Abstract
The transferrin receptor, CD71, is an attractive target for drug development because of its high expression on a number of cancer cell lines and the blood brain barrier. To generate serum-stabilized aptamers that recognize the human transferrin receptor, we have modified the traditional aptamer selection protocol by employing a functional selection step that enriches for RNA molecules which bind the target receptor and are internalized by cells. Selected aptamers were specific for the human receptor, rapidly endocytosed by cells and shared a common core structure. A minimized variant was found to compete with the natural ligand, transferrin, for receptor binding and cell uptake, but performed ~twofold better than it in competition experiments. Using this molecule, we generated aptamer-targeted siRNA-laden liposomes. Aptamer targeting enhanced both uptake and target gene knockdown in cells grown in culture when compared to nonmodified or nontargeted liposomes. The aptamer should prove useful as a surrogate for transferrin in many applications including cell imaging and targeted drug delivery.
Keywords: aftamer, liposome, siRNA, SNALPs, transferrin
Introduction
The transferrin receptor, CD71 (TfR), is one of the most widely targeted receptors for development of targeted cancer diagnostics and therapeutics.1 This type II transmembrane glycoprotein is responsible for cellular iron transport and is found at low levels on the surface of many normal cell types. However, the receptor is highly expressed on cells with increased proliferation rates. In particular, increased expression is observed across a wide range of cancer cells where increased expression is associated with poor prognosis.2 Perhaps most interestingly, the receptor is also expressed at high levels on the blood-brain barrier where it has been shown to be a route for ferrying cargoes across the blood-brain barrier.3
The natural ligand of this receptor, transferrin, has been used extensively for the generation of numerous targeted drug delivery strategies including the development of targeted toxins, some of which have gotten as far as clinical trials.4,5,6,7 Aside from anti-integrin binding RGD peptides, transferrin is perhaps the most commonly used targeting ligand for functionalizing nanoparticles for nanoparticle-based diagnostics and therapeutics.8,9 Nanoparticles in and of themselves have a tendency to accumulate in the fenestrations of tumors through a phenomenon known as the enhanced permeability and retention effect;10 however, greater gains, in particular extravasation and tissue penetration, can be achieved by including targeting ligands on the surface of nanoparticles. Specifically, transferrin has previously been shown to enhance the effect of nanoparticle-based siRNA delivery in vivo.11
In addition to the natural ligand, transferrin, a variety of antibodies1 and even peptides12 have been identified which target the transferrin receptor. More recently, Chen et al. reported the development of both RNA and DNA aptamers which target the murine transferrin receptor and demonstrated that a DNA aptamer-α-l-iduronidase conjugate taken up by α-L-iduronidase-deficient mouse fibroblasts cells was transported to lysosomes where it alleviated glycosaminoglycan accumulation.13 The selected aptamers were specific for the mouse receptor but were, unfortunately, composed of natural nucleotides rendering them highly susceptible to degradation by serum nucleases and thus limiting their utility.
We report here the development of nuclease-stabilized aptamers which target the human transferrin receptor and are readily internalized by human cells. Aptamers were selected using a two-stage selection scheme which combines stringent selection against the recombinant protein followed by a “functional” selection on cells grown in media.14 Using this approach, we have identified a family of aptamers which are readily and robustly internalized by a variety of human cancer lines known to express this receptor. A sequence analysis of functional clones revealed a conserved core motif from which we developed a minimized aptamer. To demonstrate their potential utility, minimized aptamers were used to functionalize siRNA-containing stable nucleic acid lipid particles (SNALPs). Anti-TfR targeted SNALPs showed enhanced SNALP uptake and target gene knockdown in cells grown in culture when compared to nonmodified SNALPs or those bearing a nontargeting aptamer control.
Results
Selection of anti-TfR aptamers
In order to identify nuclease-stabilized aptamers that target the human transferrin receptor and are readily endocytosed by cells, we used a hybrid in vitro selection protocol which combined stringent rounds of selection against recombinant protein with a function-based selection against cells. Selections were conducted using a 2′-fluoro-modified (2′F) RNA pool that contained a 50-nucleotide random sequence core flanked by primer binding sites. The 2′F modification can be readily incorporated during transcription by substituting 2′F CTP and 2′F UTP instead of the natural CTP and UTP and using the mutant T7 RNA polymerase protein Y639F.15,16 The resultant modified RNA is highly resistant to nuclease degradation and thus suitable for in vivo use.17
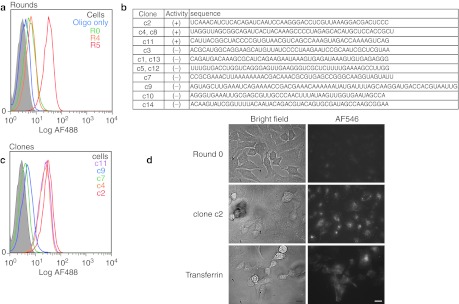
An initial four rounds of selection were conducted using a His-tagged recombinant protein produced from Sf9 cells. For the initial round, we utilized ~three copies of a library composed of ~1014 unique RNA sequences with ~2 µg recombinant hTfR immobilized on 20 µl Ni-NTA agarose. Following stringent washing, bound RNA was recovered by elution with imidazole and subsequently amplified. Following the first round, we employed a negative selection step in which the library was preincubated with Ni-NTA agarose prior to the positive selection step. For rounds 2, 3, and 4, the stringency of the selection was increased by dropping the concentration of the target protein 100-fold. However, when we labeled the Round 4 population using a fluorescently labeled oligonucleotide complementary to the 3′ constant end of the library and analyzed the population's ability to stain Jurkat cells by flow cytometry, little or no improvement in binding was observed over the starting Round 0 library (Figure 1a).
Figure 1.
Selection of aptamer which bind the human transferrin receptor. (a) Progress of the selection. The aptamer library from indicated rounds was hybridized to an AF488-labeled reverse primer and assayed by flow cytometry for binding to Jurkat cells. “Oligo only” indicates cells treated with only the AF488-labeled reverse primer. (b) Sequence of clones isolated from Round 5 of the selection. Clones capable of binding Jurkats cells are indicated with a (+) whereas those which showed no staining are indicated with a (−). Sequences are shown without the 5′ and 3′ constant regions. (c) Analysis of selected clones by flow cytometry on Jurkat cells. Aptamer labeling was performed as in a. The identity of the clones is as indicated. (d) Bright field and fluorescent microscopy images of HeLa cells following incubation with RNA from Round 0 or the full-length clone 2 (100 nmol/l). RNAs were labeled by hybridization to AF546-labeled reverse primer. For comparison, cells were also imaged following treatment with biotinylated transferrin complexed to AF546-labeled streptavidin (100 nmol/l). Bars = 10 µm.
In order to ensure the identification of aptamers which could bind hTfR in the context of the cell surface and be readily endocytosed while potentially minimizing nonspecific binding to the cell surface, following Round 4, we performed an “internalization selection” on live HeLa cells, a human cervical cancer cell line known to express TfR.14 In short, HeLa cells were incubated with the Round 4 aptamer library in media for 1 hour after which the cells were extensively washed and trypsinized. Following trypsinization, the recovered cells were treated with ribonuclease to remove any cell surface-bound RNA. Following an additional stringent wash, we used Trizol to extract total RNA from the cells, recovering any aptamers that were internalized by the cells and thus protected from nuclease treatment. The recovered RNA was subsequently amplified, and the Round 5 population was assayed by flow cytometry on Jurkat cells. As shown in Figure 1a, the Round 5 population shows almost a tenfold improvement in binding when compared to Round 4 or Round 0.
We screened 13 clones (Figure 1b) from the round 5 population by flow cytometry on Jurkat cells and identified three variants which bound robustly to Jurkat cells (Figure 1c). Of note, while some sequences appeared multiple times in the population, only one of these (c4) turned out to be functional based on our cytometric analysis. Importantly, when we incubated a fluorescently labeled aptamer (clone c2) with HeLa cells grown in culture for 1 hour, the fluorescence could be clearly seen as punctate foci in the cell cytoplasm, consistent with the biology of trafficking of the transferrin receptor.
