Abstract
Malignant glioma is a severe cancer with a poor prognosis. Local occurrence and rare metastases of malignant glioma make it a suitable target for gene therapy. Several studies have demonstrated the importance of Src kinase in different cancers. However, these studies have focused mainly on Src-deficient mice or pharmacological inhibitors of Src. In this study we have used Src small hairpin RNAs (shRNAs) in a lentiviral backbone to mimic a long-term stable treatment and determined the role of Src in tumor tissues. Efficacy of Src shRNAs was confirmed in vitro demonstrating up to 90% target gene inhibition. In a mouse malignant glioma model, Src shRNA tumors were almost 50-fold smaller in comparison to control tumors and had significantly reduced vascularity. In a syngenic rat intracranial glioma model, Src shRNA-transduced tumors were smaller and these rats had a survival benefit over the control rats. In vivo treatment was enhanced by chemotherapy and histone deacetylase inhibition. Our results emphasise the importance of Src in tumorigenesis and demonstrate that it can be efficiently inhibited in vitro and in vivo in two independent malignant glioma models. In conclusion, Src is a potential target for RNA interference-mediated treatment of malignant glioma.
Keywords: glioma, lentivirus, MRI, RNA interference, Src
Introduction
Malignant glioma is the most common brain tumor in adults and has a very poor prognosis: patients diagnosed with the most malignant form of the tumor live ~14 months after diagnosis.1,2 Although traditional treatments, such as surgical removal of the malignant tissue, chemotherapy, and radiation therapy often improve the outcome, additional therapies are still needed.3,4 Despite the significant development of therapies for several other types of cancer, the median survival of malignant glioma patients has shown little improvement over the past few decades.5 Malignant glioma, as with most brain tumors, is a localized, rarely metastasizing tumor surrounded by nondividing cells, which makes it a well-defined target for gene therapy.6,7
Tumorigenesis is a complex process which involves several molecular and cellular mechanisms. One of the central molecules is the nonreceptor protein tyrosine kinase Src, which is a multifunctional signalling molecule located downstream of several different growth factor receptors.8,9 It has a major role in cellular processes such as migration, invasion and survival which are necessary for tumorigenesis.10 Src has been shown to play a role in several different cancers at varying stages of disease, including malignant glioma, which makes it an attractive target for therapy.11 However, most of the studies conducted so far have addressed the role of Src in tumorigenesis either in Src-deficient mice or with pharmacological inhibitors of Src.12,13,14
Our approach was to specifically inhibit Src expression in the tumor tissue. Our aims were (i) to show that we are able to inhibit tumor cell growth with Src small hairpin RNA (shRNA), (ii) to demonstrate that the growth inhibition was specifically related to the inhibition of Src protein and mRNA and, (iii) to translate the in vitro findings to in vivo showing inhibition of tumor growth in two independent malignant glioma models. Lentivirus-mediated gene delivery was chosen because of its long-term expression capability and high tropism for the central nervous system.15,16 ShRNAs, the mediators of RNA interference,17,18,19 enabled us to specifically inhibit Src kinase in tumor cells.
Gene transfer vectors used in this study were first characterized in vitro for their efficacy. In vivo experiments were started in subcutaneous glioma model for preliminary screening of treatment responses. Results were further confirmed in a syngenic orthotopic rat glioma model closely resembling human malignant glioma. Src shRNA approach was also combined with standard chemotherapy and histone deacetylase inhibition to enhance the treatment response. As a result, we describe an efficient inhibition of Src expression in vitro and in vivo, which led to impaired tumor cell growth in two different glioma models. We conclude that Src is a potential target for RNA interference-based gene therapy for malignant glioma.
Results
In vitro functionality
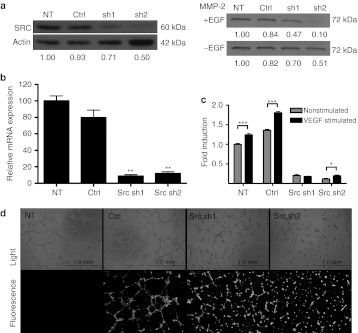
To establish efficient tools for further experiments, the functionality of Src shRNA constructs were analyzed on mRNA as well as on protein level for target gene knockdown (Figure 1a,b). Both selected shRNAs were able to reduce Src gene expression up to 90% in comparison to nontransduced (NT) and control-transduced (Ctrl) cells. In addition, Src downstream target and tumor microenvironment molecule matrix metalloproteinase 2 (MMP-2) was downregulated in cell culture supernatants after Src inhibition (Figure 1a). To assay the biological functionality of shRNA constructs, cell viability was measured by MTT assay and angiogenesis mimicked in vitro by tubulogenesis on Matrigel. The viability of NT and Ctrl cells enhanced significantly after vascular endothelial growth factor-stimulation, whereas Src-inhibited cells gained only little viability benefit (Figure 1c). Furthermore, the overall levels of cell viability in Src shRNA-treated groups were remarkably lower compared to the controls. Control cells cultured on Matrigel started to form tube-like structures within a few hours after the cells were plated. Tubules were most prominent 6 hours post-plating (Figure 1d, top panel) and started to slowly disintegrate thereafter. Tubulogenesis in Src-inhibited cells was clearly disturbed as demonstrated by cells remaining apart from each other throughout the observation time and failing to make tube-like structures. Fluorescence microscopy was used to confirm the expression of shRNAs during the experiment (Figure 1d, bottom panel).
Figure 1.
