Abstract
Epidermal growth factor receptor (EGFR) is a member of the ErbB family of receptor tyrosine kinases (RTK). EGFR overexpression or mutation in many different forms of cancers has highlighted its role as an important therapeutic target. Gefitinib, the first small molecule inhibitor of EGFR kinase function to be approved for the treatment of non-small cell lung cancer (NSCLC) by the FDA, demonstrates clinical activity primarily in patients with tumors that harbor somatic kinase domain mutations in EGFR. Here, we compare wild-type EGFR autophosphorylation kinetics to the L834R (also called L858R) EGFR form, one of the most common mutations in lung cancer patients. Using rapid chemical quench, time resolved electrospray mass spectrometry (ESI-MS) and western blot analyses, we examined the order of autophosphorylation in wild-type (WT) and L834R EGFR and the effect of gefitinib (Iressa ™) on the phosphorylation of individual tyrosines. These studies establish that there is a temporal order of autophosphorylation of key tyrosines involved in downstream signaling for WT EGFR and a loss of order for the oncogenic L834R mutant. These studies also reveal unique signature patterns of drug sensitivity for inhibition of tyrosine autophosphorylation by gefitinib; distinct for WT and oncogenic L834R mutant forms of EGFR. Fluorescence studies show that for WT EGFR, the binding affinity for gefitinib is weaker for the phosphorylated protein while for the oncogenic mutant, L834R EGFR, the binding affinity of gefitinib is substantially enhanced and likely contributes to the efficacy observed clinically. This mechanistic information is important in understanding the molecular details underpinning clinical observations as well as to aid in the design of more potent and selective EGFR inhibitors.
The epidermal growth factor receptor (EGFR) is a receptor tyrosine kinase (RTK) that plays essential roles in both normal and oncogenic signaling pathways. Wild-type EGFR is normally activated by binding of the epidermal growth factor (EGF) to the extracellular domain of EGFR. This stimulates dimerization of EGFR, bringing the cytoplasmic domains in close contact such that autophosphorylation of specific tyrosine residues on each monomer can occur. These phosphorylated tyrosines serve as specific recruitment sites for downstream signaling molecules that are involved in various cellular events such as proliferation, differentiation and apoptosis (1, 2). Activation mechanism of EGFR has received much attention in recent years. Mutational studies together with structural studies demonstrated that juxtamembrane domain, which is located in between transmembrane domain and kinase domain, acts as a latch in helping formation of active asymmetric dimer (3) (4),(5-8). Asymmetric dimer formation is a key mechanism of activation where one monomer (the donor) has an activated kinase domain and the other (the receiver) is being phosphorylated.(9) (10) The timing and regulation of these phosphorylation events and recruitment of specific signaling molecules is important for the engagement of downstream pathways. Accordingly, overexpression and/or activating mutations in EGFR have been associated with various human cancers. Moreover, the majority of activating EGFR mutations identified thus far occur in the kinase domain (11).
Gefitinib (Iressa™) is a small inhibitor that targets EGFR and was approved by the FDA in 2004 for the treatment of chemotherapy-refractory advanced NSCLC. It binds in the ATP binding cleft, inhibiting EGFR’s kinase activity. During clinical trials it was discovered that a subset of patients who harbored mutations impacting the EGFR kinase domain showed a better response profile than patients with wild-type EGFR. The majority of these mutations included either a point mutation in exon 21 (L834R being the most common one) 1 or a deletion mutation in exon 19 (12, 13). Both of these mutations in EGFR result in constitutive activation of the receptor resulting in activation of downstream signaling pathways in the absence of ligand binding.
Previous studies with FGFR 1 (fibroblast growth factor receptor 1) in this laboratory have shown that there is a sequential order in which specific FGFR1 tyrosines are autophosphorylated and that this order is perturbed in an oncogenic form of FGFR 1 (14, 15). This study suggested the order of autophosphorylation is important in providing both temporal and spatial resolution to receptor signaling and this could be a general feature of all RTK signaling. This finding prompted us to study EGFR, a receptor tyrosine kinase in a RTK family distinct from FGFR1. Even though recent developments in the field of proteomics have allowed for a global assessment of EGFR phosphorylation and the determination of EGFR binding partners with temporal and spatial resolution (16, 17), (18, 19), detailed biochemical studies on the autophosphorylation kinetics of individual tyrosines of EGFR are lacking. This is due, in part, to the fact that most structural and kinetic studies have focused on characterizing constructs of EGFR that lack the complete cytoplasmic domain containing the C-terminal tyrosines known to be sites of receptor autophosphorylation.
A more comprehensive view of the dynamics of phosphorylation with RTKs suggests that phosphorylation and dephosphorylation can result in the transient engagement of subsets of phosphorylation sites that may regulate the subsequent recruitment of various downstream adaptors and effectors to modulate distinct signaling outputs. It is possible that alterations in this dynamic regulation of RTK signaling may be an important determinant of oncogenic processes. Therefore, the ability to precisely interrogate the kinetics of RTK autophosphorylation may reveal “activation signatures” that provide further insights into the differences between normal and oncogenic forms of EGFR, and an expanded functional understanding of the emerging therapeutic class of targeted kinase inhibitors. Profiling the temporal dynamics of autophosphorylation may provide a unique molecular fingerprint for distinguishing normal and cancer cells and the responsiveness of targeted inhibitors. An initial study of EGFR autophosphorylation suggested that it may proceed with a “preferred” temporal order and that there may be distinct differences between normal and oncogenic form of EGFR (20). In the current study, we take a closer look by examining the kinetics and order of autophosphorylation in constructs containing the complete cytoplasmic domain (residues 672-1186) for wild type (WT) EGFR and comparing the kinetic profile to the oncogenic L834R EGFR (MT). We also examined the effect of gefitinib on autophosphorylation at each tyrosine residue to obtain a detailed mechanistic understanding of how gefitinib may modulate downstream signaling. Lastly, we determined the binding constants for gefitinib interaction with unphosphorylated and phosphorylated forms of wild-type and L834R EGFR using fluorescence spectroscopy. Our results showed that there are clear differences in autophosphorylation kinetics between wild type and L834R EGFR. In addition, gefitinib inhibits autophosphorylation of each tyrosine residue to differing degrees such that there are unique sensitivity patterns for WT versus L834R. Moreover, an assessment of the impact of tyrosine autophosphorylation state on inhibitor binding reveals the tighter binding of gefitinib to a phosphorylated form of L834R compared to WT EGFR that contributes to the better efficacy of this drug toward tumors expressing L834R EGFR that has been observed clinically. This information is important both to understand the molecular details underpinning clinical observations as well as to inform the design of more effective EGFR inhibitors.
EXPERIMENTAL PROCEDURES
Chemical reagents. Adenosine 3′-triphosphate (ATP), manganese chloride (MnCl2), ammonium bicarbonate (NH4HCO3), ethylendiaminetetraacetate (EDTA), and AMP-PNP were purchased from Sigma (St. Louis, MO). Digestion enzymes, trypsin gold and glutamic-C, mass spectrometry grade were obtained from Promega (Madison, WI) and Princeton Separation (Adelphia, NJ), respectively. HPLC grade water and acetonitrile were from J.T. Baker (Phillipsburg, NJ). Trifluoroacetic acid (TFA) is from Pierce (Rockford, IL). Antibodies against phosphorylated tyrosine residues 1045, 1068, 1148, 1173, and total EGFR antibody are from Cell Signaling (Danvers, MA). Antibodies against phosphorylated tyrosine residues 845 and 992 are from Millipore (Billerica, MA). Secondary antibody Alexa Fluor® 680 goat anti-mouse IgG (H+L) was from Invitrogen (Carlsbad, CA).