Characterization and minimization of anti-TfR aptamers
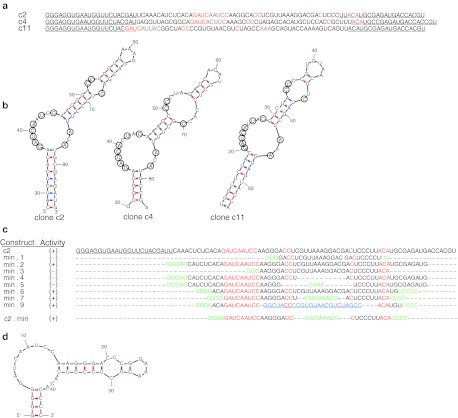
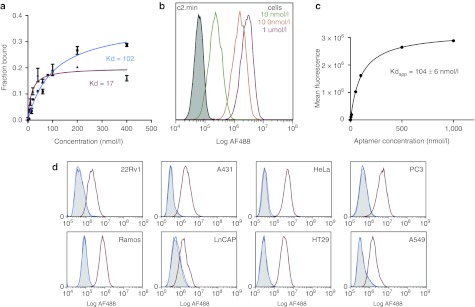
A closer inspection of the sequences of individual functional clones identified from the selection revealed short regions of sequence similarity (Figure 2a; red and blue). Strikingly, the three aptamers could be folded into a set of similar structures in which the conserved regions occupied nearly identical positions along the stem loop comprising a large asymmetric bulge of 12 nucleotides containing a conserved nonamer (GAUCAYUMC; where Y = U or C and M = A or C) and trimer (AMA) as well as a second smaller, cytosine-rich bulge situated 5–6 base pairs away (Figure 2b). Using these structures as a guide, we generated and tested truncated versions of clone c2 to investigate the role of the conserved regions (Figure 2c). As expected, deletion of the conserved 5′-nonamer (min.3) or modifications which converted the cytosine-rich bulge to a loop (min.4) or removed it entirely (min.5) resulted in a total loss of activity as determined by flow cytometry (Supplementary Figure S1). Of note, when we replaced the stem loop containing the cytosine-rich bulge from c11 on the large asymmetric bulge from c2, the clone retained full activity further supporting the requirement of both of these regions (min.9). More importantly, our analysis allowed us to identify a minimal aptamer comprised of 42 nucleotides, c2.min, in which we shortened the terminal stem formed between the 3′ and 5′ ends and replaced the terminal loop with a stable GNRA tetraloop (Figure 2d). The chemically synthesized aptamer bound recombinant protein with a Kd of 102 nmol/l, ~fivefold worse than the full length clone c2 (Kd = 17 nmol/l; Figure 3a).
Figure 2.
Comparison of anti-TfR binding clones c2, c4, and c11. (a) Sequences of clones c2, c4, and c11 with constant regions (underlined). The additional 3′ constant region used for hybridization of a fluorescent probe for flow cytometry is not shown. Invariant residues shared between the clones are colored red. (b) Functional clones share a common fold. mfold predicted structure of c2, c4, and c11. Conserved residues highlighted in red (a) have been circled. The 5′ and 3′ ends of the sequences have been removed for clarity. (c) Minimization of c2. Truncations were made by runoff transcription from the corresponding dsDNA templates. All sequences share an additional 3′ constant region used for hybridization to a fluorescent probe for flow cytometry (data not shown). The conserved regions are highlighted in red. Residues added to start transcription, force pairing or introduce a tetraloop are indicated in green. The stem loop taken from c11 is indicated in blue. (d) mfold predicted structure of minimized anti-TfR aptamer c2.min. TfR, transferrin receptor.
Figure 3.
Binding analysis of anti-TfR clone c2. (a) Binding constant for the full-length aptamer c2 (red) or the minimized variant c2.min (blue) were determined by dual filter binding assay using 32P-labled RNA. The binding constants are as indicated. (b) Representative data for the determination of the apparent binding constant for the AF488-c2.min on Jurkat cells using flow cytometry. (c) Plot of the mean fluorescence from data collected in b versus the aptamer concentration used to determine the apparent binding constant for c2.min on Jurkat cells. The binding constant is as indicated. (d) c2.min binds multiple different cancer cell lines. Flow cytometry experiments were performed at a final concentration of 100 nmol/l aptamer using either an AF488-c2.min (red) or a nonbinding control aptamer, AF488-cntrl.36 (blue). Unstained cells are shown in grey. The cell lines used for each experiment are as indicated. Each panel represents two or more experiments. Kd, binding constant; TfR, transferrin receptor.
We synthesized c2.min bearing a 3′inverted dT residue for added stability and a 5′ AlexaFluor 488 fluorescent tag to assess the molecule's ability to bind cells. When incubated with Jurkat cells in media for 1 hour, dramatic staining was observed by flow cytometry at concentrations as low as 10 nmol/l (Figure 3b). A plot of aptamer concentration versus mean fluorescence signal observed by cytometry yielded a Kdapparent on cells of 104 nmol/l, a value similar to the binding constant observed with the recombinant protein in solution (Figure 3c). Importantly, when we performed similar control experiments using a scrambled aptamer sequence, little or no background staining was observed (Figure 3c; cntrl.36).
Because the transferrin receptor is known to be overexpressed on numerous cancer cell lines, we assessed the ability of c2.min to bind different cells line. c2.min bound to every cancer cell line we tested including HeLa, 22Rv1, LnCAP, PC3, Ramos, A431, A549, and HT29 cells, as shown in Figure 3d (red). Experiments conducted with murine cell lines including mouse fibroblast 3T3 cells, mouse myoblast C2C12 cells, and T lymphocyte EL4 cells failed to show any staining, demonstrating that the aptamer is specific for the human receptor (data not shown). Importantly, no significant binding was observed when the same cell lines were tested using our control aptamer, cntrl.36 (Figure 3d; blue).
Efficient staining requires endocytosis
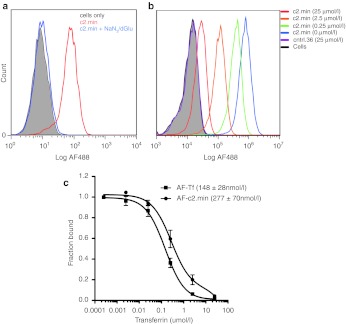
Endocytosis of the transferrin receptor is an energy-dependent process reliant on ATP.18 We therefore assessed the ability of c2.min to bind arrested cells. Of note, arrested cells (Figure 4a; blue trace) showed no staining, suggesting that the off-rate for aptamer binding is quite rapid, and it does not remain bound during the subsequent washing steps prior to cytometric analysis. No improvement in the staining of the arrested cells was observed when similar experiments were performed with the full-length aptamer clone c2 (data not shown), which displayed a lower apparent binding constant against recombinant protein. This result suggests that our selection method may have yielded “functional” aptamers more specifically tuned for internalization instead of aptamers that simply have a slow off-rate needed for high-affinity binding.
Figure 4.
c2.min and transferrin binding by flow cytometry. (a) Endocytosis is required for efficient cell staining by c2.min. Untreated Jurkat cells (red) or those arrested by treatment with a mixture of sodium azide and deoxyglucose (blue) were treated with an AF488-c2.min, for 1 hour in media at 100 nmol/l. Cells were subsequently washed and analyzed by flow cytometry. Unstained cells are shown in grey. (b) Binding and uptake of AF488-c2.min by Jurkat cells in the presence of increasing concentrations of free transferrin; 100 nmol/l aptamer was incubated with cells for 1 hour in media. The concentration of free transferrin added to the media is indicated in parentheses. Cells were washed and analyzed by flow cytometry. (c) Determination of the IC50 for binding and uptake of AF488-c2.min and DY488-Tf. Experimental conditions were as in b and described in the Methods and Materials. IC50 values are as indicated. Data represent the average of two independent trials.
C2 competes with transferrin for binding the receptor
The natural ligand for the transferrin receptor, transferrin (hTf), is present in blood at relatively high concentrations, ~25 µmol/l with ~10–50% of this in the high-affinity monoferric or diferric state.19,20 Therefore, we assessed whether or not our selected aptamer would compete with this natural ligand for receptor binding and uptake. Using flow cytometry, we assessed the uptake of AF488-labeled c2.min on Jurkat cells in the presence of increasing concentrations of diferric hTf. As shown in Figure 4b, as the concentration of diferric hTf was increased to 25 µmol/l, c2.min showed a decrease in binding with a plot of fraction aptamer bound versus the concentration of free transferrin yielding an IC50 of ~280 nmol/l (Figure 4c). A similar assay performed using fluorescently labeled iron-loaded transferrin instead of the aptamer yielded a slightly lower IC50 (~150 nmol/l) demonstrating that the aptamer binds to the receptor slightly better than the natural ligand.