Functionality of small hairpin RNAs (shRNAs) in vitro. (a) Inhibitory effects of shRNAs against Src kinase were analysed in human umbilical vein endothelial cells (HUVECs) on a protein level by western blotting. β-Actin was used as a normalization control for sample loading. Effect of Src inhibition on the cell secretion of matrix metalloproteinase 2 (MMP-2) was analyzed from +/− EGF-stimulated U118MG cell supernatants. Quantifications of western blots are shown below each lane. (b) Src inhibition in HUVECs was also measured on mRNA level by quantitative real-time reverse transcription (RT)-PCR. Target gene expression was normalized to GAPDH mRNA expression. **P < 0.01 versus nontransduced cells. (c) The effect of vascular endothelial growth factor (VEGF)-stimulation (50 ng/ml) on cell viability was measured by MTT assay in cells transduced with shRNAs. *P < 0.05; ***P < 0.001 versus nonstimulated cells. (d) To mimic angiogenesis in vitro, transduced cells were plated on growth factor reduced Matrigel and monitored over a period of several hours. Light and fluorescence microscopic pictures were taken. Representative pictures are from 6 hours timepoint. 40× magnification, scale = 1 mm. +, stimulated; −, nonstimulated; Ctrl, transduced with a control vector expressing shRNA against luciferase; EGF, epidermal growth factor; NT, nontransduced; sh1, shRNA sequence 1 against Src; sh2, shRNA sequence 2 against Src. Error bars = SEM.
Tumor growth and marker gene expression in mouse xenograft
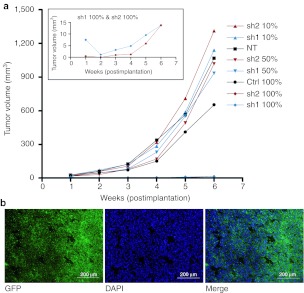
To study the efficacy of Src shRNA in vivo, we established a xenograft from the U118MG cell line. Tumor growth was measured weekly over 6 weeks of follow-up. After 3 weeks, growth differences between groups 5 and 8 (Table 1) in comparison to other groups started to emerge (Figure 2a). This difference increased toward the end of the experiment. At the end of the study (6 weeks postimplantation) the smallest tumors were <20 mm3 in size in comparison to the control tumors, which were almost 50-fold bigger. The tumors having Src shRNAs expressed in close to 100% of the cells were unable to grow. The groups having 10% and 50% cells transduced with Src shRNAs did not show reduced growth in comparison to NT tumors. The mean tumor volume and corresponding standard error of mean for each group at 6 weeks timepoint was: NT 1,068 (±168), Ctrl 100% 654 (±141), sh1 10% 1,141 (±199), sh1 50% 934 (±146), sh1 100% 13.7 (±4.2), sh2 10% 1,313 (±251), sh2 50% 1,018 (±172), sh2 100% 13.8 (±3.7). Transduction efficiency was confirmed by green fluorescent protein (GFP) marker gene expression both preimplantation (data not shown) and postsacrifice (Figure 2b). Tumor morphology assessed by hematoxylin–eosin staining showed no marked differences between the treatment groups (data not shown).
Table 1. Study groups.
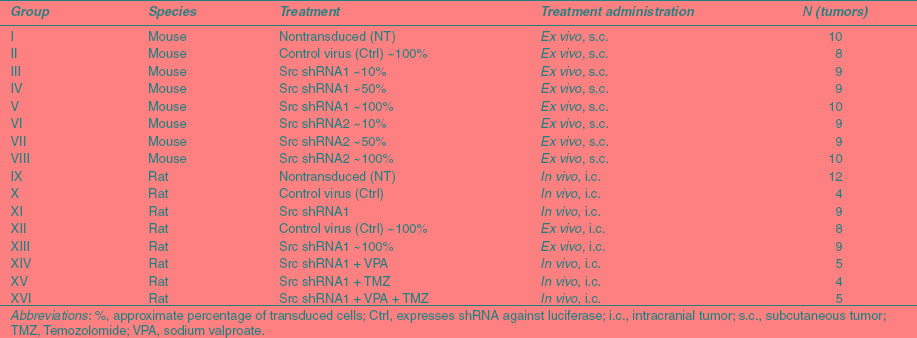
Figure 2.
Mouse tumor growth kinetics and marker gene expression. (a) Tumor growth kinetics were measured over 6 weeks follow-up. U118MG glioma cells were transduced with lentiviral vectors encoding small hairpin RNAs (shRNAs) against Src kinase and luciferase as a control. Different proportions of transduced cells were implanted subcutaneously on flanks of nude mice and tumor growth was measured weekly in different treatment groups. The insert shows tumor growth of sh1 100% and sh2 100% groups over the same study period with a more detailed scale of y axis. P < 0.05 Ctrl versus sh1/sh2 100%. (b) Maintenance of stable transduction throughout the experiment was confirmed by green fluorescent protein (GFP) marker gene expression in frozen tumor sections. Representative sections are from a tumor formed by control virus transduced cells. 100× magnification, scale = 200 μm. Ctrl, transduced with a control vector expressing shRNA against luciferase; NT, nontransduced; sh1, shRNA sequence 1 against Src; sh2, shRNA sequence 2 against Src; shRNA, small hairpin RNA.
Src expression and interferon response
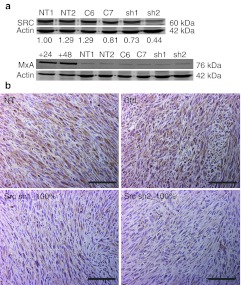
Src expression was studied from tumors by western blotting and immunohistochemistry. Tumors in groups 5 and 8 had reduced Src expression in western blot shown on lanes labelled sh1 and sh2 (Figure 3a) as well as in immunohistochemistry (Figure 3b) in comparison to NT tumors and in tumors transduced with a control vector. Tumor lysates were analyzed for MxA, a central interferon-responsive gene,20 in order to exclude this interferon response pathway as a possible mediator of the differences in tumor sizes. None of the tumor lysates expressed MxA when compared to the extracts from positive control cells (Figure 3a).
Figure 3.
Expression of Src and interferon-responsive MxA in mouse tumors. (a) Western blot was used to analyze Src and MxA protein expression from tumor lysates. β-Actin was used as a loading control. Normalization of Src by actin is shown below each lane of the corresponding blot. Lanes: NT1-2 = nontransduced tumors, C6−C7 = control virus transduced tumors, sh1 = tumor transduced with shRNA1 against Src, sh2 = tumor transduced with shRNA2 against Src, +24 and +48 = interferon-induced cell lysates collected at 24 and 48 hours postinduction (positive controls). (b) Immunostaining against Src, 200× magnification, scale = 100 µm. Ctrl, transduced with a control vector expressing shRNA against luciferase; NT, nontransduced; sh1, shRNA sequence 1 against Src; sh2, shRNA sequence 2 against Src; shRNA, small hairpin RNA.
