is generally out of reach for many, especially in an industrial setting. Starting around 8–10 years ago, a number of software platforms began to become available to address these issues (Supplementary Table S1). These software advancements have changed data processing from weeks to hours, and many software platforms include visualization tools for easier interpretation. All of the available software are not created equal however, so caution is still required. Just as one would test-drive and compare mass spectrometers to see which is a good fit, one should test-drive and compare HX MS software platforms, many of which are freely available.
We think that HX MS is not too difficult and that it does not take too long. Many of the issues that led to this reputation have been resolved and we think it is now a practical and robust method.
Can a complete HX MS experiment be done in one day?
One goal would be complete labeling, data acquisition and analysis within a single day, or even within the span of the longest time point of labeling. Is this feasible? The answer is it depends on multiple factors such as the size of the protein, digestion efficiency, operators’ experience, prior knowledge about the protein(s) that is/are interrogated, software availability, etc. In well understood protein systems with which the analyst has some experience, one could do an HX MS experiment and data analysis in a single day. But realistically speaking it generally does not happen that a completely new protein that has never been worked with before can be completely analyzed in a single day including replicate analyses. Given the technological progress in HX MS, we think it is currently realistic for the average laboratory to do an entire set of HX MS experiments (including 3+ replicates) within a week for proteins of modest (<30–50 kDa) size. More sophisticated setups and researchers will be able to realize much more in a shorter amount of time.
Is single amino acid resolution possible?
In HX detected by NMR, exchange can be measured for individual backbone amide hydrogens [87,88]. The quality of the NMR spectra dictates if every amide hydrogen can be measured but in general, the spatial resolution of HX NMR is significantly higher than HX MS that relies solely on pepsin digestion. A long standing goal has been to improve the spatial resolution of HX MS experiments in order to locate the deuterium with, at best, single backbone amide hydrogen resolution. Two ways to improve the resolution are to make shorter and shorter pepsin fragments (including perhaps overlapping fragments), or to use MS/MS to fragment deuterium labeled peptides in the gas-phase inside the mass spectrometer and locate the deuterium in the resulting fragment ions.
The aspiration of the HX MS community to achieve single-residue resolution with MS/MS started early on, with the first papers appearing in 1999 [89,90]. It was found that collision induced dissociation (CID) led to deuterium scrambling whereas ECD and ETD fragmentation did not, under the right experimental conditions (see Ref. [91–94] and references therein for a comprehensive summary). The issue at hand now for ETD fragmentation as it applies to HX MS experiments is widespread implementation, software development to ease data processing, and better defining the expectations that non-experts should have about this methodology. Although seemingly straightforward, especially as presented for model peptides and small proteins [95], routine ETD implementation is far from trivial [96]. Fragmentation efficiency, precursor ion selection in complex samples, electrospray ionization parameters and lack of software for ETD data processing are issues that need to be addressed [96]. It seems to us, and to others, that a targeted approach wherein only selected peptides are interrogated by ETD after HX and LC is the most viable option.
Conclusions
We hope that with this critical insight, readers will have a better idea where HX MS stands now as a method and what can be expected today when implementing HX MS as a routine technique for interrogating protein structure and dynamics. Compared with even a few years ago, things have greatly progressed in the field and many groups now employ the methodology on a routine basis. Some practical concerns remain, as do other ways to further improve the technique. Obtaining the protein of interest in its highest purity possible is an ongoing challenge and is protein dependent, dealing with interpreting data for heavily post translational modified proteins (such as heterogeneity due to glycosylation) remains challenging, ETD fragmentation to obtain amino acid resolution needs to become routine, and complete automation (including easy-to-use but reliable software) for higher-throughput is an ongoing endeavor. Overall, we can without a doubt acknowledge that HX MS has overcome many of its most stringent challenges and can currently be applied as a routine technique in a multitude of environments. From our perspective, we are out of the quicksand.