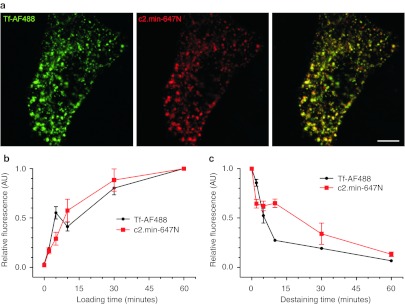
We have also investigated the uptake kinetics of hTf and of the c2.min aptamer. Cells were coincubated with AF488-labeled hTf and atto647N-labeled c2.min. Following a 30-minute incubation, cells displayed the characteristic endosomal punctuated pattern obtained with labeled transferrin. Moreover, c2.min exhibits a vast colocalization with transferrin (Figure 5a). When uptake was monitored over the course of a 60-minute period, the rates of uptake of the fluorescent cargoes showed no differences (Figure 5b). Similar experiments in which cells were first loaded with either Tf-AF488 or c2.min-647N, washed and then placed in fresh media for different time points revealed that labeled c2.min washed off more slowly than labeled hTf (Figure 5c). This finding is consistent with the relative binding constants for iron free apo-transferrin (Kd = ~4 µmol/l)21 and the aptamer (Kd = ~100 nmol/l).
Figure 5.
Labeled c2.min and Tf were followed by fluorescent microscopy. (a) Confocal images of HeLa cells treated for 30 minutes with Tf-AF488 (50 µg/ml) and c2.min-647N (250 nmol/l) show subcellular colocalization. (b) Time courses of the uptake of Tf-AF488 or c2.min-647N were monitored over 60 minutes. HeLa cells were loaded for 5 minutes with labeled c2.min or Tf, washed and transferred to fresh media for different time points. (c) The loss of fluorescence was followed for 60 minutes. Data represent the average of three independent experiments with 15–20 images per time point each. Error bars represent the SEM from three independent experiments. Bars = 5 µm. SEM, standard error of the mean; Tf, transferrin.
Enhanced delivery of siRNA using anti-TfR aptamers
In order to assess the potential to deliver molecular cargoes using anti-transferrin receptor aptamers, we generated siRNA-encapsulated liposomes containing a Cy5-labeled anti-enhanced green fluorescent protein (EGFP) siRNA and investigated gene knockdown in HeLa cells engineered to constitutively express EGFP (HeLa-EGFP).
Using a scalable, extrusion-free approach to making nucleic acid–loaded liposomes,22 we generated aptamer functionalized stable nucleic acid lipid particles (SNALPs) utilizing a lipid formulation previously shown to be effective at delivering siRNA both in mice and in nonhuman primates,23 but replacing the surface PEG-2000 molecules with a thiol-reactive PEG-2000-malemide. Following siRNA encapsulation and dialysis, the thiol-reactive liposomes were incubated with either a thiol-modified c2.min, a nonfunctional control aptamer (cntrl.36) or no aptamer at all (β-mercaptoethanol (BME)). Aptamer conjugations typically proceeded to ~90% within 4 hours as determined by gel electrophoresis (Supplementary Figures S2, S3, and S4). The liposomes were further characterized by dynamic light scattering and, consistent with previous reports, found to be ~180 nm in diameter with a polydispersity of ~20%.8,22,24 Aptamer conjugation resulted in a slight increase in liposome size (Table 1). Based on their size, the total mass of lipid used, and the concentration of aptamer used in the conjugation, we estimate that each liposome displayed ~60 aptamers per particle.
Table 1. Characterization of liposome size by dynamic light scattering.

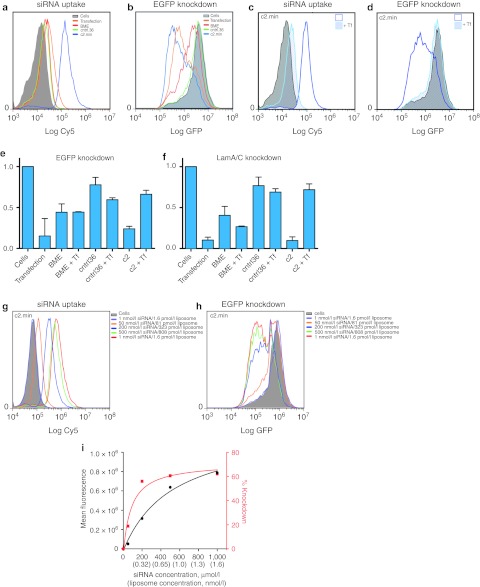
Liposome uptake (Cy5) and gene knockdown (EGFP) were assessed by flow cytometry using HeLa-EGFP cells 48 hours after treatment with aptamer-targeted or control SNALPs containing a Cy5-labeled anti-EGFP siRNA. As shown in Figure 6a, the c2.min-targeted SNALPs showed increased levels of uptake by cells when compared to both nontargeted SNALPs (BME) and those bearing the nonfunctional aptamer control (cntrl.36). More importantly, the enhanced uptake by the c2.min-conjugated SNALPs also translated to an increased level of gene knockdown, as assessed by monitoring the decrease in the expression levels of the target protein EGFP (Figure 6b).
Figure 6.
Analysis of liposome /siRNA uptake and gene knockdown using aptamer-targeted SNLAPs. (a) Uptake of SNALPs loaded with Cy5-labeled anti-EGFP siRNA and (b) knockdown of EGFP in HeLa-EGFP cells as determined by flow cytometry. Cells only (cells), targeted (c2.min), or nontargeted SNALPs (BME and cntrl.36) are as indicated. Transfection of the same Cy5-labeled anti-EGFP siRNA using HiPerfect was included for comparison (Transfection). Cells were treated as indicated for 24 hours, after which the media was changed. Cells were analyzed 48 hours after initial exposure to SNALPs. (c) Uptake of Cy5-labeled siRNA and (d) knockdown of EGFP in HeLa-EGFP cells in the absence or presence of 25 µmol/l diferric human transferrin as determined by flow cytometry. Similar experiments using nontargeted SNALPs (BME and cntrl.36) can be found in Supplementary Figure S5. Data are representative of a minimum of three independent experiments. (e,f) qPCR analysis monitoring mRNA levels in HeLa-EGFP cells treated with SNALPs containing an anti-EGFP siRNA (e) or anti-Lamin A/C siRNA (f) in the absence or presence (+ Tf) of 25 µmol/l transferrin. Cells only (cells), targeted (c2.min), or nontargeted liposomes (BME and cntrl.36) are as indicated. Cells were treated as indicated for 24 hours, after which the media was changed. Cells were analyzed 48 hours after initial exposure to SNALPs. All data represent the average of two or more independent trials using independent preparations of liposomes. (g,h) Concentration dependence of c2.min-targeted uptake and gene knockdown in HeLa-EGFP cells. Experiments were conducted as described in a. Concentrations are as indicated. Untreated cells (cells) are shown in grey. Similar experiments using nontargeted SNALPs (BME and cntrl.36) can be found in Supplementary Figure S6. (i) Plot of liposome concentration versus mean fluorescence of Cy5 uptake (black) and EGFP knockdown (red). SNALP, stable nucleic acid lipid particle; Tf, transferrin.
To confirm that uptake by c2.min-targeted SNALPs was mediated by binding and endocytosis via the transferrin receptor, we performed similar experiments using HeLa-EGFP cells and SNALPs loaded with a Cy5-labeled anti-EGFP siRNA but in the presence of free transferrin. Consistent with our competition studies performed with the free aptamer (Figure 4b,c), c2.min-targeted SNALPs displayed reduced uptake when 25 µmol/l diferric transferrin was added to the cell culture media (Figure 6c). When the same cells were analyzed for EGFP knockdown, c2.min-targeted SNALPs showed a significant decrease in the level of knockdown of EGFP (Figure 6d). The added transferrin had no effect on uptake or gene knockdown when treated with nontargeted (BME) or nonfunctional aptamer controls (Supplementary Figure S5). Gene knockdown and the mechanism of uptake were further confirmed by using real-time PCR to assess the level of EGFP mRNA degradation (Figure 6e).