Tumor vascularity and proliferation
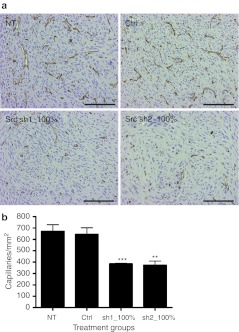
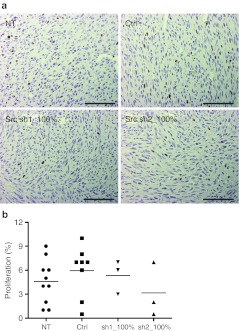
Tumor capillaries were visualized using an anti-CD34 staining for endothelial cells. Microvessel density, namely the number of capillaries/mm2, was significantly reduced in both Src shRNA groups 5 and 8 (Figure 4). Capillaries in Src-inhibited tumors appeared smaller in size and stained less intensively compared to NT and control vector-transduced tumors. Tumor sections were also stained for anti-α-smooth muscle actin for the detection of pericytes to distinguish whether the composition of vessel wall was different after the treatments. Only a few pericyte-covered vessels were detected at the tumor borders and there were very few positive vessels in any of the study groups (data not shown). Tumors were also analyzed for lymphatic vessels, which showed no consistent differences after Src inhibition (data not shown). No significant differences were found in tumor proliferation indexes between the different treatment groups (Figure 5) although the group 8 with all cells transduced with Src shRNA2 showed a decreased trend.
Figure 4.
Tumor vessels. (a) CD34 immunostaining for capillary endothelium and (b) capillary quantification (capillaries/mm2) in mouse tumors, 200× magnification, scale = 100 μm. **P < 0.01; ***P < 0.001 versus nontransduced tumors, error bars = SEM. Ctrl, transduced with a control vector expressing shRNA against luciferase; NT, nontransduced; sh1, shRNA sequence 1 against Src; sh2, shRNA sequence 2 against Src.
Figure 5.
Tumor proliferation. (a) Ki-67 immunostaining for tumor proliferation and (b) proliferation indexes in mouse tumors, 200× magnification, scale = 100 μm. Indexes as percentage (%) of proliferating cells from all the cells in different treatment groups are shown. Ctrl, transduced with a control vector expressing shRNA against luciferase; NT, nontransduced; sh1, shRNA sequence 1 against Src; sh2, shRNA sequence 2 against Src.
Rat malignant glioma model
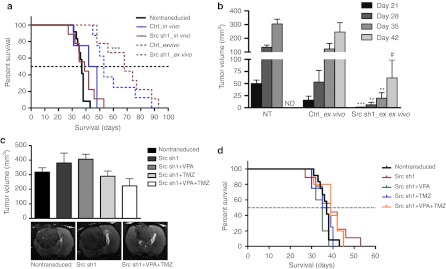
Both in vivo and ex vivo gene transduction approaches were studied in an intracranial rat glioma model. Ex vivo administrated Src shRNA1 gave rats a significant (***P < 0.001) survival benefit in comparison to rats having NT tumors (Figure 6a). Median survival for Src shRNA1 rats was 68 days and for rats with NT tumors 37 days. Overall, rats treated with ex vivo gene transductions had increased survival compared to in vivo-treated rats. Tumor volumes were measured every week by MRI. Src inhibited tumors (Src sh1_ex vivo) were significantly smaller than NT or control virus transduced (Ctrl_ex vivo) tumors at corresponding timepoints (Figure 6b). For example at day 35 mean tumor volumes were 20 mm3 (Src sh1), 123 mm3 (Ctrl), and 305 mm3 (NT). At that timepoint Src sh1 transduced tumors were 6.6% of the corresponding NT tumors. Gene transfer efficacy was also studied in order to elucidate survival differences between in vivo and ex vivo approaches (Supplementary Materials and Methods). In vivo gene transfer efficacy was 6.0% 5 days after gene transfers (day 19 postimplantation). Corresponding ex vivo transduction efficacy was 9.7% 20 days after the tumor cell implantation (Supplementary Figure S1).
Figure 6.
Rat glioma survival and tumor volumes. (a) Survival proportions in days after tumor inoculation, ***P < 0.001 Src sh1_ex vivo versus nontransduced. (b) Ex vivo tumor volumes at magnetic resonance imaging (MRI) follow-up postinoculation, **P < 0.01; ***P < 0.001 versus nontransduced tumors, #P < 0.05 versus control vector-transduced tumors. ND, not detected (no survivors left). (c) Combination treatment tumor volumes at day 35 postinoculation and representative MRI data (9.4 T MRI scanner) for some of the groups. (d) Combination treatment survival proportions in days after tumor inoculations. Error bars = SEM. Ctrl, transduced with a control vector expressing shRNA against luciferase; NT, nontransduced; sh1, shRNA sequence 1 against Src; TMZ, Temozolomide; VPA, sodium valproate.
Src shRNA treatment was also combined with sodium valproate (VPA) and/or temozolomide (TMZ) treatments. We have previously shown that VPA enhances gene expression intensity followed by lentiviral transduction (data not shown). Combinations of Src shRNA with VPA and/or TMZ were first tested in vitro by MTS viability assay (Supplementary Materials and Methods). Full combination of Src sh1, VPA, and TMZ decreased cell survival the most compared to single or dual treatments (Supplementary Figure S2). Full combination of all three treatments resulted in cell viability of 8.1% of Src shRNA alone treatment. In vivo full combination of Src sh1, VPA, and TMZ had a clearly reduced tumor volume being 59% of Src sh1 alone (Figure 6c). TMZ added to Src sh1 was able to decrease mean tumor volume to 76% of Src sh1 alone. Addition of VPA to Src sh1 did not differ from Src sh1 alone. Representative day 35 MRI data is shown for some of the treatment groups (Figure 6c). Survival of rats was also studied after combination treatment. There was no survival benefit with combination treatment in comparison to single or dual treatments (Figure 6d).