Supplementary Material
Acknowledgments
The authors would like to specifically thank Prof. Thomas E. Wales and Dr. Damian Houde for helpful discussions and suggestions related to this article. We are also grateful to the many researchers in the fields of HX MS and protein structural MS that provide continuous inspiration through their enlightening studies of proteins and protein systems. Our work has been graciously supported by the NIH (R01-GM086507) and the Waters Corporation.
References
- 1.Katta V, Chait BT. Conformational changes in proteins probed by hydrogen-exchange electrospray-ionization mass spectrometry. Rapid Commun Mass Spectrom. 1991;5:214–217. doi: 10.1002/rcm.1290050415. [DOI] [PubMed] [Google Scholar]
- 2.Anfinsen CB, Redfield RR, Choate WL, Page J, Carroll WR. Studies on the gross structure, cross-linkages, and terminal sequences in ribonuclease. J Biol Chem. 1954;207:201–210. [PubMed] [Google Scholar]
- 3.Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Z, Smith DL. Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure elucidation. Protein Sci. 1993;2:522–531. doi: 10.1002/pro.5560020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miranker A, Robinson CV, Radford SE, Dobson CM. Investigation of protein folding by mass spectrometry. FASEB J. 1996;10:93–101. doi: 10.1096/fasebj.10.1.8566553. [DOI] [PubMed] [Google Scholar]
- 6.Smith DL, Deng Y, Zhang Z. Probing the non-covalent structure of proteins by amide hydrogen exchange and mass spectrometry. J Mass Spectrom. 1997;32:135–146. doi: 10.1002/(SICI)1096-9888(199702)32:2<135::AID-JMS486>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 7.Engen JR, Smith DL. Investigating protein structure and dynamics by hydrogen exchange MS. Anal Chem. 2001;73:256A–265A. doi: 10.1021/ac012452f. [DOI] [PubMed] [Google Scholar]
- 8.Hoofnagle AN, Resing KA, Ahn NG. Protein analysis by hydrogen exchange mass spectrometry. Annu Rev Biophys Biomol Struct. 2003;32:1–25. doi: 10.1146/annurev.biophys.32.110601.142417. [DOI] [PubMed] [Google Scholar]
- 9.Wales TE, Engen JR. Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom Rev. 2006;25:158–170. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- 10.Yan X, Maier CS. Hydrogen/deuterium exchange mass spectrometry. Methods Mol Biol. 2009;492:255–271. doi: 10.1007/978-1-59745-493-3_15. [DOI] [PubMed] [Google Scholar]
- 11.Mayne L, Paterson Y, Cerasoli D, Englander SW. Effect of antibody binding on protein motions studied by hydrogen-exchange labeling and two-dimensional NMR. Biochemistry. 1992;31:10678–10685. doi: 10.1021/bi00159a006. [DOI] [PubMed] [Google Scholar]
- 12.Kim KS, Woodward C. Protein internal flexibility and global stability: Effect of urea on hydrogen exchange rates of bovine pancreatic trypsin inhibitor. Biochemistry. 1993;32:9609–9613. doi: 10.1021/bi00088a013. [DOI] [PubMed] [Google Scholar]
- 13.Engen JR, Smithgall TE, Gmeiner WH, Smith DL. Identification and localization of slow, natural, cooperative unfolding in the hematopoietic cell kinase SH3 domain by amide hydrogen exchange and mass spectrometry. Biochemistry. 1997;36:14384–14391. doi: 10.1021/bi971635m. [DOI] [PubMed] [Google Scholar]
- 14.Maier CS, Schimerlik MI, Deinzer ML. Thermal denaturation of Escherichia coli thioredoxin studied by hydrogen/deuterium exchange and electrospray ionization mass spectrometry: monitoring a two-state protein unfolding transition. Biochemistry. 1999;38:1136–1143. doi: 10.1021/bi981938w. [DOI] [PubMed] [Google Scholar]
