Abstract
Reactive oxygen species (ROS) are a family of molecules that are continuously generated, transformed and consumed in all living organisms as a consequence of aerobic life. The traditional view of these reactive oxygen metabolites is one of oxidative stress and damage that leads to decline of tissue and organ systems in aging and disease. However, emerging data show that ROS produced in certain situations can also contribute to physiology and increased fitness. This Perspective provides a focused discussion on what factors lead ROS molecules to become signal and/or stress agents, highlighting how increasing knowledge of the underlying chemistry of ROS can lead to advances in understanding their disparate contributions to biology. An important facet of this emerging area at the chemistry-biology interface is the development of new tools to study these small molecules and their reactivity in complex biological systems.
Biomolecules can be modified by oxidation-reduction (redox) reactions in a temporal and sequence-specific manner to adjust their three-dimensional structure and function. Perhaps nowhere is this fact better illustrated than in the first enzyme crystal structure, which revealed that hen egg lysozyme forms four disulfide bonds through oxidation of eight cysteine side chains to stabilize its folded form1. Despite the rich history of redox chemistry and its broad consequences for health, aging and disease, researchers are now only beginning to scratch the surface in understanding of the multitude of redox-regulated chemistries that can occur in biological systems. In this context, ROS formed from electrontransfer reactions at oxygen are major molecular sources of redox equivalents at the cell and organism level. ROS are often the small molecules responsible for mediating redox modifications of various biomolecules and are prevalent in diseases ranging from cancer to neurodegenerative diseases to diabetes. The overproduction and/or mismanagement of ROS leads to the general phenomenon of oxidative stress that is implicated in aging and death2,3. However, a more sophisticated and nuanced view of ROS is emerging that goes beyond a simple story of stress and disease, as organisms have also evolved a growing number of increasingly well understood, diverse mechanisms to harness the reactivity of ROS for a wide variety of essential physiological processes4–10.
This Perspective, in the context of the theme of sensors and switches, will highlight the signal and stress dichotomy of ROS chemistry and how it affects biological systems from a molecular, cellular and organismal level. In this spirit, the purpose of this Perspective is not to collate citations for a comprehensive review but to provide an introduction to how cells and organisms sense and use ROS as signal and/or stress agents through the exquisite control of chemistry. Although beyond the scope of this review, the related reactive nitrogen species (RNS) such as nitric oxide ([NO]•) are also important for human physiology11,12 and can react with ROS to form oxidants such as peroxynitrite13. We begin by describing biologically relevant ROS molecules and where they are generated, move on to address what and how biomolecules are modified by ROS and how they are controlled at a cellular level and close by highlighting select examples of biological processes that are mediated by ROS and the chemical tools that are available for studying the functions of these small molecules in complex systems. The common theme of this Perspective is that the chemistry of ROS is the key feature in determining the downstream biological outcome.
Biologically relevant ROS
The term ROS remains useful for global descriptions of downstream phenotypes, but because ROS encompass a family of molecules, and not one discrete chemical entity, the molecular identity of each ROS is often of critical importance in determining both its chemical reactivity and the biological response(s) to those reactions. Superoxide ([O2]•−), hydrogen peroxide (H2O2), hypochlorous acid (HOCl), singlet oxygen (1O2), lipid peroxides (ROOH), ozone (O3) and hydroxyl radical ([OH]•) are some of the major ROS in living systems, and determining whether an individual ROS is present at sufficient concentrations to participate in productive chemistry in a given situation is crucial to elucidating its biology.
Where ROS are generated
Another key consideration for ROS chemistry and biology is the subcellular location where a particular metabolite is generated, as microenvironments can dictate what targets these ROS molecules will potentially encounter in a spatial and temporal manner. The classic examples of organelles with localized ROS generation for physiology include phagosomes within specialized cells of the immune system used for pathogen killing14, and peroxisomes15, which mediate catabolic oxidation reactions for energy metabolism. In addition to these canonical ROS sources, we highlight three other main locales for ROS production in cells under physiological conditions (mitochondria, the endoplasmic reticulum (ER) and cell membranes), noting that other organelles (such as the nucleus and Golgi) as well as ROS cross-talk between subcellular regions are also open fields for study.
Mitochondria and electron transfer
Mitochondria house the electron transport chain (ETC), which transfers electrons from NADH and succinate along a controlled redox path that ends in the four-electron reduction of O2 to H2O during respiratory ATP synthesis. However, either by accident or for a purpose, the flow of electrons through the ETC is an imperfect process, and occasionally oxygen molecules undergo one- or two-electron reduction reactions to form ROS, particularly H2O2 and [O2]•− (ref. 16). Mice with mitochondria-targeted overexpression of catalase, an enzyme that quickly and specifically destroys H2O2, live longer17 and show protection against age-related decline in mitochondrial function and insulin resistance18. For this reason, it has been commonly assumed that mitochondrial leakage of ROS is an inevitable consequence of aerobic respiration, similar to pollution from an automobile, and that cells are constantly combating these aberrant ROS fluxes. However, newer data suggest that cells have evolved exquisite mechanisms to use mitochondrial ROS in a controlled fashion for physiological benefits. For example, ETC-generated H2O2 can regulate neuronal dopamine release through ATP-sensitive potassium channels19,20. In addition, the Src homology and collagen homolog protein (p66shc) is an adaptor protein for receptor protein tyrosine kinase signaling but also has a redox signaling and stress role by catalyzing electron transfer from cytochrome c to reduce O2 to [O2]•− and H2O2 (ref. 21). In response to cellular stress, p66shc mediates ROS generation, usually signaling for apoptosis22, but certain cell types use p66shc-generated ROS for growth signaling23, suggesting that this redox protein can serve to generate ROS for either proapoptotic or proliferative processes.
Endoplasmic reticulum and oxidative protein folding
The primary source of ROS from the ER results from oxidative protein folding—secreted proteins often undergo disulfide bond formation as a post-translational modification as they fold in the ER lumen. The vast majority of protein disulfide formation reactions in the ER are initiated by the glycoprotein Ero1, which triggers a two-electron oxidation of the thioredoxin protein disulfide isomerase (PDI)24. In its oxidized disulfide form, PDI is then used to introduce disulfide bonds into protein targets through thiol-disulfide exchange. Ero1 uses O2 as a two-electron acceptor to form one equivalent of H2O2 for each disulfide bond formation catalyzed, providing a strong flux of ROS in this cellular locale. In addition to the oxidative folding machinery, the ER can also house an isoform of NADPH oxidase (Nox4) that primarily generates H2O2 from O2 by a two-electron reduction25.
Cell membranes and NADPH oxidases
Another major source of physiological ROS, in the form of either [O2]•− or H2O2, are NADPH oxidases (Nox) and their dual oxidase relatives (Duox), which are localized to various cellular membranes4,26,27. Nox proteins are classically known as important ROS producers for the phagocytic killing of pathogens during the immune response. However, the discovery of a vast array of different Nox and Duox isoforms in virtually every cell type throughout the body suggests more general roles for these ROS-producing enzymes28. Indeed, beginning with the first report that receptor tyrosine kinase signaling activates Nox-derived ROS production at the plasma membrane29, the list of receptor-ligand interactions connected to Nox-regulated redox signaling continues to expand at a rapid pace6. Because of the widespread yet differential expression of Nox and Duox isoforms across organelles, cell types and organisms, the use of H2O2 and [O2]•− signaling in this manner can potentially be placed in the same class as other ubiquitous small-molecule messengers such as calcium ions (Ca2+) and [NO]•.
