Abstract
Asthma is a multifactorial disorder, primarily resulting from interactions between genetic and environmental factors. ADAM33 gene (located on chromosome 20p13) has been reported to play an important role in asthma. This review article is intended to include all of the publications, to date, which have assessed the association of ADAM33 gene polymorphisms as well as have shown the role of ADAM33 gene in airway remodeling and their expression with asthma. A PubMed search was performed for studies published between 1990 and 2010. The terms “ADAM33,” “ADAM33 gene and asthma,” and “ADAM33 gene polymorphisms” were used as search criteria. Based on available literature we can only speculate its role in the morphogenesis and functions of the lung. Fourteen studies conducted in different populations were found showing an association of ADAM33 gene polymorphisms with asthma. However, none of the single nucleotide polymorphisms (SNPs) of ADAM33 gene had found association with asthma across all ethnic groups. Because higher expression of ADAM33 is found in the fibroblast and smooth muscle cells of the lung, over- or underexpression of ADAM33 gene may result in alterations in airway remodeling and repair processes. However, no SNP of ADAM33 gene showed significant associations with asthma across all ethnic groups; the causative polymorphism, if any, still has to be identified.
Keywords: ADAM33, airway remodeling, association studies, asthma, bronchial hyperresponsiveness, chronic inflammatory disorder, multifactorial disorder, pathogenesis of asthma, positional cloning, single-nucleotide polymorphism
Asthma is a complex, chronic inflammatory disorder of airways of the lungs resulting in airflow obstruction; bronchial hyperresponsiveness (BHR) to a variety of stimuli; and symptoms of wheeze, cough, and breathlessness. It is a major global public health problem. It is estimated that there are about 300 million asthmatic patients worldwide.1 Chronic asthma is a result of abnormal repair and remodeling processes of the airways, characterized by epithelial damage, smooth muscle hyperplasia, and matrix depositions.2 It has been postulated that the aforementioned processes are a result of complex interactions between genes and the environment, often beginning in utero and early infancy.3–5 The exact role of ADAM33 in causing asthma is unclear.6Various studies have reported a cluster of single nucleotide polymorphisms (SNPs) in the ADAM33 gene that have significant association with asthma and related phenotypes.7,8 Selective expression of the ADAM33 gene in mesenchymal cells suggests its potential to affect the epithelial mesenchymal tropic unit (EMTU) along with TH2 cytokines.5,9
PATHOGENESIS OF ASTHMA: GENE AND ENVIRONMENT INTERACTION
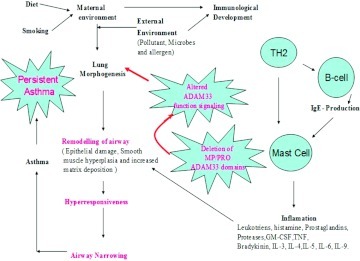
Increasing prevalence of asthma is thought to be linked with genetic changes and gene–environment interactions.4 The activation of tissue-specific susceptibility genes provides a basis for explaining environmental factors that may be more closely associated with asthma.10 Possible exposure to environmental factors such as pollutants, microbes, allergens, and oxidant stimuli reactivates the EMTU, which is involved in morphogenesis during fetal lung development (Fig. 1).9,11 It has been shown that epithelium of patients with asthma is structurally and functionally abnormal and more susceptible to oxidant-induced apoptosis.12 Apoptotic cell loss is accompanied by increased expression of epidermal growth factor (EGF) receptors and transforming growth factors (TGF) β1 and β2.13 These growth factors play an important role in promoting differentiation of fibroblasts into myofibroblasts that secrete other growth factors such as edothelin1, which act as mitogens for smooth muscles and endothelial cells.13,14 Continuous epithelial repair results in abnormal functioning of growth factor pathways and disrupts cell cycle. The altered epithelium communicates with the underlying mesenchyme to create a tropic unit that propagates and induces remodeling signals in deeper layers of submucosa.5,14
Figure 1.
Role of the gene–environment interaction and ADAM33 in development of asthma.