- 15.Deng Y, Zhang Z, Smith DL. Comparison of continuous and pulsed labeling amide hydrogen exchange/mass spectrometry for studies of protein dynamics. J Am Soc Mass Spectrom. 1999;10:675–684. doi: 10.1016/S1044-0305(99)00038-0. [DOI] [PubMed] [Google Scholar]
- 16.Hoofnagle AN, Resing KA, Goldsmith EJ, Ahn NG. Changes in protein conformational mobility upon activation of extracellular regulated protein kinase-2 as detected by hydrogen exchange. Proc Natl Acad Sci U S A. 2001;98:956–961. doi: 10.1073/pnas.98.3.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iacob RE, Zhang J, Gray NS, Engen JR. Allosteric interactions between the myristate- and ATP-site of the Abl kinase. PLoS One. 2011;6:e15929. doi: 10.1371/journal.pone.0015929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hvidt A, Nielsen SO. Hydrogen exchange in proteins. Adv Protein Chem. 1966;21:287–386. doi: 10.1016/s0065-3233(08)60129-1. [DOI] [PubMed] [Google Scholar]
- 19.Englander SW, Downer NW, Teitelbaum H. Hydrogen exchange. Annu Rev Biochem. 1972;41:903–924. doi: 10.1146/annurev.bi.41.070172.004351. [DOI] [PubMed] [Google Scholar]
- 20.Woodward CK, Hilton BD. Hydrogen exchange kinetics and internal motions in proteins and nucleic acids. Annu Rev Biophys Bioeng. 1979;8:99–127. doi: 10.1146/annurev.bb.08.060179.000531. [DOI] [PubMed] [Google Scholar]
- 21.Woodward C, Simon I, Tuchsen E. Hydrogen exchange and the dynamic structure of proteins. Mol Cell Biochem. 1982;48:135–160. doi: 10.1007/BF00421225. [DOI] [PubMed] [Google Scholar]
- 22.Roder H, Wagner G, Wuthrich K. Amide proton exchange in proteins by EX1 kinetics: Studies of the basic pancreatic trypsin inhibitor at variable pH 2 and temperature. Biochemistry. 1985;24:7396–7407. doi: 10.1021/bi00346a055. [DOI] [PubMed] [Google Scholar]
- 23.Raschke TM, Marqusee S. Hydrogen exchange studies of protein structure. Curr Opin Biotechnol. 1998;9:80–86. doi: 10.1016/s0958-1669(98)80088-8. [DOI] [PubMed] [Google Scholar]
- 24.Zhu MM, Rempel DL, Du Z, Gross ML. Quantification of protein-ligand interactions by mass spectrometry, titration, and H/D exchange: PLIMSTEX. J Am Chem Soc. 2003;125:5252–5253. doi: 10.1021/ja029460d. [DOI] [PubMed] [Google Scholar]
- 25.Xiao H, Kaltashov IA, Eyles SJ. Indirect assessment of small hydrophobic ligand binding to a model protein using a combination of ESI MS and HDX/ESI MS. J Am Soc Mass Spectrom. 2003;14:506–515. doi: 10.1016/S1044-0305(03)00135-1. [DOI] [PubMed] [Google Scholar]
- 26.Brier S, Lemaire D, DeBonis S, Kozielski F, Forest E. Use of hydrogen/deuterium exchange mass spectrometry and mutagenesis as a tool to identify the binding region of inhibitors targeting the human mitotic kinesin Eg5. Rapid Commun Mass Spectrom. 2006;20:456–462. doi: 10.1002/rcm.2329. [DOI] [PubMed] [Google Scholar]
