Abstract
The cellular response to DNA damage signaling by MMR proteins is incompletely understood. It is generally accepted that MMR-dependent apoptosis pathway in response to DNA damage detection is independent of MMR's DNA repair function. In this study we investigate correlated motions in response to the binding of mismatched and PCL DNA fragments by MutSα, as derived from 50 ns molecular dynamics simulations. The protein dynamics in response to the mismatched and damaged DNA recognition suggests that MutSα signals their recognition through independent pathways providing evidence for the molecular origin of the MMR-dependent apoptosis. MSH2 subunit is indicated to play a key role in signaling both mismatched and damaged DNA recognition; localized and collective motions within the protein allow identifying sites on the MSH2 surface possible involved in recruiting proteins responsible for downstream events. Unlike in the mismatch complex, predicted key communication sites specific for the damage recognition are on the list of known cancer causing mutations or deletions. This confirms MSH2's role in signaling DNA-damage induced apoptosis and suggests that defects in MMR alone is sufficient to trigger tumorigenesis, supporting the experimental evidence that MMR-damage response function could protect from the early occurrence of tumors. Identifying these particular communication sites may have implications for the treatment of cancers that are not defective for MMR, but are unable to function optimally for MMR-dependent responses following DNA damage such as the case of resistance to cisplatin.
Keywords: correlated motions, collective motions, signal transduction by MSH2, MMR-dependent apoptosis, DNA-damage signaling, MMR repair function
1. Introduction
It has been long recognized that proteins are dynamic systems (1-5). Functional proteins are not static structures, but rather generally stable soft mechanical constructs that allow certain types of internal motion to enable their biological function (1,6-8). Recently, in biophysics this concept lead to an extension of structure-function paradigm to include dynamics (3,8,9). Within the protein dynamics, correlated motions have a large contribution to the net atomic displacements (4). Localized or long-range, correlated motions are likely to be critical for function of large molecular complexes, and they are considered to play an important functional role in recognition, binding, signal transduction or mechanical/thermodynamic energy transport in biomolecules, and have been the subject of extensive experimental and theoretical studies (2,4,7,10,11).
Functional dynamics in complex biomolecular systems is difficult to probe experimentally, as seen in recent reviews surveying advances in investigations of conformational fluctuations of proteins in solutions using NMR structural and relaxation techniques (2,12-14). A profound insight into the molecular nature of these motions and their relevance for biological function can be obtained by molecular dynamics simulation, advancing the path for protein engineering or rational drug design. NMR spectroscopy techniques are typically used to study of individual atoms in a protein with respect to their surroundings, but in large complexes they usually provide little information regarding whether and how atoms dynamically communicate with each other. Molecular dynamics simulations, with their unique property of probing the space and timescales simultaneously, allow for sampling of motions of biological interest (3,7,9,15) and provide information regarding the correlated atomic motions inside proteins (16-19).
Currently there are two main approaches to performing molecular simulations of protein motions. The first is all-atom molecular dynamics (15,20-22), in which atoms and molecules are allowed to interact over time at a given temperature following the laws of classical mechanics and which provide a detailed description of atomic motions. The second approach is normal mode analysis, in which harmonic motions of the molecule about a local energy minimum are calculated (23-25).
In this study we investigate correlations between atomic positional fluctuations in response to the binding of mismatched and PCL double stranded DNA fragments by MutSα (MSH2-MSH6 heterodimer), as derived from 50 ns molecular dynamics simulations. The treatment is different than harmonic or quasi-harmonic normal mode analysis, since we do not analyze the motion but rather the positional fluctuations without involving the atomic masses in the analysis (6). The differences between the localized and long-range correlated displacements in the two systems will be highlighted and possible links between correlated motions and MutSα‘s function will be proposed.
For the same system, we has been previously examined the conformational effects of binding of DNA damaged by a chemotherapeutic on MutSα(26), providing a refined protein-DNA binding mode and identifying specific conformational changes induced by the recognition of mismatched and damaged DNAs, which concluded that MutSα proteins recognize the mismatched and the damaged DNA substrates in significantly different modes. Here we found evidence for localized and collective motions that connect the DNA binding and protein recruitment surfaces in this complex and also found key residues that are involved in cancer-causing mutations that are implicated in communication pathways. This complements our previous work by suggesting that there are classes of residues responsible for dynamic perturbations and other responsible for conformational perturbations.
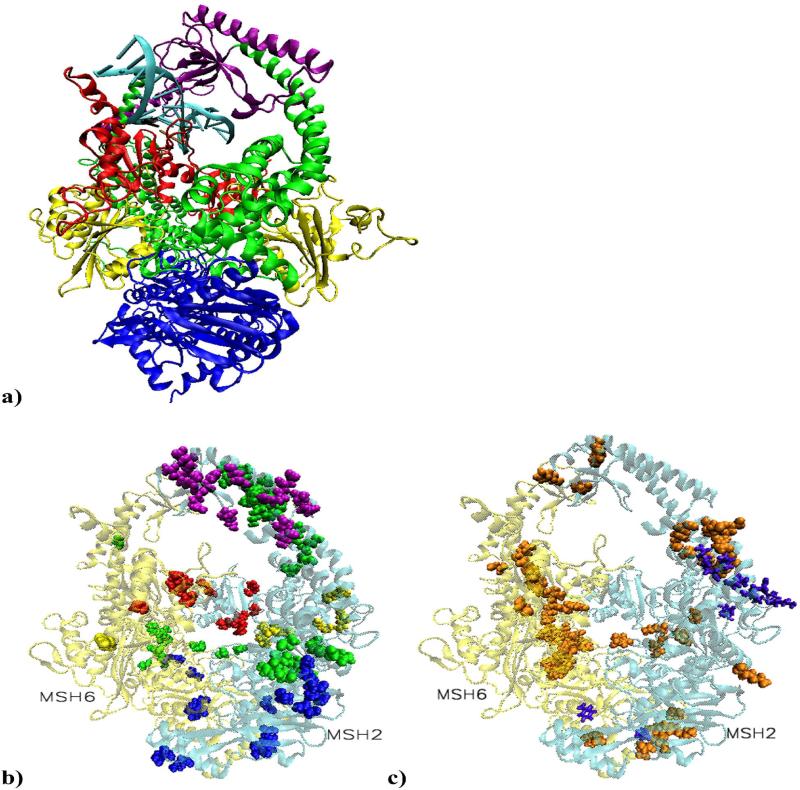
DNA mismatch repair proteins maintain genetic stability in both prokaryotes and eukaryotes by detecting and repairing mismatched bases and insertion/deletion loops mistakenly incorporated during DNA replication. In addition, MMR proteins initiate cellular response to certain types of DNA damage, such as those produced by chemotherapeutic agent cisplatin. Mammalian cells with defective MMR show partial or complete failure to undergo apoptosis following specific types of DNA damage and the human mismatch-binding factor MutSα promotes apoptosis in normal cells (27). It is considered that cells deficient in MMR proteins have a “mutator phenotype” in which the rate of spontaneous mutations is greatly elevated causing predicposition to cancer (28). Inactivation of MMR in human cells is associated with hereditary and sporadic cancers, with hereditary non-polyposis colon cancer (HNPCC) being the most prominent (29,30). In our simulations, the structural model for DNA mismatch recognition by the MMR machinery was the solved structure of MutSα with two ADP molecules bound in the ATPase sites of the heterodimer, bound to a 15 base pair duplex DNA containing a central G-T mispair causing distortion in the orientation of the bases (31). The structural model for DNA damage recognition was based on the crystal structure of DNA containing a (1, 2)GpG cisplatin intra-strand crosslink (32,33) built into the crystal structure of MutSα (31). (1, 2)GpG cisplatin intra-strand DNA adduct is the most prevalent of those made by cisplatin and it is recognized by human mismatch repair proteins (34,35). Since MutSα purified from cell extracts carries ADP (28,31), the model with two ADP molecules was chosen in both cases. The mismatched or the damaged DNA fragment is fully encircled by the mismatch binding (red) and clamp (purple) domains from both MutSα subunits (Figure 1a). Note that in our system, residue 1 of MSH6 corresponds to residue 362 in the solved structure, (31). Residues sequence and numbering for MSH2 and MSH6 are included in SM, Figures S3 and S4.