To ensure specificity, we performed similar experiments using an alternate siRNA which targets the housekeeping gene lamin A/C. HeLa-EGFP cells treated with SNALPs containing a Cy5-labeled anti-Lamin A/C siRNA showed similar levels of siRNA uptake but failed to show a decrease in EGFP signal (data not shown). In contrast, real-time PCR analysis of the treated cells revealed enhanced knockdown of lamin A/C mRNA when treated with c2.min-targeted SNALPs as compared with nontargeted SNALPs or those bearing the nonfunctional aptamer control (Figure 6f). Importantly, when these experiments were performed in the presence of 25 µmol/l diferric transferrin, only the samples treated with c2.min-targeted SNALPs were affected, returning to near background levels of mRNA knockdown.
Nanoparticles and liposomes often benefit from the fact that targeting ligands are displayed in a polyvalent manner which can lead to significant enhancements in binding affinity through avidity effects.24,25 Indeed, at 200 nmol/l siRNA, the concentration used in our delivery experiments, the concentration of liposomes is actually ~300 pmol/l, which is far below the observed binding constant measured for our aptamer (100 nmol/l). To explore this effect further, we performed uptake (Figure 6g) and knockdown experiments (Figure 6h) over a range of concentrations from 1 nmol/l to 1 µmol/l siRNA (1.6 pmol/l to 1.6 nmol/l liposome). Liposome binding/uptake displayed an apparent Kd of 310 ± 130 pmol/l (Figure 6i), ~300-fold less than the aptamer alone. Gene knockdown displayed an EC50 of 74 ± 64 pmol/l (Figure 6i) demonstrating that the siRNA pathway saturates long before the cell uptake pathway does. Consistent with our previous results, little background uptake and almost no change in EGFP expression is observed when similar experiments are performed with nontargeted SNALP controls (Supplementary Figure S6).
Finally, the delivery of siRNA, especially via endosomal routes has the potential to lead to nonspecific immune activation.25 Therefore, we assessed the cellular immune response to cells treated with aptamer targeted SNALPs by monitoring the expression of OAS1, CDKL2, and interferon B by real-time PCR. No increase in mRNA levels of these genes was observed indicating that aptamer-mediated SNALP delivery did not induce any adverse cellular effects (Supplementary Figures S7 and S8).
Discussion
Aptamers that target cell surface receptors have great potential for development of both targeted diagnostics and therapeutics and have been a focus of many recent selection experiments (reviewed in refs (26,27)). One very promising target for targeted delivery is the transferrin receptor, a transmembrane protein that is highly overexpressed across a number of different cancer lines. Indeed, CALAA-0, a targeted siRNA nanoparticle formulation which utilizes the natural ligand for this receptor, transferrin, has previously been shown to function in humans28,29 and is currently undergoing clinical trials.30 The transferrin receptor has also been shown to be expressed on the endothelium of the blood-brain barrier, where targeting has been demonstrated as a way to enhance delivery to the brain.3,31
While both DNA and RNA aptamers to the murine transferrin receptor have previously been reported, neither of these can be easily adapted for therapeutic or diagnostic purposes in humans, as they were selected against the mouse variant and do not cross react.13 Additionally, because these aptamers are nonmodified RNA and DNA, they are not serum stable and will require additional modifications and optimizations before use in vivo. In experiments conducted in our laboratory, a FITC-labeled version of the anti-murine transferrin receptor DNA aptamer did not stain murine fibroblasts (3T3) or T cells (EL4) in media (data not shown). However, the apparent lack of observed binding might be the result of differences in valency and our cell staining approach, as Chen et al. reported staining using biotinylated aptamers and fluorescently labeled streptavidin.
We have generated nuclease-stabilized aptamers which bind the human transferrin receptor and are readily internalized by cells, thereby providing a means to specifically deliver cargoes to human cells which express this receptor. To do this, we utilized a two-stage selection strategy in which we first enriched a nuclease-stabilized RNA library for molecules which could bind to recombinant protein. We then enriched for molecules which had the desired function—namely, to bind the receptor on the surface of cells grown in tissue culture and to be internalized. Analysis of functional clones from the 5th round of the selection allowed for the identification of a core anti-TfR binding motif and a minimized aptamer, c2.min, that is specific for the human receptor and is robustly endocytosed by multiple cancer cell lines grown in culture. Of note, inhibition of endocytosis resulted in almost a complete loss of cell staining as determined by flow cytometry (Figure 4b) suggesting that the off-rate for bound aptamers is relatively fast and that surface-bound molecules are lost during sample workup. This unexpected result may, in fact, be a direct consequence of the selection method which we employed. That is, by performing a functional selection for uptake, rather than just binding, we appear to have enriched for molecules which likely would not have been identified as they would have been lost from the cell surface during the subsequent wash steps of the selection process.
Because the natural ligand for the transferrin receptor, transferrin, is present in blood at the relatively high concentration of ~25 µmol/l, we assessed whether our selected aptamers would compete with this ligand for binding. In serum, transferrin exists in three major forms: the low affinity apo-form (Kd = ~4 µmol/l) and the high-affinity monoferric (Kd = ~200 nmol/l) and diferric forms (Kd = ~10 nmol/l).21 These high affinity forms of the protein are typically present at levels ranging from ~ 2 to10 µmol/l depending on the level of iron in the blood.19,20 In our titration experiments in which we utilized the high-affinity diferric transferrin as a competitor, significant aptamer binding and uptake were observed even at superphysiological concentrations of free ligand. When compared to fluorescently labeled transferrin, our aptamer displayed a more favorable IC50 (~twofold) than the natural ligand suggesting that these molecules would serve as superior in vivo targeting agents. However, because our aptamer does not bind the mouse receptor, care will have to be taken in evaluating this aptamer in vivo using human xenograft tumor models, as the aptamer will not likely bind to any tissues other than the tumor. Additionally, in vivo results may be further complicated by the fact that mouse transferrin binds the human transferrin receptor with weaker affinity and, as expected, shows a sixfold higher IC50 when utilized in competition assays with c2.min (Supplementary Figure S9).
Because of its ability to be internalized into TfR-expressing cells, we explored the possibility of using this aptamer for delivery of potential therapeutics. Of particular interest is the delivery of siRNA, which can be designed to target almost any gene. One attractive strategy for aptamer-mediated siRNA delivery is the design of aptamer-siRNA chimeras.32,33,34,35,36,37 Therefore, based on the designs reported by Dassie et al. (A10-SWAP),37 we generated aptamer-siRNA chimeras targeting the gene PLK1 using our minimized aptamer c2.min as the targeting molecule. Unfortunately, while our aptamer-siRNA chimeras were readily taken up by HeLa, LNCaP, and 22Rv1 cells at essentially the same levels as the free aptamer, the efficacy of the delivered siRNA proved very poor with no effect on cell viability observed (data not shown). Control experiments using the constructs reported by Dassie et al., which target the prostate-specific membrane antigen on LnCAP and 22Rv1 cells (two cell lines known to express prostate-specific membrane antigen) also failed to elicit any effect on cell viability even when delivered at 100 times the published minimal effective dose (400 nmol/l). The lack of efficacy of this approach prompted us to explore alternate methods for the delivery of siRNA.
Nanoparticle-based delivery systems are gaining much interest both in the laboratory and in the clinic, and aptamers have previously been shown capable of enhancing the delivery of nanoparticle and liposomal small molecule drug formulations.38,39,40,41,42 With an interest in targeting siRNA, we tested our aptamer for its ability to enhance delivery of nanoparticle-based siRNA formulations. In particular, we chose to investigate a lipid nanoparticle formulation called SNALPs (stable nucleic acid lipid particles), which have previously been used to deliver siRNA to liver cells as well as solid tumors43 and are currently undergoing clinical trials.44,45 While these neutral liposomes have been shown to associate with Apo-E leading to efficient liver uptake, more recent work has demonstrated that the particles and their siRNA cargoes can be specifically targeted to other receptors by functionalizing the liposomal surface with ligands.46 To this end, we synthesized maleimide functionalized SNALPs using an extrusion-free approach.47 This yielded liposomes approximately ~200 nm in diameter which we subsequently derivatized with a thiol-modified, minimized anti-TfR aptamer, c2.min. While the utility of liposomes this size may be somewhat limited in vivo, they serve as a convenient platform to demonstrate the potential utility of our aptamers, at least in vitro. In this regard, the ease with which aptamers can be chemically synthesized and site specifically modified makes them ideal candidates for chemical conjugation and functionalization of nanoparticles.