Discussion
Src was the first kinase to be characterized in late 1970s21 and the knowledge about it has vastly expanded since then.11,22 It is crucial for various cellular responses downstream of several important growth factor receptors.9,10 These properties make its inhibition a potential cancer therapy approach. On the other hand, these same properties make it a less desirable target elsewhere in the body when considering potential adverse effects of systemically administrated treatments. Importantly, a need for high systemic concentrations in order to reach effective treatment levels in a target tissue is likely to cause unwanted side effects.23 By using local gene therapy one could circumvent some systemic adverse effects.
To address these issues we studied lentiviral RNA interference-mediated inhibition of Src in order to restrict the growth of malignant glioma. For preliminary in vitro screening, human umbilical vein endothelial cells (HUVECs) were used to evaluate the functionality of the lentiviral RNA interference-mediated Src inhibition since Src has been shown to be biologically important and highly expressed in endothelial cells.10,24 Being primary cells, HUVECs also resemble in vivo conditions better than immortalized cell lines. ShRNAs targeting Src were shown to be functional and highly efficient when measured by mRNA and protein levels. According to requirements for RNA interference-based studies,25 shRNAs were further tested in suitable functionality assays, namely cell viability and tubulogenesis assays. Sequences were searched for homology to unrelated targets and an irrelevant vector-matched control shRNA against luciferase was used in every experiment. Upstream from Src are several growth factors out of which vascular endothelial growth factor mediated cell viability was shown to decrease after Src inhibition. Reduced MMP-2 levels confirmed impaired downstream signalling mediated through Src kinase.9 MMP-2 has also shown to be a crucial factor for tumor invasiveness and metastasis further emphasising the importance of its inhibition.2
In a nude mouse glioma model, both Src shRNAs were able to almost completely abolish the tumor growth when expressed by all tumor cells. Cells were not sorted by fluorescence-activated cell sorting, which resulted in the presence of some nontransduced cells. In practice, the proportions of transduced tumor cells were >97% in these groups measured by fluorescence-activated cell sorting. In mice having only 50% or 10% of tumor cells transduced, tumor growth was comparable to NT tumors. Based on these results, it could be estimated that the threshold portion of the transduced cells needed to retain the growth block was somewhere between 50% and 100% taking into account the timeline of the experiment. Slower growth and smaller volumes of the control vector-transduced tumors were likely caused by having all tumor cells transduced by the vector occupying cellular machinery and slowing down the growth. MxA, a central interferon-responsive protein, was analyzed from the mouse tumor lysates in order to exclude its influence in this approach. It is well known that double-stranded RNA and RNA interference can trigger interferon response and thus cause inefficient response to shRNA therapy.20,26 Since MxA was only expressed in positive control cell lysates, it can be concluded that neither shRNAs themselves nor lentiviral vectors induced the interferon response pathway, which could explain the differences in tumor growth. However, it is possible that some other signalling pathways not examined here could have contributed to the reduced growth rate. Other explanations for the off-target effect in shRNA approaches could be related to unintentional miRNA inhibition caused by shRNA overexpression and perturbation of gene regulation through miRNAs.27,28 If this is the case, the shRNA therapy that we are proposing could be improved by regulating the intracellular shRNA levels for example with regulated or tissue specific expression systems.27,29
Src protein expression was analyzed from mouse tumor lysates by western blot and shown to be reduced in Src-inhibited tumors. This was verified with immunohistochemistry by staining corresponding tumor sections with anti-Src antibody. Intensity of the staining was clearly reduced in Src-inhibited tumors. In Src-inhibited tumors there were significantly less vessels and they were smaller in size compared to NT and control vector-transduced tumors. These vessels were confirmed to be small capillaries based on nonexistent pericyte coverage after α-smooth muscle actin staining. Lymphatic vessels were scarce and showed no differences between different treatments. The smaller size and reduced capillary number of Src inhibited tumors is in agreement with the general notion that a tumor cannot grow bigger than ~1 mm in diameter without new capillaries to support its nutrient and oxygen demands.30 Tumor proliferation did not significantly vary between different treatments, although Src sh2_100% group showed a decreased trend. In support of this, current data from many tumor cell types and clinical trials of Src inhibitors have not shown objective responses on proliferation.31,32
The maintenance of high transduction efficacies throughout the nude mouse glioma experiment was confirmed postsacrifice by GFP fluorescence microscopy. This confirmed the stable long-term transgene expression characteristic of lentiviral vectors.33 In addition, lentiviral vectors have shown to have high tropism for neural tissues.15,16 They can be targeted or delivered locally into tumor mass therefore reducing off-target effects and systemic toxicity. One of the pitfalls of in vivo gene therapy has been relatively low transduction efficacy. This aspect needs further optimization and several strategies are currently under investigation.6,34 With lentiviral vectors, viral purification has been shown to increase transduction efficacy35,36 and is expected to cause less immune response and may therefore enable consecutive viral administrations, if necessary.
The growth deterioration following Src inhibition was further confirmed in an orthotopic rat glioma model, which has been shown to closely resemble human malignant glioma.34,37 Rats treated with ex vivo Src inhibition had significant survival benefit and decreased tumor volumes in comparison to rats having NT tumors. Similar benefit was not seen with in vivo gene transfer. In vivo gene transfer efficacy was 6.0%, which supports the observation from our mouse glioma model that over 50% transduction efficiency is needed in order to see Src-mediated inhibition of tumor growth. Ex vivo gene transfer efficacy was ~94% preimplantation. However, only 9.7% of cells were GFP-positive 20 days postimplantation. This difference is likely explained by overgrowth of nontransduced cell population and possibly immune response against nonmammalian GFP protein included in the viral construct.38 Similar reduction of gene transfer efficacy was not seen in nude mouse glioma model possibly due to the compromised immune response in these mice. To reach high gene transfer efficiency and improved treatment response, one needs to either transduce ex vivo and sort the cells to be 100% transgene positive, use replication competent vectors or systems with a bystander effect.6,39
Another way to enhance the efficacy is to combine gene therapy with standard treatments. In this study, we combined Src inhibition with TMZ and/or VPA. Rational for this was that TMZ is being currently used for standard therapy of glioblastoma multiforme and VPA (being a histone deacetylace inhibitor) has shown its potential in enhancing gene expression.40 When combined all together, we were able to clearly reduce the tumor volume to 59% of Src sh1 alone. However, this did not result in corresponding benefit in rat survival. It is known that antiangiogenic compounds can interact with other cancer drugs in several ways making it critical to evaluate dosing and scheduling in detail. Some studies have shown that pretreating with an antiangiogenic compound will normalize the vasculature and thus the chemotherapeutic agent uptake into tumor will be enhanced.41 This increase in vascular patency is transient and thus the “window of opportunity” needs to be carefully determined.