Figure 1.
a) Structural model of MutSα in complex with a 15 base pair duplex DNA containing a central G-T mismatch. DNA is shown in light blue with the mismatch pair marked: black for guanine and ochre for thymine. The color code for the heterodimer domains is: red for the mismatch binding domain, residues 1 to 124 in MSH2 and 1 to 157 in MSH6; yellow for the connector domain, residues 125 to 297 in MSH2 and 158 to 356 in MSH6; green for the lever domain, residues 300 to 456 and 554 to 619 in MSH2, and 357 to 573 and 648 to 714 in MSH6; purple for the clamp domain, residues 457 to 553 in MSH2 and 574 to 647 in MSH6; blue for the ATPase domain, residues 620 to 855 in MSH2 and 715 to 974 in MSH6. Note that in our system, residue 1 of MSH6 corresponds to residue 362 in the solved structure. b) Residues on the list of cancer causing mutations or deletions involved in highly significant correlated atom displacements. The large majority is unique for the platinum cross-linked complex and located on the surface of MSH2 subunit, covering all five domains.
c) Proposed cancer related highly significant correlations. In violet are residues associated with the mismatched system and in orange those associated with the damaged system. The predicted repair signaling residues are on the surface of the connector, lever and ATPase domains of MSH2. In the damaged complex, distinctively, most of MSH6's predictions are on the heterodimer interface (mismatch binding, connector, lever and ATPase domain), while MSH2's predictions are on its surface (connector, lever, clamp and ATPase domains).
2. Methods
2.1. Molecular Dynamics Simulations
The simulations of the G-T mismatch are performed using the same basic protocol as in our previous work (36,37) based on the X-ray structure of human MSH2-MSH6 protein complex with heteroduplex DNA (31). The bases at the mismatch remain in the DNA helix but do not stack, and as a result a bending of ~60° occurs in this region. Coordination of the cis-{Pt(NH3)2}2+ fragment to DNA , in which platinum atom links N7 atoms of two adjacent guanine residues, Gua8 and Gua9, alters the duplex DNA. The structural alterations induced in DNA by forming adducts with platinum have been extensively studied (38). NMR studies led to the conclusion that the (1, 2)GpG cisplatin intra-strand adduct bends the DNA helix by 78° towards the major groove and also unwinds it by 25° at the site of the platination, resulting in de-stacking of the bases (39). Hydrogen atoms were added using the hbuild facility of CHARMM (40). The CHARMM force field was used for the entire complex with additional parameters based on preexisting cisplatin parameters (41-43). This force field has been extensively parameterized for a wide range of biologically important molecules, including nucleic acids, amino acids, lipids and some small-molecule ligands. The platinum cross-linked DNA structure was built using the mismatch as a template. The cross-linked structure was fitted into the binding pocket to maximize the structural overlap with the mismatched DNA structure, followed by rotations and translations to minimize the energy of the unrelaxed structure using the coordinate manipulation and energy minimization facilities of CHARMM. The platinum atom cross-links two adjacent guanines. The structure was fully solvated with TIP3P water (44) in a cubic box using the visual molecular dynamics (VMD) package (45). Although there are increasingly accurate implicit-solvent models, e.g., (46-48), they have yet to be thoroughly vetted on large DNA/protein complexes such as the ones simulated herein. The water molecules were briefly minimized for 100 cycles of conjugate gradient minimization with a small harmonic force constant on all protein atoms. The entire system then underwent 250 ps of molecular dynamics simulation to achieve a thermal equilibration using Berendsen pressure regulation with isotropic position scaling(49). The system's temperature was equilibrated by reassigning atom velocities from a Boltzmann distribution for a given temperature every 1000 cycles, in 25 K increments, from an initial temperature of 0 K to a target temperature of 300 K. Following the equilibration, a 10 ns production simulation was performed in NAMD package (50), under NPT ensemble, using standard parameters: a 2.0 fs time step using SHAKE on all bonds to hydrogen atoms (51), a 12 Å cutoff, Particle Mesh Ewald with a 128 grid points on a side (52), Langevin temperature control with a damping coefficient of 5/ps, Berendsen's constant pressure algorithm with a target pressure of 1.01325 bar, a compressibility of 45.7 mbar, a relaxation time of 1 ps, and a pressure frequency of 40 fs, and a coordinate save frequency of 200 fs; all as implemented in NAMD. A total of ten simulations were performed, five for each system. For each of the five trajectories the same protocol was employed with different initial velocities and the same coordinates. The initial coordinates, velocities, and system dimensions were taken from the final state of the corresponding equilibration simulation. There are 855 residues in MSH2, 974 residues in MSH6, 30 nucleotides in the DNA fragment, and two ADP molecules, a total of 30048 atoms in the platinum cross-linked complex and 30039 in the mismatched system.
Cα root mean square deviations and total energies are provided in SM, Figure S5. These data show there are two different relaxation timescales, a fast one on the 10s-100s of picosecond time scale, and a slow one on the nanoscale. Data show that most of the relaxation to equilibrium occurs within the first 2ns, and that while there may be additional long-time relaxation, starting the simulation analysis at 5ns allows for a conservative removal of the majority of the non-equilibrium effects. Since our different simulations started from different initial conditions, it is expected they to show different pathways to equilibration, and they show the expected variation in relaxation.
2.2. Covariance Analysis
Cα normalized variance-covariance matrixes, or Pearson correlation coefficients, were calculated from the simulated trajectories of the mismatched and platinum cross-linked MutSα-DNA complexes using CHARMM program. The covariance of two sets of atoms, A and B, of XA and XB coordinates sets is given by the expectation value:
where 〈XA〉 and 〈XB〉 are the expected values. The correlation of the two variables is the dimensionless normalized covariance defined as . σA and σB are the standard deviations of XA and XB. Variance is the special case of covariance when the two variables are identical.
Pearson correlation coefficient, also known as product-moment coefficient of correlation or cross-correlation coefficient, quantifies the linear dependence between variable, in our case atom displacements of pairs of atoms. Given the size of the system non-linear correlations were not investigated. The correlation coefficients of the matrix are symmetrical about the diagonal and range from -1 to 1. A value of 1 implies that a linear equation describes the relationship and the atom displacements are in the same direction, denoting correlated motions. A value of -1 implies that all data points lie on a line but the atom displacements are in the opposite direction, denoting anti-correlated motions. Zero corresponds to the orthogonal atomic motions which are not detected by this method. In our analysis, the atom motions are considered strongly correlated/anti-correlated for correlation coefficients higher/lower than 0.5/-0.5 and weakly correlated/anti-correlated for the rest.
To prevent the equilibration period for adding rare events to the correlations, the first half of each simulation was discarded during this analysis. The covariances were calculated about the average equilibrated structure and were calculated as covariances among the protein's Cα atomic fluctuations (within MSH2, within MSH6 and between MSH2 and MSH6), as well as between the protein and the heavy-atom residue-averaged fluctuations of the DNA base pairs. Fluctuation correlations depend on the time scale over which data are analyzed since different correlations may arise on different time scales. In this study they are based on an ensemble of one structure per picosecond, which has been shown in other studies to provide a good sampling of the trajectory (17) .
Correlated motions can occur over a wide-range of timescales, depending on the molecular events involved. This is especially true for protein folding and large-scale conformational rearrangements. For such studies, longer time-scale simulations would need to be performed, although such timescale are not yet readily accessible by all-atom MD simulations. The focus of this paper is on the correlated motions that occur around the native states of two different complexes, where the differences between the complexes are solely due to slightly different binding events.