Anti-TfR targeted SNALPs displayed both enhanced uptake and enhanced target gene knockdown when compared to SNALPs bearing a maleimide group which had been quenched with BME or those bearing a nontargeting control aptamer (Figure 6a). SNALPs bearing the control aptamer performed worse than those quenched with BME suggesting that the negatively charged molecules on the surface of the liposome may inhibit interactions with serum ApoE and hinder uptake by this mechanism. To confirm the route of uptake of our anti-TfR targeted SNALPs, we performed experiments in the presence of 2 mg/ml human holotransferrin (diferric). As observed, with the free aptamer, at this concentration of free ligand, liposome binding was significantly decreased as was the observed gene knockdown, confirming the link between receptor targeting and the observed enhanced function.
In summary, by combining a “traditional” selection approach using recombinant protein with a functional selection for internalization, we have identified aptamers that bind the human transferrin receptor and are readily internalized by cells. We envision that this hybrid selection approach may prove useful for other cell surface targets which are known to be internalized or where the desired function is aptamer-induced internalization. Perhaps more importantly, using a minimized anti-TfR aptamer identified by this selection approach, we generated aptamer-targeted SNALPs and have shown that receptor targeting enhances cell uptake and more importantly the efficiency of gene knockdown on cells grown in tissue culture. Aptamer-targeted liposomes may therefore play an important role in the future development of nanoparticle-based siRNA therapies.
Materials and Methods
Cell lines and tissue culture. All cells were obtained from the American Tissue Culture Collection (ATTC, Manassas, VA). HeLa, A431, and HT29 cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS) (Invitrogen). Jurkat, Ramos, 22Rv1, and LnCap cells were cultured in RPMI-1640 (Invitrogen) supplemented with 10% FBS. PC3 and A549 cells were cultured in F12K (Invitrogen) supplemented with 10% FBS. All cells were grown at 37 °C with 5% CO2 and 99% humidity. Sf9 insect cells were grown at 27 °C in Grace's Insect Media (Invitrogen).
Protein production and purification. The plasmid pAcGP67A-TfR (Addgene, Cambridge, MA) was used to express human transferrin receptor (hTfR) in insect cells (Sf9). The His-tagged protein was purified on Ni-NTA agarose, dialyzed to remove imidazole and concentrated to ~1 mg/ml for storage.
Selection of nuclease-stabilized anti-TfR aptamers. The sequence of the N50 library used for selection was 5′GGGAGGTGAATGGTTCTACGAT-N50- TTACATGCGAGATGACCACGTAATTGAATTAAATGCCCGCCATGACCAG-3′. The single-stranded DNA library was synthesized such that N regions contained an equal probability of containing A, T, G, or C, as previously described.48 Following deprotection, the library was gel purified by denaturing (7 mol/l urea) gel electrophoresis on an 8% polyacrylamide gel. The single-stranded DNA library was amplified by PCR to generate double-stranded DNA bearing a T7 promoter. The double-stranded pool was transcribed in vitro using the Y639F mutant of T7 RNA polymerase15,16 and 2′-fluoro (2′F) pyrimidines. The RNA was purified on a denaturing (7 mol/l urea) 8% polyacrylamide gel.
For the initial round of selection, we utilized ~3 copies of a library composed of ~1014 unique RNA sequences. Prior to each round, the library was thermally equilibrated in 30 µl HBSS (Hank's Buffered Saline Solution; Invitrogen) for 3 minutes at 70 °C and allowed to cool on the benchtop for at least 15 minutes. The sample was subsequently supplemented with 0.1% bovine serum albumin, 1 µg/µl ssDNA and tRNA in a final volume of 50 µl of HBSS.
The thermally equilibrated aptamer library was incubated with 2 µg of recombinant hTfR immobilized on 20 µl of Ni-NTA agarose. Following a 30-minute incubation at room temperature, the resin was washed five times with 500 µl of HBSS, and then protein-bound RNA was eluted by the addition of 400 µl HBSS containing 200 mmol/l imidazole. Eluted RNA was recovered by ethanol precipitation, reverse transcribed, PCR amplified and retranscribed into RNA for the subsequent round. Following the first round, we employed a negative selection step in which the library was preincubated with 200 µl of Ni-NTA agarose prior to the positive selection step. For rounds 2, 3, and 4, we utilized 5 µg of RNA, and the stringency of the selection was increased by dropping the number of target protein by 100-fold. This was achieved by resuspending 2 µg of protein immobilized on 20 µl of Ni-NTA agarose in 200 µl of HBSS and using 2 µl of the well-mixed slurry. For these rounds, washing was facilitated by using a 0.45-micron spin filter to capture the trace amount of Ni-NTA resin. All incubations and washes (6 × 400 µl) were performed at 37 °C.
For Round 5, we performed an “internalization selection” using HeLa cells. To ensure that selected aptamers could easily be assayed by flow cytometry, the pool was combined with a 1.5-fold molar excess of reverse primer bearing a 5′ fluorescent dye (AF488-CTGGTCATGGCGGGCATTTAATTC). Following thermal equilibration in 50 µl of HBSS, the library was added to one well of a 24-well plate containing ~105 adherent HeLa cells in 300 µl DMEM supplemented with 10% FBS and 1 mg/ml tRNA and ssDNA (Sigma, St. Louis, MO) as blocking agents. Following a 1-hour incubation at 37 °C, the cells were washed three times with 1 ml HBSS containing 0.1% sodium azide, once with 1 ml cold 200 mmol/l glycine and 150 mmol/l NaCl at pH 4, and three more times with HBSS. Cells were then lifted with 500 µl trypsin-EDTA containing 0.1% sodium azide, removed from the plate, washed with 1 ml HBSS, and resuspended in 100 µl HBSS containing 5 µl Riboshredder RNAse cocktail (Epicentre, Madison, WI). Following a 5-minute incubation at room temperature, the cells were washed an additional three times with 1 ml of HBSS, and the total cellular RNA was recovered using Trizol extraction according to manufacturer's protocol (Invitrogen). The recovered RNA was reverse transcribed, amplified by PCR, and transcribed back into RNA.
Minimized aptamers were generated by runoff transcription using the Y639F RNA polymerase and 2′-fluoro (2′F) pyrimidines. The RNA was purified on a denaturing (7 mol/l urea) 8% polyacrylamide gel. The minimized 2′F RNA variants all bore the same 3′ constant region (GAATTAAATGCCCGCCATGACCAG), which allow for hybridization of reverse primer bearing a 5′ fluorescent dye. Binding assays were conducted using flow cytometry as described above.
Chemical synthesis of RNA aptamers. The minimized aptamer was synthesized in our laboratory on an Expedite 8909 DNA synthesizer (Applied Biosystems, Carlsbad, CA) using 2′-fluoro-deoxycytidine and 2′-fluoro-deoxyuridine phosphoramidites (Metkenin, Kuusisto, Finland). Unless noted otherwise, all reagents were purchased from Glen Research (Sterling, VA). The aptamer was synthesized bearing a 5′ thiol modification using a thiol-modifier C6 S-S phosphoramidite and a 3′ inverted dT residue for added serum stability. The sequences of the minimized aptamer, c2.min, and a nonbinding aptamer, cntrl.36, were: 5SGGGGGAUCAAUCCAAGGGACCCGGAAACGCUCCCUUACACCCCt and 5SGGCGUAGUGAUUAUGAAUCGUGUGCUAAUACACGCCt, respectively, where “t” is a 3′inverted dT and “5S” is the 5′ thiol. All aptamers were synthesized with the final dimethoxytrityl group on to facilitate purification. Following deprotection, aptamers were purified by reversed-phase HLPC on a 10 × 50 mm Xbridge C18 column (Waters, Milford, MA) using a linear gradient of acetonitrile in 0.1 mol/l triethylammonium acetate at pH 7.0.