GENETIC BASIS OF ASTHMA
Although ∼120 genes have been identified to be associated with asthma, no clear pattern of inheritance is known. Heritability estimates of asthma lie between 36 and 79%.15–17 Recently, positional cloning and candidate genes approach have been widely used to identify genes for asthma and associated phenotypes.17,18A large number of asthma candidate genes have been found on chromosomal regions 2q33, 5q23-31, 6p24-21, 11q 21-13, 12q24-12, and 13q14-12.19 It has been established that inflammation of airways leads to its remodeling and is presumedly related to asthma and associated phenotypes.9 Since the 1990s, various genes associated with inflammation have also been identified using new molecular techniques.15 Several genomewide screens have identified linkage of certain chromosomal regions with both asthma and inflammation such as 5q23-31, 5p15, 6p21.3-23, 11p15, 12q14-24.2, 13q21.3, 14q11.2-13, 17p11.1-q11.2, 19q13, and 21q21.15,20–22 Among them, 5q23-31, 5p15, and 12q14-24.2 are constantly replicated, which contain genes such as IL-3, IL-4, IL-5, IL-9, IL-12b, IL-13, interferon (IFN) γ, iNOS, and FcεRIβ.21,23 Most of these genes influence cytokine regulation and activity of eosinophils, mast cells, and neutrophils.21,23,24
Linkage analysis and genomewide screening of 260 families in the United Kingdom and United States led to a region on chromosome 20p13 containing a putative asthma susceptibility gene, ADAM33.20,25 It is postulated that ADAM33 is one of many genes that influence the onset and progression of asthma. Identification of ADAM33 gene changed the belief that airway remodeling was solely the result of persistent inflammation and established that it is influenced by other genetic factors also. Its potential in shaping multiple phenotypes such as early life lung functions and chronic obstructive pulmonary disorder makes ADAM33 an important asthma susceptibility gene.26 Genetic origin of BHR has also been linked with ADAM33. Likewise, another gene, dipeptidyl peptidase 10 (DPP10), was found associated with BHR and IgE.25,27,28 Expression of various genes, such as G-protein coupled receptor for asthma (GPRA) and serine protease inhibitors of the Kazal type (SPINK5), have also been found to have a role in airway remodeling and asthma.29
ABOUT ADAM33
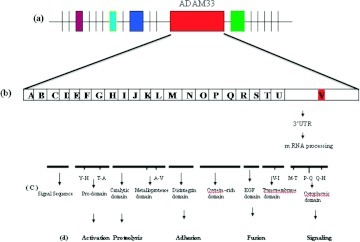
ADAM molecules are members of the disintegrin and metalloprotease family, which are type I transmembrane zymogen glycoproteins that typically contain an N-terminal secretion signal domain, pro, catalytic, disintegrin, cysteine-rich, EGF-like transmembrane part, and a cytoplasmic (C-terminal) domain (Fig. 2, c).30,31 Its complex organization involves eight domains that facilitate their participation in many cellular processes.31,32 Their adhesion domain as well as protease domain makes it exclusive among cell surface proteins. The autocatalytic removal of the prodomain is activation signal for ADAM proteins.33 It is presumed that ADAM proteins have a critical role in cell adhesion, proliferation differentiation, signaling, and apoptosis, as well as the inflammatory responses.34,35 Because ADAM33 belongs to the ADAM12, ADAM15, ADAM19, and ADAM28 subfamily, it is expected to have similar activities.36 From a wide range of substrates for ADAM33 only four, viz., stem cell factor (C- kit), β-amyloid precursor protein, TNF-related activation–induced cytokine, and insulin β, were cleaved. However, it is not certain if any of these are natural substrates.37 To date, no natural substrate has been identified for ADAM33.
Figure 2.
(a) ADAM33 and other flanking regions; (b) exon–intron structure of ADAM33 gene; (c) different domains of ADAM33 protein; (d) probable function of different domains.
A crystallization study of catalytic domain of ADAM33 revealed that the catalytic site shares the feature of other matrix metalloproteinases having a Zn2+ binding site but differs in structure of substrate pocket.38 It has been speculated that ADAM33 may have cytokine stimulating effects, given that other ADAM proteins (ADAM10 and ADAM17) also appear to interact with inflammatory cytokines.13 Cysteine-rich and EGF domains of ADAM33 have been identified to have a role in cell adhesion and membrane fusion events (Fig. 2 d).39 These properties of ADAM33 suggest that it might play a role in progression of asthma.40,41
Objective
This article is intended to include all of the publications to date, which have assessed the association of ADAM33 gene polymorphisms as well as have shown a role of ADAM33 gene in airway remodeling and their expression with asthma.
METHODS
Identification and Selection of Studies
N. Sharma and P. Tripathi contributed equally to this study. A comprehensive literature search was conducted to collect data from all of the studies that investigated the association of genes with asthma in worldwide populations. All of the studies that were published between 1990 and 2010 were included. We did extensive computer-based searches of PubMed to identify the studies evaluating the role of the ADAM33 gene in relation to asthma. The terms “ADAM33,” “ADAM33 gene and asthma,” and “ADAM33 gene polymorphisms” were used as search criteria. The search results were limited to humans.