3. Results and Discussion
3.1. Correlated motions within the protein
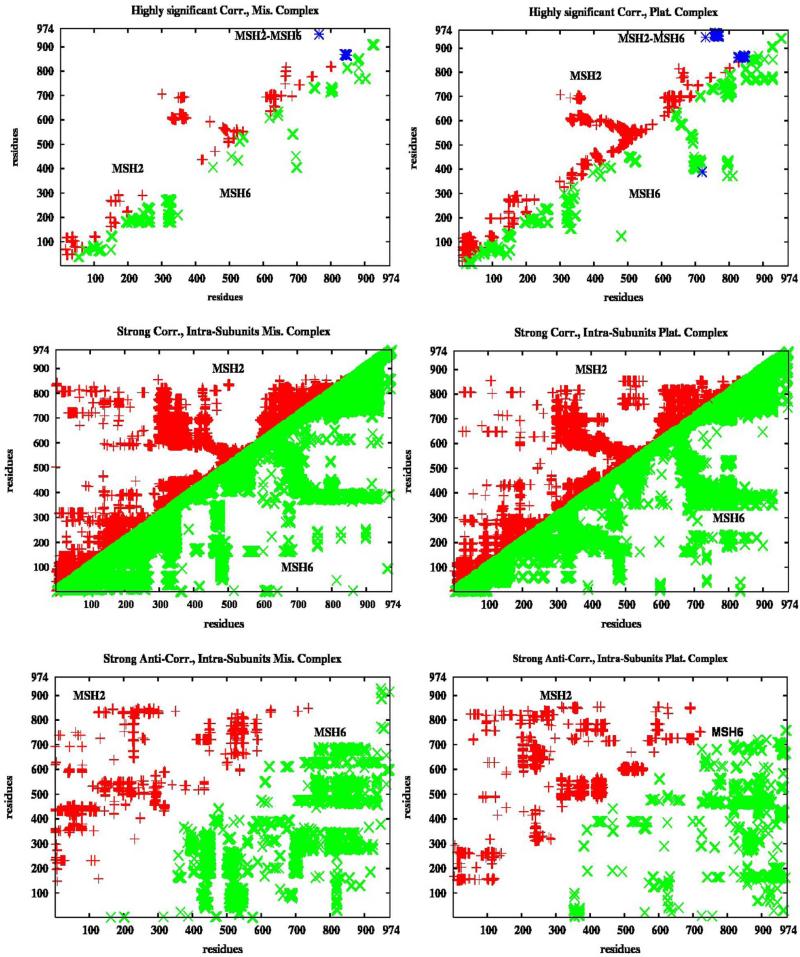
Considering the size of the system, a large number of correlations is expected: about 3*106 correlations within the heterodimer protein and 5.5 *104 protein-DNA correlations. The standard deviation of the former is 0.18 for the mismatched system and 0.17 for the platinum cross-linked (PCL) system. Highly significant were considered correlations 3.5 standard deviations above the mean (which is 0.24 in the mismatched and 0.22 in the PCL system) and the binding to cisplatin almost double their number. In Figure 2, upper plots, are presented highly significant correlations between residues at least ten units apart within MSH2 (data in red), MSH6 (data in green), as well as between the two subunits of the protein complex (data in blue; mostly between their ATPase domains). The localized and long-range correlated and collective motions specific for the mismatched and PCL MutSα-DNA complexes will be discussed subsequently. Almost continuous strong correlations (higher than 0.5 but not highly significant) are seen within each domain of the protein's subunits, with small gaps at the end of the mismatch binding, connector, and lever domains of MSH2 (Figure 2, middle plots). Notice that the inter-subunits strong correlations are mimicking at a larger scale the pattern of the highly significant long-range collective motions. Strong anti-correlations (negative values lower than -0.5 but not highly significant) within the protein complex's subunits, the bottom plots in Figure 2, display different patterns in the two systems. Profoundly different patterns in the inter subunits correlations and anti-correlations (SM, Figure S9) are associated with change of motion directionality in the two complexes, as it will be addressed later. The complete variance-covariance correlation maps for the two systems are presented in SM, Figure S1, and, despite their differences at the highly correlated level, they are quite similar in general. Additional long-time relaxation, which simulations suggest, may affect the overall cross-correlations, but not the highly significant correlations which are the focus of our correlation analysis in the two systems.
Figure 2. Intra-subunits Correlations.
In red and green are represented correlations within MSH2 and MSH6, respectively. The highly significant correlations (only covariance between residues at least ten units apart) are twice as many in the platinum cross-linked complex than in the mismatched complex. Almost continuous strong correlations are seen throughout the subunits. Different patterns are seen in the anti-correlations in the two complexes. There is a limited number of highly significant correlations between the two subunits, data in blue. High correlations between the atomic displacements of adjacent residues are expected, but when considered those between residues at least ten units apart there are 2429 such correlated motions within
MutSα sensor and signal DNA damage
Highly significant correlations within MSH2
Presented in Figure 3a-b, they involve extensive and unique regions in the damaged DNA binding response. The dynamics of both system exhibits highly significant correlations within the mismatch binding domain, between the lever and the ATPase domains, as well as between the lever and the clamp domains. Unique in MSH2's response to the protein complex binding to the damaged DNA are the highly significant correlations between the mismatch binding and the connector domains.
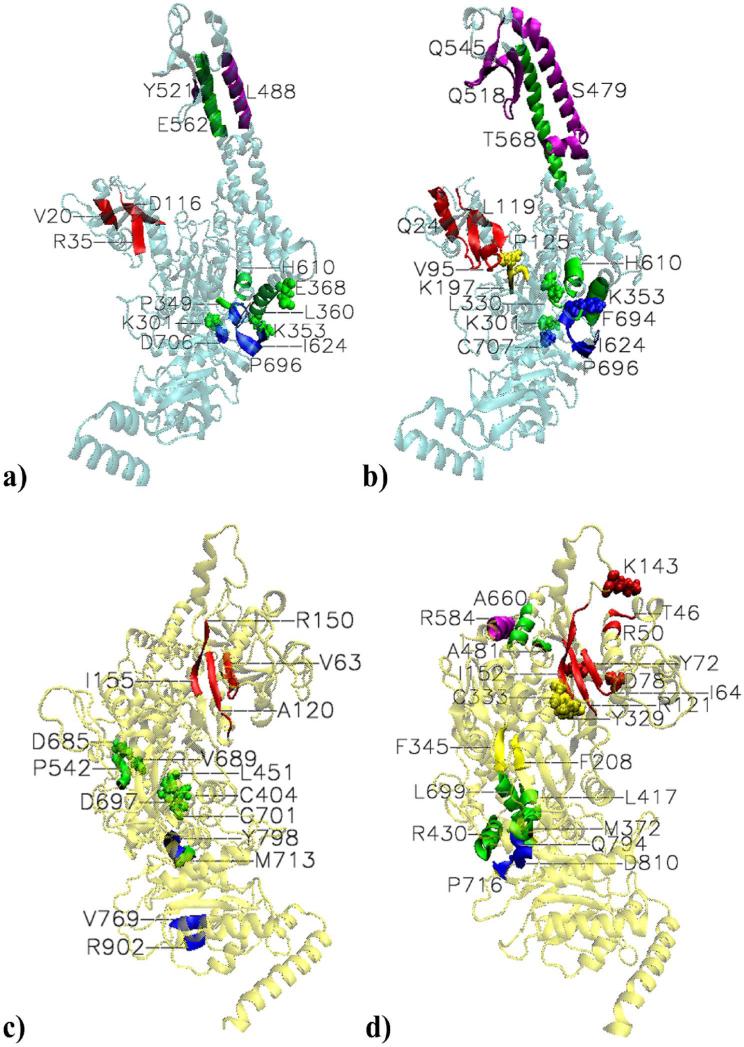
Figure 3. Highly significant long range communications within the subunits.