Thiolated aptamers were used to generate the AlexaFluor488 (AF488)- or Atto-647-labeled aptamers used in cytometry or microscopy. Labeling was performed using AF488-C5-malemide (Invitrogen) or Atto-647-malemide (ATTO-TEC GmbH, Siegen, Germany) as follows: 10 nmoles thiolated aptamer was reduced using 10 mmol/l tricarboxyethylphosphine in 100 µl of 0.1 mol/l TEAA. Samples were heated at 70 °C for 3 minutes followed by incubation at room temperature for 1 hour. The reduced aptamers were desalted using Biospin 6 columns (BioRad, Hercules, CA) into phosphate-buffered saline (PBS) supplemented with 50 mmol/l phosphate pH 7.5. To this, 10 µl of dimethyl sulfoxide was added containing the fluorescent maleimide. Following an overnight reaction at 4 °C, the aptamer was recovered by ethanol precipitation, resuspended in PBS, and desalted an additional time using a Biospin 6 column. Dye to aptamer ratios were determined at 260 and 650 nm and were typically ~1. The absence of free dye in the final product was confirmed by reversed-phase HLPC.
Aptamer binding by flow cytometry. Aptamer binding and uptake was assessed by flow cytometry. Rounds from each selection or isolated clones were first hybridized to the AF488-labeled reverse primer complementary to the 3′ end of the library. Rounds or individual aptamers were incubated at 1 µmol/l with 1.5 µmol/l biotinylated oligonucleotide in Dulbecco's phosphate-buffered saline (DPBS) heated to 70 °C for 3 minutes and then allowed to cool on the bench top for 15 minutes. Following hybridization, the aptamers were added to cells in the appropriate media supplemented with 1 µg/ml tRNA and sheared salmon sperm DNA (ssDNA) at final concentration of 100 nmol/l.
For assays conducted on suspension cells, ~150,000 cells were incubated in a final reaction volume of 50 µl. For adherent cells, ~100,000 cells were incubated per well in a 24-well plate in 100 µl final volume. Unless otherwise noted, rounds/aptamers were incubated with cells for 1 hour at 37 °C. Following incubation, adherent cells were trypsinized with 100 µl trypsin-EDTA for 5 minutes. The reaction was quenched by the addition of 1 ml fluorescence-activated cell sorting (FACS) buffer, and the cells were pelleted and washed an additional time with FACS buffer before being resuspended in 500 µl FACS buffer containing 2 µg/ml 7-aminoactinomycin D stain (7AAD) to exclude dead cells in the analysis. Flow cytometry was performed on a FACScan or FACScalibur flow cytometer (Beckton Dickinson, Franklin Lakes, NJ).
Fluorescence microscopy. For microscopic analysis of Round 0 and full-length c2 on HeLa cells, RNA was labeled by hybridization to an AF546-labeled oligonucleotide as described above for the flow cytometry experiments. As a control, 1 µmol/l biotinylated transferrin (Invitrogen) was preincubated with 1 µmol/l AF546-labeled streptavidin (Invitrogen). HeLa cells were grown as adherent monolayers in eight-chamber glass slide systems (Labtek; Nunc Nalgene International, Rochester, NY). Thirty minutes prior to aptamer treatment, cells were blocked with 0.1 µg/ml tRNA and ssDNA in DMEM supplemented with 10% FBS at 37 °C. Labeled Round 0, c2 or transferrin was added to the media to a final concentration of 100 nmol/l, and the samples were incubated at 37 °C for 1 hour, after which the media was changed, and the cells were imaged on an Olympus IX71 inverted microscope.
Colocalization images (Figure 5a) of hTf-AF488 and c2.min-647N were obtained using a true confocal scanner SP5 fluorescence microscope from Leica Microsystems GmbH, with a 1.4 N.A., ×100 objective (Leica Microsystems GmbH). HeLa cells were exposed simultaneously to Tf-AF488 (50 µg/ml) and c2.min-647N (250 nmol/l) for 30 minutes in Ringer buffer containing 10 µg/ml each of yeast tRNA and ssDNA as blocking agents. Coverslips were then taken out, washed thoroughly with Ringer buffer, and placed in ice-cold paraformaldehyde (4%) for 30 minutes. Samples were quenched for 10 minutes with PBS containing 100 mmol/l of glycine and NH4Cl. Finally, cells were mounted in Mowiöl.
Uptake and washout experiments of transferrin or aptamer were imaged with an Olympus IX71 microscope equipped with 1.45 NA/×100 objective. Images were captured with an Olympus F-View II CCD camera (all from Olympus, Hamburg, Germany). For uptake experiments, HeLa cells were washed in Ringer buffer (124 mmol/l NaCl, 5 mmol/l KCl, 2 mmol/l CaCl2, 1 mmol/l MgCl2, 30 mmol/l D-glucose, and 25 mmol/l HEPES, pH 7.4) and exposed to a mixture of Tf-AF488 (50 µg/ml) and c2.min-647N (250 nmol/l) prepared in Ringer buffer containing 10 µg/ml each of yeast tRNA and ssDNA as blocking agents. After the specified times, coverslips were washed thoroughly, fixed in ice-cold paraformaldehyde (4%) for 30 minutes and then quenched for 10 minutes with PBS containing 100 mmol/l of glycine and NH4Cl. Finally, cells were mounted in Mowiöl.
For monitoring the loss of fluorescence, HeLa cells were washed in Ringer buffer and exposed for 5 minutes to a mixture of Tf-AF488 (50 µg/ml) or c2.min-647N (250 nmol/l) prepared in Ringer buffer containing 10 µg/ml each of yeast tRNA and ssDNA as blocking agents. Cells were then briefly washed with a large volume (100 ml) of Ringer buffer and placed back into their complete medium inside the incubator (37 °C and 5% CO2). After the specified times, coverslips were taken out, washed with Ringer buffer, and placed in ice-cold paraformaldehyde (4%) for 30 minutes. Samples were quenched for 10 minutes with PBS containing 100 mmol/l of glycine and NH4Cl. Finally, cells were mounted in Mowiöl.
Graphs represent the average data obtained from three independent experiments with 15–20 images per time point each. Error bars represent standard error of the mean (SEM) from three independent experiments.
Determination of binding constants. Binding constants were determined by nitrocellulose filter binding assays.49 Aptamers were radiolabeled using T4 PNK (Optikinase; USBiologicals, Swampscott, MA) and γ-32P-ATP (Perkin Elmer, Waltham, MA) following standard protocols. The full-length c2 aptamer was generated by transcription as described above, and the terminal triphosphate was removed by treatment with alkaline phosphatase (FastAP; Fermentas, Glen Burnie, MD) prior to kinasing.
For each assay, the aptamer was thermally equilibrated in tissue culture grade DPBS containing Mg2+ and Ca2+ (Invitrogen) by heating at 70 °C for 3 minutes followed by incubation at room temperature for 15 minutes. Ten fmoles of radiolabeled aptamer was combined with varying concentrations of protein in a total volume of 50 µl in DPBS. Binding was allowed to come to equilibrium for 30 minutes at room temperature, and bound species were partitioned from unbound species by passing through nitrocellulose and nylon filters under vacuum followed by three successive washes with 500 µl DPBS. Filters were exposed to phosphor screens overnight which were then imaged using a Storm Molecular Imager Phosphorimager (GE Healthcare, Piscataway, NJ). The fraction of bound aptamer was determined using the ImageQuant software. Kinetic constants were determined by plotting the fraction of bound aptamer against concentration of hTfR, and data were fit to equation (1).
where f = fraction of bound aptamer, F = maximum fraction of bound aptamer, A = concentration of human transferrin receptor, and Kd = affinity dissociation constant.
For determination of apparent binding constants by flow cytometry, 100,000 Jurkat cells were incubated with increasing concentrations of AF488-labled c2.min in RPMI-1640 supplemented with 10% FBS and 1 mg/ml tRNA and ssDNA in 50 µl. Following a 1-hour incubation at 37 °C, the cells were washed twice with 1 ml FACS buffer and then resuspended in 500 µl FACS buffer containing 7AAD to exclude dead cells in the analysis. The apparent binding constant was determined from a fit of the mean fluorescence versus the aptamer concentration using the equation (1).