As generally acknowledged, no single gene therapy is likely to be sufficient for a complex disease like cancer.4 As a combination therapy, Src inhibition could be a useful tool, not only by directly restricting the tumor growth, but also by normalizating the tumour vasculature and controlling tumor invasiveness and thereby metastasis in tumor types prone to metastasise.31,42 As the treatment regimens are moving toward combination therapies, Src inhibition could be beneficial in several combinations. However, to improve therapeutic responses, combination of several treatments requires careful optimization of dosing, scheduling and interactions between the individual treatments.43
Materials and Methods
shRNAs and lentiviral vectors. Lentiviral vectors containing shRNA inserts against Src kinase (LV.shSRC1 and LV.shSRC2) and luciferase (LV.shGL3) as a control were selected from shRNA lentivirus library provided by the Genomics Institute of Novartis Research Foundation (San Diego). All vector constructs included a sense-loop-antisense hairpin structure driven by H1 promoter and a GFP marker gene driven by CMV promoter. Target sequences of shRNAs were: 5′-gctgttcggaggcttcaac-3′ (shSRC1), 5′-gggcctcaacgtgaagcac-3′ (shSRC2) and 5′-gacgaacacttcttcatcg-3′ (shGL3). Third generation, self-inactivating lentiviruses were prepared by standard calcium phosphate transfection method in 293T cells as described.44 Viral preps were titered in HeLa cells with biological titers ranging form 2.0 × 108 to 3.7 × 108 TU/ml.
Cell culture and viral transduction. Original screening of Src shRNAs for target inhibition efficiency was conducted in low passage HUVECs. HUVECs were isolated from umbilical cords obtained from the maternity unit of the Kuopio University Hospital, by the approval of the Kuopio University Hospital Ethics Committee. HUVECs were cultured on plastic ware coated with 0.05% gelatin/10 µg/ml fibronectin and maintained in endothelial basal medium with growth supplements (Lonza, Cologne, Germany). For glioma experiments U118MG cells (ATCC: HTB-15) were used in vitro as well as for mouse ex vivo implantations. BT4C cells45 were used in rat malignant glioma model. Glioma cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin. For MMP-2 analysis U118MG cells were serum starved (0.5% fetal bovine serum) and stimulated with 100 ng/ml epidermal growth factor (R&D Systems, Minneapolis, MN). Lentiviral transductions were done with MOI 5 for HUVECs, MOI 10 for U118MG cells and MOI 20 for BT4C cells. Basal growth medium was changed for the cells the following day. In each experiment transduction efficiency was monitored by fluorescence microscopy (Olympus IX71, Olympus, Tokyo, Japan) and fluorescence-activated cell sorter (FACScalibur; BD Biosciences, Franklin Lakes, NJ) measuring GFP.
Real-time reverse transcription-PCR. Total RNA was extracted from cells by Tri Reagent (Invitrogen, Carlsbad, CA) according to manufacturer's protocol. RNA was reverse transcribed to cDNA by M-MuLV reverse transcriptase (Fermentas, Leon-Rot, Germany). Target gene mRNA levels were measured by real-time PCR (ABI PRISM 7700 detection system; Applied Biosystems) using specific Taqman gene expression assays (Applied Biosystems, Foster City, CA) for human Src (Hs00178494_m1) and GAPDH (4333764F) for normalization.
Western blot. Cultured cells were lysed and protein concentrations measured by BCA assay (Thermo Scientific, Rockford, IL). 20–30 µg of total proteins were run on polyacrylamide gel electrophoresis and transferred on nitrocellulose membrane. Blocked blots were incubated with a primary antibody (Src: Cell Signalling Technology, Danvers, MA; MMP-2: R&D Systems Inc.; MxA: a kind gift from professor Ilkka Julkunen, The National Institute for Health and Welfare, Helsinki, Finland; β-actin: Abcam, Cambridge, UK) followed by a corresponding secondary antibody (goat anti-rabbit IgG-HRP, goat anti-mouse IgG-HRP: Thermo Fisher Scientific, Rockford, IL). Blots were visualized using ECL Plus western Blotting Detection System with Typhoon 9400 (GE Healthcare, Piscataway, NJ).
Cell viability assay. HUVECs were transduced with lentiviral vectors and 4 days post-transduction 5,000 cells were replated on 96-well plates. Cells were let to adhere and serum-deprived medium containing 0.5% fetal bovine serum was changed to the wells the following day. After 24 hours, stimulation medium containing 50 ng/ml recombinant human vascular endothelial growth factor in serum-deprivation medium was added on cells. MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrasodium bromide) assay, CellTiter-Glo Luminescent Cell Viability Assay (Promega, Madison, WI) was performed at 48 and 72 hours poststimulation according to the manufacturer's protocol.
In vitro angiogenesis. HUVECs were transduced with lentiviral constructs and 4 days post-transduction 40,000 cells were replated on 48-well plates coated with growth factor reduced Matrigel (BD Biosciences) diluted 1:1 with basal medium, all together 150 µl/well. Cells were monitored and pictures were taken during the next 24 hours with light as wells as fluorescence microscopy. Representative pictures are from 6 hours timepoint.