a) MSH2. Mismatched MutSα-DNA complex. Special case within the mismatch binding domain. 18-23 (V20) and 34-37 (R35) coupled with 115-121 (D116). Couplings between lever and ATPase domains. K301 is coupled with D706. K353 is coupled with region 624-625 (I624). 349-350 (P349), E368 and 356-365 (L360) are coupled with 689-696 (labeled with X for I691); the latter region of the lever domain is also coupled with 621-624. 608-612 (H610), and Y619 are coupled with 693-699 (P696). Couplings between clamp and lever domains. From the former, 481-495 (L488) and 521-522 (Y521) are coupled with 555-570 (E562). b)MSH2. Platinum cross-linked MutSα-DNA complex. Special case within the mismatch binding domain:11-38(E24) is paired with 114 to 124 (L119). Couplings between mismatch binding and connector domains: 91-99 (V95) of the former is paired with P125 and G126 of the latter. Aditioanlly, 93 and 96-98 are paired with196-198 (K197). Couplings between lever and ATPase domains: K301- C707 and L330-F694 coupled pairs. 49-364 (K353) is paired with 689-704 (P696). 607-612 (H610) and 618-619 are paired with 693-695. Couplings between clamp and lever domains. 457-502 (S479) of the former is paired with 555-582 (T568); 512-524 (Q518) is paired with 555-563; and 538 to 553 (Q545) is paired with 554-564 of the lever domain. c) MSH6. Mismatched MutSα-DNA complex. Coupled β-stands within the mismatch binding domain: 60-67 (V63) is paired with 117-123 (A120); the latter is also paired with 150 -155 region, and K67 is paired with R150. Couplings between lever and ATPase domains. 713-714 (M713) of the former is paired with798-799 (Y798) of the latter. Communications within the lever domain: C404 and H405 are paired with C701, and L451 is paired with D697; 541-543 (P542) is paired with D685 and A689. Communication within the ATPase domain: 768-770 (V769) is paired with R902 and L903. d) MSH6. Platinum cross-linked MutSα-DNA complex.
Extensive communication within the mismatch binding domain: paired with the end of the mismatch binding domain, K143 and β strand150-153 (I152), are 46-47 (T46), 50-51 (R50), and 64-67 (I64) from the mismatch binding site; β strand 61-67, Y72, D78, and I81 are paired with β strand 117-124 (R121); the latter is also paired with150-157. Communication between mismatch binding and lever domains: 123-125 of the former is paired with 481-482 (A481) of the latter. Communication between mismatch binding and connector domains: R154 and I155 are paired with 329-335 (Y329) and C333, respectively. Communications between lever and ATPase domains: 372 -373( M372) is paired with 794-795 (Q794) and 810-81 (D810); 417-421 (L417) is paired with 792-796 (Q794); 424-437 (R430) is paired with both regions 715-718 (P716) and 793-799 (see Q794); 697-706 and 713-714 are paired with R715 (see P716 ) and 795-799 (see Q794), and 789-805, respectively. Communication between the clamp and lever domains: 581-588 (R585) of the former is paired with 656-664 (A660) of the latter. Communication within the connector domain: 206-210 ( F208) is paired with 344-347 (F345. Communication within the lever domain: 400-416 (see L417) and 427-437(R430) are paired with 694-706 (L699).
The special case of long-range correlations within the mismatch binding domain in the mismatched complex (Figure 3a) includes couplings of 18-23 α-helix (labeled by V20) and 34-37 β-sheet (labeled by R35) with the end of it, 115-121 β-sheet (labeled by D116). On the contrary, in the PCL complex (Figure 3b) it involves extended regions from the beginning and the end of the mismatch binding domain: the collective highly correlated long-range motions between 11-38 region (labeled by Q24) and 114-124 region (labeled by L119).
In the mismatched complex, long-range highly significant correlations between the first section of the lever and the ATPase domains within MSH2 include couplings between key sites (depicted in Figure 3a by van der waals representations), such as K301- D706, couplings between key sites and small or large regions, the case of K353-region 624-625 (labeled by I624) and E368-region 689-696 (labeled with X for I691), as well as multiple collective motions between region 356-365 (labeled by L360) and regions 689-696 and 621-624. From the second section of the lever domain, region 608-612 (labeled by H610) and Y619 are coupled with region 693-699 (labeled by P696). Similarly, couplings between key sites are seen in the dynamics of the PCL complex (Figure 3b), the case of K301-C707 and L330-F694. However, by comparison with the dynamics of the mismatched complex, there are noticeably the collective motions between 349-364 (labeled by K353), from the first section of the lever domain, and 689-704 (labeled by P696). Of special interest is K353 which makes multiple correlations with I624, Q690, and I704. In both systems, the end of the lever domain, 607-612 (labeled by H610) and 618-619 are paired with residues form region 693-695 (see P696 for reference) of the ATPase domain.
Within non-binding subunit, highly significant correlations are also seen between the clamp domain and the second section of the lever domain. In the mismatched complex they include correlations between 481-495 α-helix (labeled by L488) and 521-522 (labeled by Y521) from the former (Figure 3b), and 555-570 α-helix (labeled by E562) from the latter. It is highly noticeable that the clamp-lever correlations in the PCL complex include extended and additional regions by comparison with the mismatched complex. It is the case of correlations between 457-502 α-helix (labeled by S479) from the clamp domain and 555-582 α-helix (labeled by T568) form the lever domain. A second region from the clamp domain, 512-524 (labeled by Q515), is paired with 555-563 region of the lever domain. In addition, a third region from the clamp domain, β-sheet 538-553 (labeled by Q545), is paired with 554-564 region of the lever domain.
Unique for the PCL complex, the dynamics of its non-binding subunit reveals highly significant correlated motions between region 91-99 (labeled by V95 in Figure 3b) of the mismatch binding domain and the beginning of the connector domain, residues P125 and G126. Additionally, highly significant correlations made by L93, R96 toY98 with region 196-198 (labeled by K197) become weak in the mismatched system, as it will be addressed later.
Highly significant correlations within MSH6
The highly significant correlations within the mismatch recognition subunit contrast significantly in the two systems: while long–range correlations within the mismatch binding domain, within the lever domain, and lever-ATPase domains are common for both systems, the binding of MutSα to the damaged DNA translates into an increase in the possible functionally relevant dynamics (Figure 3c-d). Unique for the damaged complex are highly significant mismatch binding-connector, mismatch binding-lever, and clamp-lever correlations, which, as it will be addressed subsequently, are either weak or strong without being highly significant in the mismatched complex.
As in MSH2, but on a larger scale, the mismatch binding domain of MSH6 exhibits dynamically coupled β-stands. In the mismatched complex (Figure 3c), 60-67 (labeled by V63) is coupled with 117-123 (labeled by A120); the latter is also paired with the end of the mismatch binding domain, region 150-155, a two step communication.More expansive correlations are seen within the mismatch binding domain in the damaged complex (Figure 3d): paired with the end of the mismatch binding domain, K143 and region 150-153 (labeled by I152), are regions 46-47, 50-51, and 64-67 from the mismatch binding site, labeled by T46, R50, and I64, respectively. On the other hand, the mismatch binding site residues 61-67, Y72, D78, and I81 are paired with 117-124 (labeled by R121); the latter region is also paired with the end of the mismatch binding domain, region 150-157.
Limited in the mismatched complex, highly significant correlations between lever and ATPase domains , which include 713-714 (labeled by M713) from the end of the former with 798-799 (labeled by Y798) of the latter, are broader in the PCL complex (Figure 3d). From the first section of the lever domain, M372 and V373 are paired with regions 794-795 (labeled by Q794) and 810-811 (labeled by D810). Region 417-421 (labeled by L417) is paired with region 792-796 (labeled by Q794). Collective motions are seen between 424-437 α-helix (labeled by R430) and regions 715-718 (labeled by P716) and 793-799 (see Q794 for reference). From the end of the second section of the lever domain, region 697-706 is paired with R715 (see P716 for reference) and region 795-799 (see Q794 for reference), while 713-714 is paired with region 789-805 (see Q794 for reference).