RESULTS
Role of ADAM33 Gene in Asthma
Role of ADAM33 Gene in Airway Remodeling.
Remodeling of airways has been assumed to result from uncontrolled inflammatory/repair processes, which may occur early in development, either as an effect of a maternally dictated (in utero) environment or of genetic inheritance or a combination of both.27,42 Family-based studies of asthma have shown that there is a genetic predisposition to atopy, which alters susceptibility to asthma as well as other allergic disorders. Genetic effects also regulate susceptibility of the lungs to both allergic and other environmentally induced inflammation. Traditionally, it has been assumed that inflammation resulting in airway remodeling is the cause of asthma. However, inflammation alone does not explain many of the characteristic features of the chronic and relapsing nature of asthma.5 Selective expression of ADAM33 in mesenchymal cells of asthmatic airways strongly suggests that alteration in its activity may result in abnormalities in the function of airway smooth muscle cells and fibroblasts linked to BHR and remodeling in asthma.
ADAM33 acts as a cell surface sheddase to release growth factors and modify cell surface receptor expression. It might be linked to the action of TGF- β. TGF-β increases ADAM33 expression as a part of differentiation trajectory of common fibroblastic progenitor of components of EMTU to myofibroblasts.5,14,43,44 Studies have shown ADAM33 expression in lungs during embryogenesis.45,46 Therefore, it might be possible that ADAM33 functions in lung growth and that alteration in this gene may alter the lung morphogenesis or subsequent susceptibility of the lung to asthma.
Association of ADAM33 Gene with Asthma
ADAM33 and Their Expression.
Northern blot analysis showed two transcripts (5.0 and 3.5 kb) of ADAM33, of which only one was (3.5 kb) found in cytoplasm.15,25 In situ hybridization technique revealed that ADAM33 preferentially expressed in smooth muscles, myofibroblasts, and fibroblasts of asthmatic airway and was not found in epithelium, endothelium, and T cell or inflammatory leukocytes.46 High expression of ADAM33 is found in MRC5 (lung fibroblast), jurkat (transformed T cell), the stomach, and the small intestine. Moderate expression of ADAM33 is observed in the trachea and bronchus and low expression is shown in the lymph node and thymus.5 The potential of alternative transcript of ADAM33 gene showed marked differences in tissue expression profile of pro and protease domains, which indicates the potential for tissue-selective functions of ADAM33.9,45 However, the correlation of this phenomenon with individual SNPs is still unknown.
ADAM33 protein has two isoforms, viz., α and β. The α-form is found abundantly in airway fibroblasts, myofibroblasts, and smooth muscles cells. Secreted splice variants of ADAM33 are rarely found.17,45 Because of complex differential splicing property of the ADAM33 gene, a high degree of variability in gene products has been observed. The majority of variation is seen in the 5′ end of the gene and the 3′ domain is present in all transcripts.48 A study conducted by Powel et al.49 showed no clear difference in the expression of ADAM33 spliced variants in bronchial biopsy specimens between normal subjects and subjects with asthma. However, in another study, increased expression of ADAM33 has been reported in moderate and severe asthma.50 Down-regulation of the ADAM33 gene by IFN-γ has been observed in airway smooth muscles cells.51 Studies suggest that mRNA expression of ADAM33 in response to IFN-γ may be regulated by the promoter region. The difference between regulation of ADAM33 gene by IFN-γ in subjects with and without asthma may be caused by the presence of polymorphism in the upstream region of ADAM33.7,51
SNPs in ADAM33 as Indicator of Severe and Progressive Asthma.
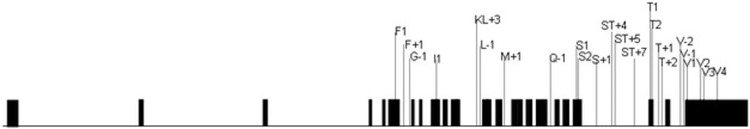
Although the exact role of ADAM33 in the pathogenesis of asthma is unclear, studies have shown an association of ADAM33 and its related SNPs with asthma (Fig. 3). Table 1 summarizes the results of studies conducted in various populations. In the original description of ADAM33 as an asthma candidate gene, Van Eerdewegh et al.25 identified a locus on the short arm of chromosome 20 and assessed 135 polymorphisms of 23 genes in this region and reported ADAM33 gene to be significantly associated with asthma association of ADAM33 SNPs with asthma as reported in published studies, are summarized in Table 1.