Mouse malignant glioma model. U118MG human malignant glioma cells were transduced with lentiviral shRNA constructs against Src and luciferase 4 days preimplantation. Transduction efficiencies were confirmed, cells were counted for viability and suspended in Optimem medium (Invitrogen). Cell suspensions were prepared to implant 250,000 cells in 50 µl volume/tumor, 2 tumors/mouse in NMRI nude mice (Taconic, Ejby, Denmark). There were five mice/group resulting in ~10 tumors/group. The treated mice were divided into eight groups as described in Table 1. In groups 3, 4, 6, and 7 cell suspensions were prepared by mixing transduced cells with nontransduced cells. Cells were injected in both flanks of sedatated mice with a 27 G needle. During the experiment tumors were weekly measured in three dimensions and volumes calculated using the formula 4/3πr3. All mice were sacrificed at 6 weeks timepoint.
Rat malignant glioma model. Rat malignant glioma cells were inoculated intracranially as described previously34 in inbred male BDIX rats (Charles River Laboratories, Lille, France). Briefly, 10,000 BT4C cells in 5 µl Optimem medium were inoculated over right corpus callosum under stereotactic guidance. Coordinates were 1 mm caudal to bregma, 2 mm right of sagittal suture, and a depth of 2.5–3.0 mm. Inoculations were done slowly over 5 minutes to avoid backflow and needle was left in place for another 5 minutes. In ex vivo transduced tumors cells were transduced 4 days preinoculation as described above. shRNA1 against Src was used in this model due to its sequence homology to rat Src mRNA. All rats were imaged with MRI at day 12 postinoculation to confirm tumor existence and in vivo gene transfers were done at days 13 and 14 as described by Tyynelä et al.37 A total volume of 20 µl of virus (LV.shSRC1 or LV.shGL3) was administrated stereotactically at multiple sites to cover the tumor area. Thereafter rats were imaged once a week for tumor growth follow-up. For rats receiving VPA subcutaneous injections of 200 mg/kg Deprakine (Sanofi Winthrop Industrie, Ambarès, France) were given twice a day for 14 days starting at day 19 postinoculation. VPA was also administered once preceeding gene transfers at days 13 and 14. For rats receiving TMZ i.p. injections of 20 mg/kg Temodal (SP Europe, Bruxelles, Belgium) were given once a day for 5 days starting at day 28 postinoculation. Temodal capsules were dissolved 1:9 in DMSO:0.9% saline to yield final concentration of 20 mg/ml. All animal procedures were approved by the Experimental Animal Committee of the University of Kuopio.
MRI.Magnetic resonance imaging (MRI) was used in rat malignant glioma model for confirmation of tumor existence (day 12 postinoculation) and weekly tumor growth follow-up after the gene transfers. For rats undergoing MRI, anesthesia was induced with 5% isoflurane gas, in a gas chamber having a 70:30% mixture of N2O:O2 and anesthesia was maintained at 1.5% isoflurane. MRI scans were acquired on a 4.7 T small animal MRI scanner (Magnex Scientific, Abington, UK) interfaced to a Varian Unity Inova (Palo Alto, CA) console or a 9.4 T small animal MRI scanner (Varian Medical Systems, Palo Alto, CA) interfaced to a Varian DirectDrive console. Noncontrast enhanced, T2-weighted fast spin-echo images of 17 coronal images (1 mm thickness, no gap between slices) were positioned to cover the tumor area in the rat brain. The details for the MRI contrast parameters for the 4.7 T MRI scanner were as follows: spin-echo preparation with 80 ms echo time and repetition time 2.5 seconds. The field of view of 40 × 40 mm with 256 × 256 data matrices resulted in pixel size of (0.16 mm)2. For the 9.4 T MRI scanner parameters were as follows: spin echo preparation with 40 ms echo time, followed a segmented fast spin echo with four echoes per excitation and echo time of 8 ms, repetition time of 3 seconds. Field of view of 30 × 30 mm with 256 × 128 data matrices (zerofilled to 256 × 256) resulted in pixel size of (0.12 mm)2. The total tumor volumes were calculated by delineating the tumor area in all the image slices using a home-built Matlab program Aedes (http://aedes.uku.fi; MathWorks, Natick, MA).
Immunohistochemistry. Isopentane-frozen tumor sections (10 µm) were used to check the maintenance of tumor transduction by GFP. PFA-fixed paraffin-embedded tumor sections (4 µm) were used for all other immunohistochemical stainings. General morphology of tumors was analyzed from hematoxylin–eosin stainings. Tumor sections were also immunostained for Src (1:50; Cell Signalling Technology), mouse CD34 (1:10; HyCult Biotechnology b.v., Uden, the Netherlands), LYVE-1 (1:1,000; ReliaTech, Braunschweig, Germany), Ki-67 (1:50; Dako, Glostrup, Denmark) and anti-Muscle Actin (HHF35) (1:50; Enzo Life Sciences, Farmingdale, NY). Control immunostainings were conducted without the primary antibody. Histological sections were assessed using Olympus AX-70 microscope (Olympus) with analySIS software (Soft Imaging System, Münster, Germany). Ki-67 quantifications were done by an experienced pathologist in a blinded-manner. For tumor capillaries, CD34 stained microscopic slides were evaluated and vascular “hot-spots” from five microscopic fields46 were counted.
Statistical analysis. All experiments were carried out with three or more replicates. Results are expressed as mean ± SEM. Statistical analyses were performed with GraphPad Prism version 4.03 (GraphPad Software, San Diego, CA). Kolmogorov–Smirnov normality test was used to assess sample normality. Kruskall–Wallis test, followed by Dunn's multiple comparison test, was used as a nonparametric test and unpaired t-test as a parametric test when relevant. P < 0.05 was considered statistically significant.
Acknowledgments
The authors thank Riina Liikala, Anne Martikainen, Anneli Miettinen, Mervi Nieminen, and Seija Sahrio for their skillful technical assistance. Doctor Johanna Närväinen (Biomedical NMR research group, A.I. Virtanen Institute for Molecular Sciences, University of Eastern Finland) is acknowledged for her assistance in MRI issues. American Cancer Society Professor of Molecular Biology, Inder Verma and the staff in his laboratory (Laboratory of Genetics, Salk Institute for Biological Studies, La Jolla, CA) are warmly acknowledged for their valuable research collaboration. This study was supported by grants from Cancer Foundation of Northern Savo, Finnish Academy, Finnish Cultural Foundation, Finnish Foundation for Cardiovascular Research, Leducq Foundation, Paavo Koistinen Foundation, Research Foundation of Orion Corporation, University of Kuopio and Ark Therapeutics Ltd. Haritha Samaranayake, Jere Pikkarainen, and Ann-Marie Määttä are employees of Ark Therapeutics Ltd. The authors declared no conflict of interest.
