Abstract
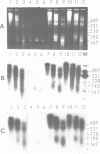
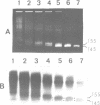
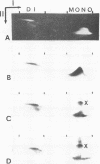
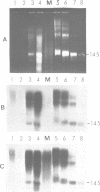
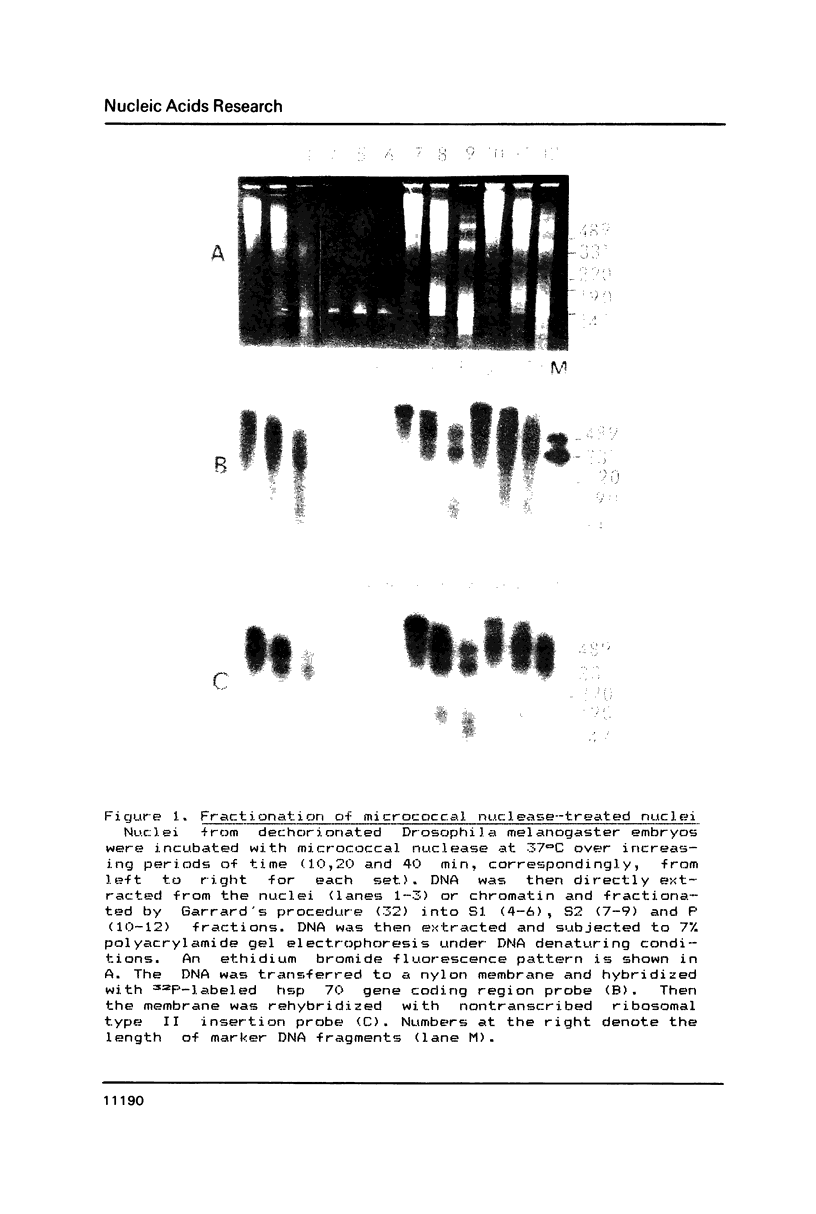
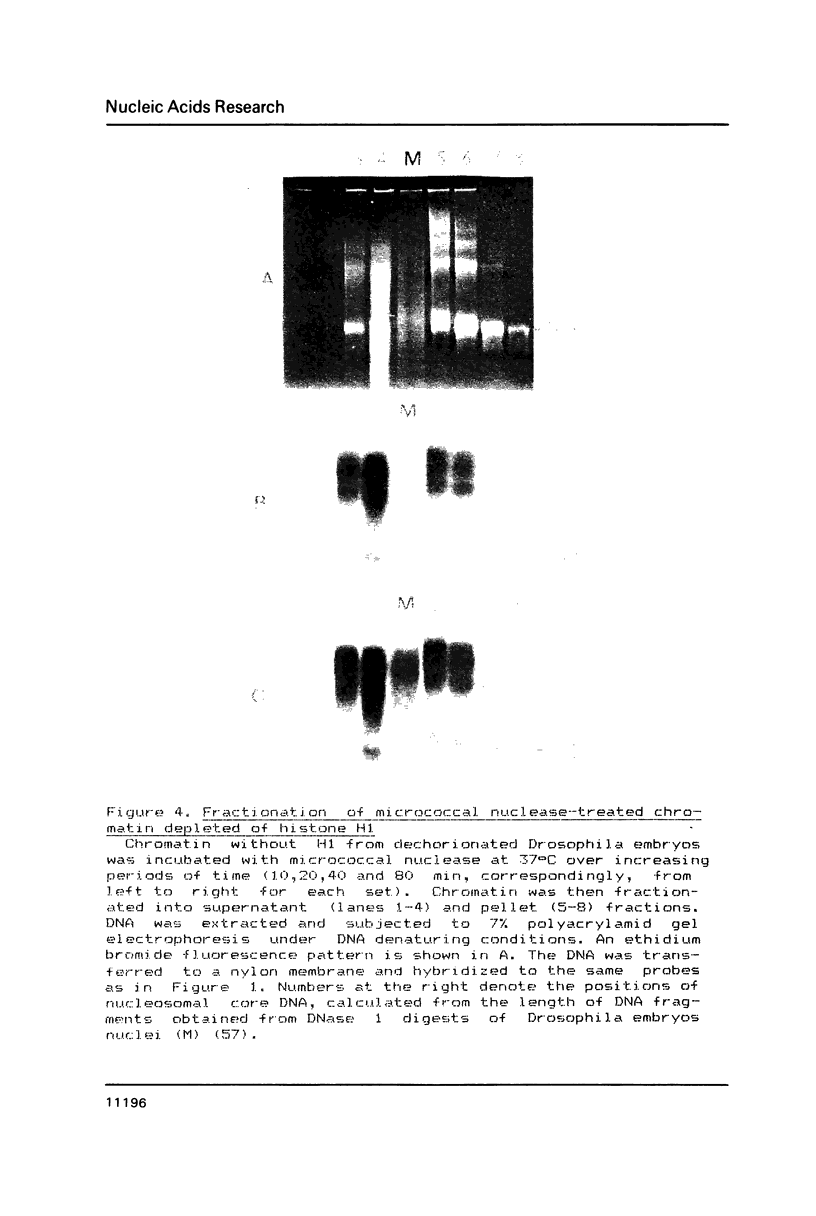
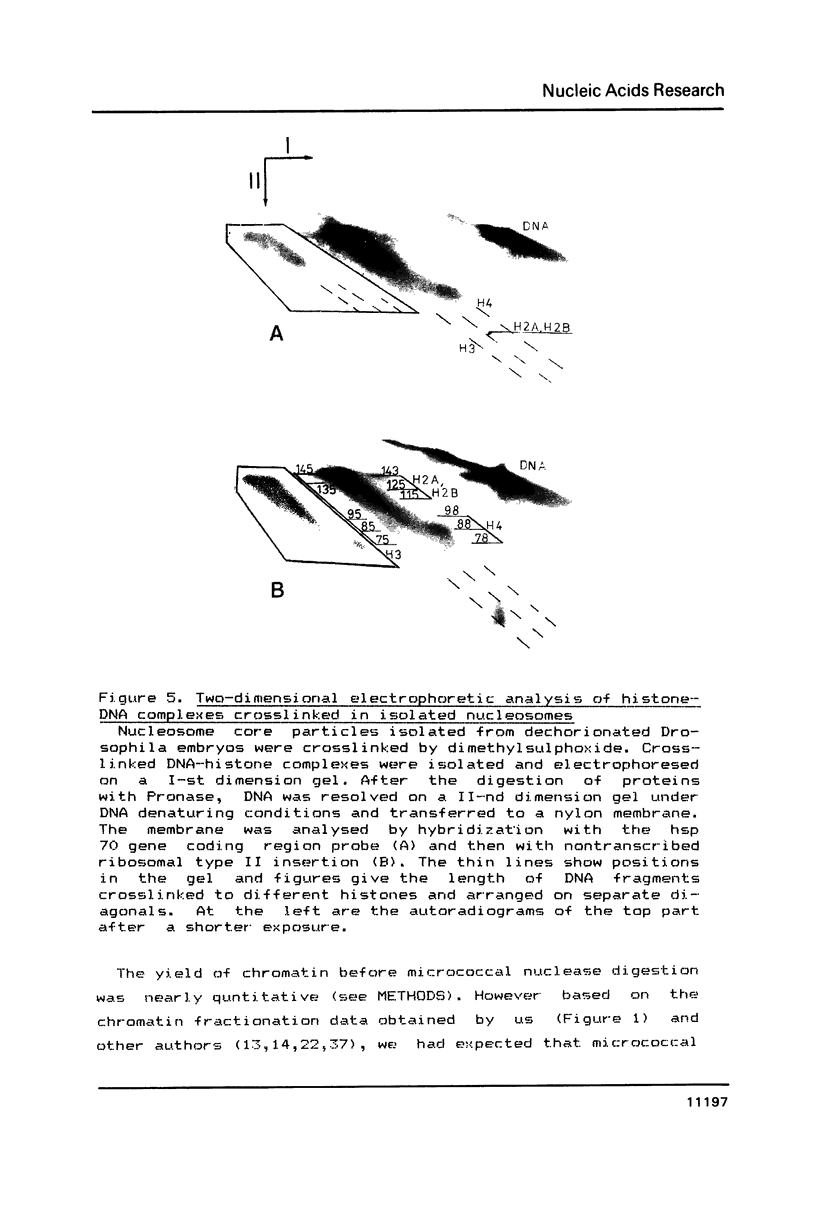
The arrangement of histones along DNA in nucleosomal core particles within transcribed heat shock gene (hsp 70) region and repressed insertion within ribosomal genes of Drosophila was analysed by using protein-DNA crosslinking methods combined with hybridization tests. In addition, two-dimensional gel electrophoresis was employed to compare the overall nucleosomal shape and the nucleosomal DNA size. The arrangement of histones along DNA and general compactness of nucleosomes were shown to be rather similar in transcriptionally active and inactive genomic regions. On the other hand, nucleosomes within transcriptionally active chromatin are characterized by a larger size of nucleosomal DNA produced by micrococcal nuclease digestion and some peculiarity in electrophoretic mobility.