Figure 3.
Reported single nucleotide polymorphisms in the ADAM33 gene. Exons (black rods) and introns of the ADAM33 gene are shown. The association results of a case–control study in different populations shown in Table 1.
Table 1.
Previously reported association results with ADAM33 gene polymorphisms with asthma and BHR
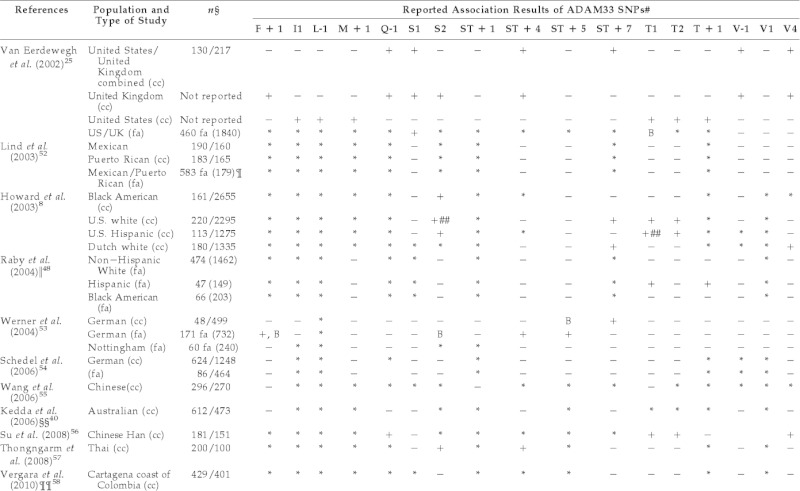
#The SNP designations are derived from the exon labeling (see Fig. 2).
Association with Asthma (+); BHR (B); SNPs that are not significantly associated (−); SNPs not studied (*).
§In case–control studies number of cases and controls and in family studies number of families and individuals.
¶Two hundred sixty-five Mexican families and 318 Puerto Rican families, no association in single or in pooled populations.
‖In the study of Raby et al., eight additional SNPs were investigated: G1, I1, KL + 3, N1, S + 1, T + 2, V-2, and V3. None of the additional SNPs showed an association with asthma.
##Only trend (0.05 < p ≤ 0.06).
§§In the study of Kedda et al., three additional SNPs were investigated: V-2, V2, and V5.
¶¶Haplotype of ADAM33 SNPs showed association with asthma.
BHR = bronchial hyperresponsiveness; cc = case control; fa = family study; n = number; SNP = single nucleotide polymorphism.
In another case–control study, Howard et al.8 assessed association of eight SNPs of ADAM33 gene, namely S1, S2, ST + 4, ST + 7, T1, T2, V-1, and V4, with asthma in four unique ethnic groups with asthma viz: U.S. white, Dutch white, black American, and Hispanic. Significant associations of SNPs were observed in all four studied population, but no single SNP was consistently associated with a specific asthma phenotype across all of the ethnicities. They observed significant associations with asthma in the Dutch population with SNPs ST + 7 and V4 by using a codominant model (p = 0.0093 and p = 0.0009, respectively). Associations with asthma were also observed in black Americans with SNP S2 (p = 0.03), U.S. white populations with SNPs ST + 7 (p = 0.017), T1 (p = 0.03), T2 (p = 0.02) and in U.S. Hispanic populations with SNPs S2 (p = 0.04) and T2 (p = 0.04).
Lind et al.52 working on six SNPs, viz., S1, T1, T2, V-1, V1, and V4, of the ADAM33 gene used the transmission disequilibrium test to analyze associations between the ADAM33 gene variants and asthma, asthma severity, bronchodilator responsiveness, and total IgE levels using single SNPs, two to six SNP combinations, specific haplotypes but were unable to establish an association of any SNPs either with asthma or other outcomes. Raby et al.48 assessed 17 SNPs of ADAM33 gene, viz., G-1, Il, KL + 3, M + 1, N1, S2, S + 1, ST + 4, ST + 5, T1, T2, V-2, V-1, V3, and V4, but failed to detect association in a family-based study of white, black Americans, and Hispanic trios representative of North American children with mild-to-moderate asthma. However, two SNPs in strong linkage disequilibrium (T1 and T + 1) were marginally associated with asthma in the Hispanic cohort only (p = 0.04). Wernar et al.53 analyzed 15 SNPs of the ADAM33 gene, viz., F + 1, Il, M + 1, Q-1, S1, S2, S + 1, ST + 4, ST + 5, ST + 7, T1, T2, T + 1, V-1, and V4, and observed a statistically significant association of asthma with SNPs F + 1, ST + 4, and ST + 5 in a family-based study. Significant association of SNP, ST + 7, was found in the case–control study.