Highly significant correlations between the two sections of the lever domain are seen in both systems. In the mismatched complex there are localized correlations between C404, H405 and L451 from the first section and C701 and D697, respectively, from the second section; in addition, region 541-543 (labeled P542) is paired with D685 and A689. In the PCL complex there are correlated motions between two α-helix regions from the first section, 400-416 (labeled by L417) and 427-437 (labeled by R430), and 694-706 α-helix (labeled by L699) from the second section of the lever domain.
Exclusive for the damaged complex, its mismatch recognition subunit features highly significant correlations between the mismatch binding domain, which “senses” the damaged DNA, and connector and lever domains, as well as between clamp and lever domains. From the end of the mismatch binding domain, residues R154 and I155, are paired with region 329-335 (labeled by Y329) and C333, respectively, from the connector domain. Long-range correlations are also seen between residues 123-125 of the mismatch binding domain and residues 481-482 (labeled by A481) of the lever domain, as well as between residues of α-helix 581-588 (labeled by R584) of the clamp domain and residues 556-664 (labeled by A660) of the lever domain's extended α-helix. Unique for the PCL complex are also highly significant correlations between residues 206-210 (labeled by F208) and 344-347 (labeled by F345) within the connector domain.
Up to this point, the analysis of the protein dynamics in response to the mismatch and damage recognition suggests that MutSα signals the recognition of the mismatched and damaged DNAs through different if not independent pathways. Additional support for this finding derives from the analysis of inter-subunits correlations for each system, as well as from the investigation of the highly significant correlations in the PCL complex that became weak or change directionality in the mismatched complex, which will follow.
Highly significant correlations in platinum cross-linked that became weak in the mismatched
No change of directionality is seen when differences in the highly significantly correlated motions in the two systems are analyzed. However, a meaningful number of highly significant correlations in the PCL complex became weak in the mismatched complex (Figure 4), highlighting the independent DNA-damage signaling by MutSα.
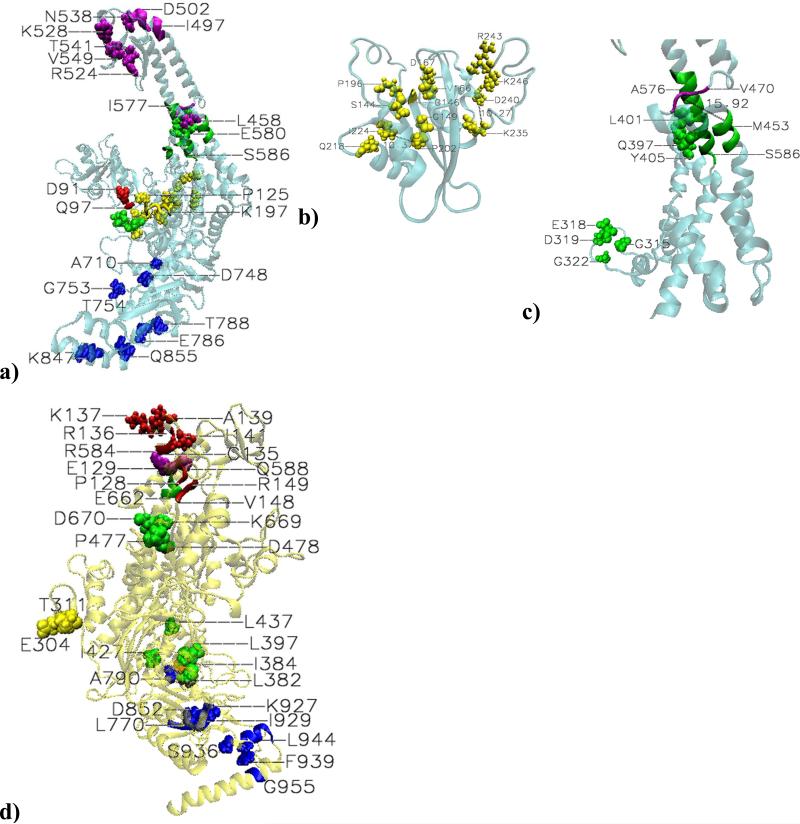
Figure 4. Highly significant correlations in platinum cross-linked system that became weak in the mismatched system.MSH2.
a) Correlations between mismatch binding and connector domain: D91 and 96-98 (Q97), from the former, are paired with P125 and 196-198 (K197) from the latter. Correlations within the connector domain (b): S144-G146-V147-D167; G149-Q218; V166-P196; P202-I224 (10.37 Å apart); K235-D240 (10.27 Å apart); R243-K246. Correlations within the lever domain (c): G315-E318; D319-G322; Q397-L401; and A398-Y405 coupled pairs. A bifurcation point in information flow: the end of the first section of the lever domain, region 451-456 (M453) is coupled with both 469-478 (V470) of the clamp domain and 574-580 (A576) of the second section of the lever domain. Additionally, high correlations between two adjacent regions, 572-583 (see A576) and 584-586 (S586) became weak correlations in the mismatched system. Correlations within the clamp domain: N538 is coupled with 496-498 (I497), 500-503 (D502), and R524, which are 18.60, 8.86, and 19.4Å apart, respectively; K528, F539, S540, T541, and V549 are paired with residues 498-511. Coupling between clamp and lever domains: L458-E580 pair, 15.54Å apart. Couplings within the ATPase domain: A710-D748; G753-T754; E786-T788; and K847-Q855 coupled pairs.
MSH6: d) Correlations within the mismatch binding domain: C135 and R136 are paired with P128; R136 is also paired with E129; K137 is paired with I141, V148 and R149; V148 is also paired with A139. Correlation within the connector domain: T311-E304 coupled pair. Correlations within the lever domain: from the first section of it, L382-I384-L397 and the coupled pairs I427-L437 and P477- D478; from the second section, D670 is paired with residues 660-663(E662). Correlations between the clamp and lever domains: R584- E661 and Q588-E662coupled pairs. Correlations between lever and ATPase domains: C714 from the end of the former, depicted in brown for clarity, is paired with A790 from the later. Correlations within the ATPase domain: the correlated pair D852-I929, 21.28 Å apart; region 769-771 (L771) is coupled with K927, about 17 Å apart; 943-947 (L944) and 955-956 (G955) are coupled with F939.
Possible key role played by the connector domain of MSH2 in DNA-damage signaling
As mentioned above, within MSH2, highly significant mismatch binding - connector correlations in the PCL complex un-correlate in the mismatched complex. Similarly, highly significant correlations within the connector domain of MSH2 (Figure 4b), often between residues in direct contact, un-correlate in the mismatch system, suggesting the following pattern for the damage information flow: S144-G146-V147-D167; G149-Q218; V166-P196; P202-I224 (10.37 Å apart); K235-D240 (10.27 Å apart); and R243-K246. These correlated motions may be responsible for opening up of the connector domain, known for its flexibility, in response to the damaged DNA binding. Anticipatory, many of these residues are on the known list of MutSα's cancer related mutations or deletions, as will be addressed latter.