Chemical targets and reactions mediated by ROS
Once specific types of ROS are generated at a given time and place, they can mediate a diverse array of reversible or irreversible redox modifications on biomolecules ranging from proteins to lipids to DNA and RNA. In this context, the chemical reactivity of an individual ROS is generally dictated by whether it prefers one- or twoelectron oxidations. Additionally, as many highly reactive oxidants have short half-lives within the cellular milieu, ROS reactivity is also regulated by which biomolecule targets are within close proximity of the site of ROS generation (see next section). With these principles in mind, we highlight a selection of ROS targets classified by their chemical functionality, using protein-based examples (Fig. 1). We primarily focus on cysteine residues owing to the greater knowledge base about the pathology and physiology of the oxidation of this amino acid, and we then extend our discussion to other amino acid targets of ROS.
Figure 1. Reactions of primary ROS with functional groups on proteins.
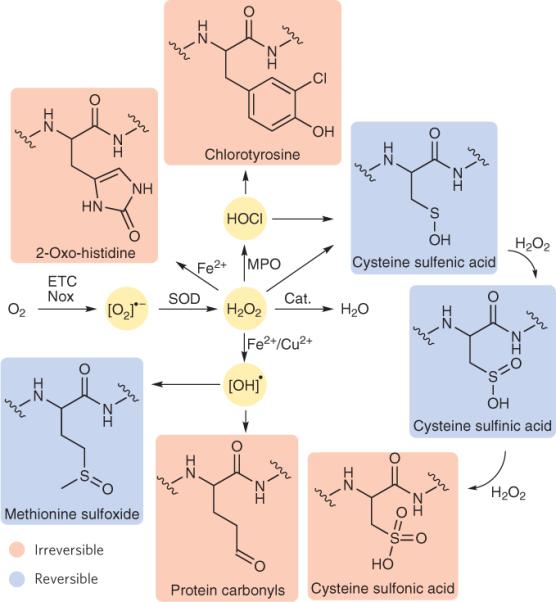
A one-electron reduction of molecular oxygen, either from the electron transport chain (ETC) or through the action of NADPH oxidases (Nox), yields superoxide ([O2]•−). Superoxide is converted to hydrogen peroxide (H2O2) by superoxide dismutase (SOD) or by dismutation in aqueous solution. H2O2 can react with various functional groups; for example, this ROS can oxidize cysteine residues to form sulfenic acids or histidine residues to form 2-oxo-histidines. Sulfenic acids can then go on to form disulfide bonds or be further oxidized to sulfinic and then sulfonic acids by a second and third equivalent of H2O2, respectively. H2O2 can also be converted to hydroxy radical ([OH]•) by catalysis with redox-cycling metals such as Fe2+ and Cu2+, which can then oxidize functional groups such as methionine residues to form methionine sulfoxides or other amino acids such as lysine, arginine, proline and histidine to form protein carbonyls. The enzyme myeloperoxidase (MPO) can convert H2O2 to the highly reactive hypochlorous acid (HOCl), which can oxidize cysteine residues to form sulfenic acids or tyrosine residues to form chlorotyrosine. Oxidized products in blue are those with known pathways to reverse the redox modification, whereas those products highlighted in red are thought to be irreversibly oxidized.
Cysteines
Oxidation of the thiol side chains of cysteine, usually by an initial two-electron oxidation mediated by H2O2 or HOCl, is the most commonly recognized and studied redox post-translational modification7 of proteins. Such reactions generally exploit the electrophilic nature of these particular ROS, as nucleophilic attack by deprotonated thiols releases either H2O or Cl− to form a sulfenic acid, which can then go on to form internal or mixed disulfides or other products. HOCl reacts rapidly with thiolates in a relatively nonspecific manner, as illustrated by the difference in reaction rate of glutathione with HOCl (3 × 107 M−1 s−1) as compared to H2O2 (0.9 M−1 s−1)14. However, the thiolate reactivity of H2O2 can be tuned and substantially accelerated by the local structure and environment of the target residue. Indeed, this fact is highlighted by the large difference in reaction rate constants of H2O2 with redox-active peroxiredoxin 2 (Prx2; 2 × 107 M−1 s−1)30 and protein tyrosine phosphatase, nonreceptor type 1B (PTP1B; 20 M−1 s−1)31, two major targets of redox signaling in cells. Thus, H2O2 can serve as a ubiquitous yet selective second messenger that can oxidize and modulate a wide array of thiol or thiolate-containing targets with kinetic control14. Sulfenic acids are reactive intermediates that can be trapped by internal thiols to form an intramolecular disulfide, react with glutathione (glutathionylation) or other external thiols to form intermolecular disulfide species or cyclize into sulfenamide structures32,33. Moreover, sulfinic and sulfonic acids can be formed by subsequent two- and four-electron oxidations of sulfenic acid congeners, and the discovery of Sulfiredoxin proteins, which can convert sulfinic acids back to thiols, presages an additional layer of redox regulation at this higher oxidation state34,35.
The number of cellular targets of H2O2 that undergo reversible cysteine oxidation is rapidly growing and encompasses a range of different biological processes; we highlight a few examples here. Phosphatases such as PTEN5,36 and PTP-1B32 can be reversibly deactivated by H2O2 production, forming an intermolecular disulfide or sulfenamide, respectively, to enhance forward kinase signaling for various receptor-ligand interactions. Transcription factors such as Yap1 (ref. 37) in yeast and FoxO4 (ref. 38) in mammals can detect H2O2 and activate genes associated with redox regulation. The activity of matrix metalloproteinase-7 can be regulated in vitro by oxidation, in which the addition of HOCl results in activation of the proenzyme39. The nucleocytoplasmic shuttling of the histone deacetylase HDAC4 is controlled by a thioredoxin 1—dependent, H2O2-mediated disulfide formation to regulate cardiac hypertrophy40.
Other amino acids
In addition to cysteine thiols, various other amino acids can be oxidized by ROS. The aberrant oxidation of lysine, arginine, proline and histidine residues, usually catalyzed by redox-cycling metal ions such as Fe2+ and Cu2+, can result in the conversion of the side chain amines to carbonyls, potentially altering a protein's function, and the protein carbonyl content of an organism or tissue often serves as a marker of general oxidative stress41. HOCl, which is generated from the activity of myeloperoxidase, can react with tyrosine residues to form 3-chlorotyrosine, 5-chlorotyrosine and 3,5-chlorotyrosine42, which are implicated in impaired protein function associated with high-density lipoprotein in atherosclerosis43 as well as diminished airway function in children with cystic fibrosis44. However, organisms have evolved mechanisms to both sense and recycle oxidation of at least some amino acids other than cysteine, suggesting that these modifications may be physiological in nature. For example, methionine thioethers can be oxidized to the corresponding sulfoxides, and methionine sulfoxide reductase, an enzyme that is crucial for normal lifespan in mammals, can reverse this modification45. PerR is a transcription factor found in Bacillus subtilis that regulates redox defense genes and can detect low levels of H2O2 by metal-catalyzed oxidation of histidine46. Protein cofactors can also serve as redox sensors, as exemplified by the SoxR transcription factor, which senses [O2]•− by oxidation of an iron-sulfur cluster47, as [O2]•— oxidizes iron-sulfur clusters at a rate that is almost diffusion limited as a result of high charge attraction8.
How cells control ROS chemistry for signaling functions
Because of their transient and reactive nature, a major question for ROS chemistry in living systems is how do cells funnel these small molecules selectively toward physiological redox signaling over uncontrolled oxidative stress pathways? Here are a few recent findings that illustrate the principles of how ROS selectivity can be regulated in a spatial and temporal manner at the subcellular level to promote kinetically competent redox reactions; it is likely that multiple layers of control are working in conjunction to mediate physiological ROS signaling (Fig. 2).