Blakey et al.6 conducted transmission disequilibrium and case–control studies in Icelandic and Nottingham study populations but did not find any association with asthma; however, after performing meta-analysis, all existing data showed either positive or negative association results with asthma and showed that F + 1 and ST + 7 SNPs were significantly associated with asthma.6 Schedel et al.54 analyzed 10 SNPs of ADAM33, viz., F + 1, M + 1, S1, S2, ST + 4, ST + 5, ST + 7, T1, T2, and V4, and found none to be significantly associated with asthma in both case–control and cohort study designs in a German population.
A study conducted on a Chinese population by Wang et al.55 did not find any association of SNPs, viz., S + 1, T1, and F + 1, with asthma. Another study on a Chinese Han population analyzing six SNPs of the ADAM33 gene, viz., V4, T + 1, T2, T1, S1, and Q-1, for association with allergic asthma, found that V4, T2, T1, and Q-1 increased risk of susceptibility.56 Kedda et al.40 did a study on an Australian population and were unable to find association of SNPs of ADAM33, viz., F + 1, Q-1, S1, ST + 4, ST + 7, V-2, V-1, V2, V4, and V5, with asthma.
Thongngarm et al.57 conducted a study in a Thai population on eight SNPs of ADAM33, viz., S1, S2, ST + 4, ST + 7, T1, V-1, and V4, and found a positive association between ADAM33 polymorphisms S2 and ST + 4 with asthma susceptibility. A study by Vergara et al.58 using six SNPs of ADAM33, viz., S2, ST + 7, T1, T2, V-1, and V4, in population of Cartagena, Colombia, were unable to find association of any SNPs with asthma. However, they found association of TT genotype of ST + 7(C/T) SNP with asthma (p = 0.05) that disappeared after correcting for multiple testing. They also identified eight common haplotypes; among them, H4 (GCAGGG) was associated with asthma in the family group (Z score, −2.049; p = 0.04).
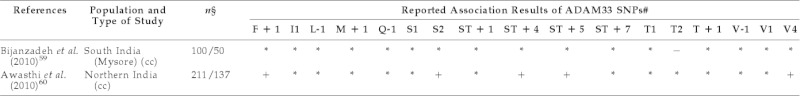
A study by Bijanzadeh et al.59 in children as well as adults, also failed to find an association between asthma and the T1 SNP of ADAM33 gene in a southern Indian population. However, another recent case–control study, conducted in Northern India to assess association of ADAM33 gene polymorphisms, viz., F + 1, S2, ST + 4, ST + 5, and V4, with asthma in children aged 1–15 years, showed significant association of all of them with the disease.60
Certain studies have shown that multiple SNPs may act together to increase the risk of asthma; haplotype analysis found a significant association which was not found during individual SNPs analysis.40,47,58 However, it is still unclear which of the identified SNPs relate causatively to asthma.
CONCLUSIONS
In the last 10 years >120 genes have been found associated with asthma and related phenotypes. Gene–gene as well gene–environment interactions are involved in the pathogenesis of asthma. This causes airway inflammation and remodeling by the action of cytokines, metalloprotease (ADAM33), and matrix metalloproteinase 9.
Identification of ADAM33 as an asthma susceptibility gene marks the entry of asthma research into the genomic area and provides new insight to the understanding of molecular basis of pathogenesis of asthma. Based on the available literature the exact biological activities of ADAM33 are still unknown. We can only speculate its role in the morphogenesis and functions of the lungs. Because higher expression of ADAM33 is found in the fibroblast and smooth muscle cells of the lung, over- or underexpression of the ADAM33 gene may result in alterations in airway remodeling and repair processes.
Probable associations of various polymorphisms of the ADAM33 gene with asthma and related phenotypes have been found in literature review. However, no single SNP of the ADAM33 gene has been found associated with asthma across all of the studies published so far. Therefore, we conclude that the causative polymorphisms of ADAM33 gene, if any, resulting in asthma and its related phenotypes, either alone or as a result of gene–gene or gene–environment interactions, have not been identified and additional in-depth research is needed in this area.