Lever-clamp bifurcation point in DNA-damage information flow
Highly correlated motions within the lever domain that un-correlate in the mismatched complex (Figure 4a,c) include localized motions between G315-E318 and D319-G322 pairs from the interconnected disorganized loop, Q397-L401 and A389-Y405 pairs from the long α-helix, as well as collective motions of two adjacent regions of the long α-helix, 572-583 (see A576 for reference) and 584-586 (labeled by S586). Additionally, several localized and collective highly significant correlated motions within the clamp domain (the couplings of N538 with 496-498, 500-503, and R524, as well as the couplings of K528, F539, S540, T541, and V549 with residues from region 498-511) and within the ATPase domain of MSH2 in the damaged complex (pairs A710-D748, G753-T54, and E786-T788, and K847-Q855) became weak in the mismatched complex. Specific to the damaged DNA recognition are also the highly significant correlations between the end of the first section of the lever domain, region 451-456 (labeled by M453) and region 459-478 (labeled by V470) of the clamp domain, as well as between the former and region 574-580 (labeled by A576) of the second section of the lever domain, suggesting a clamp-lever bifurcation point specific to damage recognition information flow.
Additionally, MSH6's highly significant correlations that became weak in the mismatched complex (Figure 4d) enhance the differences in the mismatch versus damage signaling. Specific to MSH6's response to the damage recognition are certain highly significant correlations within the mismatch binding domain (the couplings of P128 and E129 with C135 and R136, and K137-I141, V148-R149,and A139-V148 dynamically coupled pairs), within the connector domain (T311-E304 pair), within the lever domain (from the first section L382-I384-L397 and from the second section I427-L437 and P477-D478 pairs), and within ATPase domain (the couplings of 769-771, labeled by L771, with K927, 17.11 Å apart, D852-I929 pair, 21.28 Å apart, and the couplings of F939 with 943-947and 955-956). Furthermore, the above described clamp-lever highly significant correlations in the PCL complex mismatch binding subunit are weak in the mismatched complex, and so is the coupling of C714 from the end of the lever domain (depicted in Figure 4d in brown for clarity) and A790 from the ATPase domain.
Possible MSH2 key role in mismatched DNA signaling
On the other hand, a limited number of highly significant correlations in the mismatched complex became weak in the PCL complex (Figure S6, SM). It is suggested a possible key role in signaling mismatched DNA recognition by N412, P415, N416, V417, I418, L421 and E422 on MSH2‘s lever domain surface. By comparison with the possible involvement of MSH2's lever domain in signaling damaged DNA recognition documented above, a different region of the long α-helix seems to signal the mismatched DNA recognition in a “molecular wire” like pattern involving adjacent residues.
In summary, the analysis of the intra-subunits highly significant correlations that could have function implications in the two systems concludes that MutSα sensor and signal mismatched and damaged DNA differently. There are more than double correlations at this level in response to the damaged DNA binding, and the meaningful difference arises from the highly correlated motions between extended regions of the lever domain and the clamp domain of the non-binding subunit, MSH2, as well as from those within the mismatch binding, connector and lever domains of both subunits (as summarized in Figure S9a-b, SM). While is well known that MSH6 subunit plays a key role in the mismatched / damaged DNA recognition , this analysis suggests that MSH2 subunit may play a key role in signaling it through independent channels, and possibly in recruiting the downstream events’ proteins.
Inter-subunits correlations
There is a limited number of highly significant inter-subunit correlations (data in blue in Figure 2, upper level plots), with about 90% more in the PCL complex than in the mismatched complex. In both systems they are carried out mainly by residues from the ATPase domains (Figure S6c-d, SM), with several of those specific to the damaged complex being on the list of cancer related mutations (Table I). No inter-subunits highly significant anti-correlations are seen in either system. However, strong inter-subunits correlations and anti-correlations exhibit different patterns in the two complexes (Figure S9, SM), further substantiating the above argument that MutSα sensor and signal mismatched and damaged DNA differently.
Table I.
Cancer related highly significant communications within the protein and protein-DNA1.
| On the list | To be tested | |
|---|---|---|
| MSH2 | S13, V17,E28, T33, L92, L93, R95, R96, Q97, Y98, Y118, A123, G146, G149, P196, E198, I224, K235, R243, K246, K301, G315, G322, P349, R359, D361, Q397, V417, L421, L458, V470, S473, S479, R482, E483, Q493, L503, G504, D506, G508, Q510, S516, E518, F523, R524, S554, L556, L559, E561, E562, Y563, T564, G566, K567, T568, E569, I577, E580, N583, A609, R612, Y619, P622, E690, G692, C697, S699, V702, T788 | 144, 166, 197, 202, 204, 218, 318, 319, 353, 401, 404, 447, 451, 452, 455, 458, 528, 538, 539, 549, 585, 586, 588, 786; 249, 252, 253, 290, 291, 412, 415, 416, 418, 420 422, 668, 817 |
| MSH6 | I81, I155, F345, R400, R411, W416, A426, L431, A433, L437, A660, D697, G796, G798, A801, E802, E832, D852, R902, R943 | 117-124, 333, 427, 670, 695, 696, 698-706, 715-718, 793-795, 797; 830 |
| MSH2-MSH6 | MSH2: W764, E842,Q846 MSH6: T864, I866, I952, Q953, A959, R960, F962 |
MSH6: 862, 865, 951, 954-956, 958, 959; MSH2: 757-769, 843-845, 848 |
| DNA-MSH2 | - | - |
| DNA-MSH6 | L74, R107 | - |
Based on the present analysis we propose the following residues for mutagenesis studies to assess mutations in MSH2 that would affect the DNA-damage induced apoptosis but not the MMR repair function: S144, V166, K197, P202, G204, Q218, E318, D319, K353, L401, C404, F447, Q451, E452, E455, L458, K528, N538, F539, V549, E580, S585, S586, Y588 and E786.
In italic are residues associated with mismatched complex, or proposed for the repair signaling. Unmarked are residues found involved in correlations unique for the PCL complex, or proposed for apoptosis signaling. Underlined are the common residues. Certain residues on the list were found as correlated pairs. The proposed residues were predicted as correlated with residues on the list, found to be involved in multiple correlations (the case of K353 and N538 from MSH2, as well as 715-718, 695, 696, 698-706, 793-795 and 797 from MSH6), or the case of highly significant correlations in the repair system that became weak.
Possible structural changes by the change of directionality in long-range inter and intra subunits correlated motions
Of particular interest in the comparison of the strong inter-subunits correlations in the two complexes is their change of directionality (Figure S9). Strongly correlated atom displacements in the damaged complex that change directionality in the mismatched complex include couplings between the connector domain of MSH2 and ATPase domain of MSH6, as well as between the lever domain of MSH2 and the clamp domain of MSH6. On the hand, strongly correlated atom displacements in the mismatched complex that change directionality in the damaged complex include correlations between the mismatch binding domain of MSH2 and the ATPase domain of MSH6, between the connector domain of MSH2 and both the clamp domain and the ATPase domain of MSH6, as well as couplings between the clamp domain of MSH2 and the mismatch binding and the connector domains of MSH6. Additionally, several strong intra-subunits correlations in the PCL complex change directionality becoming strong anti-correlations in the mismatched complex and vice-versa (details in Figure S7c, SM). Since the method employed here can not establish the causality of these correlated motions, this analysis does not reveal how these long-range inter and intra subunits correlated motions that change directionality in response to mismatched and damaged DNA binding transmit information or the direction of information flow. However, they not only support the already established meaningful difference between mismatch and damage signaling, but heir location suggests that certain structural changes in MutSα's upper and lower channels may be associated with mismatched and damaged DNA signaling.
3.2. Protein-DNA correlated motions
MutSα's response upon binding to the mismatched and PCL DNA fragments was investigated. Again, granted the size of the systems, were considered functionally relevant or highly significant those correlations with absolute value exceeding the mean plus 3.5 standard deviations. In both systems, highly significant protein-DNA correlations (Figure S8, upper plots) are comparative in number and, naturally, they involve mainly the binding subunit, MSH6, particularly the mismatch binding domain. However, the major difference between the two systems resides in the fact that while mismatched DNA displays highly significant and strong correlations with the clamp domain of MSH6, residues E619, C642, and K643, the damaged DNA displays similar correlations but with the lever domain of MSH6, residues I482 and M483. (Details of the differences in the highly significant correlations in the two systems are depicted in SM, Figure S6c-d). There are 39% more strong MSH2-DNA anti-correlations in the mismatched complex than in the PCL complex. In the mismatched complex, residues from all but clamp domain of MSH2 are strongly anti-correlated with most of the DNA bases, suggesting a coherent displacement of MSH2 away from the DNA fragment upon its mismatch recognition. On the contrary, in the PCL complex, binding to the damaged mismatched base triggers an ambiguous protein-DNA anti-correlations pattern.