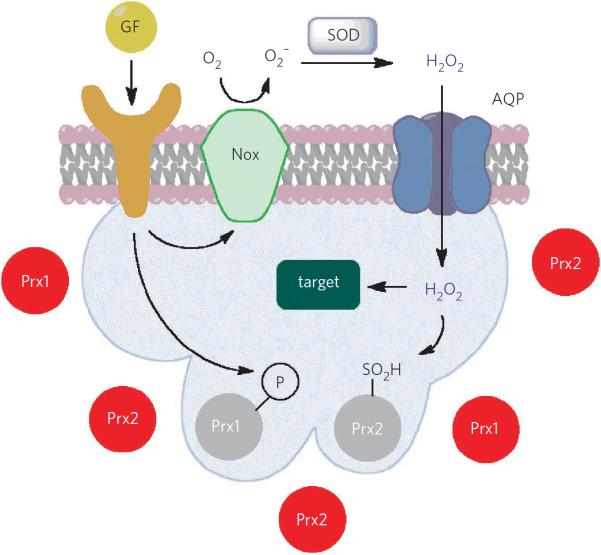
Figure 2. Potential layers of regulation for membrane-localized H2O2 signaling.
Receptor activation, often by growth factors (GF) or other ligands, leads to superoxide ([O2]•−) generation at the cellular membrane by Nox proteins, with subsequent production of H2O2 by dismutation or action of SOD. H2O2 can then pass through specific aquaporins (AQP) to reach the intracellular cytosol. Concomitantly, receptor activation also leads to localized Prx1 phosphorylation and deactivation, decreasing the redox-buffering capacity near the cell membrane. Localized rises in intracellular H2O2 levels can cause further deactivation of Prx2 by overoxidation. These various points of regulation can work together to lead to transient rises in H2O2 concentrations and the subsequent oxidation of local redox targets.
Colocalization of ROS sources and targets
Many types of ROS will not migrate far from their source of production because of their inherent instability and reactivity, and because of the redox-buffering capacity of a cell. For example, the cellular half-life of [OH]• is only about 10−9 s because of its reactivity (the reduction potential of the [OH]•, H+−H2O couple is 2.31 V) compared to about 1 ms for H2O2 (ref. 8). This means that [OH]• will react with or very near to the biomolecule that produced it, whereas H2O2 can diffuse away from its source. Moreover, as shown by the reactivity of H2O2 with various cysteine thiols, the wide range of observed reaction rates between ROS sources and targets affords another level of discrimination. As such, a primary layer of control for ROS signaling is the colocalization of sources and targets of ROS by generation of the small-molecule oxidant in proximity to a given substrate. This form of regulation directly influences the kinetics of a putative chemical signaling reaction by controlling the local concentrations of the participating molecular reactants. For example, Nox proteins that influence receptor tyrosine kinase signaling via H2O2 and [O2]•− are often colocalized with their putative physiological targets, such as phosphatases and kinases, at the plasma membrane. This also prevents oxidation of pathological targets such as nucleotides that are confined to other parts of the cell48,49. Indeed, recent data show that ROS generation is localized for signaling in various cell types50. Other examples of colocalization of ROS signaling sources and targets is ER localization of the H2O2 generator Nox4 and its phosphatase target PTP1B51 and the localized generation of HOCl by myeloperoxidase in phagosomes for pathogen defense14.
Modulation of local redox buffer capacity
Through various measurements and calculations, the intracellular concentration of H2O2 has been determined to fluctuate between the low-nanomolar to low-micromolar range6. These estimates, however, assume an even distribution of H2O2 throughout the cell. As previously explained, the sources of each ROS are localized to specific regions, suggesting that ROS fluxes may not be homogenous in concentration across a living cell and that the concentration of ROS near a source of generation can reach a high local concentration.
In addition to localizing a transient increase in ROS concentrations in proximity to a given target, another layer of physiological ROS control occurs through alterations in local redox buffering capacity. As described above, the millimolar concentrations of cellular glutathione provide a substantial redox buffer for many ROS such as HOCl, but they react too slowly with H2O2 to provide much buffering capacity. Peroxiredoxins, however, have remarkably fast reaction rates with H2O2 and provide a prime example of local redox control of H2O2 owing to two different mechanisms that can modulate peroxiredoxin activity.
Peroxiredoxin proteins typically cycle between reduced dithiol and oxidized disulfide forms mediated by glutathione and H2O2, respectively52. However, the Prx2 isoform is also susceptible to overoxidation by reaction of the sulfenic acid form of the protein with a second equivalent of H2O2, resulting in a transient catalytically inactive protein. In this way, low levels of H2O2 are quickly and efficiently quenched by the redox-buffering capacity of Prx2. However, H2O2 generation at specific subcellular locales can cause a localized overoxidation and deactivation of Prx2, thereby allowing the redox signal to build up in a defined and controlled region as dictated by the source. This so-called `floodgate' model illustrates an elegant and powerful mechanism by which a cell can control localized fluxes of ROS for selective cellular chemistry52,53.
A second example of localized redox buffer control has been identified recently, in which the Prx1 isoform, which is substantially less susceptible to overoxidation than its Prx2 congener, can be selectively deactivated by phosphorylation in cells stimulated with growth factors or in mice during cutaneous wound healing54. In this model, receptor activation is directly coupled to local changes in redox-buffering capacity through discrete kinase signaling cascades.
Membrane transport and sequestration
We recently discovered a third form of physiological ROS regulation that involves membrane ROS transport and sequestration, providing a physical barrier to off-site redox reactions. Building on previous work in plant and yeast models55,56, we showed that the transport of H2O2 across mammalian cell membranes can be controlled by specific classes of aquaporins, integral membrane proteins originally identified as transporters of water and other small-molecule metabolites. Members of the aquaglyceroporin and unorthodox families of aquaporins, but not classical aquaporins, can enhance the permeability of mammalian cell membranes to H2O2, and models with both Nox and aquaporin use the latter to regulate transport of extracellularly generated H2O2 across the plasma membrane to mediate intracellular signaling cascades57. This work suggests that individual cell and tissue types can potentially be tuned for their susceptibility to H2O2-mediated cellular signaling or stress, depending on which aquaporins or related channels are displayed on their cell surfaces.
Physiological processes mediated by ROS signaling
Various redox-regulated physiological processes have been identified that cement ROS signaling as a diverse, important and widespread biological phenomenon. Owing to space limitations, the discussion here is limited to three recent examples in which ROS signaling contributes to physiology and shows the breadth of redox regulation at the cellular and organismal scale (Fig. 3).
Figure 3. ROS signaling in physiology.
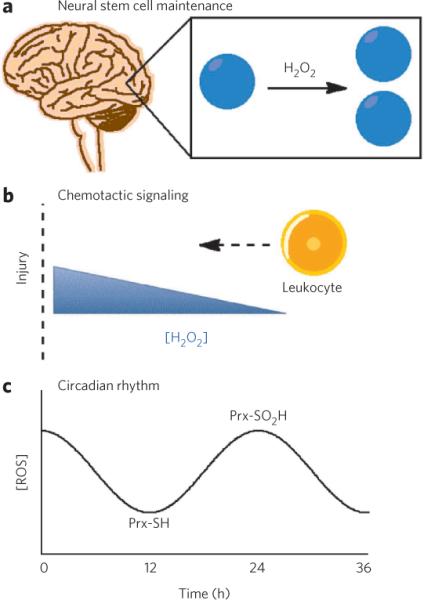
(a) ROS have recently been discovered as second-messenger signaling agents used to control growth and maintenance of neural stem cells located in both the subgranular zone of the hippocampus as well as the subventricular zone of the lateral ventricles. (b) ROS have also been discovered as signaling agents at both the biochemical and whole-organism level to trigger chemotaxis and recruitment of leukocytes to damaged tissue. (c) Finally, the oxidation state of peroxiredoxins (Prx) have been shown to be modulated between reduced (Prx-SH) and oxidized (Prx-SO2H) forms to regulate circadian rhythms in the absence of transcription or translation.