ACKNOWLEDGMENTS
The authors thank Avivar Awasthi for editing this article.
Footnotes
The authors have no conflicts of interest to declare pertaining to this article
REFERENCES
- 1. Bousquet J, Bousquet PJ, Godard P, et al. The public health implications of asthma. Bull World Health Organ 83:548–554, 2005 [PMC free article] [PubMed] [Google Scholar]
- 2. Elias JA, Zhou Z, Chupp G, et al. Airway remodeling in asthma. J Clin Invest 104:1001–1006, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ball TM, Castro-Rodriguez JA, Griffith KA, et al. Siblings, day-care attendance, and the risk of asthma and wheezing during childhood. N Engl J Med 343:538–543, 2000 [DOI] [PubMed] [Google Scholar]
- 4. Koppelman GH. Gene by environment interaction in asthma. Curr Allergy Asthma Rep 6:103–111, 2006 [DOI] [PubMed] [Google Scholar]
- 5. Davies DE, Holgate ST. Asthma: The importance of epithelial mesenchymal communication in pathogenesis. Inflammation and the airway epithelium in asthma. Int J Biochem Cell Biol 34:1520–1526, 2002 [DOI] [PubMed] [Google Scholar]
- 6. Blakey J, Halapi E, Bjornsdohir US, et al. Contribution of ADAM33 polymorphism to the risk of asthma. Thorax 60:274–276, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chae SC, Yoon KH, Chung HT. Identification of novel polymorphisms in the ADAM33 gene. J Hum Genet 48:278–281, 2003 [DOI] [PubMed] [Google Scholar]
- 8. Howard TD, Postma DS, Jongepier H, et al. Association of disintegrin and metalloprotease 33 (ADAM33) gene with asthma in ethnically diverse populations. J Allergy Clin Immunol 112:717–722, 2003 [DOI] [PubMed] [Google Scholar]
- 9. Cakebread JA, Haitchi HM, Holloway JW, et al. The role of ADAM33 in the pathogenesis of asthma. Semin Immunol 25:361–375, 2004 [DOI] [PubMed] [Google Scholar]
- 10. Holgate ST. Genetic and environmental interaction in allergy and asthma. J Allergy Clin Immunol 104:1139–1146, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chou KT, Huang CC, Chen YM, et al. Asthma and risk of erectile dysfunction—A nationwide population-based study. J Sex Med 8:1754–1760, 2011 [DOI] [PubMed] [Google Scholar]
- 12. Bucchieri F, Puddicombe SM, Lordan JL, et al. Asthmatic bronchial epithelium is more susceptible to oxidant-induced apoptosis. Am J Respir Cell Mol Biol 27:179–185, 2002 [DOI] [PubMed] [Google Scholar]
- 13. Redington AE, Roche WR, Holgate ST, et al. Co-localization of immunoreactive transforming growth factor-beta 1 and decorin in bronchial biopsies from asthmatic and normal subjects. J Pathol 186:410–415, 1998 [DOI] [PubMed] [Google Scholar]
- 14. Richter A, Puddicombe SM, Lordan JL, et al. The contribution of interleukin IL-4 and IL-13 to the epithelial-mesenchymal tropic unit in asthma. Am J Respir Cell Mol Biol 25:385–391, 2001 [DOI] [PubMed] [Google Scholar]
- 15. Duffy DL. Genetics of asthma and hay fever in Australian twins. Am Rev Respir Dis 142:1351–1358, 1990 [DOI] [PubMed] [Google Scholar]
- 16. Cookson WOCM, Faux JA, Sharp PA, et al. Linkage between immunoglobulin E responses underlying asthma and rhinitis and chromosome 11q. Lancet 333:1292–1295, 1989 [DOI] [PubMed] [Google Scholar]
- 17. Zhang J, Pare PD, Sandford JA. Recent advances in asthma genetics. Respir Res 9:4, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wick J. Transient up regulation of ADAM33 by TGF-β precedes myofibroblast differentiation. Am J Respir Crit Care Med 167:157, 2003 [Google Scholar]
- 19. Hoffjan S, Ober C. Present status on the genetic studies of asthma. Curr Opin Immunol 14:709–714, 2002 [DOI] [PubMed] [Google Scholar]
- 20. Malebra G, Pignatti PF. A review of asthma genetics: Gene expression studies and recent candidates. J Appl Genet 46:93–104, 2005 [PubMed] [Google Scholar]