Expression and Purification of EGFR
Viral stocks containing cytoplasmic constructs of wild type and L834R EGFR (from residues 672-1186) were prepared as previously described (20). Sf9 cells were transfected with viral stocks of wild type or L834R EGFR containing a FLAG affinity tag. Cells were harvested at 2000x g for 10 min after 72 h transfection. Pellets were rinsed in PBS (phosphate buffered saline) and resuspended in a buffer containing 50mM Tris-HCl (pH 7.4), 150 mM NaCl, 10% glycerol, 1% Tween-20, and Complete, an EDTA-free protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN). Cell lysates were sonicated for 1 min at 10 sec intervals, and centrifuged for 40 min at 16000x g at 4 °C. The cleared supernatant was incubated with M2 agarose beads (Sigma, St. Louis, MO) and lambda phosphatase (New England Biolabs, UK) overnight at 4°C. M2 beads were pelleted and washed with a buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 10% glycerol and 0.05% Tween-20. EGFR proteins were eluted from the beads using 1x FLAG peptide (100 Og/mL) (Sigma, St. Louis, MO). Eluted protein was concentrated to 5 mg/mL and loaded onto Superdex 200 10/300 (GE Healthcare, Piscataway, NJ) using ACTA Purifier FPLC system (GE Healthcare, Piscataway, NJ). Column was equilibrated with a buffer containing 50mM TrisCl (pH 7.4), 150 mM NaCl2, 0.01 % Tween-20. Flow-rate was 0.4 mL/min. Absorbance at 280 nm was monitored and elution peaks were analyzed by SDS-PAGE. Eluted EGFR fractions were pooled and concentrated to 10 mg/mL using Amicon ultra centrifugal filter (10K MWCO, Millipore, Billerica, MA), aliquoted and stored in a −80° C freezer. (see Supporting Information for details)
Radiolabeled autophosphorylation assay
Increasing concentrations of EGFR (2-65 μM for wild-type and 1-20 μM for L834R EGFR) were autophosphorylated in the presence of γ32P-ATP and MnCl2 in a buffer containing 20 mM TrisCl (pH 7.4) and 150 mM NaCl. The reaction was quenched with 100 mM EDTA at different reaction times (5-150 sec). Final concentrations of ATP and MnCl2 in the reaction were 500 μM and 10 mM, respectively. Quenched samples were separated by SDS-PAGE and autophosphorylated EGFR was visualized using Molecular Imager FX (Bio-Rad, Hercules, CA) and quantified using Imagequant software (Bio-Rad, Hercules, CA). Data were fitted to following equation to determine the rate of autophosphorylation. The autophosphorylation rate obtained from different concentrations of EGFR was plotted against EGFR concentration to determine the saturating concentration of EGFR where catalytic activity is maximal and receptor dimerization is not rate-limiting. Experiments were carried out in triplicate.
| (equation 1) |
ESI-TOF MS to identify the level of EGFR autophosphorylation
The ESI-TOF MS (electrospray ionization-time of flight mass spectrometry) analysis was performed on intact protein EGFR samples to assess phosphorylation state using an Ettan ESI-TOF mass spectrometer (Amersham Biosciences, Uppsala, Sweden). Prior to MS analysis, EGFR samples were desalted by HPLC using C4 column (Vydac, Deerfield, IL) (Solvent A: 5% Acetonitrile, 0.1% Formic Acid, 0.02 % TFA in water; Solvent B: 90% Acetonitrile, 0.1% Formic Acid, 0.02 % TFA in water). Twenty μg of EGFR was loaded onto the C4 column and EGFR protein was eluted at 63% B. Eluted peak was diluted 3x with HPLC water and concentrated back to 25 μL for spraying.
In vitro autophosphorylation assay by western blotting
Autophosphorylation of EGFR was initiated by adding 500 μM ATP and 10 mM MnCl2 to 25 μM EGFR. The autophosphorylation reaction was quenched with 100 mM EDTA at different times using rapid chemical quench (Kintek Instruments, Austin, TX). Quenched samples were separated by SDS-PAGE and blotted with phosphospecific antibodies that specifically recognize 6 tyrosine residues (845, 992, 1045, 1068, 1148, and 1173) in EGFR. Standard curves for each antibody were generated to determine the dilution factor necessary to obtain a signal that is linear in response to the amount of protein loaded. Adjusted antibody dilution was as follows: 1:2000 for 845, 1:5000 for 992, 1:200 for 1045, 1:1000 for 1068, 1:250 for 1148, 1:5000 for 1173, and 1:2000 for EGFR. Phosphorylated EGFR bands were visualized upon addition of secondary antibody Alexa Fluor® 680 goat antimouse IgG (H+L) using Odyssey Infrared Imaging System (LI-COR Biosciences, Lincoln, NE) and quantified using Odyssey v3.0 software (LI-COR Biosciences, Lincoln, NE). The intensity of bands was plotted against increasing times and the data were fitted to equation 1 to determine the rate of autophosphorylation. Experimental analyses were carried out in triplicate.
Inhibition of autophosphorylation by gefitinib
Wild-type and L834R EGFR (25 μM) were incubated with different concentrations of gefitinib for 5 minutes at room temperature. The autophosphorylation reaction times were examined over a range of 2 to 10 min. Phosphospecific analysis of autophosphorylation in the presence of gefitinib was examined as described earlier. The intensity of phosphorylated EGFR bands was quantified using Molecular Imager FX and plotted against gefitinib concentration. Data were fitted to the following equation to determine the concentration of gefitinib that results in 50 % EGFR activity. Experimental analyses were carried out in triplicate.
| (equation 2) |
Determining the Kd of gefitinib using fluorimetry
Phosphorylated wild-type and L834R EGFR were prepared by incubating the constructs with ATP and MnCl2 for 2 minutes. Excess ATP was removed using Amicon centrifugal filter devices 10K concentrator (Millipore, Billerica, MA). Both unphosphorylated and phosphorylated EGFR constructs at 50 nM concentration were titrated with gefitinib at concentrations ranging from 0 – 7 μM. Quenching of the intrinsic tryptophan fluorescence signal of EGFR was measured using a fluorimeter (Photon Technology International, Lawrenceville, NJ). Both excitation and emission slits were set to 4 nm, and excitation and emission wavelengths were 285 nm and 325 nm, respectively. Fluorescence emission was measured for 20 seconds at 1 point/second (Felix software, version 1.41, Photon Technology International, Lawrenceville, NJ). Twenty points of emission intensity were averaged and plotted against total concentration of gefitinib. Percent EGFR bound was calculated according to the fluorescence signal of EGFR alone being 0% bound. The % bound fraction was plotted against the total concentration of gefitinib in the reaction and fit to the following equation.
| (equation 3) |
RESULTS
The architecture of RTKs consists of an extracellular ligand binding domain, a single transmembrane region, and an intracellular cytoplasmic domain that includes the core kinase catalytic activity. In vitro studies of individual domains of various recombinant forms of RTKs including the EGFR have played a key role in defining our current understanding of the structure, function, and biological function for these proteins. Moreover, these studies have been essential for establishing the nature of downstream signaling pathways that mediate their cellular functions. The EGFR constructs used in the following studies contained residues 672-1186 encompassing the complete cytoplasmic domain, including the C-terminal end of the juxtamembrane segment, the kinase domain, and the C-terminal tail. They were prepared using a baculovirus Sf9 protein expression system (20). The importance of the juxtamembrane segment of EGFR in asymmetric dimer formation and activation of the kinase domain has been reported (4, 7, 8). A mechanistic understanding of the autophosphorylation kinetics of the intracellular cytoplasmic domain and the role of these tyrosines in the absence of other cellular factors (cell-free) is a key step in establishing the molecular mechanism underlying EGFR protein function, and serves as a foundation for designing and interpreting more complex studies conducted at a cellular level.