Cell migration
Redox signaling can regulate cell migration at both the molecular and whole-organism levels. At a molecular level, cells respond to various stimuli by generation of Nox-derived H2O2, which can then modulate the local cytoskeleton organization and hence cell migration. This process is mediated by cofilin, an important regulator of cellular actin dynamics, through redox modulation of its opposing phosphatase Slingshot-1L. Slingshot-1L is activated by its release from a regulatory complex through H2O2-mediated oxidation, which in turn induces cofilin-mediated membrane ruffling and cell motility58. Specifically, colon cancer cells rely on c-Src tyrosine kinase–induced, Nox1-generated H2O2 to form functional invadopodia for normal cell migration59. At the whole-organism level, zebrafish have been shown to produce tissue-scale fluxes of H2O2 generated from the Nox isoform Duox upon tail lacerations60. The H2O2 signal traverses hundred of micrometers through the zebrafish epithelium to recruit leukocytes to the wounded area.
Circadian rhythm
Another exciting new area of physiological redox signaling is the circadian rhythm, with the recent discovery that the oxidation state of peroxiredoxin proteins can provide a way for cells to keep time without transcription or translation. Red blood cells, which lack a nucleus and most other organelles, including mitochondria, use 24-h redox cycles of peroxiredoxin proteins that persist for many days, under constant conditions that are entrainable and temperature compensated61. Similar findings were reported for Ostreococcus tauri, a green algae that suspends all transcription when kept in the dark but does not reset its clock upon reintroduction into light. O. tauri keeps track of time in the dark, without transcription, through similar peroxiredoxin redox cycling events62. Although the molecular basis and ROS responsible for these redox fluctuations are yet to be identified, the correlations between fluctuations in ATP and NADPH suggest a link between peroxiredoxin oxidation, central metabolism and circadian rhythm. This fascinating finding could potentially define a general and conserved regulatory mechanism for controlling circadian rhythms in various contexts.
Stem cell proliferation and neurogenesis
Finally, recent studies have established that physiological H2O2 signaling is essential for stem cell proliferation, as illustrated in neural stem cell models, and can also influence subsequent neurogenesis. Using a newly developed H2O2-responsive fluorophore, Peroxyfluor-6 (PF6), in combination with in vitro biochemistry and cellular assays and in vivo knockout mice studies, we recently discovered that adult neural hippocampal progenitors use Nox2-derived H2O2 to regulate growth signaling and maintain normal stem cell population sizes and levels of neurogenesis63. Concurrent with our work, another report documented that a population of neural stem cells located in the subventricular zone also use Nox2-derived ROS to modulate stem and progenitor pools and neurogenesis64. Taken together, these two studies show that brain-derived ROS are not solely detrimental to the fitness of a living organism and can in fact provide tangible benefits. These results also provide a molecular theory for the cognitive deficits observed in mice and humans lacking Nox2, and they suggest a link between H2O2, brain health and memory formation. Moreover, these findings provide one physiological mechanism to explain why nonspecific administering of antioxidants is generally a poor therapeutic.
chemical tools for studying ROS biology
The broad physiological and pathological consequences of ROS biology and the chemical complexities associated with these reactive small molecules provide a need for new and better methods to monitor the origins and fates of ROS, particularly those that can be used in intact living specimens and give real-time information. We present a subset of the most current chemical tools for studying ROS biology (Fig. 4), highlighting new opportunities for innovation.
Figure 4. Chemical tools to study redox biology.
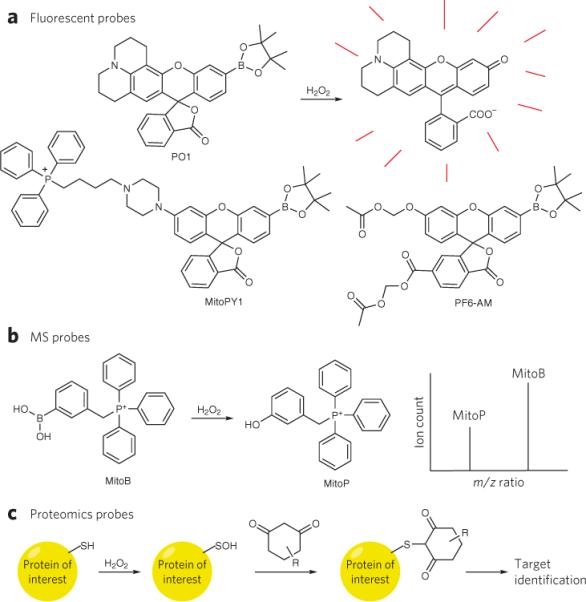
(a) The conversion of boronates to phenols by H2O2 has been used to create a suite of novel fluorescent probes with various properties, such as red-shifted emission (Peroxy Orange 1, PO1), mitochondrial localization (Mitochondria Peroxy Yellow 1, MitoPY1) and enhanced sensitivity through cytosolic trapping groups (Peroxyfluor-6 acetoxymethyl ester, PF6-AM). (b) A mitochondrial-targeted MS probe, which similarly uses the conversion of a boronic acid to a phenol, allows ratiometric detection and quantification of H2O2 in vivo by analysis of the ion count ratios between the protected and deprotected form of the probe, which can be distinguished by differences in mass to charge (m/z) ratios. (c) Dimedone-based reactivity probes can trap oxidized cysteine residues from a sulfenic acid and when coupled to purification or labeling groups, allow the identification of the redox-modified target.
ROS detection
As emphasized in the previous sections, each type of ROS molecule will have its own distinct reactivity in terms of selectivity and kinetics in a given biological context, so a primary need is to devise methods that allow detection of specific ROS metabolites in living cells and organisms. Traditional probes such as dichlorodihydrofluorescein remain useful as global ROS indicators, but because the chemistry for C-H oxidation in this dye and related fluorophores is not selective, one must be cautious about overinterpreting data and attributing biological effects to a single ROS65. To address this concern, a growing number of small-molecule and protein-based detectors have been introduced for monitoring various ROS, including [O2]•−, H2O2, HOCl, 1O2 and O3, as well as global redox changes and related reactive nitrogen species such as [NO]• and peroxynitrite (ONOO−)66–79. Protein-based ROS detectors take advantage of redox-active domains, often transcription factors or antioxidant defense proteins, tethered to fluorescent proteins, which generally rely on cysteine oxidations to modulate a fluorescent response. Two notable examples include HyPer73, a H2O2-specific protein sensor that uses the bacterial transcription factor OxyR, as well as circularly permuted yellow fluorescent protein (cpYFP), which was fortuitously discovered to respond to reaction with [O2]•− (ref. 77). Small-molecule probes generally attempt to selectively detect a single ROS by targeting a unique reactivity of that particular ROS. For example, [NO] probes have taken advantage of the [NO]•-mediated conversion of diamines into triazoles68 as well as the redox reaction between [NO]• and Cu(II)69.