- 21. Vereceill D. Discovering susceptibility genes for asthma and allergy. Nat Rev Immunol 8:169–182, 2008 [DOI] [PubMed] [Google Scholar]
- 22. Guerra S, Martinez FD. Asthma genetics: From linear to multifactorial approaches. Annu Rev Med 59:327–341, 2008 [DOI] [PubMed] [Google Scholar]
- 23. Boss Y, Hudson TJ. Toward a comprehensive set of asthma susceptibly genes. Annu Rev Med 58:171–184, 2007 [DOI] [PubMed] [Google Scholar]
- 24. Anderson GG, Morrison JF. Molecular biology and genetics of allergy and asthma. Arch Dis Child 78:488–496, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Van Eerdewegh P, Little RD, Dupuis J, et al. Association of the ADAM 33 gene with asthma and bronchial hyperresponsives. Nature 418:426–430, 2002 [DOI] [PubMed] [Google Scholar]
- 26. Raby BA, Weiss ST. ADAM 33 Where are we now? AM J Respir Cell Mol 31:1–2, 2004 [DOI] [PubMed] [Google Scholar]
- 27. Davies DE. The role of the epithelium in airway remodeling in asthma. Proc Am Thorac Soc 6:678–682, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Allen M, Heinzmann A, Noguchi E, et al. Positional cloning of a novel gene influencing asthma from chromosome 2q14. Nat Genet 35:258–263, 2003 [DOI] [PubMed] [Google Scholar]
- 29. Laitinen T, Polvi A, Rydman P, et al. Characterization of a common susceptibility locus for asthma related traits. Science 304:300–304, 2004 [DOI] [PubMed] [Google Scholar]
- 30. Black RA, White JM. ADAMs: Focus on the protease domain. Curr Opin Cell Biol 10:654–659, 1998 [DOI] [PubMed] [Google Scholar]
- 31. Blobel CP. ADAMs: Key components in EGFR signaling and development. Nat Rev Mol Cell Biol 6:32–43, 2005 [DOI] [PubMed] [Google Scholar]
- 32. Primakoff P, Myles DG. The ADAM gene family: Surface proteins with adhesion and protease activity. Trends Genet 16:83–87, 2000 [DOI] [PubMed] [Google Scholar]
- 33. Stone AL, Kroenger M, Sang QXA. Structure-function analysis of the ADAM family of disintegrin-like and metallo-proteinase-containing proteins. J Protein Chem 18:447–465, 1999 [DOI] [PubMed] [Google Scholar]
- 34. Wolfsberg TG, Straight PD, Gerena RL, et al. ADAM, a family wide distributed and developmentally regulated gene family encoding membrane proteins with a disintegrin and metalloprotease domain. Dev Biol 131:275–248, 1995 [DOI] [PubMed] [Google Scholar]
- 35. Wolfsberg TG, Primak off P, Myles DG, et al. ADAM novel family of membrane proteins containing a disintegrin and metalloprotease domain: Multipotential function in cell–matrix interactions. J Cell Bio 131:275–278, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Howard L, Maciewicz RA, Blobel CP. Cloning and characterization of ADAM28: Evidence for autocatalytic pro-domain removal and for cell surface localization of mature ADAM28. Biochem J 348:21–27, 2000 [PMC free article] [PubMed] [Google Scholar]
- 37. Zou J, Zhu F, Liu J, et al. Catalytic activity of human ADAM 33. J Biol Chem 279:9818–9830, 2004 [DOI] [PubMed] [Google Scholar]
- 38. Orth P, Reichert P, Wang W, et al. Crystal structure of the catalytic domain of human ADAM33. J Mol Biol 335:129–137, 2004 [DOI] [PubMed] [Google Scholar]
- 39. Iba K, Albrechtsen R, Gilpin B, et al. The cysteine-rich domain of human ADAM 12 supports cell adhesion through syndecans and triggers signaling events that lead to beta1 integrin-dependent cell spreading. J Cell Biol 149:1143–1156, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kedda MA, Duffy DL, Bradley B, et al. ADAM33 haplotypes are associated with asthma in a large Australian population. Eur J Hum Genet 14:1027–1036, 2006 [DOI] [PubMed] [Google Scholar]
- 41. Holloway JW, Keith TP, Davies DE, et al. The discovery and role of ADAM 33 a new candidate gene for asthma. Expert Rev Mol Med 6:12, 2004 [DOI] [PubMed] [Google Scholar]