- 27.Kong L, Huang CC, Coales SJ, Molnar KS, Skinner J, Hamuro Y, Kwong PD. Local conformational stability of HIV-1 gp120 in unliganded and CD4-bound states as defined by amide hydrogen/deuterium exchange. J Virol. 2010;84:10311–10321. doi: 10.1128/JVI.00688-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chalmers MJ, Busby SA, Pascal BD, West GM, Griffin PR. Differential hydrogen/deuterium exchange mass spectrometry analysis of protein-ligand interactions. Expert Rev Proteomics. 2011;8:43–59. doi: 10.1586/epr.10.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seckler JM, Barkley MD, Wintrode PL. Allosteric suppression of HIV-1 reverse transcriptase structural dynamics upon inhibitor binding. Biophys J. 2011;100:144–153. doi: 10.1016/j.bpj.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tiyanont K, Wales TE, Aste-Amezaga M, Aster JC, Engen JR, Blacklow SC. Evidence for increased exposure of the Notch1 metalloprotease cleavage site upon conversion to an activated conformation. Structure. 2011;19:546–554. doi: 10.1016/j.str.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi SH, Wales TE, Nam Y, O’Donovan DJ, Sliz P, Engen JR, Blacklow SC. Conformational Locking upon Cooperative Assembly of Notch Transcription Complexes. Structure. 2012;20:340–349. doi: 10.1016/j.str.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee CT, Graf C, Mayer FJ, Richter SM, Mayer MP. Dynamics of the regulation of Hsp90 by the co-chaperone Sti1. Embo J. 2012 doi: 10.1038/emboj.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Creighton TE. Pathways and mechanisms of protein folding. Adv Biophys. 1984;18:1–20. doi: 10.1016/0065-227x(84)90004-2. [DOI] [PubMed] [Google Scholar]
- 34.Chothia C. Principles that determine the structure of proteins. Annu Rev Biochem. 1984;53:537–572. doi: 10.1146/annurev.bi.53.070184.002541. [DOI] [PubMed] [Google Scholar]
- 35.Matthews CR. Pathways of protein folding. Annu Rev Biochem. 1993;62:653–683. doi: 10.1146/annurev.bi.62.070193.003253. [DOI] [PubMed] [Google Scholar]
- 36.Thévenon-Emeric G, Kozlowski J, Zhang Z, Smith DL. Determination of amide hydrogen exchange rates in peptides by mass spectrometry. Anal Chem. 1992;64:2456–2458. doi: 10.1021/ac00044a027. [DOI] [PubMed] [Google Scholar]
- 37.Berger A, Linderstrom-Lang K. Deuterium exchange of poly-DL-alanine in aqueous solution. Arch Biochem Biophys. 1957;69:106–118. doi: 10.1016/0003-9861(57)90478-2. [DOI] [PubMed] [Google Scholar]
- 38.Hvidt A. A Discussion of the pH Dependence of the Hydrogen-Deuterium Exchange of Proteins. C R Trav Lab Carlsberg. 1964;34:299–317. [PubMed] [Google Scholar]
- 39.Kagi JH, Ulmer DD. Hydrogen-deuterium exchange of cytochrome c. II. Effect of pH. Biochemistry. 1968;7:2718–2723. doi: 10.1021/bi00848a004. [DOI] [PubMed] [Google Scholar]
- 40.Molday RS, Englander SW, Kallen RG. Primary structure effects on peptide group hydrogen exchange. Biochemistry. 1972;11:150–158. doi: 10.1021/bi00752a003. [DOI] [PubMed] [Google Scholar]
- 41.Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burkitt W, O’Connor G. Assessment of the repeatability and reproducibility of hydrogen/deuterium exchange mass spectrometry measurements. Rapid Commun Mass Spectrom. 2008;22:3893–3901. doi: 10.1002/rcm.3794. [DOI] [PubMed] [Google Scholar]
- 43.Houde D, Berkowitz SA, Engen JR. The utility of hydrogen/deuterium exchange mass spectrometry in biopharmaceutical comparability studies. J Pharm Sci. 2011;100:2071–2086. doi: 10.1002/jps.22432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chalmers MJ, Pascal BD, Willis S, Zhang J, Iturria SJ, Dodge JA, Griffin PR. Methods for the analysis of high precision differential hydrogen deuterium exchange data. Int J Mass Spectrom. 2011;302:59–68. doi: 10.1016/j.ijms.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chalmers MJ, Busby SA, Pascal BD, He Y, Hendrickson CL, Marshall AG, Griffin PR. Probing protein ligand interactions by automated hydrogen/deuterium exchange mass spectrometry. Anal Chem. 2006;78:1005–1014. doi: 10.1021/ac051294f. [DOI] [PubMed] [Google Scholar]