Full text
PDF















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ausio J., van Holde K. E. Histone hyperacetylation: its effects on nucleosome conformation and stability. Biochemistry. 1986 Mar 25;25(6):1421–1428. doi: 10.1021/bi00354a035. [DOI] [PubMed] [Google Scholar]
- Cohen R. B., Sheffery M. Nucleosome disruption precedes transcription and is largely limited to the transcribed domain of globin genes in murine erythroleukemia cells. J Mol Biol. 1985 Mar 5;182(1):109–129. doi: 10.1016/0022-2836(85)90031-2. [DOI] [PubMed] [Google Scholar]
- Eissenberg J. C., Cartwright I. L., Thomas G. H., Elgin S. C. Selected topics in chromatin structure. Annu Rev Genet. 1985;19:485–536. doi: 10.1146/annurev.ge.19.120185.002413. [DOI] [PubMed] [Google Scholar]
- Levina E. S., Bavykin S. G., Shick V. V., Mirzabekov A. D. The method of crosslinking histones to DNA partly depurinated at neutral pH. Anal Biochem. 1981 Jan 1;110(1):93–101. doi: 10.1016/0003-2697(81)90117-2. [DOI] [PubMed] [Google Scholar]
- Lohr D. The chromatin structure of an actively expressed, single copy yeast gene. Nucleic Acids Res. 1983 Oct 11;11(19):6755–6773. doi: 10.1093/nar/11.19.6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorch Y., LaPointe J. W., Kornberg R. D. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell. 1987 Apr 24;49(2):203–210. doi: 10.1016/0092-8674(87)90561-7. [DOI] [PubMed] [Google Scholar]
- Losa R., Brown D. D. A bacteriophage RNA polymerase transcribes in vitro through a nucleosome core without displacing it. Cell. 1987 Aug 28;50(5):801–808. doi: 10.1016/0092-8674(87)90338-2. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Felsenfeld G. Nucleosome structure. Annu Rev Biochem. 1980;49:1115–1156. doi: 10.1146/annurev.bi.49.070180.005343. [DOI] [PubMed] [Google Scholar]
- Nicolas R. H., Wright C. A., Cockerill P. N., Wyke J. A., Goodwin G. H. The nuclease sensitivity of active genes. Nucleic Acids Res. 1983 Feb 11;11(3):753–772. doi: 10.1093/nar/11.3.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olins A. L., Olins D. E., Levy H. A., Durfee R. C., Margle S. M., Tinnel E. P. DNA compaction during intense transcription measured by electron microscope tomography. Eur J Cell Biol. 1986 Mar;40(1):105–110. [PubMed] [Google Scholar]
- Razin S. V., Yarovaya O. V., Georgiev G. P. Low ionic strength extraction of nuclease-treated nuclei destroys the attachment of transcriptionally active DNA to the nuclear skeleton. Nucleic Acids Res. 1985 Oct 25;13(20):7427–7444. doi: 10.1093/nar/13.20.7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves R. Transcriptionally active chromatin. Biochim Biophys Acta. 1984 Sep 10;782(4):343–393. doi: 10.1016/0167-4781(84)90044-7. [DOI] [PubMed] [Google Scholar]
- Shick V. V., Belyavsky A. V., Mirzabekov A. D. Primary organization of nucleosomes. Interaction of non-histone high mobility group proteins 14 and 17 with nucleosomes, as revealed by DNA-protein crosslinking and immunoaffinity isolation. J Mol Biol. 1985 Sep 20;185(2):329–339. doi: 10.1016/0022-2836(85)90407-3. [DOI] [PubMed] [Google Scholar]
- Smith R. D., Yu J., Annunziato A., Seale R. L. beta-Globin gene family in murine erythroleukemia cells resides within two chromatin domains differing in higher order structure. Biochemistry. 1984 Jun 19;23(13):2970–2976. doi: 10.1021/bi00308a019. [DOI] [PubMed] [Google Scholar]
- Stein A., Townsend T. HMG 14/17 binding affinities and DNAase I sensitivities of nucleoprotein particles. Nucleic Acids Res. 1983 Oct 11;11(19):6803–6819. doi: 10.1093/nar/11.19.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeponteau B., Martinson H. G. Gamma rays and bleomycin nick DNA and reverse the DNase I sensitivity of beta-globin gene chromatin in vivo. Mol Cell Biol. 1987 May;7(5):1917–1924. doi: 10.1128/mcb.7.5.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Wu R. S., Panusz H. T., Hatch C. L., Bonner W. M. Histones and their modifications. CRC Crit Rev Biochem. 1986;20(2):201–263. doi: 10.3109/10409238609083735. [DOI] [PubMed] [Google Scholar]
- Xu M., Barnard M. B., Rose S. M., Cockerill P. N., Huang S. Y., Garrard W. T. Transcription termination and chromatin structure of the active immunoglobulin kappa gene locus. J Biol Chem. 1986 Mar 15;261(8):3838–3845. [PubMed] [Google Scholar]