Concentration-dependence of wild type (WT) and L834R EGFR activity
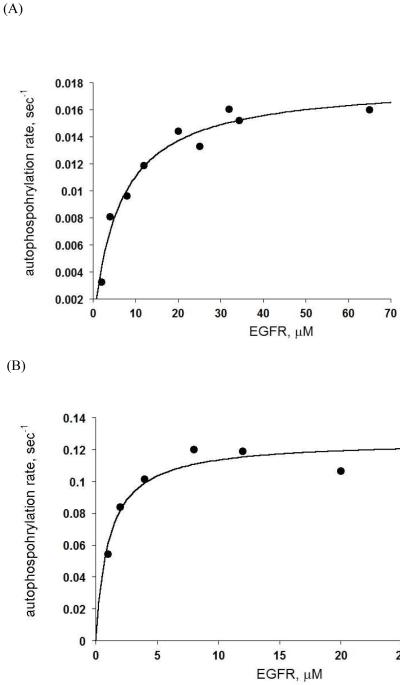
To begin to compare the enzyme kinetic properties of these proteins, it was initially important to determine how kinase activity, in the context of an autophosphorylation assay, may vary as a function of protein concentration. This information would provide an indication of the relative propensity for dimerization. In order to address this question, the WT and L834R EGFR proteins were dephosphorylated with lambda phosphatase and Yersinia tyrosine phosphatase and evaluated using ESI-TOF MS to confirm that the proteins were in a homogeneous and unphosphorylated state. Recent cellular studies have shown that EGFRs can spontaneously form dimers that are primed for ligand binding and activation (21). For our in vitro studies, to ensure proper dimerization and activation of the constructs, we examined the kinetics of tyrosine autophosphorylation as a function of wild-type or L834R EGFR concentration (see Supporting Information, Fig. S2). The overall autophosphorylation rate was first measured using a radioactivity assay via incorporation of γ-32P ATP over time as a function of protein concentration (see Experimental Procedures). An examination of autophosphorylation rates reveals that L834R EGFR autophosphorylation proceeds > 6 times faster than for the wild type enzyme, and the Kd for saturation of dimerization/maximal catalytic efficiency is ~6-fold tighter (Figure 1). Fitting the data to a hyperbolic equation yielded Kd and kcat values of 6.3 ± 1 μM and 0.018 ± 0.001 sec−1, respectively, for wild type and 1 ± 0.3 μM and 0.125 ± 0.007 sec−1 for L834R EGFR.
Figure 1.
Concentration dependence of (A) wild type and (B) L834R EGFR. The rate of autophosphorylation was plotted versus increasing concentration of EGFR for each construct.
As illustrated in Figure 1, the rate of autophosphorylation increases upon increasing wild-type EGFR concentration, reaching a plateau at a protein concentration ~25 μM. The rate did not change even at concentrations up to 65 μM. For the L834R EGFR mutant, the rate of autophosphorylation was saturated at ~5 μM enzyme. To make sure that dimerization does not limit the rate of autophosphorylation for either construct, an enzyme concentration of 25 μM was used in all of the experiments. We also prepared his-tagged EGFR which was used to attach to DOGS-NTA-Ni lipids at 5mole percent. At 5 mole percent of DOGS-NTA-Ni, the local protein concentration is ~3.7mM according to Zhang X. et al (3) (see materials and methods in Supporting info for details). The rate of autophosphorylation measured with the EGFR construct on lipid vesicles was 0.02 s−1 (Supplemental Figure S3) that is analogous to the rate we obtained with the original WT EGFR (containing a FLAG tag) in solution at concentrations 25-65 OM (Fig 1).
Comparison of in vitro intrinsic tyrosine autophosphorylation for WT and L834R EGFR using ESI-TOF MS
Our previous studies with FGFR1 have established electrospray time-of-flight (ESI-TOF) mass spectrometry (MS) as a valuable tool for monitoring protein phosphorylation (14, 15). With this technique, the formation of individual phosphorylation states in WT and L834R EGFR was followed as a function of time by ESI-TOF MS (Figure 2). For both constructs, spectra representing increasing reaction times showed the sequential addition of phosphate groups (~80 Da). The spectra further support the kinetic differences between WT and MT as autophosphorylation of L834R EGFR is much faster than that seen with wild type EGFR. For example, at 1 second, wild type EGFR shows mainly 0P and small amount of 1P and 2P species formed. In comparison, there are up to 4P species formed in the same time period for L834R EGFR.
Figure 2.
Analysis of wild type (A) and L834R (B) EGFR autophosphorylation by ESI-TOF MS.
Temporal control of autophosphorylation
Our previous studies on FGFR revealed a specific pattern of autophosphorylation that was altered in oncogenic forms of FGFR (14, 15). For EGFR, there are several tyrosine residues within the kinase domain and the C-terminal tail that have been suggested to play important roles in recruitment of downstream signaling proteins (22). Among those tyrosines, we examined the autophosphorylation pattern of Y845, Y992, Y1045, Y1068, Y1148, and Y1173 for WT and L834R MT constructs of EGFR using phosphospecific antibodies and mass spectrometry to determine whether there was a preferred temporal order. The results shown in Figure 3 reveal there is indeed a molecular order for autophosphorylation for WT EGFR. As illustrated in Fig. 3A, analysis of the reaction time course by gel electrophoresis and western blotting with phosphospecific antibodies for each tyrosine reveals qualitatively that these tyrosines are phosphorylated in different time domains. As exemplified in 3C, quantitation of the data to determine the individual rate constants shows there is a clear temporal pattern of tyrosine autophosphorylation for WT EGFR: the preferred molecular order is 1173 > 1148,1068 >992 > 845 >1045. The ordered pattern of autophosphorylation was also confirmed by ESI/LC/MS/MS and phosphopeptide mapping. Representative data for phosphopeptide mapping is shown in Supporting Information (see Figure S4). For example, autophosphorylation reaction times of 5 and 30 seconds were digested with trypsin and/or glutamic-C and phosphorylation of tyrosine residues were analyzed by LC-MS/MS (see Materials and Methods in Supporting Information). The samples with the 5-second reaction time showed phosphorylation at tyrosines 1148, 1173, and 1068. In contrast, in the 30-second reaction time, all six tyrosine residues were phosphorylated.
Figure 3.
A-B. In vitro autophosphorylation of wild type (A) and L834R (B) EGFR mapped with phospho-specific antibodies. 25 μM EGFR was phosphorylated in the presence of 0.5 mM ATP and 10 mM MnCl2 and the reaction was quenched by 100 mM EDTA using chemical quench at indicated times. Phosphorylated EGFR was analyzed by SDS-PAGE and probed with phospho-specific antibodies. C-D. In vitro autophosphorylation of wild type (C) and L834R (D) EGFR mapped with phospho-specific antibodies. The intensity of phosphorylated EGFR bands were quantified and plotted against reaction time. Data were fit to equation 1 as in Experimental Procedures. E. Bar graph representation of wild type and L834R EGFR autophosphorylation rates at specific tyrosine residues.