- 46.Colon LA, Baird LJ. Detectors in Modern Gas Chromatography. In: Grob RL, Barry EF, editors. Modern Practice of Gas Chromatography. 4. John Wiley & Sons; Hoboken: 2004. [Google Scholar]
- 47.Peale NV. The power of positive thinking. Random House, Inc; New York: 1952. [Google Scholar]
- 48.Woods VL, Jr, Hamuro Y. High resolution high-throughput amide deuterium exchange-mass spectrometry (DXMS) determination of protein binding site structure dynamics: utility in pharmaceutical design. J Cell Biochem Suppl. 2001;37:89–98. doi: 10.1002/jcb.10069. [DOI] [PubMed] [Google Scholar]
- 49.Hamuro Y, Coales SJ, Southern MR, Nemeth-Cawley JF, Stranz DD, Griffin PR. Rapid analysis of protein structure and dynamics by hydrogen/deuterium exchange mass spectrometry. J Biomol Tech. 2003;14:171–182. [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Q, Willison LN, Tripathi P, Sathe SK, Roux KH, Emmett MR, Blakney GT, Zhang HM, Marshall AG. Epitope mapping of a 95 kDa antigen in complex with antibody by solution-phase amide backbone hydrogen/deuterium exchange monitored by Fourier transform ion cyclotron resonance mass spectrometry. Anal Chem. 2011;83:7129–7136. doi: 10.1021/ac201501z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mandell JG, Falick AM, Komives EA. Measurement of amide hydrogen exchange by MALDI-TOF mass spectrometry. Anal Chem. 1998;70:3987–3995. doi: 10.1021/ac980553g. [DOI] [PubMed] [Google Scholar]
- 52.Andersen MD, Shaffer J, Jennings PA, Adams JA. Structural characterization of protein kinase A as a function of nucleotide binding. Hydrogen-deuterium exchange studies using matrix-assisted laser desorption ionization-time of flight mass spectrometry detection. J Biol Chem. 2001;276:14204–14211. doi: 10.1074/jbc.M011543200. [DOI] [PubMed] [Google Scholar]
- 53.Turner BT, Jr, Maurer MC. Evaluating the roles of thrombin calcium in the activation of coagulation factor XIII using H/D exchange and MALDI-TOF MS. Biochemistry. 2002;41:7947–7954. doi: 10.1021/bi025630n. [DOI] [PubMed] [Google Scholar]
- 54.Li X, Hood RJ, Wedemeyer WJ, Watson JT. Characterization of peptide folding nuclei by hydrogen/deuterium exchange-mass spectrometry. Protein Sci. 2005;14:1922–1928. doi: 10.1110/ps.051458905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Woofter RT, Maurer MC. Role of calcium in the conformational dynamics of factor XIII activation examined by hydrogen-deuterium exchange coupled with MALDI-TOF MS. Arch Biochem Biophys. 2011;512:87–95. doi: 10.1016/j.abb.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Emmett MR, Kazazic S, Marshall AG, Chen W, Shi SD, Bolanos B, Greig MJ. Supercritical fluid chromatography reduction of hydrogen/deuterium back exchange in solution-phase hydrogen/deuterium exchange with mass spectrometric analysis. Anal Chem. 2006;78:7058–7060. doi: 10.1021/ac060693n. [DOI] [PubMed] [Google Scholar]
- 57.Neue UD. HPLC Columns: Theory, Technology and Practice. John Wiley and Sons; New York: 1997. [Google Scholar]