Atomic displacements of the mismatched Gua8-Thy23 base pair are highly correlated with those of residues K67, V68, G69 and F71 from the mismatch “probing” loop of MSH6 (Figure S7a, SM). Damaged Gua8 (Figure S7b, SM) displays no highly significant correlations, but it is strongly correlated mainly with residues from MSH6’ “probing” loop (G69, K70 and F71), as well as P101 also at the binding site. Its mispaired base makes extensive highly significant correlations with residues F71-P101 from the binding site. Strong anti-correlations are predicted between the mismatched Gua8-Thy23 pair and the connector domain of MSH2: residues 143 and 187-190, and 197 and 222-223, respectively. The damaged pair makes only one strong anti-correlation, which is with E585 from the clamp domain of MSH6.
4. Conclusions
MutSα proteins signal DNA-damage induced programmed cell death and mismatched DNA repair through different channels
The predicted highly significant localized and long-range atom displacement correlations may describe motions which are relevant to MutSα's function. The internal motions in response to the recognition of a mismatched and damaged DNA are significantly different, “preparing” the protein for very different pathways following the mismatch or the damage recognition, which complements the rigorous hydrogen bonding analysis (26) showing that MutSα proteins recognize the mismatched and damaged DNA fragments in significantly different modes. Most of the predicted key communication sites found on the list of cancer causing mutations (InSight database: http://www.insight-group.org/; residues named on Table I and depicted in Figure 1b) are unique for the damage recognition by MMR. This is not only supporting the hypothesis of very different mechanisms for repair and DNA-damage cell death, but also suggesting that if those highly correlated regions are altered the damage recognition may not take place and the apoptosis mechanism is not triggered, leading to genomic instability.
Regarding the first issue, there is a large body of experimental evidence supporting the MMR-dependent damage signaling. Mutsα directly recognizes the damage and, excision and processing of the DNA damage is not required for an MMR-dependent apoptotic signal (36,53-56). Here, strong correlated motions in MutSα, often involving large regions of the protein, and their unique connection to the cancerous tumors when altered, without being linked to the mismatched DNA recognition, support the hypothesis that recognition of DNA damage is sufficient to signal apoptosis. Regarding the second issue, this suggest that defects in MMR alone is sufficient to trigger tumorigenesis, indicating that MMR-damage response function could protect from the early occurrence of tumors, and there is experimental evidence to support it (53,57,58).
Cancer related long range communications confirm MSH2's major role in signaling DNA-damage induced apoptosis
The entire analysis shows an overall increase in the protein's dynamics upon binding to the damaged DNA than to the mismatched DNA. The large majority of the predicted correlated sites related to cancer causing mutations or deletions (Figure 1b) belong to the non-mismatch binding subunit, MSH2, covering all five domains (marked with * in Table I). Most of these key residues are at the surface of the protein, suggesting that MSH2 may play a significant role in downstream events for MMR-mediated cellular response to DNA damage. And there is experimental evidence to support it: cells expressing reduced levels of MSH2 have almost normal levels of MMR activity, do not display microsatellite instability, but are more tolerant to DNA damaging agents (59,60). This has possible implications for the treatment of cancers that are not defective for MMR (normal DNA repair), but are unable to function optimally for MMR-dependent responses following DNA damage.
Of special interest may be the observation in the damaged complex of sites involved in multiple long-range correlated motions: in MSH6, region 427-437 from the first part of the lever domain is coupled with region 695-706 from the second part of the lever domain, as well as with residues 715-718 and 793-798 from the ATPase domain (details in Figure 3d). Several of these residues are on the list of known cancer causing mutations or deletions, recommending for the investigation of similar effects on the following residues: 715-718, 695, 696, 698-706, 793-795, and 797.
It must be noted that all of the presently proposed communication cites associated with known MutSα's cancer causing mutations are independent of the previously predicted essential contacts at the heterodimer interface found among the observed MutSα's cancer causing mutations(61), suggesting their defined role in the mismatched and damage recognition, for the later, and signaling, for the former.
Putative drug discovery approach in targeting MSH2 damage signaling
Remarkably, it has also been suggested that the second monomer plays a dynamic role in the function of eukaryotes homologue MutS complex (62). Recent efforts have focused on mimicking the structural effects of cisplatin (37). Given the evidence for MSH2's role in mismatch and damage recognition signaling, a novel drug discovery approach in discovering and designing inhibitors that interact with the mismatch signaling regions on the surface of MSH2 (residues K249, Q252, M253, E290, L291, N412, P415, N416, I418, A420, E422, T668 and V817, which are highly significant in the repair system and became weak in the damaged system) so as to severe these communications but to promote the damage signaling can be explored. Notice that the predicted repair signaling residues are on the surface of the connector, lever and ATPase domains of MSH2 (Figure 1c). Distinctively, in the damaged complex, most of MSH6's predictions are on the heterodimer interface (mismatch binding, connector, lever and ATPase domains), while MSH2's predictions are on its surface (connector, lever, clamp and ATPase domains). These suggest that the predicted cancer related highly significant correlations in MSH6, same of them arising from inter-subunit communications (MSH2-MSH6 in Table I), are responsible for allosteric transduction of conformational adjustments upon damage recognition, while MSH2 is responsible for damage signaling.
Supplementary Material
Acknowledgements
This research was partially supported by NIH R01CA129373 to FRS. The computations herein were performed on the WFU DEAC cluster; we thank WFU's Provost's office and Information Systems Department for their generous support.
Footnotes
Supplementary Material
Supplementary material dealing with details on protein-DNA and inter-subunit correlations, simulations convergence data, dynamic cross-correlation maps, protein’ residues sequence and numbering, is available at no charge from the authors directly; It can also be downloaded for free of charge from the author's server at http://bob.olin.wfu.edu/~web/.