Our laboratory has developed reaction-based approaches to selective H2O2 detection that exploit the H2O2-mediated conversion of aryl boronates to phenols or oxidative decarboxylation of α-ketoacids and incorporated these organic switches into fluorescent, bioluminescent and magnetic resonance imaging (MRI) modalities80–87. Recently, we have used the simple and versatile boronate switch to create H2O2-selective fluorescent turn-on probes with color modularity for dual imaging of multiple ROS simultaneously during the phagocytic respiratory burst85, targeting groups for imaging of mitochondrial-localized ROS production in disease states83 and methyl ester or acetoxymethyl ester functionalities as cytosolic trapping groups for sensitive detection of H2O2 during growth factor signaling in colon cancer and neural stem cells57,63 (Fig. 4a). In addition, nanoparticle- and luciferase-based systems have proven useful for in vivo imaging of H2O2 (refs. 88,89). More recently, a quantitative mass spectrometry approach using a mitochondrial-targeted boronate was developed that allows the measurement of mitochondrial H2O2 levels in various tissues in vivo to address issues in aging and longevity10 (Fig. 4b).
As the identity of the ROS generated in a given system is crucial to dictating the chemistry and, hence, downstream biology, extreme care must be taken to clearly delineate which molecules are involved in initiating these processes. Although many creative and useful specific ROS detection systems have been developed, specifically those for [O2]•−, H2O2, HOCl and [NO]•, improved technologies are still in great demand. In particular, the discovery of new chemical reactions that can specifically detect other ROS, the creation of probes with faster reaction rates and the creation of reversible probes that can detect transient ROS fluxes would all help to decipher the complex redox processes that take place in biological systems.
Controlled ROS production
Because the timing and location of ROS chemistry is tightly regulated in living systems, exogenous addition of reactive molecules to whole-cell cultures or animal models cannot, in many cases, accurately mimic true physiological situations. As such, another potentially powerful set of tools to dissect ROS biology includes reagents that can locally produce a particular ROS on demand and in a controlled fashion. One technology for chemically controlled H2O2 generation uses D-amino acid oxidase, an enzyme that produces H2O2 upon reaction with the substrate N-acetyl-D-alanine90. In this way, local H2O2 generation from a genetically encodable system can be initiated using a chemical trigger, and this method has been applied to astrocyte H2O2 production. General oxidative stress photosensitizers that target nuclei and mitochondria were created using organelle-specific peptide delivery systems91. In parallel, our laboratory has developed a caged small-molecule H2O2 generator that rapidly produces H2O2 on demand upon cleavage of a photolabile protecting group92. We used this new reagent to produce H2O2 in living cells by light activation and trigger downstream redox regulation of cofilin, leading to actin polymerization and cell migration.
ROS target identification
A final key area in ROS chemical biology is to develop new approaches to measure and detect ROS-mediated modifications in living systems. Classically, oxidative stress conditions are globally assessed by measuring oxidized DNA or RNA (for example, 8-oxoG) and/or oxidized proteins (for example, protein carbonyl content). Additionally, hyperoxidized protein cysteines can be detected by western blot analysis with an antibody that detects sulfinic and sulfonic acids in proteins. However, technologies for detecting the transient and reversible oxidations associated with physiological events, particularly those that can be used in live-cell or even live-animal settings, are more informative to ROS signaling. For example, proteomics approaches that interrogate entire populations offer unbiased assessments of redox-active proteins. In this context, isotope-coded affi nity tags have been used to distinguish oxidant-sensitive cysteines in complex protein mixtures by using reactivity differences between free and oxidized thiols93,94, and computational approaches comparing homologous proteins to identify sporadic incorporation of selenocysteine at cysteine active sites have been used to predict putative redox-active cysteine residues95. In addition, recent advances in activity-based protein profiling allow the quantification of nucleophilic, reactive cysteine residues that are more likely to be oxidized by ROS such as H2O2 (ref. 96). In terms of specific chemical modifications, elegant tools that exploit the selectivity of dimedone for sulfenic acid–modified cysteines coupled to fluorophores, affinity tags or both can be used to mark and identify proteins that have undergone this particular redox modification97–99, and an antibody has been produced against a dimedone-modified cysteine residue for analysis by western blotting or pull-down assays100 (Fig. 4c). This area is a particularly fruitful one as chemical biologists become more sophisticated in their thinking of redox chemistry and discrete reactions in the context of more complex systems.
Summary and prospects
The chemistry of a given ROS, which is influenced by its identity, concentration and local environment, is the key determinant of its downstream biological responses. As such, the study of ROS is an inherently well-suited area for the intellectual and practical approaches of chemical biology, as the invention of new chemical technologies that enable the assignment of sources, identities, concentrations and targets of ROS in complex living systems will greatly aid in elucidating the basic principles that control redox biology at the molecular, cellular and organismal levels.
Proteomics and other unbiased screening approaches should allow the identification of many new redox-sensing biomolecules and, when coupled with ROS-specific detection methods, allow chemical biologists to uncover many new processes mediated by ROS signaling and stress. Indeed, recent discoveries of ROS signaling in chemotaxis, stem cell proliferation, neurogenesis and circadian rhythm presage that redox chemistry can regulate a diverse array of biological processes. However, the molecular basis for much of this regulation is still largely unexplored. For example, it is clear that cell migration is regulated by specific redox modifications of proteins directly involved in cytoskeletal rearrangements and that external ROS cues can also signal cells to move toward an ROS source over relatively large distances. However, many of the links between the biochemical and whole-organism responses are still missing, and this will be a rich area for exploration. Studying various modes of ROS production, sensing and signaling at both the cellular and organismal levels is crucial for providing a coherent picture of how ROS are used by biological systems and offers an exciting set of challenges for understanding the contributions of these small molecules to health, aging and disease.
Acknowledgments
We thank the Packard Foundation, Amgen, Astra Zeneca and Novartis and the US National Institutes of Health (NIH; GM 79465) for providing funding for this work. C.J.C. is an Investigator with the Howard Hughes Medical Institute. B.C.D. was partially supported by a Chemical Biology Training Grant from the NIH (T32 GM066698). Due to space limitations, we apologize if we inadvertently omitted citations of major contributions to this area.
Footnotes
Competing financial interests The authors declare no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html. Correspondence and requests for materials should be addressed to C.J.C.