Transferrin competition binding assays. Assays were performed by flow cytometry on Jurkat cells. Aptamers were thermally equilibrated in tissue culture grade DPBS containing Mg2+ and Ca2+ by heating at 70 °C for 3 minutes followed by incubation at room temperature for 15 minutes prior to addition to cells. For each reaction, 105 cells were preblocked by incubation in 50 µl RPMI containing 10% FBS and 1 mg/ml ssDNA and tRNA at 37 °C for 15 minutes prior the addition of 100 nmol/l labeled aptamer (c2minAF488 or cntrl.36AF488) or 100 nmol/l labeled iron-loaded transferrin (Dy488-Tf; Jackson ImmunoResearch West Grove, PA) and unlabeled competitor (25 µmol/l, 2.5 µmol/l, 250 nmol/l, 25 nmol/l, or 2.5 nmol/l) in a final volume of 100 µl RPMI containing 10% FBS and 0.5 mg/ml ssDNA and tRNA. Cells were incubated at 37 °C for 1 hour in 96 well tissue culture plates after which cells were washed three times with 1 ml FACS buffer before being resuspended in 500 µl FACS buffer containing 2 µg/ml 7-aminoactinomycin D stain (7AAD) to exclude dead cells in the analysis. Data shown are the average of two independent trials. To ensure iron loading of the labeled transferrin, we performed similar experiments using Dy488-transferrin which had been pretreated by incubation in 400 µg/ml ferric ammonium citrate 10 mmol/l NaHCO3/20 mmol/l HEPES pH 7.7 for 10 minutes followed by desalting.50 Similar results were observed (data not shown).
Liposome preparation. SNALPs were prepared using the spontaneous vesicle formation by ethanol dilution method.22 1,2-Distearoyl-sn-glycero-3-phosphocoline (DSPC), cholesterol, and 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[maleimide(polyethylene glycol)-2000] (DSPE-PEG-Mal) were purchased from Avanti Polar Lipids (Alabaster, AL). 1,2-dilinoleyloxy-N,N-dimethyl-3-aminopropane (DLinDMA) was synthesized following the method described by Heyes et al.24 siRNA was purchased from Integrated DNA Technologies (Coralville, IA).
A 90% vol/vol ethanol solution was prepared containing 10 µmole of total lipid consisting of DSPC/DLinDMA/Cholesterol/DSPE-PEG-Mal (10:40:48:2 mol%). A second solution of equal volume was prepared containing siRNA at a 1:10 molar ratio of Cy5-labeled siRNA to unlabeled siRNA in 20 mmol/l citrate buffer pH 5.0. The amount of anti-GFP or antiLamin A/C siRNA added maintained a 2:1 (+/−) charge ratio based on the molar amounts of negatively charged siRNA and positively charged DLinDMA. Both solutions were heated to 37 °C prior to vesicle formation. The siRNA solution was then added to the lipid solution with rapid mixing. The final mixture was diluted with an equal volume of 20 mmol/l citrate pH 6.0 containing 300 mmol/l NaCl and mixed well by rapid pipetting. The resulting mixture was incubated at 37 °C for 30 minutes followed by dialysis with a 10 kDa membrane into PBS, pH 8.0 with 1 mmol/l EDTA overnight at 4 °C. siRNA encapsulation efficiency typically exceeded 90% and was determined using Quant-iT RiboGreen (Invitrogen) as previously described24 or by gel electrophoresis as described below.
Thiol-modified aptamers were reduced prior to SNALP conjugation by adding Tris-(2-carboxyethyl phosphine) (TCEP) at a final concentration of 20 mmol/l, heating at 70 °C for 3 minutes, and incubating at room temperature for 2 hours. TCEP was removed by passing the sample through a Micro Bio-Spin 6 column (BioRad). After dialysis, thiol-modified aptamers were added to the liposomes at a final concentration of 1.5 µmol/l and incubated for 4 hours at 4 °C. Free maleimide groups were quenched after aptamer conjugation with 1 mmol/l BME and incubated at room temperature for 3 hours. SNALPs used for transfection purposes were BME-quenched (nontargeted with no aptamer), cntrl.36-conjugated (nontargeted with a conjugated control aptamer), and c2-conjugated (targeted).
Liposome characterization. Particle size distribution was assessed by dynamic light scattering (DynaPro Plate Reader, Wyatt Technology). SNALPs were diluted 1:40 in PBS and analyzed in a 384 well plate after removing bubbles by centrifugation at 1,000g for 3 minutes.
Aptamer conjugation and siRNA encapsulation were analyzed by running each sample on a denaturing (7 mol/l urea) 20% acrylamide gel. Samples were diluted 1:1 in 7 mol/l urea loading buffer and subsequently loaded on the gel without heating. Under these conditions, the liposomes remained intact and trapped in the well while any unencapsulated siRNA or unconjugated aptamer ran into the gel. Gels were stained with SYBR Gold and imaged on a Storm 840 phosphorimager (GE Health Sciences). The amount of free aptamer and siRNA was determined by comparison to aptamer or siRNA standard curve (Supplemental Figures S2, S3, and S4). Aptamer functionalized and nonfunctionalized liposomes were stable for at least 1 week when stored at 4 °C (Supplementary Figure S10).
The number of aptamers per liposome was estimated based on the observed conjugation efficiency and the concentration of liposomes. Liposome concentration was estimated based on the average diameter of these liposomes (~180 nm), the lipid surface area for packing of a phospholipid, 0.6 nm2, and by assuming that the liposomes are unilamellar and have spherical geometry.51
Cell transfection. HeLa-EGFP cells (20,000 cells/well) were seeded in a 24-well plate in DMEM/High Glucose culture medium supplemented with 5% FBS and antibiotics (D5 media). Twenty-four hours later, D5 media was replaced with D5 supplemented with 10 mg/ml ssDNA in DPBS and 10 mg/ml tRNA in DPBS, as blocking agents. BME-quenched, cntrl.36 nontargeted or c2 targeted SNALPs containing either GFP or LaminA/C siRNA were added to cells at a final siRNA concentration in each well of 200 nmol/l and cultured for 24 hours at 37 °C. As a positive control, cells were transfected with either 7.5 ng GFP or 7.5 ng LaminA/C siRNA and 3 µl HiPerFect Transfection Reagent (QIAGEN, Valencia, CA) following the manufacturer's protocol. As a negative control, 3 µl HiPerFect in 100 µl Opti-MEM was added to cells growing in 500 µl D5. After a 24-hour incubation with liposomes or transfection reagent, all media was replaced with fresh D5 media and cells were further incubated for 24 hours before assaying.
Flow cytometry and RNA isolation. Transfected cells were washed with PBS and trypsinized. Half of the cells in each sample were centrifuged at 300g for 3 minutes and resuspended in flow cytometry buffer consisting of HBSS with 1% bovine serum albumin and 0.1% sodium azide. Bisbenzimide was added to the buffer to assess cell viability. These samples were analyzed for knockdown of GFP expression and uptake of Cy5-siRNA on a Sony iCyt Eclipse Analyzer. The second half of each sample was passed through a QIAshredder cell homogenizer (QIAGEN). Total cell RNA for each sample was isolated from the cell lysate according to QIAGEN's RNAeasy Plus Mini Kit protocol.
Real-time PCR. cDNA synthesis was performed by first heating the following components to 70 °C for 3 minutes: 100 ng of total RNA isolated as described above, 25 ng of random hexamer primers (Invitrogen), and 2 µl of 5× First Strand RT buffer (Invitrogen). Mixtures were cooled to 4 °C, and 2 µl of 4 mmol/l dNTP mix, 1 µl of 0.1 mol/l DTT, and 1 µl of Moloney murine leukemia virus reverse transcriptase (Invitrogen) were added to each mixture. Distilled water was added to each sample to make 10 µl final volume reactions. Samples were heated to 37 °C for 50 minutes followed by a 15-minute incubation at 75 °C.
Expression levels of GFP mRNA and LaminA/C mRNA were analyzed by quantitative real-time PCR on an Applied Biosystems 7300 Real Time PCR machine using TaqMan Univeral Master Mix II (Invitrogen) and specific primer sets. GFP expression was measured using the following primers: GFP-For 5′GACAACCACTACCTGAGCAC, GFP-Rev 5′CAGGACCATGTGATCGCG, and GFP-probe 5′FAM-CCTGAGCAAAGACCCCAACGAGAA-IBFQ. LaminA/C expression was measured using the following primers: Lam-For 5′ ATGATCGCTTGGCGGTCTAC, Lam-Rev 5′, GCCCTGCGTTCTCCGTTT and Lam-probe 5′ FAM-TCGACCGTGTGCGCTCGCTG-IBFQ. RPLP0 expression was used as a standard control with primers: RPLP0-For 5′GGCGACCTGGAAGTCCAACT, RPLP0-Rev 5′CCATCAGCACCACAGCCTTC and RPLP0-probe 5′FAM-ATCTGCTGCATCTGCTTGGAGCCCA-IBFQ.52 Reverse transcription reactions were diluted 1:40, and 10 µl of each sample was used in each 25 µl quantitative real-time PCR reaction according to the TaqMan Universal Master Mix II protocol. Each sample was analyzed in triplicate.