The rates of tyrosine autophosphorylation fall into three groups: a fast rate, an intermediate rate, and a slow rate of phosphorylation. The individual rates of autophosphorylation for each tyrosine residue are summarized in Table 1. These data show that Y1173 falls into the fast rate category (~0.055 s−1) and is the first tyrosine to be phosphorylated. In the context of downstream signal propagation, this residue is associated with the recruitment of PLCγ and the activation of the DAG/ IP3 kinase pathway. There are two additional residues that also fall into this category that are phosphorylated at similar rates, Y1148 and Y1068 (~0.04 s−1). Two tyrosine residues fall into the intermediate rate category: Y845 and Y992 (~0.007-0.016 s−1). The final tyrosine, Y1045, is phosphorylated at a significantly slower rate (0.002 s−1) -- almost 30- fold slower than that of Y1173, the first tyrosine to be phosphorylated.
Table 1.
Autophosphorylation rate of wild type and L834R EGFR at tyrosine residues.
| tyrosine residue | wild type (sec-1) | L834R (sec-1) |
|---|---|---|
| 845 | 0.007 ± 0.002 | 0.250 ± 0.069 |
| 992 | 0.016 ± 0.002 | 0.240 ± 0.120 |
| 1045 | 0.002 ± 0.001 | 0.131 ± 0.065 |
| 1068 | 0.043 ± 0.003 | 0.300 ± 0.081 |
| 1148 | 0.045 ± 0.010 | 0.330 ± 0.078 |
| 1173 | 0.055 ± 0.007 | 0.220 ± 0.077 |
The temporal order observed for WT EGFR is in contrast to results for L834R EGFR. The gel analysis and western blotting with phosphospecific antibodies for L834R EGFR, shown in Figure 3B, indicate that the overall rates for tyrosine autophosphorylation are much faster than WT EGFR, consistent with results from the radiolabeled assay described in Figure 1. A quantitation of the data for the L834R EGFR as shown in Figure 3D reveals there is loss of temporal control and little or no distinct order. A summary of the individual rates of tyrosine autophosphorylation for L834R is shown in Table 1. For this mutant EGFR, the tyrosine with the fastest rate of autophosphorylation, Y1148 (0.33 s−1), is only 2.5-fold that of the slowest tyrosine, Y1045 (0.13 s−1). A graphical comparison of the data for WT and L834R autophosphorylation for individual tyrosines is shown in Figure 3E. The findings with the L834R mutant, illustrated in dark bars, reveal a complete lack of temporal control such that the five tyrosines: Y845, Y992, Y1068, Y1148, and Y1173, are autophosphorylated with similar rates, and the sixth, Y1045, is only slightly slower. On the other hand, the distinct, three set pattern of tyrosine autophosphorylation is clear with the WT EGFR shown in open bars. It is also interesting to note that the L834R mutation has affected the enhancement of the rate of autophosphorylation at each tyrosine in a disproportionate manner. The three rates of tyrosine autophosphorylation for the fastest residues for WT EGFR: Y1173, Y1148, and Y1068, have rates in the L834R mutant EGFR that are enhanced 4 to 8-fold and Y992 is 15-fold. This is in contrast to Y845 and Y1045 where those rates are enhanced >35 and 65-fold, respectively.
Effect of gefitinib on autophosphorylation of wild type and L834R EGFR
It has been shown that patients with mutations in the kinase domain of EGFR have a more favorable response to treatment with gefitinib (Iressa ™) than do patients with wild type EGFR, although the underlying mechanism(s) for this differential response are not completely understood (13, 23). It is possible that gefitinib may elicit unique signature patterns of drug sensitivity for individual tyrosines and that the patterns may be distinct for WT and L834R forms of EGFR.
To determine whether there are disparate effects of gefitinib inhibition on the autophosphorylation of the individual tyrosine residues, the relative drug inhibition was examined for the complete cytoplasmic domain constructs of WT and L834R forms of EGFR described above. A protein concentration of 25 μM EGFR as described above was used to ensure dimerization/catalytic activity of the enzyme was saturated. Both wild type and L834R EGFR were incubated with increasing concentrations of gefitinib and autophosphorylation at each of the six tyrosine residues (Y845, Y992, Y1045, Y1068, Y1148, and Y1173) was monitored using phosphospecific antibodies as described in the experimental procedures. It is important to note that since the concentration of protein used in these experiments is in the micromolar range, a relative or apparent inhibition constant Ki,app is obtained. Therefore, gefitinib concentrations for relative IC50 inhibition of L834R EGFR autophosphorylation in Table 2 indicate upper limit and fold differences indicate lower limit. The goal of this experiment was to determine if there was differential inhibition of autophosphorylation for individual tyrosine residues and to see if the patterns of drug sensitivity were unique for WT versus MT (L834R) forms of EGFR.
Table 2.
Gefitinib concentration for relative IC50 inhibition of WT and L834R EGFR autophosphorylation at individual tyrosine residues.
| tyrosine residue | wild type (μM) | L834R (μM) a | fold difference a |
|---|---|---|---|
| 845 | 13.12 ± 3.51 | 0.54 ± 0.24 | 24.3 |
| 992 | 10.21 ± 2.52 | 0.26 ± 0.10 | 39.3 |
| 1045 | 5.46 ± 0.40 | 0.1 | 50 |
| 1068 | 28.87 ± 6.29 | 3.29 ± 1.08 | 8.8 |
| 1148 | 33.34 ± 4.72 | 4.94 ± 1.58 | 6.7 |
| 1173 | 32.11 ± 6.67 | 3.64 ± 1.38 | 8.8 |
Gefitinib concentrations for relative IC50 inhibition of L834R EGFR autophosphorylation indicate upper limit.
Fold differences indicate lower limit.
The gel analysis for determination of relative inhibition of autophosphorylation for WT and L834R EGFR is shown in Supporting Information (Figures S5A & B). A quantitation of the data (Figures 4A & B) with increasing concentrations of gefitinib revealed that autophosphorylation of the tyrosines for both EGFR constructs does indeed occur to differing degrees. The relative Ki,app of gefitinib for each phosphorylation site of both constructs was determined as described in the experimental procedures. The bar graph shown in Figure 4C compares the Ki,app values for the autophosphorylation of wild type (open bars) and L834R (dark bars) EGFR at different tyrosine residues. The numbers in parentheses over each set of bars represent the relative difference in inhibition for WT versus L834R mutant forms of EGFR indicating lower limit. A complete summary of the Ki,app values is shown in Table 2.
Figure 4.
A-B. Gefitinib effect on autophosphorylation of wild type (C) and L834R (D) EGFR. Data were fit to equation 2 as in Experimental Procedures to determine the concentration of C. Bar graph representation of the effect of gefitinib on wild type and L834R EGFR autophosphorylation. The number over the bars indicate fold difference in relative IC50 values between wild type and L834R EGFR.
The data show several striking features regarding the range and pattern of relative gefitinib sensitivities (Figure 4 and Table 2). First, the data reveal that for both WT and L834R MT EGFR, the relative range of drug sensitivity for gefitinib varies substantially for each individual tyrosine residue. The range of relative drug sensitivities for gefitinib inhibition of autophosphorylation at these individual tyrosines for WT EGFR is approximately 6-fold. In general, gefitinib is more potent for the L834R MT EGFR and the range of drug sensitivities is much more pronounced. In this case, for the oncogenic MT form of EGFR, the gefitinib potency for individual tyrosines varies >50-fold.