- 58.Jorgenson JW. Capillary liquid chromatography at ultrahigh pressures. Annu Rev Anal Chem (Palo Alto Calif) 2010;3:129–150. doi: 10.1146/annurev.anchem.1.031207.113014. [DOI] [PubMed] [Google Scholar]
- 59.Plumb R, Castro-Perez J, Granger J, Beattie I, Joncour K, Wright A. Ultra-performance liquid chromatography coupled to quadrupole-orthogonal time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 2004;18:2331–2337. doi: 10.1002/rcm.1627. [DOI] [PubMed] [Google Scholar]
- 60.Wu Y, Engen JR, Hobbins WB. Ultra performance liquid chromatography (UPLC) further improves hydrogen/deuterium exchange mass spectrometry. J Am Soc Mass Spectrom. 2006;17:163–167. doi: 10.1016/j.jasms.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 61.Wales TE, Fadgen KE, Gerhardt GC, Engen JR. High-speed and high-resolution UPLC separation at zero degrees Celsius. Anal Chem. 2008;80:6815–6820. doi: 10.1021/ac8008862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chalmers MJ, Busby SA, Pascal BD, Southern MR, Griffin PR. A two-stage differential hydrogen deuterium exchange method for the rapid characterization of protein/ligand interactions. J Biomol Tech. 2007;18:194–204. [PMC free article] [PubMed] [Google Scholar]
- 63.Jones LM, Zhang H, Vidavsky I, Gross ML. Online, high-pressure digestion system for protein characterization by hydrogen/deuterium exchange and mass spectrometry. Anal Chem. 2010;82:1171–1174. doi: 10.1021/ac902477u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Houde D, Demarest SJ. Fine details of IGF-1R activation, inhibition, and asymmetry determined by associated hydrogen/deuterium-exchange and peptide mass mapping. Structure. 2011;19:890–900. doi: 10.1016/j.str.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 65.Iacob RE, Murphy JP, 3rd, Engen JR. Ion mobility adds an additional dimension to mass spectrometric analysis of solution-phase hydrogen/deuterium exchange. Rapid Commun Mass Spectrom. 2008;22:2898–2904. doi: 10.1002/rcm.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ruotolo BT, Gillig KJ, Stone EG, Russell DH. Peak capacity of ion mobility mass spectrometry: separation of peptides in helium buffer gas. J Chromatogr B Analyt Technol Biomed Life Sci. 2002;782:385–392. doi: 10.1016/s1570-0232(02)00566-4. [DOI] [PubMed] [Google Scholar]
- 67.Hilderbrand AE, Myung S, Barnes CA, Clemmer DE. Development of LC-IMS-CID-TOFMS techniques: analysis of a 256 component tetrapeptide combinatorial library. J Am Soc Mass Spectrom. 2003;14:1424–1436. doi: 10.1016/j.jasms.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 68.Ruotolo BT, McLean JA, Gillig KJ, Russell DH. Peak capacity of ion mobility mass spectrometry: the utility of varying drift gas polarizability for the separation of tryptic peptides. J Mass Spectrom. 2004;39:361–367. doi: 10.1002/jms.592. [DOI] [PubMed] [Google Scholar]
- 69.Valentine SJ, Liu X, Plasencia MD, Hilderbrand AE, Kurulugama RT, Koeniger SL, Clemmer DE. Developing liquid chromatography ion mobility mass spectometry techniques. Expert Rev Proteomics. 2005;2:553–565. doi: 10.1586/14789450.2.4.553. [DOI] [PubMed] [Google Scholar]
- 70.Rosa JJ, Richards FM. An experimental procedure for increasing the structural resolution of chemical hydrogen-exchange measurements on proteins: application to ribonuclease S peptide. J Mol Biol. 1979;133:399–416. doi: 10.1016/0022-2836(79)90400-5. [DOI] [PubMed] [Google Scholar]