Supplementary Material
Transduction efficacy in rat malignant glioma model.
Combination treatment efficacy in vitro.
References
- Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, European Organisation for Research and Treatment of Cancer Brain Tumour and Radiation Oncology Groups; National Cancer Institute of Canada Clinical Trials Group et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- Louis DN. Molecular pathology of malignant gliomas. Annu Rev Pathol. 2006;1:97–117. doi: 10.1146/annurev.pathol.1.110304.100043. [DOI] [PubMed] [Google Scholar]
- Sathornsumetee S, Reardon DA, Desjardins A, Quinn JA, Vredenburgh JJ., and, Rich JN. Molecularly targeted therapy for malignant glioma. Cancer. 2007;110:13–24. doi: 10.1002/cncr.22741. [DOI] [PubMed] [Google Scholar]
- Rich JN., and, Bigner DD. Development of novel targeted therapies in the treatment of malignant glioma. Nat Rev Drug Discov. 2004;3:430–446. doi: 10.1038/nrd1380. [DOI] [PubMed] [Google Scholar]
- Pulkkanen KJ., and, Yla-Herttuala S. Gene therapy for malignant glioma: current clinical status. Mol Ther. 2005;12:585–598. doi: 10.1016/j.ymthe.2005.07.357. [DOI] [PubMed] [Google Scholar]
- Määttä AM, Samaranayake H, Pikkarainen J, Wirth T., and, Ylä-Herttuala S. Adenovirus mediated herpes simplex virus-thymidine kinase/ganciclovir gene therapy for resectable malignant glioma. Curr Gene Ther. 2009;9:356–367. doi: 10.2174/156652309789753365. [DOI] [PubMed] [Google Scholar]
- Lawler SE, Peruzzi PP., and, Chiocca EA. Genetic strategies for brain tumor therapy. Cancer Gene Ther. 2006;13:225–233. doi: 10.1038/sj.cgt.7700886. [DOI] [PubMed] [Google Scholar]
- Olsson AK, Dimberg A, Kreuger J., and, Claesson-Welsh L. VEGF receptor signalling - in control of vascular function. Nat Rev Mol Cell Biol. 2006;7:359–371. doi: 10.1038/nrm1911. [DOI] [PubMed] [Google Scholar]
- Park CM, Park MJ, Kwak HJ, Lee HC, Kim MS, Lee SH.et al. (2006Ionizing radiation enhances matrix metalloproteinase-2 secretion and invasion of glioma cells through Src/epidermal growth factor receptor-mediated p38/Akt and phosphatidylinositol 3-kinase/Akt signaling pathways Cancer Res 668511–8519. [DOI] [PubMed] [Google Scholar]
- Werdich XQ., and, Penn JS. Src, Fyn and Yes play differential roles in VEGF-mediated endothelial cell events. Angiogenesis. 2005;8:315–326. doi: 10.1007/s10456-005-9021-x. [DOI] [PubMed] [Google Scholar]
- Summy JM., and, Gallick GE. Treatment for advanced tumors: SRC reclaims center stage. Clin Cancer Res. 2006;12:1398–1401. doi: 10.1158/1078-0432.CCR-05-2692. [DOI] [PubMed] [Google Scholar]
- Lund CV, Nguyen MT, Owens GC, Pakchoian AJ, Shaterian A, Kruse CA.et al. (2006Reduced glioma infiltration in Src-deficient mice J Neurooncol 7819–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criscuoli ML, Nguyen M., and, Eliceiri BP. Tumor metastasis but not tumor growth is dependent on Src-mediated vascular permeability. Blood. 2005;105:1508–1514. doi: 10.1182/blood-2004-06-2246. [DOI] [PubMed] [Google Scholar]
- Weis S, Cui J, Barnes L., and, Cheresh D. Endothelial barrier disruption by VEGF-mediated Src activity potentiates tumor cell extravasation and metastasis. J Cell Biol. 2004;167:223–229. doi: 10.1083/jcb.200408130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer O., and, Verma IM. Applications of lentiviral vectors for shRNA delivery and transgenesis. Curr Gene Ther. 2008;8:483–488. doi: 10.2174/156652308786848067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consiglio A, Gritti A, Dolcetta D, Follenzi A, Bordignon C, Gage FH.et al. (2004Robust in vivo gene transfer into adult mammalian neural stem cells by lentiviral vectors Proc Natl Acad Sci USA 10114835–14840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanotto D., and, Rossi JJ. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457:426–433. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K., and, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE., and, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Rautsi O, Lehmusvaara S, Salonen T, Häkkinen K, Sillanpää M, Hakkarainen T.et al. (2007Type I interferon response against viral and non-viral gene transfer in human tumor and primary cell lines J Gene Med 9122–135. [DOI] [PubMed] [Google Scholar]
- Oppermann H, Levinson AD, Varmus HE, Levintow L., and, Bishop JM. Uninfected vertebrate cells contain a protein that is closely related to the product of the avian sarcoma virus transforming gene (src) Proc Natl Acad Sci USA. 1979;76:1804–1808. doi: 10.1073/pnas.76.4.1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez RH, Kantarjian HM., and, Cortes JE. The role of Src in solid and hematologic malignancies: development of new-generation Src inhibitors. Cancer. 2006;107:1918–1929. doi: 10.1002/cncr.22215. [DOI] [PubMed] [Google Scholar]
- Yuan SY. Protein kinase signaling in the modulation of microvascular permeability. Vascul Pharmacol. 2002;39:213–223. doi: 10.1016/s1537-1891(03)00010-7. [DOI] [PubMed] [Google Scholar]