Second, the pattern of drug sensitivity is distinctive for WT and L834R forms of EGFR. For the WT EGFR, the pattern of gefitinib drug sensitivity reveals that there are two distinct sets of tyrosines. The pattern of drug sensitivity for WT EGFR shows that the C-terminal residues, Y1173, Y1148, and Y1068 are the least vulnerable to gefitinib inhibition. For the second set of tyrosines, comprised by Y845, Y992, and Y1045, gefitinib exhibits a 3-6 fold higher potency.
In contrast, the pattern of gefitinib drug sensitivity for the L834R EGFR shows three distinct sets of tyrosines. The first set, comprised by Y1173, Y1148, and Y1068, are the least sensitive toward gefitinib inhibition, however, these potencies are ~10-fold greater than those observed with WT EGFR. As illustrated in Figure 4B, the gefitinib inhibition of Y1148 plateaus at approximately 45%. These data suggest that autophosphorylation of this tyrosine residue cannot be completely inhibited. This type of kinetic inhibition behavior implies that gefitinib may be a noncompetitive inhibitor of ATP in the autophosphorylation of Y1148. There are earlier studies in the literature that have suggested that under some conditions, gefitinib may function as a noncompetitive inhibitor of ATP. (24)
The second set of tyrosines, comprised by Y845 and Y992 reveal an intermediate potency with a 20-40-fold higher level of drug sensitivity for L834R EGFR relative to WT EGFR. The most potent inhibition of tyrosine autophosphorylation for this MT EGFR was observed for Y1045. In this case, the gefitinib drug sensitivity was in the nanomolar range and was > 50-fold more efficiently inhibited relative to the Y1045 for WT EGFR.
Gefitinib binding to unphosphorylated and phosphorylated forms of wild type and L834R EGFR
Structural and biochemical studies with other RTKs and TKs, most notably BCR-ABL (bcr gene product fused to Abelson TK) have shown that the phosphorylation state of the protein affects the affinity of TKIs such as imatinib (Gleevec™) (25). These studies showed that imatinib binds to a form of the TK in which the activation loop has adopted an inactive confirmation and it has a higher potency on the unphosphorylated form of the kinase.
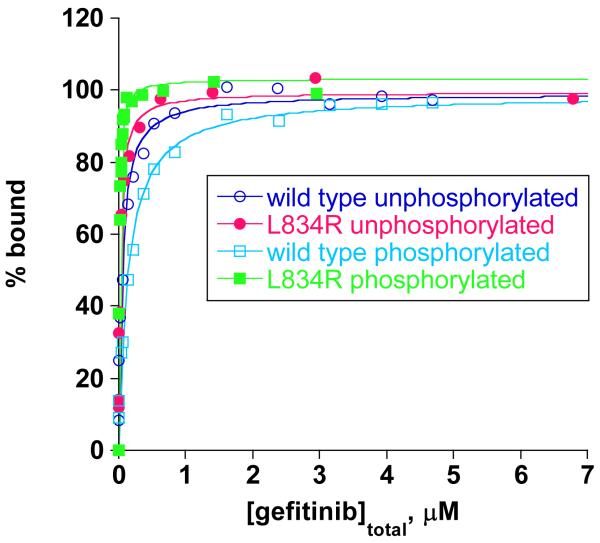
Several studies have examined the structures of the kinase domain of WT and L834R EGFR and the effects of gefitinib using unphosphorylated forms of EGFR containing only the Y845 and Y992 tyrosines (26, 27). Additional structural and biochemical studies have examined the effects of a closely related TKI, erlotinib (Tarceva™) on EGFR (28, 29). While the majority of these studies suggest that gefitinib binds more tightly to L834R, one study indicates that the drug binds WT and MT EGFR with similar affinities (30). Important caveats in reconciling these data are that most experiments were performed with the kinase domain and phosphorylated forms were not examined. Furthermore, the C-terminal domain, containing the tyrosines important for propagation of downstream signaling, was absent. To examine the effect of phosphorylation state as well the potential role of the C-terminal tyrosine residues in modulating gefitinib sensitivity, we determined the dissociation constants for gefitinib binding to unphosphorylated and fully phosphorylated forms of WT and L834R MT EGFR containing the complete intracellular cytoplasmic domain using fluorescence spectroscopy. The dissociation constants of gefitinib for both unphosphorylated and phosphorylated forms of WT and L834R EGFR are shown in Figure 5 and summarized in Table 3. The data in Figure 5 and Table 3 represent a plot of % bound versus total gefitinib concentration and the dissociation constant values from the fit, respectively. The dissociation constant for gefitinib binding to the unphosphorylated wild type EGFR (50 ± 6 nM) was 2.7-fold lower than for the phosphorylated wild-type protein (140± 16 nM), indicating that an increase in phosphorylation state of the protein weakens the drug interaction. The results of gefinitib binding to the L834R EGFR are in direct contrast to that observed for WT. In this case, the Kd for gefitinib binding to the unphosphorylated forms of L834R EGFR (24± 3 nM) was 2.4-fold weaker than for the phosphorylated form of the mutant (10± 1 nM), showing that the increase in phosphorylation state of the protein results in a higher affinity interaction. A comparison of gefitinib interaction with the unphosphorylated forms of WT and L834R EGFR shows that there is only a 2-fold difference in the Kd values (50 nM versus 24 nM). However, in the case of the fully phosphorylated forms of the EGFR proteins, the difference is much more dramatic, such that the Kd for gefitinib interaction for wild type and L834R EGFR is a 14-fold (140 nM versus 10 nM for WT and MT, respectively).
Figure 5.
Gefitinib binding to unphosphorylated and phosphorylated forms of wild type and L834R EGFR. 50 nM EGFR was titrated with increasing concentration of gefitinib and the intrinsic fluorescence signal of EGFR was quenched. The percent EGFR bound was calculated according to procedures outlined in Experimental Procedures.
table 3.
Kd values for gefitinib binding to wild type and L834R EGFR.
| unphosphorylated (nm) | phosphorylated (nm) | ||
|---|---|---|---|
| wild type | L834R | wild type | L834R |
| 51 ± 6 | 24 ± 3 | 140 ± 16 | 10 ± 1 |
DISCUSSION
Initiation of protein signaling events involves the autophosphorylation of key tyrosines on RTKs such as EGFR. These phosphorylation sites serve as docking sites for the binding of signaling partner proteins and downstream signal propagation. Dynamic control of multiple phosphorylation modifications of a single RTK like EGFR can manifest critical control on multiple downstream signal transduction pathways. Alterations of this sensitive dynamic in growth factor networks and aberrant activities in EGFR have severe biological consequences and are linked to oncogenic processes. One such example is the L834R mutant form of EGFR that is found in about 43% of NSCLC patients with EGFR mutation. About 10 % of NSCLC patients have mutations in EGFR (31). The current study was designed to investigate the molecular mechanisms of the early dynamic autophosphorylation events that may hold the key to understanding and predicting the nature of oncogenic behavior and the effect of precisely guided cancer therapy such as the TKI, gefitinib.
This work focused on several key questions regarding how early tyrosine autophosphorylation events in protein signaling may be governed during normal and oncogenic signal transduction for the EGFR tyrosine kinase pathway. Among the questions we explored were: (1) Are there differences in the overall tyrosine autophosphorylation kinetics and dimerization/catalytic efficiency for WT and L834R MT forms of EGFR? (2) Is temporal control a common feature of various RTKs such as EGFR and is temporal control altered with MT forms of EGFR such as the L834R? (3) Is there preferential inhibition of specific tyrosine residues by TKIs such as gefitinib? (4) Are there differences in drug sensitivity for gefitinib inhibition of WT versus L834R and is that drug sensitivity influenced by phosphorylation state of the protein?