References
- 1.Blake CC, et al. Structure of hen egg-white lysozyme. A three-dimensional Fourier synthesis at 2 Angstrom resolution. Nature. 1965;206:757–761. doi: 10.1038/206757a0. [DOI] [PubMed] [Google Scholar]
- 2.Barnham KJ, Masters CL, Bush AI. Neurodegenerative diseases and oxidative stress. Nat. Rev. Drug Discov. 2004;3:205–214. doi: 10.1038/nrd1330. [DOI] [PubMed] [Google Scholar]
- 3.Finkel T, Serrano M, Blasco MA. The common biology of cancer and ageing. Nature. 2007;448:767–774. doi: 10.1038/nature05985. [DOI] [PubMed] [Google Scholar]
- 4.Lambeth JD. NOX enzymes and the biology of reactive oxygen. Nat. Rev. Immunol. 2004;4:181–189. doi: 10.1038/nri1312. [DOI] [PubMed] [Google Scholar]
- 5.Rhee SG. H2O2, a necessary evil for cell signaling. Science. 2006;312:1882–1883. doi: 10.1126/science.1130481. [DOI] [PubMed] [Google Scholar]
- 6.Stone JR, Yang S. Hydrogen peroxide: a signaling messenger. Antioxid. Redox Signal. 2006;8:243–270. doi: 10.1089/ars.2006.8.243. [DOI] [PubMed] [Google Scholar]
- 7.Veal EA, Day AM, Morgan BA. Hydrogen peroxide sensing and signaling. Mol. Cell. 2007;26:1–14. doi: 10.1016/j.molcel.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 8.D'Autréaux B, Toledano MB. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell Biol. 2007;8:813–824. doi: 10.1038/nrm2256. [DOI] [PubMed] [Google Scholar]
- 9.Paulsen CE, Carroll KS. Orchestrating redox signaling networks through regulatory cysteine switches. ACS Chem. Biol. 2010;5:47–62. doi: 10.1021/cb900258z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cochemé HM, et al. Measurement of H2O2 within living Drosophila during aging using a ratiometric mass spectrometry probe targeted to the mitochondrial matrix. Cell Metab. 2011;13:340–350. doi: 10.1016/j.cmet.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ignarro LJ. Nitric oxide as a unique signaling molecule in the vascular system: a historical overview. J. Physiol. Pharmacol. 2002;53:503–514. [PubMed] [Google Scholar]
- 12.Bryan NS, Bian K, Murad F. Discovery of the nitric oxide signaling pathway and targets for drug development. Front. Biosci. 2009;14:1–18. doi: 10.2741/3228. [DOI] [PubMed] [Google Scholar]
- 13.Szabó C, Ischiropoulos H, Radi R. Peroxynitrite: biochemistry, pathophysiology and development of therapeutics. Nat. Rev. Drug Discov. 2007;6:662–680. doi: 10.1038/nrd2222. [DOI] [PubMed] [Google Scholar]
- 14.Winterbourn CC. Reconciling the chemistry and biology of reactive oxygen species. Nat. Chem. Biol. 2008;4:278–286. doi: 10.1038/nchembio.85. [DOI] [PubMed] [Google Scholar]
- 15.del Río LA, Sandalio LM, Palma JM, Bueno P, Corpas FJ. Metabolism of oxygen radicals in peroxisomes and cellular implications. Free Radic. Biol. Med. 1992;13:557–580. doi: 10.1016/0891-5849(92)90150-f. [DOI] [PubMed] [Google Scholar]
- 16.Murphy MP. How mitochondria produce reactive oxygen species. Biochem. J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schriner SE, et al. Extension of murine life span by overexpression of catalase targeted to mitochondria. Science. 2005;308:1909–1911. doi: 10.1126/science.1106653. [DOI] [PubMed] [Google Scholar]
- 18.Lee HY, et al. Targeted expression of catalase to mitochondria prevents age-associated reductions in mitochondrial function and insulin resistance. Cell Metab. 2010;12:668–674. doi: 10.1016/j.cmet.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Avshalumov MV, Rice ME. Activation of ATP-sensitive K+ (KATP) channels by H2O2 underlies glutamate-dependent inhibition of striatal dopamine release. Proc. Natl. Acad. Sci. USA. 2003;100:11729–11734. doi: 10.1073/pnas.1834314100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bao L, et al. Mitochondria are the source of hydrogen peroxide for dynamic brain-cell signaling. J. Neurosci. 2009;29:9002–9010. doi: 10.1523/JNEUROSCI.1706-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giorgio M, et al. Electron transfer between cytochrome c and p66Shc generates reactive oxygen species that trigger mitochondrial apoptosis. Cell. 2005;122:221–233. doi: 10.1016/j.cell.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 22.Migliaccio E, et al. The p66shc adaptor protein controls oxidative stress response and life span in mammals. Nature. 1999;402:309–313. doi: 10.1038/46311. [DOI] [PubMed] [Google Scholar]
- 23.Veeramani S, Yuan TC, Lin FF, Lin MF. Mitochondrial redox signaling by p66Shc is involved in regulating androgenic growth stimulation of human prostate cancer cells. Oncogene. 2008;27:5057–5068. doi: 10.1038/onc.2008.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gross E, et al. Generating disulfides enzymatically: reaction products and electron acceptors of the endoplasmic reticulum thiol oxidase Ero1p. Proc. Natl. Acad. Sci. USA. 2006;103:299–304. doi: 10.1073/pnas.0506448103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen K, Kirber MT, Xiao H, Yang Y, Keaney JF., Jr. Regulation of ROS signal transduction by NADPH oxidase 4 localization. J. Cell Biol. 2008;181:1129–1139. doi: 10.1083/jcb.200709049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suh YA, et al. Cell transformation by the superoxide-generating oxidase Mox1. Nature. 1999;401:79–82. doi: 10.1038/43459. [DOI] [PubMed] [Google Scholar]
- 27.Geiszt M, Kopp JB, Várnai P, Leto TL. Identification of Renox, an NAD(P)H oxidase in kidney. Proc. Natl. Acad. Sci. USA. 2000;97:8010–8014. doi: 10.1073/pnas.130135897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bedard K, Krause KH. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol. Rev. 2007;87:245–313. doi: 10.1152/physrev.00044.2005. [DOI] [PubMed] [Google Scholar]
- 29.Sundaresan M, Yu ZX, Ferrans VJ, Irani K, Finkel T. Requirement for generation of H2O2 for platelet-derived growth factor signal transduction. Science. 1995;270:296–299. doi: 10.1126/science.270.5234.296. [DOI] [PubMed] [Google Scholar]
- 30.Peskin AV, et al. The high reactivity of peroxiredoxin 2 with H2O2 is not reflected in its reaction with other oxidants and thiol reagents. J. Biol. Chem. 2007;282:11885–11892. doi: 10.1074/jbc.M700339200. [DOI] [PubMed] [Google Scholar]
- 31.Denu JM, Tanner KG. Specific and reversible inactivation of protein tyrosine phosphatases by hydrogen peroxide: evidence for a sulfenic acid intermediate and implications for redox regulation. Biochemistry. 1998;37:5633–5642. doi: 10.1021/bi973035t. [DOI] [PubMed] [Google Scholar]
- 32.Salmeen A, et al. Redox regulation of protein tyrosine phosphatase 1B involves a sulphenyl-amide intermediate. Nature. 2003;423:769–773. doi: 10.1038/nature01680. [DOI] [PubMed] [Google Scholar]
- 33.van Montfort RL, Congreve M, Tisi D, Carr R, Jhoti H. Oxidation state of the active-site cysteine in protein tyrosine phosphatase 1B. Nature. 2003;423:773–777. doi: 10.1038/nature01681. [DOI] [PubMed] [Google Scholar]
- 34.Biteau B, Labarre J, Toledano MB. ATP-dependent reduction of cysteine sulphinic acid by S. cerevisiae sulphiredoxin. Nature. 2003;425:980–984. doi: 10.1038/nature02075. [DOI] [PubMed] [Google Scholar]
- 35.Chang TS, et al. Characterization of mammalian sulfiredoxin and its reactivation of hyperoxidized peroxiredoxin through reduction of cysteine sulfinic acid in the active site to cysteine. J. Biol. Chem. 2004;279:50994–51001. doi: 10.1074/jbc.M409482200. [DOI] [PubMed] [Google Scholar]