- Gavard J, Patel V., and, Gutkind JS. Angiopoietin-1 prevents VEGF-induced endothelial permeability by sequestering Src through mDia. Dev Cell. 2008;14:25–36. doi: 10.1016/j.devcel.2007.10.019. [DOI] [PubMed] [Google Scholar]
- Whither RNAi. Nat Cell Biol. 2003;5:489–490. doi: 10.1038/ncb0603-490. [DOI] [PubMed] [Google Scholar]
- Bridge AJ, Pebernard S, Ducraux A, Nicoulaz AL., and, Iggo R. Induction of an interferon response by RNAi vectors in mammalian cells. Nat Genet. 2003;34:263–264. doi: 10.1038/ng1173. [DOI] [PubMed] [Google Scholar]
- Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR.et al. (2006Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways Nature 441537–541. [DOI] [PubMed] [Google Scholar]
- Khan AA, Betel D, Miller ML, Sander C, Leslie CS., and, Marks DS. Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat Biotechnol. 2009;27:549–555. doi: 10.1038/nbt.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjunath N, Wu H, Subramanya S., and, Shankar P. Lentiviral delivery of short hairpin RNAs. Adv Drug Deliv Rev. 2009;61:732–745. doi: 10.1016/j.addr.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain RK, di Tomaso E, Duda DG, Loeffler JS, Sorensen AG., and, Batchelor TT. Angiogenesis in brain tumours. Nat Rev Neurosci. 2007;8:610–622. doi: 10.1038/nrn2175. [DOI] [PubMed] [Google Scholar]
- Brunton VG., and, Frame MC. Src and focal adhesion kinase as therapeutic targets in cancer. Curr Opin Pharmacol. 2008;8:427–432. doi: 10.1016/j.coph.2008.06.012. [DOI] [PubMed] [Google Scholar]
- Huang F, Reeves K, Han X, Fairchild C, Platero S, Wong TW.et al. (2007Identification of candidate molecular markers predicting sensitivity in solid tumors to dasatinib: rationale for patient selection Cancer Res 672226–2238. [DOI] [PubMed] [Google Scholar]
- Mäkinen PI, Koponen JK, Kärkkäinen AM, Malm TM, Pulkkinen KH, Koistinaho J.et al. (2006Stable RNA interference: comparison of U6 and H1 promoters in endothelial cells and in mouse brain J Gene Med 8433–441. [DOI] [PubMed] [Google Scholar]
- Sandmair AM, Turunen M, Tyynelä K, Loimas S, Vainio P, Vanninen R.et al. (2000Herpes simplex virus thymidine kinase gene therapy in experimental rat BT4C glioma model: effect of the percentage of thymidine kinase-positive glioma cells on treatment effect, survival time, and tissue reactions Cancer Gene Ther 7413–421. [DOI] [PubMed] [Google Scholar]
- Lesch HP, Laitinen A, Peixoto C, Vicente T, Makkonen KE, Laitinen L.et al. (2011Production and purification of lentiviral vectors generated in 293T suspension cells with baculoviral vectors Gene Ther 18531–538. [DOI] [PubMed] [Google Scholar]
- Yamada K, McCarty DM, Madden VJ., and, Walsh CE. Lentivirus vector purification using anion exchange HPLC leads to improved gene transfer. BioTechniques. 2003;34:1074–8, 1080. doi: 10.2144/03345dd04. [DOI] [PubMed] [Google Scholar]
- Tyynelä K, Sandmair AM, Turunen M, Vanninen R, Vainio P, Kauppinen R.et al. (2002Adenovirus-mediated herpes simplex virus thymidine kinase gene therapy in BT4C rat glioma model Cancer Gene Ther 9917–924. [DOI] [PubMed] [Google Scholar]
- Stripecke R, Carmen Villacres M, Skelton D, Satake N, Halene S., and, Kohn D. Immune response to green fluorescent protein: implications for gene therapy. Gene Ther. 1999;6:1305–1312. doi: 10.1038/sj.gt.3300951. [DOI] [PubMed] [Google Scholar]
- Marconi P, Manservigi R., and, Epstein AL. HSV-1-derived helper-independent defective vectors, replicating vectors and amplicon vectors, for the treatment of brain diseases. Curr Opin Drug Discov Devel. 2010;13:169–183. [PubMed] [Google Scholar]
- Fan S, Maguire CA, Ramirez SH, Bradel-Tretheway B, Sapinoro R, Sui Z.et al. (2005Valproic acid enhances gene expression from viral gene transfer vectors J Virol Methods 12523–33. [DOI] [PubMed] [Google Scholar]
- Jain RK. Normalization of tumor vasculature: an emerging concept in antiangiogenic therapy. Science. 2005;307:58–62. doi: 10.1126/science.1104819. [DOI] [PubMed] [Google Scholar]
- de Groot J., and, Milano V. Improving the prognosis for patients with glioblastoma: the rationale for targeting Src. J Neurooncol. 2009;95:151–163. doi: 10.1007/s11060-009-9916-2. [DOI] [PubMed] [Google Scholar]
- Ylä-Herttuala S., and, Alitalo K. Gene transfer as a tool to induce therapeutic vascular growth. Nat Med. 2003;9:694–701. doi: 10.1038/nm0603-694. [DOI] [PubMed] [Google Scholar]
- Follenzi A., and, Naldini L. HIV-based vectors. Preparation and use. Methods Mol Med. 2002;69:259–274. [PubMed] [Google Scholar]
- Laerum OD, Rajewsky MF, Schachner M, Stavrou D, Haglid KG., and, Haugen A. Phenotypic properties of neoplastic cell lines developed from fetal rat brain cells in culture after exposure to ethylnitrosourea in vivo. Z Krebsforsch Klin Onkol Cancer Res Clin Oncol. 1977;89:273–295. doi: 10.1007/BF00283783. [DOI] [PubMed] [Google Scholar]
- Gupta K, Radotra BD, Banerjee AK., and, Nijhawan R. Quantitation of angiogenesis and its correlation with vascular endothelial growth factor expression in astrocytic tumors. Anal Quant Cytol Histol. 2004;26:223–229. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transduction efficacy in rat malignant glioma model.
Combination treatment efficacy in vitro.