- 71.Englander JJ, Rogero JR, Englander SW. Protein hydrogen exchange studied by the fragment separation method. Anal Biochem. 1985;147:234–244. doi: 10.1016/0003-2697(85)90033-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ehring H. Hydrogen exchange/electrospray ionization mass spectrometry studies of structural features of proteins and protein/protein interactions. Anal Biochem. 1999;267:252–259. doi: 10.1006/abio.1998.3000. [DOI] [PubMed] [Google Scholar]
- 73.Wang L, Pan H, Smith DL. Hydrogen exchange-mass spectrometry: optimization of digestion conditions. Mol Cell Proteomics. 2002;1:132–138. doi: 10.1074/mcp.m100009-mcp200. [DOI] [PubMed] [Google Scholar]
- 74.Cravello L, Lascoux D, Forest E. Use of different proteases working in acidic conditions to improve sequence coverage and resolution in hydrogen/deuterium exchange of large proteins. Rapid Commun Mass Spectrom. 2003;17:2387–2393. doi: 10.1002/rcm.1207. [DOI] [PubMed] [Google Scholar]
- 75.Mazon H, Marcillat O, Forest E, Vial C. Local dynamics measured by hydrogen/deuterium exchange and mass spectrometry of creatine kinase digested by two proteases. Biochimie. 2005;87:1101–1110. doi: 10.1016/j.biochi.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 76.Brier S, Maria G, Carginale V, Capasso A, Wu Y, Taylor RM, Borotto NB, Capasso C, Engen JR. Purification and characterization of pepsins A1 and A2 from the Antarctic rock cod Trematomus bernacchii. Febs J. 2007;274:6152–6166. doi: 10.1111/j.1742-4658.2007.06136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rey M, Man P, Brandolin G, Forest E, Pelosi L. Recombinant immobilized rhizopuspepsin as a new tool for protein digestion in hydrogen/deuterium exchange mass spectrometry. Rapid Commun Mass Spectrom. 2009;23:3431–3438. doi: 10.1002/rcm.4260. [DOI] [PubMed] [Google Scholar]
- 78.Marcoux J, Thierry E, Vives C, Signor L, Fieschi F, Forest E. Investigating alternative acidic proteases for H/D exchange coupled to mass spectrometry: plasmepsin 2 but not plasmepsin 4 is active under quenching conditions. J Am Soc Mass Spectrom. 2010;21:76–79. doi: 10.1016/j.jasms.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 79.Woods VL., Jr Methods for the high-resolution identification of solvent-accessible amide hydrogens in polypeptides or proteins and for characterization of the fine structure of protein binding sites. U.S. Patent 6,291,189. 2001
- 80.Iacob RE, Pene-Dumitrescu T, Zhang J, Gray NS, Smithgall TE, Engen JR. Conformational disturbance in Abl kinase upon mutation and deregulation. Proc Natl Acad Sci U S A. 2009;106:1386–1391. doi: 10.1073/pnas.0811912106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang HM, McLoughlin SM, Frausto SD, Tang H, Emmett MR, Marshall AG. Simultaneous reduction and digestion of proteins with disulfide bonds for hydrogen/deuterium exchange monitored by mass spectrometry. Anal Chem. 2010;82:1450–1454. doi: 10.1021/ac902550n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Johnson RS, Walsh KA. Mass spectrometric measurement of protein amide hydrogen exchange rates of apo- and holo-myoglobin. Protein Sci. 1994;3:2411–2418. doi: 10.1002/pro.5560031224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dharmasiri K, Smith DL. Mass spectrometric determination of isotopic exchange rates of amide hydrogens located on the surfaces of proteins. Anal Chem. 1996;68:2340–2344. doi: 10.1021/ac9601526. [DOI] [PubMed] [Google Scholar]