References
- 1.Debrunner PG, Frauenfelder H. Ann Rev Phys Chem. 1982;33:283–299. [Google Scholar]
- 2.Kay LE. Nature Struct Biol NMR Supplement. 1998:513–516. doi: 10.1038/755. [DOI] [PubMed] [Google Scholar]
- 3.Livesay DR. Curr Opin Pharmacol. 2010;10:1. doi: 10.1016/j.coph.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCammon JA. Rep Prog Phys. 1984;47:1–64. [Google Scholar]
- 5.Salsbury FR. Curr Opin Pharmacol. 2010;10:738–744. doi: 10.1016/j.coph.2010.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amadei A, Linssen ABM, Berendsen HJC. Proteins. 1993;17:412–425. doi: 10.1002/prot.340170408. [DOI] [PubMed] [Google Scholar]
- 7.Karplus M, Kurian J. Proc Natl Acad Sci U S A. 2005;102:6679–6685. doi: 10.1073/pnas.0408930102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruschweiler R. Nature Chem. 2011;3:665–666. doi: 10.1038/nchem.1124. [DOI] [PubMed] [Google Scholar]
- 9.Henzler-Wildman K, Kern D. Nature. 2007;450:964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- 10.Faure P, Micu A, Perahia D, Doucet J, Smith JC, Benoit JP. Nature Struct Biol. 1994;1:124–128. doi: 10.1038/nsb0294-124. [DOI] [PubMed] [Google Scholar]
- 11.Lange OF, Grubmuller H. Proteins. 2007;70:1294–1312. doi: 10.1002/prot.21618. [DOI] [PubMed] [Google Scholar]
- 12.Ishima R, Torchia DA. Nature Struct Biol. 2000;7:740–743. doi: 10.1038/78963. [DOI] [PubMed] [Google Scholar]
- 13.Mittermaier A, Kay LE. Science. 2006;312:224–227. doi: 10.1126/science.1124964. [DOI] [PubMed] [Google Scholar]
- 14.Ishima R. Top Curr Chem. 2011:1–24. [Google Scholar]
- 15.Dodson GG, Lane DP, Verma CS. EMBO. 2008;9:144–150. doi: 10.1038/sj.embor.7401160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamberaj H, van der Vaart A. Biophys J. 2009;97:1747–1755. doi: 10.1016/j.bpj.2009.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kormos BL, Baranger AM, Beveridge DL. J Struct Biol. 2007;157:500–513. doi: 10.1016/j.jsb.2006.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arnold GE, Ornstein RL. Biophys J. 1997;73:1147–1159. doi: 10.1016/S0006-3495(97)78147-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horstink LM, Abseher R, Nilges M, Hilbers CW. J Mol Biol. 1999;287:569–577. doi: 10.1006/jmbi.1999.2629. [DOI] [PubMed] [Google Scholar]
- 20.Knaggs MH, Salsbury FR, Edgell MH, Fetrow JS. Biophys J. 2007;92:2062–2079. doi: 10.1529/biophysj.106.081950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doruker P, Atilan AR, Bahar I. Proteins. 2000;40:512–524. [PubMed] [Google Scholar]
- 22.Salsbury FR, Crowder MW, Kingsmore SF, Huntley JA. J Mol Model. 2009;15:133–145. doi: 10.1007/s00894-008-0410-0. [DOI] [PubMed] [Google Scholar]
- 23.Go N, Noguti T, Nishikawa T. Proc Natl Acad Sci U S A. 1983;80:3696–3700. doi: 10.1073/pnas.80.12.3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levy RM, Karplus M. Biopolymers. 1979;18:2465–2495. [Google Scholar]
- 25.Levy RM, Perahia D, Karplus M. Proc Natl Acad Sci U S A. 1982;79:1346–1350. doi: 10.1073/pnas.79.4.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Negureanu L, Salsbury FR. J Biomol Struct Dyn. 2012;29:757–776. doi: 10.1080/07391102.2012.10507412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Young L, et al. J Invest Dermatol. 2003;121:876–880. doi: 10.1046/j.1523-1747.2003.12486.x. [DOI] [PubMed] [Google Scholar]
- 28.Jiricny J. Nat Rev. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 29.Peltomaki P. J Clin Oncol. 2003;21:1174–1179. doi: 10.1200/JCO.2003.04.060. [DOI] [PubMed] [Google Scholar]
- 30.Peltomaki P. Hum Mol Genetics. 2001;10:735–740. doi: 10.1093/hmg/10.7.735. [DOI] [PubMed] [Google Scholar]
- 31.Warren J, et al. Mol Cell. 2007;26:579–592. doi: 10.1016/j.molcel.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 32.Takahara PM, Rosenzweig AC, Frederick CA, Lippard SJ. Nature. 1995;377:649–652. doi: 10.1038/377649a0. [DOI] [PubMed] [Google Scholar]
- 33.Takahara PM, Frederick CA, Lippard SJ. J Am Chem Soc. 1996;118:12309–12321. [Google Scholar]
- 34.Duckett DR, Drummond JT, Murchie AI, Reardon JT, Sancar A, Lilley DM, Modrich, P. P. Proc Natl Acad Sci U S A. 1996;93:6443–6447. doi: 10.1073/pnas.93.13.6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamada M, O'Regan E, Brown R, Karran, P. P. Nucleic Acids Res. 1997;25:491–496. doi: 10.1093/nar/25.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salsbury FR, et al. Nucleic Acids Res. 2006;34:2173–2185. doi: 10.1093/nar/gkl238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vasilyeva A, Clodfelter JE, Gorczynski MJ, Gerardi AR, King SB, Salsbury FR, Scarpinato KD. J Nucleic Acids. 2010 doi: 10.4061/2010/162018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kartalou M, Essigmann JM. Mutat Res. 2001;478:1–21. doi: 10.1016/s0027-5107(01)00142-7. [DOI] [PubMed] [Google Scholar]
- 39.Gelasco A, Lippard SJ. Biochemistry. 1998;37:9230–9239. doi: 10.1021/bi973176v. [DOI] [PubMed] [Google Scholar]
- 40.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 41.Scheeff ED, Briggs JM, Howell SB. Mol Pharmacol. 1999;56:633–643. [PubMed] [Google Scholar]
- 42.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fisher S, Gao J, Guo H, Ha S, et al. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 43.MacKerell DAJ, Banavali N, Foloppe N. Biopolymers. 2001;56:257–265. doi: 10.1002/1097-0282(2000)56:4<257::AID-BIP10029>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 44.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, R.W., Klein ML. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 45.Humphrey W, Dalke A, Schulten K. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 46.Lee MS, Salsbury FR, Brooks CL. J Chem Phys. 2002;116:10606–10615. [Google Scholar]
- 47.Lee MS, Salsbury FR, Olson MA. J Comp Chem. 2004;25:1967–1978. doi: 10.1002/jcc.20119. [DOI] [PubMed] [Google Scholar]
- 48.Salsbury FR. Mol Phys. 2006 [Google Scholar]
- 49.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- 50.Kale L, Skeel R, Bhandarkar M, Brunner R, Gursoy A, Krawetz N, Phillips J, Shinozaki A, Varadarajan K, Schulten K. J Comp Phys. 1999;15:1283–1312. [Google Scholar]
- 51.van Gunsteren WF, Berendsen HJC. Mol Phys. 1977;34:1311–1327. [Google Scholar]
- 52.Darden T, York D, Pedersen L. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 53.Lin PD, Wang Y, Scherer SJ, Clark AB, Yang K, Avdievich E, Jin, B. B, et al. Cancer Res. 2004;64:517–522. doi: 10.1158/0008-5472.can-03-2957. [DOI] [PubMed] [Google Scholar]
- 54.O'Brien V, Brown R. Carcinogenesis. 2006;27:682–692. doi: 10.1093/carcin/bgi298. [DOI] [PubMed] [Google Scholar]
- 55.Zhang H, Richard B, Wilson T, Lloyd M, Cranston A, Thorburm A, Fishel R, Meuth M. Cancer Res. 1999;59:3021–3027. [PubMed] [Google Scholar]
- 56.Stojic L, Brun R, Jiricny J. DNA Repair. 2004;3:1091–1101. doi: 10.1016/j.dnarep.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 57.Yang G, Scherer SJ, Shell SS, Yang M, Lipkin M, Kucherlapati R, Kolodner RD, Edelman W. Cancer Cell. 2004;6:139–150. doi: 10.1016/j.ccr.2004.06.024. [DOI] [PubMed] [Google Scholar]
- 58.Yanamadala S, Ljungman M. Mol Cancer Res. 2003;1:747–754. [PubMed] [Google Scholar]
- 59.Tomlinson IP, Hampson R, Karran P, Bodmer WF. Mutat Res. 1997;383:177–182. doi: 10.1016/s0921-8777(96)00058-4. [DOI] [PubMed] [Google Scholar]
- 60.Marra G, D'Atri S, Corti C, Bonmassar L, Cattaruzza MS, Schweizer P, Heinimann K, Bartosova Z, Nystrom-Lahti M, Jiricny J. Proc Natl Acad Sci U S A. 2001;98:7164–7169. doi: 10.1073/pnas.121136498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Negureanu L, Salsbury FR. J Biomol Struct Dyn. 2011;29:757–776. doi: 10.1080/07391102.2012.10507412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Salsbury FR. Protein Pept Lett. 2010;17:744–750. doi: 10.2174/092986610791190318. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.