- 42. Cokugras H, Akcakaya N, Seckin I, et al. Ultrastructural examination of bronchial biopsy specimens from children with moderate asthma. Thorax 56:25–29, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang S, Smartt H, Holgate ST, et al. Growth factors secreted by bronchial epithelial cell control myofibroblast proliferation: An in vitro co-culture model of airway remodeling in asthma. Lab Invest 79:395–405, 1999 [PubMed] [Google Scholar]
- 44. Puddicombe SM, Polosa R, Richter A, et al. Involvement of the epidermal growth factor receptor in epithelial repair in asthma. FASEB J 14:1362–1374, 2000 [DOI] [PubMed] [Google Scholar]
- 45. Powell RM, Wicks J, Holloway JW. Identification and quantification of novel spliced variants of a disintegrin and metalloprotease (ADAM) 33 reveal distinct tissue expression profile. Am J Respir Crit Care Med 167:A440, 2003 [Google Scholar]
- 46. Umland SP, Garlisi CG, Shah H, et al. Human ADAM 33 messenger RNA expression profile and post transcriptional regulation. Am J Respir Cell Mol Biol 29:571–582, 2003 [DOI] [PubMed] [Google Scholar]
- 47. Holgate ST, Yang Y, Haitchi HM, et al. The genetic asthma ADAM 33 as an example of a susceptibility gene. Am Thorac Soc 3:440–443, 2006 [DOI] [PubMed] [Google Scholar]
- 48. Raby BA, Silverman EK, Kwiatkowski DJ, et al. ADAM33 polymorphisms and phenotype associations in childhood asthma. J Allergy Clin Immunol 113:1071–1078, 2004 [DOI] [PubMed] [Google Scholar]
- 49. Powell RM, Wicks J, Holloways JW, et al. The splicing and fate of ADAM33 transcripts in primary human airways fibroblasts. Am J Respir Cell Mol Biol 31:13–21, 2004 [DOI] [PubMed] [Google Scholar]
- 50. Foley SC, Mogas AK, Olivenstein R, et al. Increased expression of ADAM 33 and ADAM 8 with disease progression in asthma. J Allergy Clin Immunol 119:863–871, 2007 [DOI] [PubMed] [Google Scholar]
- 51. Ito I, Laporte JD, Fiset PO, et al. Down regulation of a disintegrin and metalloproteinase 33 by IFNγ in human airway smooth muscles cells. J Allergy Clin Immunol 119:89–97, 2007 [DOI] [PubMed] [Google Scholar]
- 52. Lind DL, Choudhry S, Ung N, et al. ADAM33 is not associated with asthma in Puerto Rican or Mexican populations. Am J Respir Crit Care Med 168:1312–1316, 2003 [DOI] [PubMed] [Google Scholar]
- 53. Werner M, Herbon N, Gohlke H, et al. Asthma is associated with single-nucleotide polymorphisms in ADAM 33. Clin Exp Allergy 34:26–36, 2004 [DOI] [PubMed] [Google Scholar]
- 54. Schedel M, Depner M, Schoen C, et al. The role of polymorphisms in ADAM33, a disintegrin and metalloprotease 33, in childhood asthma and lung function in two German populations. Respir Res 7:91, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang P, Liu QJ, Li JS, et al. Lack of association between ADAM33 gene and asthma in a Chinese population. Int J Immunogenet 33:303–306, 2006 [DOI] [PubMed] [Google Scholar]
- 56. Su D, Zhang X, Sui H, et al. Association of ADAM33 gene polymorphisms with adult allergic asthma and rhinitis in a Chinese Han population. BMC Med Genet 9:82, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Thongngarm T, Jameekornrak A, Limwongse C, et al. Association between ADAM33 polymorphisms and asthma in a Thai population. Asian Pac J Allergy Immunol 26:205–211, 2008 [PubMed] [Google Scholar]
- 58. Vergara CI, Acevedo N, Jiménez S, et al. A six SNP haplotype of ADAM33 is associated with Asthma in a population of Cartagena, Colombia. Int Arch Allergy Immunol 152:32–40, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bijanzadeh M, Ramachandra NB, Mahesh PA, et al. Association of IL-4 and ADAM33 gene polymorphisms with asthma in an Indian population. Lung 188:415–422, 2010 [DOI] [PubMed] [Google Scholar]
- 60. Awasthi S, Tripathi P, Ganesh S, et al. Association of ADAM33 gene polymorphisms with asthma in Indian children. J Hum Genet 56:188–195, 2011 [DOI] [PubMed] [Google Scholar]