- 36.Kwon J, et al. Reversible oxidation and inactivation of the tumor suppressor PTEN in cells stimulated with peptide growth factors. Proc. Natl. Acad. Sci. USA. 2004;101:16419–16424. doi: 10.1073/pnas.0407396101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wood MJ, Storz G, Tjandra N. Structural basis for redox regulation of Yap1 transcription factor localization. Nature. 2004;430:917–921. doi: 10.1038/nature02790. [DOI] [PubMed] [Google Scholar]
- 38.Dansen TB, et al. Redox-sensitive cysteines bridge p300/CBP-mediated acetylation and FoxO4 activity. Nat. Chem. Biol. 2009;5:664–672. doi: 10.1038/nchembio.194. [DOI] [PubMed] [Google Scholar]
- 39.Fu X, Kassim SY, Parks WC, Heinecke JW. Hypochlorous acid oxygenates the cysteine switch domain of pro-matrilysin (MMP-7). A mechanism for matrix metalloproteinase activation and atherosclerotic plaque rupture by myeloperoxidase. J. Biol. Chem. 2001;276:41279–41287. doi: 10.1074/jbc.M106958200. [DOI] [PubMed] [Google Scholar]
- 40.Ago T, et al. A redox-dependent pathway for regulating class II HDACs and cardiac hypertrophy. Cell. 2008;133:978–993. doi: 10.1016/j.cell.2008.04.041. [DOI] [PubMed] [Google Scholar]
- 41.Stadtman ER. Protein oxidation and aging. Free Radic. Res. 2006;40:1250–1258. doi: 10.1080/10715760600918142. [DOI] [PubMed] [Google Scholar]
- 42.Winterbourn CC, Kettle AJ. Biomarkers of myeloperoxidase-derived hypochlorous acid. Free Radic. Biol. Med. 2000;29:403–409. doi: 10.1016/s0891-5849(00)00204-5. [DOI] [PubMed] [Google Scholar]
- 43.Shao B, Oda MN, Oram JF, Heinecke JW. Myeloperoxidase: an oxidative pathway for generating dysfunctional high-density lipoprotein. Chem. Res. Toxicol. 2010;23:447–454. doi: 10.1021/tx9003775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomson E, et al. Identifying peroxidases and their oxidants in the early pathology of cystic fibrosis. Free Radic. Biol. Med. 2010;49:1354–1360. doi: 10.1016/j.freeradbiomed.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 45.Moskovitz J, et al. Methionine sulfoxide reductase (MsrA) is a regulator of antioxidant defense and lifespan in mammals. Proc. Natl. Acad. Sci. USA. 2001;98:12920–12925. doi: 10.1073/pnas.231472998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee JW, Helmann JD. The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature. 2006;440:363–367. doi: 10.1038/nature04537. [DOI] [PubMed] [Google Scholar]
- 47.Gaudu P, Weiss B. SoxR, a [2Fe-2S] transcription factor, is active only in its oxidized form. Proc. Natl. Acad. Sci. USA. 1996;93:10094–10098. doi: 10.1073/pnas.93.19.10094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hilenski LL, Clempus RE, Quinn MT, Lambeth JD, Griendling KK. Distinct subcellular localizations of Nox1 and Nox4 in vascular smooth muscle cells. Arterioscler. Thromb. Vasc. Biol. 2004;24:677–683. doi: 10.1161/01.ATV.0000112024.13727.2c. [DOI] [PubMed] [Google Scholar]
- 49.Ushio-Fukai M. Localizing NADPH oxidase–derived ROS. Sci. STKE. 2006;2006:re8. doi: 10.1126/stke.3492006re8. [DOI] [PubMed] [Google Scholar]
- 50.Mishina NM, et al. Does cellular hydrogen peroxide diffuse or act locally? Antioxid. Redox Signal. 2011;14:1–7. doi: 10.1089/ars.2010.3539. [DOI] [PubMed] [Google Scholar]
- 51.Wu RF, Ma Z, Liu Z, Terada LS. Nox4-derived H2O2 mediates endoplasmic reticulum signaling through local Ras activation. Mol. Cell. Biol. 2010;30:3553–3568. doi: 10.1128/MCB.01445-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wood ZA, Poole LB, Karplus PA. Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science. 2003;300:650–653. doi: 10.1126/science.1080405. [DOI] [PubMed] [Google Scholar]
- 53.Woo HA, et al. Reversing the inactivation of peroxiredoxins caused by cysteine sulfinic acid formation. Science. 2003;300:653–656. doi: 10.1126/science.1080273. [DOI] [PubMed] [Google Scholar]
- 54.Woo HA, et al. Inactivation of Peroxiredoxin I by phosphorylation allows localized H2O2 accumulation for cell signaling. Cell. 2010;140:517–528. doi: 10.1016/j.cell.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 55.Bienert GP, et al. Specific aquaporins facilitate the diffusion of hydrogen peroxide across membranes. J. Biol. Chem. 2007;282:1183–1192. doi: 10.1074/jbc.M603761200. [DOI] [PubMed] [Google Scholar]
- 56.Dynowski M, Schaaf G, Loque D, Moran O, Ludewig U. Plant plasma membrane water channels conduct the signalling molecule H2O2. Biochem. J. 2008;414:53–61. doi: 10.1042/BJ20080287. [DOI] [PubMed] [Google Scholar]
- 57.Miller EW, Dickinson BC, Chang CJ. Aquaporin-3 mediates hydrogen peroxide uptake to regulate downstream intracellular signaling. Proc. Natl. Acad. Sci. USA. 2010;107:15681–15686. doi: 10.1073/pnas.1005776107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim JS, Huang TY, Bokoch GM. Reactive oxygen species regulate a slingshot-cofilin activation pathway. Mol. Biol. Cell. 2009;20:2650–2660. doi: 10.1091/mbc.E09-02-0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gianni D, Taulet N, DerMardirossian C, Bokoch GM. c-Src-mediated phosphorylation of NoxA1 and Tks4 induces the reactive oxygen species (ROS)-dependent formation of functional invadopodia in human colon cancer cells. Mol. Biol. Cell. 2010;21:4287–4298. doi: 10.1091/mbc.E10-08-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Niethammer P, Grabher C, Look AT, Mitchison TJ. A tissue-scale gradient of hydrogen peroxide mediates rapid wound detection in zebrafish. Nature. 2009;459:996–999. doi: 10.1038/nature08119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O'Neill JS, Reddy AB. Circadian clocks in human red blood cells. Nature. 2011;469:498–503. doi: 10.1038/nature09702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.O'Neill JS, et al. Circadian rhythms persist without transcription in a eukaryote. Nature. 2011;469:554–558. doi: 10.1038/nature09654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dickinson BC, Peltier J, Stone D, Schaffer DV, Chang CJ. Nox2 redox signaling maintains essential cell populations in the brain. Nat. Chem. Biol. 2011;7:106–112. doi: 10.1038/nchembio.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Le Belle JE, et al. Proliferative neural stem cells have high endogenous ROS levels that regulate self-renewal and neurogenesis in a PI3K/Akt-dependant manner. Cell Stem Cell. 2011;8:59–71. doi: 10.1016/j.stem.2010.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hempel SL, Buettner GR, O'Malley YQ, Wessels DA, Flaherty DM. Dihydrofluorescein diacetate is superior for detecting intracellular oxidants: comparison with 2′,7′-dichlorodihydrofluorescein diacetate, 5(and 6)-carboxy-2′,7′-dichlorodihydrofluorescein diacetate, and dihydrorhodamine 123. Free Radic. Biol. Med. 1999;27:146–159. doi: 10.1016/s0891-5849(99)00061-1. [DOI] [PubMed] [Google Scholar]
- 66.Ostergaard H, Henriksen A, Hansen FG, Winther JR. Shedding light on disulfide bond formation: engineering a redox switch in green fluorescent protein. EMBO J. 2001;20:5853–5862. doi: 10.1093/emboj/20.21.5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hanson GT, et al. Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. J. Biol. Chem. 2004;279:13044–13053. doi: 10.1074/jbc.M312846200. [DOI] [PubMed] [Google Scholar]