Autophosphorylation kinetics and catalytic efficiency
Earlier studies using the complete cytoplasmic domains (but lacking the extracellular ligand-binding domain) for a number of RTKs have shown that autophosphorylation occurs at higher protein concentrations and is therefore a good model system for examining kinetic behavior. A combination of rapid chemical quench with radioactivity detection coupled with time-resolved ESI-MS/TOF allowed us to monitor the concentration dependence for formation of phosphorylated proteins. This assessment reveals that the rate (kcat) for the L834R mutant is > 6-fold faster, and the Kd for saturation of dimerization/catalytic activation is ~6-fold tighter than WT EGFR (Fig. 1). Therefore, the catalytic efficiency (kcat/ Kd) for the oncogenic (L834R) form of the enzyme is enhanced relative to WT EGFR. Our study suggests that the Leu to Arg mutation not only promotes dimerization but also increases the intrinsic catalytic efficiency for autophosphorylation.
Two other recent studies have examined the catalytic efficiency of WT and L834R EGFR proteins to phosphorylate a peptide substrate. In one study, a comparison of the core kinase domains of WT and L834R revealed that the rate of peptide substrate phosphorylation for L834R mutant EGFR was 2.5-fold faster than WT (32). Another study examined WT and L834R EGFR protein constructs that contained extracellular, transmembrane, and kinase domains that lacked the C-terminal tail containing four of the tyrosines that are autophosphorylated (Y1045, Y1068, Y1148, and Y1173). In this study, the WT EGFR had similar catalytic activity for peptide substrate phosphorylation as L834R EGFR when purified in the presence of EGF (33). These differences in kinetic behavior could be due to differences in the protein constructs or experimental conditions employed such as the concentration of ATP.
It is instructive to consider how the kinase kinetic behavior might correlate with structure and function. Under normal signaling conditions, EGFR exists in an equilibrium between inactive monomer and inactive dimer (22, 34). Recent cellular studies with EGFR have shown that spontaneous dimers may form and the binding of ligand shifts the equilibrium toward an active dimer for complete activation (21).
These kinetic data for the L834R EGFR is consistent with a mechanism in which the mutant is constitutively active such that dimerization and activation may readily occur in the absence of ligand binding. Structural studies with the kinase domain of L834R EGFR revealed that this single residue mutation is not compatible with the inactive conformation of EGFR in which the activation loop is closed over the active site. Rather, the leucine to arginine mutation locks the activation loop in an open conformation, which renders L834R EGFR constitutively active (26). Recent molecular dynamic simulation studies have examined the core kinase domain structures for WT and L834R EGFR. These studies suggest that the N lobe dimerization interface of WT EGFR is intrinsically disordered and only become ordered upon dimerization. These simulations also suggest that oncogenic mutants such as L834R may facilitate EGFR dimerization by suppressing the local disorder (32).
A defined temporal order of autophosphorylation for WT EGFR and loss of temporal control for an oncogenic mutant
Our previous studies of FGFR1 revealed that tyrosine autophosphorylation was precisely ordered and that order was lost for an oncogenic form of FGFR1 associated with glioblastoma (14, 15). In the present study, we examined another distinct receptor tyrosine kinase family, the EGFR RTK, to see if temporal order might be a general feature of RTKs, offering an exquisite means of amplifying or attenuating a downstream signal. We also extended the study to evaluate an oncogenic EGFR mutant, L834R.
As illustrated by the data in Fig. 3 and Table 1, there is a clear temporal pattern of autophosphorylation for WT EGFR showing three discrete sets of tyrosines. The tyrosines phosphorylated most rapidly are those located in the C-terminal tail domain of EGFR: Y1173, Y1148, and Y1068. The Y1173 has been suggested to be a preferred docking site for Shc and the N-terminal SH2 binding domain of PLC-γ (22). Y1148 is suggested to be a docking site for the adaptor proteins, Shc, Dok-R, and PTP1B while Y1068 is associated with the adaptor protein GRB-2 (22).
A second set of tyrosines
Y845 and Y992 have an intermediate rate of phosphorylation. Tyrosine 845 is located in the activation loop and Y992 is at the N-terminus of the C-terminal tail. The Y845 has been implicated in the src and STAT-5 signaling pathways while Y992 has been associated with C-terminal SH2 binding domain of PLC-γ (22). The last tyrosine to be phosphorylated (0.002 s−1) is Y1045 also located in the C-terminal tail. A number of studies have confirmed that Y1045 is a docking site for interaction with CBL proteins, E3 ubiquitin ligases that are negative regulators of EGFR signaling via ubiquitination and receptor degradation (35), (36). Thus Y1045 may play an important role in attenuating or terminating the signal.
Several interesting features of the WT EGFR signaling pathway emerge from this analysis. Our data showing a temporal pattern of EGFR tyrosine autophosphorylation suggest that this may be a general feature of RTKs that may act as a means of regulating downstream signaling. The temporal pattern for EGFR autophosphorylation is distinct compared with FGFR1. The phosphorylation of tyrosines in the activation loop are important for the optimal catalytic activity for FGFR1 kinase and Y653, located in that loop is the first tyrosine to be phosphorylated (14). The role of Y845 in the activation loop for EGFR is less clear and remains controversial. Earlier studies have suggested that Y845 is phosphorylated by Src kinase thereby influencing EGFR function although the phosphorylation is not required for EGFR activity (37-40). Later studies indicate that Y845 is phosphorylated in response to TGFα stimulation and in the case of oncogenic EGFR mutations, the autophosphorylation of Y845 was dependent upon EGFR activity and associated with STAT-5 mediated gene expression (41). Moreover, Y845 was shown to be autophosphorylated with purified truncated forms of an EGF/EGFR complex lacking the C-terminal tail following ATP addition (42). Our in vitro studies with the complete cytoplasmic domain clearly show that Y845 can be autophosphorylated upon ATP addition. A comparison of rates of tyrosine autophosphorylation shows that there is a 27-fold difference in the dynamic range for the fastest and slowest rates. We might posit that this kinetic profile for EGFR allows another level of regulation by providing a “conformational tunability” to attenuate or amplify and orchestrate, at the appropriate time, the recruitment of partner proteins and the propagation of the downstream signals.
The results for the kinetics of autophosphorylation for the oncogenic mutant form of EGFR (L834R) are in contrast to that observed with WT EGFR as illustrated in Fig. 3 and Table 1. This is consistent with previous results from our laboratory and others (20) (26). In this case of L834R EGFR, the reaction kinetics reveal a mutant kinase that lacks temporal control. A similar heterogeneity and loss of temporal control was observed for an oncogenic form of FGFR1 (15).
Interestingly, the autophosphorylation rates of Y845 and Y1045 are most affected by the L834R mutation and this kinetic behavior may, in part, explain results at a cellular level. Previous cellular studies have implicated an aberrant role for Y845 in L834R EGFR signaling and STAT-5 activation (41). In addition, L834R EGFR displayed ligand independent phosphorylation at Y1045, the CBL binding site, stable association with CBL and impaired ligand mediated receptor ubiquitination and downregulation (43).
The question of drug sensitivity patterns for gefitinib for WT and L834R EGFR, and the underlying molecular mechanism(s) of clinical responses for NSCLC expressing mutant EGFRs, is somewhat unclear. Some studies suggested the mutations such as L834R do not affect the binding affinity for gefitinib and other TKIs (30), while other laboratories including ours have shown that that drug sensitivity is higher for the mutant L834R EGFR (20) (29, 44) (45). In the current study, we have addressed this question in a more detailed manner for WT and L834R EGFR by determining gefitinib relative drug sensitivity patterns for inhibition of autophosphorylation at individual tyrosines and examining the effect of receptor phosphorylation upon gefitinib binding affinity.