- 84.Zhang Z, Post CB, Smith DL. Amide hydrogen exchange determined by mass spectrometry: application to rabbit muscle aldolase. Biochemistry. 1996;35:779–791. doi: 10.1021/bi952227q. [DOI] [PubMed] [Google Scholar]
- 85.Deng Y, Smith DL. Identification of unfolding domains in large proteins by their unfolding rates. Biochemistry. 1998;37:6256–6262. doi: 10.1021/bi972711o. [DOI] [PubMed] [Google Scholar]
- 86.Chen J, Walter S, Horwich AL, Smith DL. Folding of malate dehydrogenase inside the GroEL-GroES cavity. Nat Struct Biol. 2001;8:721–728. doi: 10.1038/90443. [DOI] [PubMed] [Google Scholar]
- 87.Schmid FX, Baldwin RL. Detection of an early intermediate in the folding of ribonuclease A by protection of amide protons against exchange. J Mol Biol. 1979;135:199–215. doi: 10.1016/0022-2836(79)90347-4. [DOI] [PubMed] [Google Scholar]
- 88.Roder H, Wuthrich K. Protein folding kinetics by combined use of rapid mixing techniques and NMR observation of individual amide protons. Proteins: Structure, Function, and Genetics. 1986;1:34–42. doi: 10.1002/prot.340010107. [DOI] [PubMed] [Google Scholar]
- 89.Deng Y, Pan H, Smith DL. Selective isotope labeling demonstrates that hydrogen exchange at individual peptide amide linkages can be determined by collision-induced dissocation mass spectrometry. J Am Chem Soc. 1999;121:1966–1967. [Google Scholar]
- 90.Akashi S, Naito Y, Takio K. Observation of hydrogen-deuterium exchange of ubiquitin by direct analysis of electrospray capillary-skimmer dissociation with Fourier transform ion cyclotron resonance mass spectrometry. Anal Chem. 1999;71:4974–4980. doi: 10.1021/ac990444h. [DOI] [PubMed] [Google Scholar]
- 91.Rand KD, Adams CM, Zubarev RA, Jorgensen TJ. Electron capture dissociation proceeds with a low degree of intramolecular migration of peptide amide hydrogens. J Am Chem Soc. 2008;130:1341–1349. doi: 10.1021/ja076448i. [DOI] [PubMed] [Google Scholar]
- 92.Zehl M, Rand KD, Jensen ON, Jorgensen TJ. Electron transfer dissociation facilitates the measurement of deuterium incorporation into selectively labeled peptides with single residue resolution. J Am Chem Soc. 2008;130:17453–17459. doi: 10.1021/ja805573h. [DOI] [PubMed] [Google Scholar]
- 93.Rand KD, Zehl M, Jensen ON, Jorgensen TJ. Protein hydrogen exchange measured at single-residue resolution by electron transfer dissociation mass spectrometry. Anal Chem. 2009;81:5577–5584. doi: 10.1021/ac9008447. [DOI] [PubMed] [Google Scholar]
- 94.Konermann L, Pan J, Liu YH. Hydrogen exchange mass spectrometry for studying protein structure and dynamics. Chem Soc Rev. 2011;40:1224–1234. doi: 10.1039/c0cs00113a. [DOI] [PubMed] [Google Scholar]
- 95.Rand KD, Pringle SD, Morris M, Engen JR, Brown JM. ETD in a Traveling Wave Ion Guide at Tuned Z-Spray Ion Source Conditions Allows for Site-Specific Hydrogen/Deuterium Exchange Measurements. J Am Soc Mass Spectrom. 2011;22:1784–1793. doi: 10.1007/s13361-011-0196-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Landgraf RR, Chalmers MJ, Griffin PR. Automated Hydrogen/Deuterium exchange electron transfer dissociation high resolution mass spectrometry measured at single-amide resolution. J Am Soc Mass Spectrom. 2012;23:301–309. doi: 10.1007/s13361-011-0298-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.