- 68.Sasaki E, et al. Highly sensitive near-infrared fluorescent probes for nitric oxide and their application to isolated organs. J. Am. Chem. Soc. 2005;127:3684–3685. doi: 10.1021/ja042967z. [DOI] [PubMed] [Google Scholar]
- 69.Lim MH, Xu D, Lippard SJ. Visualization of nitric oxide in living cells by a copper-based fluorescent probe. Nat. Chem. Biol. 2006;2:375–380. doi: 10.1038/nchembio794. [DOI] [PubMed] [Google Scholar]
- 70.Yang D, Wang HL, Sun ZN, Chung NW, Shen JG. A highly selective fluorescent probe for the detection and imaging of peroxynitrite in living cells. J. Am. Chem. Soc. 2006;128:6004–6005. doi: 10.1021/ja0603756. [DOI] [PubMed] [Google Scholar]
- 71.Robinson KM, et al. Selective fluorescent imaging of superoxide in vivo using ethidium-based probes. Proc. Natl. Acad. Sci. USA. 2006;103:15038–15043. doi: 10.1073/pnas.0601945103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kenmoku S, Urano Y, Kojima H, Nagano T. Development of a highly specific rhodamine-based fluorescence probe for hypochlorous acid and its application to real-time imaging of phagocytosis. J. Am. Chem. Soc. 2007;129:7313–7318. doi: 10.1021/ja068740g. [DOI] [PubMed] [Google Scholar]
- 73.Belousov VV, et al. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat. Methods. 2006;3:281–286. doi: 10.1038/nmeth866. [DOI] [PubMed] [Google Scholar]
- 74.Miller EW, Bian SX, Chang CJ. A fluorescent sensor for imaging reversible redox cycles in living cells. J. Am. Chem. Soc. 2007;129:3458–3459. doi: 10.1021/ja0668973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Robinson KM, Janes MS, Beckman JS. The selective detection of mitochondrial superoxide by live cell imaging. Nat. Protoc. 2008;3:941–947. doi: 10.1038/nprot.2008.56. [DOI] [PubMed] [Google Scholar]
- 76.Gutscher M, et al. Real-time imaging of the intracellular glutathione redox potential. Nat. Methods. 2008;5:553–559. doi: 10.1038/nmeth.1212. [DOI] [PubMed] [Google Scholar]
- 77.Wang W, et al. Superoxide flashes in single mitochondria. Cell. 2008;134:279–290. doi: 10.1016/j.cell.2008.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Garner AL, et al. Specific fluorogenic probes for ozone in biological and atmospheric samples. Nature Chemistry. 2009;1:316–321. doi: 10.1038/nchem.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dickinson BC, Srikun D, Chang CJ. Mitochondrial-targeted fluorescent probes for reactive oxygen species. Curr. Opin. Chem. Biol. 2010;14:50–56. doi: 10.1016/j.cbpa.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chang MC, Pralle A, Isacoff EY, Chang CJ. A selective, cell-permeable optical probe for hydrogen peroxide in living cells. J. Am. Chem. Soc. 2004;126:15392–15393. doi: 10.1021/ja0441716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Miller EW, Albers AE, Pralle A, Isacoff EY, Chang CJ. Boronatebased fluorescent probes for imaging cellular hydrogen peroxide. J. Am. Chem. Soc. 2005;127:16652–16659. doi: 10.1021/ja054474f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Miller EW, Tulyanthan O, Isacoff EY, Chang CJ. Molecular imaging of hydrogen peroxide produced for cell signaling. Nat. Chem. Biol. 2007;3:263–267. doi: 10.1038/nchembio871. [DOI] [PubMed] [Google Scholar]
- 83.Dickinson BC, Chang CJ. A targetable fluorescent probe for imaging hydrogen peroxide in the mitochondria of living cells. J. Am. Chem. Soc. 2008;130:9638–9639. doi: 10.1021/ja802355u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Srikun D, Miller EW, Domaille DW, Chang CJ. An ICT-based approach to ratiometric fluorescence imaging of hydrogen peroxide produced in living cells. J. Am. Chem. Soc. 2008;130:4596–4597. doi: 10.1021/ja711480f. [DOI] [PubMed] [Google Scholar]
- 85.Dickinson BC, Huynh C, Chang CJ. A palette of fluorescent probes with varying emission colors for imaging hydrogen peroxide signaling in living cells. J. Am. Chem. Soc. 2010;132:5906–5915. doi: 10.1021/ja1014103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lippert AR, Keshari KR, Kurhanewicz J, Chang CJ. A hydrogen peroxide-responsive hyperpolarized (13)c MRI contrast agent. J. Am. Chem. Soc. 2011;133:3776–3779. doi: 10.1021/ja111589a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Srikun D, Albers AE, Chang CJ. A dendrimer-based platform for simultaneous dual fluorescence imaging of hydrogen peroxide and pH gradients produced in living cells. Chem. Sci. 2011;2:1156–1165. [Google Scholar]
- 88.Lee D, et al. In vivo imaging of hydrogen peroxide with chemiluminescent nanoparticles. Nat. Mater. 2007;6:765–769. doi: 10.1038/nmat1983. [DOI] [PubMed] [Google Scholar]
- 89.Van de Bittner GC, Dubikovskaya EA, Bertozzi CR, Chang CJ. In vivo imaging of hydrogen peroxide production in a murine tumor model with a chemoselective bioluminescent reporter. Proc. Natl. Acad. Sci. USA. 2010;107:21316–21321. doi: 10.1073/pnas.1012864107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Haskew-Layton RE, et al. Controlled enzymatic production of astrocytic hydrogen peroxide protects neurons from oxidative stress via an Nrf2-independent pathway. Proc. Natl. Acad. Sci. USA. 2010;107:17385–17390. doi: 10.1073/pnas.1003996107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mahon KP, et al. Deconvolution of the cellular oxidative stress response with organelle-specific peptide conjugates. Chem. Biol. 2007;14:923–930. doi: 10.1016/j.chembiol.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 92.Miller EW, et al. Light-activated regulation of cofilin dynamics using a photocaged hydrogen peroxide generator. J. Am. Chem. Soc. 2010 Nov 15; doi: 10.1021/ja107783j. published online, doi: 10.1021/ja107783j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Winter J, Ilbert M, Graf PC, Ozcelik D, Jakob U. Bleach activates a redox-regulated chaperone by oxidative protein unfolding. Cell. 2008;135:691–701. doi: 10.1016/j.cell.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Leichert LI, et al. Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proc. Natl. Acad. Sci. USA. 2008;105:8197–8202. doi: 10.1073/pnas.0707723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fomenko DE, Xing W, Adair BM, Thomas DJ, Gladyshev VN. High-throughput identification of catalytic redox-active cysteine residues. Science. 2007;315:387–389. doi: 10.1126/science.1133114. [DOI] [PubMed] [Google Scholar]
- 96.Weerapana E, et al. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468:790–795. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Poole LB, et al. Fluorescent and affinity-based tools to detect cysteine sulfenic acid formation in proteins. Bioconjug. Chem. 2007;18:2004–2017. doi: 10.1021/bc700257a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Leonard SE, Reddie KG, Carroll KS. Mining the thiol proteome for sulfenic acid modifications reveals new targets for oxidation in cells. ACS Chem. Biol. 2009;4:783–799. doi: 10.1021/cb900105q. [DOI] [PubMed] [Google Scholar]
- 99.Seo YH, Carroll KS. Quantification of protein sulfenic acid modifications using isotope-coded dimedone and iododimedone. Angew. Chem. Int. Edn Engl. 2011;50:1342–1345. doi: 10.1002/anie.201007175. [DOI] [PubMed] [Google Scholar]
- 100.Seo YH, Carroll KS. Profiling protein thiol oxidation in tumor cells using sulfenic acid-specific antibodies. Proc. Natl. Acad. Sci. USA. 2009;106:16163–16168. doi: 10.1073/pnas.0903015106. [DOI] [PMC free article] [PubMed] [Google Scholar]