As our data show, the relative drug sensitivity for gefitinib inhibition of autophosphorylation for each individual tyrosine varies for WT EGFR but for the oncogenic L834R EGFR, is more enhanced. Moreover, the patterns of gefitinib drug sensitivity are unique for WT and L834R forms of EGFR and each displays a distinct molecular fingerprint.
To complement the gefitinib drug sensitivity studies, equilibrium fluorescence titrations studies were carried out to directly assess binding affinity for the complete full-length cytoplasmic domain in an unphosphorylated and fully phosphorylated state for WT and L834R EGFR. Our fluorescence studies with the entire cytoplasmic domain summarized in Figure 5 and Table 3 show clear differences in the impact of phosphorylation state upon gefitinib binding affinity. The data indicate that for unphosphorylated and phosphorylated forms of WT EGFR, the binding of gefitinib to the fully phosphorylated form of the receptor is weaker whereas, for L834R, gefitinib binds more potently to the phosphorylated form of the receptor. Thus, phosphorylation weakens gefitinib potency for WT EGFR while enhancing potency for the oncogenic L834R EGFR. Taken together, the results from gefitinib inhibition of autophosphorylation of individual tyrosines and binding as a function of receptor phosphorylation suggest that there may be multiple conformational changes that occur, which in turn affect the ligand binding affinity that are not discernible by crystallographic studies using the kinase domain.
In summary, our studies have examined the intrinsic tyrosine autophosphorylation behavior for WT and an oncogenic form of EGFR (L834R) that may be involved in normal and aberrant protein signaling. A temporal order of autophosphorylation is observed in WT EGFR that might play in important regulatory role in recruiting downstream partners and signal propagation. This temporal order and regulation are lost with the L834R EGFR. The WT and L834R EGFR display different molecular fingerprints for inhibition of autophosphorylation in terms of gefitinib drug sensitivity that offers insight into how downstream pathways may be affected.These in vitro studies lay the foundation for examining whether there is a temporal order of autophosphorylation at a cellular level, whether downstream signaling partners may be recruited in an orchestrated fashion with normal and aberrant signaling, and finally how TKIs such as gefitinib may affect these processes. This mechanistic information can, in turn, improve our understanding of the nature of drug responsiveness in a clinical setting.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Anne Roberts for helpful comments on the manuscript. This research was supported by National Institute of Health grants NCI CA127580 and CA125284 to K.S.A.
Abbreviations
- EGFR
epidermal growth factor receptor
- RTK
receptor tyrosine kinase
- MS
mass spectrometry
- FGFR
fibroblast growth factor receptor
Footnotes
1Numbering based on mature EGFR. An alternate numbering for this residue in EGFR is L858R based upon the 24 amino acid nuclear localization sequence that is absent in the mature EGFR.
This research was supported by National Institute of Health grants NCI CA127580 and CA125284 to K.S.A.
SUPPORTING INFORMATION AVAILABLE
Size exclusion chromatography of wild type and L834R EGFR, autophosphorylation assay of EGFR on vesicles, and identification of tyrosine phosphorylation sites by mass spectrometry. This material is available free of charge via the Internet at http://pubs.acs.org
REFERENCES
- 1.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 2.Schlessinger J. Common and distinct elements in cellular signaling via EGF and FGF receptors. Science. 2004;306:1506–1507. doi: 10.1126/science.1105396. [DOI] [PubMed] [Google Scholar]
- 3.Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 4.Red Brewer M, Choi SH, Alvarado D, Moravcevic K, Pozzi A, Lemmon MA, Carpenter G. The juxtamembrane region of the EGF receptor functions as an activation domain. Mol Cell. 2009;34:641–651. doi: 10.1016/j.molcel.2009.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang X, Pickin KA, Bose R, Jura N, Cole PA, Kuriyan J. Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface. Nature. 2007;450:741–744. doi: 10.1038/nature05998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jura N, Zhang X, Endres NF, Seeliger MA, Schindler T, Kuriyan J. Catalytic control in the EGF receptor and its connection to general kinase regulatory mechanisms. Mol Cell. 2011;42:9–22. doi: 10.1016/j.molcel.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thiel KW, Carpenter G. Epidermal growth factor receptor juxtamembrane region regulates allosteric tyrosine kinase activation. Proc Natl Acad Sci U S A. 2007;104:19238–19243. doi: 10.1073/pnas.0703854104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jura N, Endres NF, Engel K, Deindl S, Das R, Lamers MH, Wemmer DE, Zhang X, Kuriyan J. Mechanism for activation of the EGF receptor catalytic domain by the juxtamembrane segment. Cell. 2009;137:1293–1307. doi: 10.1016/j.cell.2009.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bessman NJ, Lemmon MA. Finding the missing links in EGFR. Nature structural & molecular biology. 2012;19:1–3. doi: 10.1038/nsmb.2221. [DOI] [PubMed] [Google Scholar]
- 11.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 12.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, Naoki K, Sasaki H, Fujii Y, Eck MJ, Sellers WR, Johnson BE, Meyerson M. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 13.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 14.Furdui CM, Lew ED, Schlessinger J, Anderson KS. Autophosphorylation of FGFR1 kinase is mediated by a sequential and precisely ordered reaction. Mol Cell. 2006;21:711–717. doi: 10.1016/j.molcel.2006.01.022. [DOI] [PubMed] [Google Scholar]
- 15.Lew ED, Furdui CM, Anderson KS, Schlessinger J. The precise sequence of FGF receptor autophosphorylation is kinetically driven and is disrupted by oncogenic mutations. Sci Signal. 2009;2:ra6. doi: 10.1126/scisignal.2000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schuchardt S, Borlak J. Quantitative mass spectrometry to investigate epidermal growth factor receptor phosphorylation dynamics. Mass Spectrom Rev. 2008;27:51–65. doi: 10.1002/mas.20155. [DOI] [PubMed] [Google Scholar]
- 17.Guo L, Kozlosky CJ, Ericsson LH, Daniel TO, Cerretti DP, Johnson RS. Studies of ligand-induced site-specific phosphorylation of epidermal growth factor receptor. J Am Soc Mass Spectrom. 2003;14:1022–1031. doi: 10.1016/S1044-0305(03)00206-X. [DOI] [PubMed] [Google Scholar]
- 18.Wu SL, Kim J, Bandle RW, Liotta L, Petricoin E, Karger BL. Dynamic profiling of the post-translational modifications and interaction partners of epidermal growth factor receptor signaling after stimulation by epidermal growth factor using Extended Range Proteomic Analysis (ERPA) Mol Cell Proteomics. 2006;5:1610–1627. doi: 10.1074/mcp.M600105-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 20.Mulloy R, Ferrand A, Kim Y, Sordella R, Bell DW, Haber DA, Anderson KS, Settleman J. Epidermal growth factor receptor mutants from human lung cancers exhibit enhanced catalytic activity and increased sensitivity to gefitinib. Cancer Res. 2007;67:2325–2330. doi: 10.1158/0008-5472.CAN-06-4293. [DOI] [PubMed] [Google Scholar]
- 21.Chung I, Akita R, Vandlen R, Toomre D, Schlessinger J, Mellman I. Spatial control of EGF receptor activation by reversible dimerization on living cells. Nature. 2010;464:783–787. doi: 10.1038/nature08827. [DOI] [PubMed] [Google Scholar]
- 22.Jorissen RN, Walker F, Pouliot N, Garrett TP, Ward CW, Burgess AW. Epidermal growth factor receptor: mechanisms of activation and signalling. Exp Cell Res. 2003;284:31–53. doi: 10.1016/s0014-4827(02)00098-8. [DOI] [PubMed] [Google Scholar]
- 23.Sordella R, Bell DW, Haber DA, Settleman J. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science. 2004;305:1163–1167. doi: 10.1126/science.1101637. [DOI] [PubMed] [Google Scholar]
- 24.Wakeling AE, Guy SP, Woodburn JR, Ashton SE, Curry BJ, Barker AJ, Gibson KH. ZD1839 (Iressa): an orally active inhibitor of epidermal growth factor signaling with potential for cancer therapy. Cancer Res. 2002;62:5749–5754. [PubMed] [Google Scholar]
- 25.Schindler T, Bornmann W, Pellicena P, Miller WT, Clarkson B, Kuriyan J. Structural mechanism for STI-571 inhibition of abelson tyrosine kinase. Science. 2000;289:1938–1942. doi: 10.1126/science.289.5486.1938. [DOI] [PubMed] [Google Scholar]
- 26.Yun CH, Boggon TJ, Li Y, Woo MS, Greulich H, Meyerson M, Eck MJ. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell. 2007;11:217–227. doi: 10.1016/j.ccr.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yun CH, Mengwasser KE, Toms AV, Woo MS, Greulich H, Wong KK, Meyerson M, Eck MJ. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci U S A. 2008;105:2070–2075. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stamos J, Sliwkowski MX, Eigenbrot C. Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. J Biol Chem. 2002;277:46265–46272. doi: 10.1074/jbc.M207135200. [DOI] [PubMed] [Google Scholar]
- 29.Carey KD, Garton AJ, Romero MS, Kahler J, Thomson S, Ross S, Park F, Haley JD, Gibson N, Sliwkowski MX. Kinetic analysis of epidermal growth factor receptor somatic mutant proteins shows increased sensitivity to the epidermal growth factor receptor tyrosine kinase inhibitor, erlotinib. Cancer Res. 2006;66:8163–8171. doi: 10.1158/0008-5472.CAN-06-0453. [DOI] [PubMed] [Google Scholar]
- 30.Fabian MA, Biggs WH, 3rd, Treiber DK, Atteridge CE, Azimioara MD, Benedetti MG, Carter TA, Ciceri P, Edeen PT, Floyd M, Ford JM, Galvin M, Gerlach JL, Grotzfeld RM, Herrgard S, Insko DE, Insko MA, Lai AG, Lelias JM, Mehta SA, Milanov ZV, Velasco AM, Wodicka LM, Patel HK, Zarrinkar PP, Lockhart DJ. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat Biotechnol. 2005;23:329–336. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- 31.Mitsudomi T, Yatabe Y. Epidermal growth factor receptor in relation to tumor development: EGFR gene and cancer. The FEBS journal. 2010;277:301–308. doi: 10.1111/j.1742-4658.2009.07448.x. [DOI] [PubMed] [Google Scholar]
- 32.Shan Y, Eastwood MP, Zhang X, Kim ET, Arkhipov A, Dror RO, Jumper J, Kuriyan J, Shaw DE. Oncogenic Mutations Counteract Intrinsic Disorder in the EGFR Kinase and Promote Receptor Dimerization. Cell. 2012;149:860–870. doi: 10.1016/j.cell.2012.02.063. [DOI] [PubMed] [Google Scholar]
- 33.Wang Z, Longo PA, Tarrant MK, Kim K, Head S, Leahy DJ, Cole PA. Mechanistic insights into the activation of oncogenic forms of EGF receptor. Nature structural & molecular biology. 2011;18:1388–1393. doi: 10.1038/nsmb.2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 35.Yokouchi M, Kondo T, Houghton A, Bartkiewicz M, Horne WC, Zhang H, Yoshimura A, Baron R. Ligand-induced ubiquitination of the epidermal growth factor receptor involves the interaction of the c-Cbl RING finger and UbcH7. J Biol Chem. 1999;274:31707–31712. doi: 10.1074/jbc.274.44.31707. [DOI] [PubMed] [Google Scholar]
- 36.Levkowitz G, Waterman H, Ettenberg SA, Katz M, Tsygankov AY, Alroy I, Lavi S, Iwai K, Reiss Y, Ciechanover A, Lipkowitz S, Yarden Y. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol Cell. 1999;4:1029–1040. doi: 10.1016/s1097-2765(00)80231-2. [DOI] [PubMed] [Google Scholar]
- 37.Margolis BL, Lax I, Kris R, Dombalagian M, Honegger AM, Howk R, Givol D, Ullrich A, Schlessinger J. All autophosphorylation sites of epidermal growth factor (EGF) receptor and HER2/neu are located in their carboxyl-terminal tails. Identification of a novel site in EGF receptor. J Biol Chem. 1989;264:10667–10671. [PubMed] [Google Scholar]
- 38.Biscardi JS, Maa MC, Tice DA, Cox ME, Leu TH, Parsons SJ. c-Src-mediated phosphorylation of the epidermal growth factor receptor on Tyr845 and Tyr1101 is associated with modulation of receptor function. J Biol Chem. 1999;274:8335–8343. doi: 10.1074/jbc.274.12.8335. [DOI] [PubMed] [Google Scholar]
- 39.Sato K, Sato A, Aoto M, Fukami Y. c-Src phosphorylates epidermal growth factor receptor on tyrosine 845. Biochem Biophys Res Commun. 1995;215:1078–1087. doi: 10.1006/bbrc.1995.2574. [DOI] [PubMed] [Google Scholar]
- 40.Gotoh N, Tojo A, Hino M, Yazaki Y, Shibuya M. A highly conserved tyrosine residue at codon 845 within the kinase domain is not required for the transforming activity of human epidermal growth factor receptor. Biochem Biophys Res Commun. 1992;186:768–774. doi: 10.1016/0006-291x(92)90812-y. [DOI] [PubMed] [Google Scholar]
- 41.Yang S, Park K, Turkson J, Arteaga CL. Ligand-independent phosphorylation of Y869 (Y845) links mutant EGFR signaling to stat-mediated gene expression. Exp Cell Res. 2008;314:413–419. doi: 10.1016/j.yexcr.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qiu C, Tarrant MK, Boronina T, Longo PA, Kavran JM, Cole RN, Cole PA, Leahy DJ. In vitro enzymatic characterization of near full length EGFR in activated and inhibited states. Biochemistry. 2009;48:6624–6632. doi: 10.1021/bi900755n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang S, Qu S, Perez-Tores M, Sawai A, Rosen N, Solit DB, Arteaga CL. Association with HSP90 inhibits Cbl-mediated down-regulation of mutant epidermal growth factor receptors. Cancer Res. 2006;66:6990–6997. doi: 10.1158/0008-5472.CAN-06-1042. [DOI] [PubMed] [Google Scholar]
- 44.Sakai K, Yokote H, Murakami-Murofushi K, Tamura T, Saijo N, Nishio K. In-frame deletion in the EGF receptor alters kinase inhibition by gefitinib. Biochem J. 2006;397:537–543. doi: 10.1042/BJ20051962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eck MJ, Yun CH. Structural and mechanistic underpinnings of the differential drug sensitivity of EGFR mutations in non-small cell lung cancer. Biochim Biophys Acta. 2010;1804:559–566. doi: 10.1016/j.bbapap.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.