Interferon response. HeLa-EGFP cells were transfected, total RNA was isolated, and cDNA was synthesized as described above. As a positive control, HeLa-EGFP cells were transfected with 100 ng of Poly(I:C) complexed with 3 µl HiPerFect in 100 µl Opti-MEM and incubated for 24 hours at 37 °C. RNA isolation and cDNA synthesis were then carried out as described above. Expression levels of interferon-β, CDKL2, and OAS1 were quantified with real-time PCR as described above using Power SYBR Green Master Mix (Invitrogen) and specific primer sets: interferon-β (IFNB)-For 5′AGACTTACAGGTTACCTCCGAA, IFNB-Rev 5′CAGTACATTCGCCATCAGTCA, OAS1-For 5′CGAGGGAGCATGAAAACACATTT, OAS1-Rev 5′GCAGA GTTGCTGGTAGTTTATGAC, CDKL2-For 5′GCCTCCTTGG GTTCGTCTATAA, CDKL2-Rev 5′CTCAGGGCCCGCTCATA GTA. RPLP0 was used for normalization purposes. Each sample was analyzed in triplicate (Supplementary Figures S5 and S6).
Acknowledgments
This work was supported by funding from Stand Up 2 Cancer, the Marion Bessin Liver Research Center at the Albert Einstein College of Medicine and the National Cancer Institute at the National Institutes of Health (1R21CA157366). M.L. and S.E.W. would like to thank Debbie Palliser, Teresa P. DiLorenzo, Gayatri Mukherjee, and Joe Katakowski for our weekly discussions on liposomes and the targeted delivery of siRNA.
Supplementary Material
Analysis of minimized c2 constructs by flow cytometry.
Denaturing polyacrylamide gel analysis of liposomes showing nonencapsulated siRNA and unconjugated aptamer.
Denaturing polyacrylamide gel analysis of liposomes used to determine amount of unconjugated c2.
Denaturing polyacrylamide gel analysis of liposomes used to determine amount of unconjugated cntrl.36.
Effect of free transferrin on cell uptake and gene knockdown using nontargeted liposomes.
Effect of liposome concentration on cell uptake and gene knockdown using nontargeted liposomes.
Interferon response of HeLa-GFP cells treated with anti-TfR-targeted (c2), nontargeted (BME), or nonspecific aptamer (cntrl.36) functionalized SNALPs.
Influence of aptamer-conjugated SNALPs on interferon-β (IFNB) expression levels in HeLa-EGFP cells.
Transferrin competition binding assay using mouse transferrin (mTf).
SNALP stability at 4 °C.
References
- Daniels TR, Delgado T, Helguera G., and, Penichet ML. The transferrin receptor part II: targeted delivery of therapeutic agents into cancer cells. Clin Immunol. 2006;121:159–176. doi: 10.1016/j.clim.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Daniels TR, Delgado T, Rodriguez JA, Helguera G., and, Penichet ML. The transferrin receptor part I: Biology and targeting with cytotoxic antibodies for the treatment of cancer. Clin Immunol. 2006;121:144–158. doi: 10.1016/j.clim.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Jones AR., and, Shusta EV. Blood-brain barrier transport of therapeutics via receptor-mediation. Pharm Res. 2007;24:1759–1771. doi: 10.1007/s11095-007-9379-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Head JF, Wang F., and, Elliott RL. Antineoplastic drugs that interfere with iron metabolism in cancer cells. Adv Enzyme Regul. 1997;37:147–169. doi: 10.1016/s0065-2571(96)00010-6. [DOI] [PubMed] [Google Scholar]
- Laske DW, Muraszko KM, Oldfield EH, DeVroom HL, Sung C, Dedrick RL.et al. (1997Intraventricular immunotoxin therapy for leptomeningeal neoplasia Neurosurgery 411039–49; discussion 1049. [DOI] [PubMed] [Google Scholar]
- Laske DW, Youle RJ., and, Oldfield EH. Tumor regression with regional distribution of the targeted toxin TF-CRM107 in patients with malignant brain tumors. Nat Med. 1997;3:1362–1368. doi: 10.1038/nm1297-1362. [DOI] [PubMed] [Google Scholar]
- Weaver M., and, Laske DW. Transferrin receptor ligand-targeted toxin conjugate (Tf-CRM107) for therapy of malignant gliomas. J Neurooncol. 2003;65:3–13. doi: 10.1023/a:1026246500788. [DOI] [PubMed] [Google Scholar]
- Mendonça LS, Firmino F, Moreira JN, Pedroso de Lima MC., and, Simões S. Transferrin receptor-targeted liposomes encapsulating anti-BCR-ABL siRNA or asODN for chronic myeloid leukemia treatment. Bioconjug Chem. 2010;21:157–168. doi: 10.1021/bc9004365. [DOI] [PubMed] [Google Scholar]
- Sahoo SK, Ma W., and, Labhasetwar V. Efficacy of transferrin-conjugated paclitaxel-loaded nanoparticles in a murine model of prostate cancer. Int J Cancer. 2004;112:335–340. doi: 10.1002/ijc.20405. [DOI] [PubMed] [Google Scholar]
- Matsumura Y., and, Maeda H. A new concept for macromolecular therapeutics in cancer chemotherapy: mechanism of tumoritropic accumulation of proteins and the antitumor agent smancs. Cancer Res. 1986;46 12 Pt 1:6387–6392. [PubMed] [Google Scholar]
- Bartlett DW, Su H, Hildebrandt IJ, Weber WA., and, Davis ME. Impact of tumor-specific targeting on the biodistribution and efficacy of siRNA nanoparticles measured by multimodality in vivo imaging. Proc Natl Acad Sci USA. 2007;104:15549–15554. doi: 10.1073/pnas.0707461104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler JA, Lee JH, Collawn JF., and, Moore BA. Receptor mediated uptake of peptides that bind the human transferrin receptor. Eur J Biochem. 2001;268:2004–2012. doi: 10.1046/j.1432-1327.2001.02073.x. [DOI] [PubMed] [Google Scholar]
- Chen CH, Dellamaggiore KR, Ouellette CP, Sedano CD, Lizadjohry M, Chernis GA.et al. (2008Aptamer-based endocytosis of a lysosomal enzyme Proc Natl Acad Sci USA 10515908–15913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalhães ML, Byrom M, Yan A, Kelly L, Li N, Furtado R.et al. (2012A general RNA motif for cellular transfection Mol Ther 20616–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla R., and, Sousa R. Mutant T7 RNA-polymerase as a DNA-polymerase. Embo Journal. 1995;14:4609–4621. doi: 10.1002/j.1460-2075.1995.tb00140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla R., and, Sousa R. Efficient synthesis of nucleic acids heavily modified with non-canonical ribose 2'-groups using a mutantT7 RNA polymerase (RNAP) Nucleic Acids Res. 1999;27:1561–1563. doi: 10.1093/nar/27.6.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieken WA, Olsen DB, Benseler F, Aurup H., and, Eckstein F. Kinetic characterization of ribonuclease-resistant 2'-modified hammerhead ribozymes. Science. 1991;253:314–317. doi: 10.1126/science.1857967. [DOI] [PubMed] [Google Scholar]
- Schmid SL., and, Carter LL. ATP is required for receptor-mediated endocytosis in intact cells. J Cell Biol. 1990;111 6 Pt 1:2307–2318. doi: 10.1083/jcb.111.6.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie RF, Palomaki GE, Neveux LM, Navolotskaia O, Ledue TB., and, Craig WY. Reference distributions for serum iron and transferrin saturation: a practical, simple, and clinically relevant approach in a large cohort. J Clin Lab Anal. 2002;16:237–245. doi: 10.1002/jcla.10048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isselbacher KJ, Braunwald E, Wilson JD, Martin JB, Fauci AS., and, Kasper DL.1994Harrison's Principles of Internal Medicine13th edn. McGraw-Hill: New York [Google Scholar]
- Young SP, Bomford A., and, Williams R. The effect of the iron saturation of transferrin on its binding and uptake by rabbit reticulocytes. Biochem J. 1984;219:505–510. doi: 10.1042/bj2190505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffs LB, Palmer LR, Ambegia EG, Giesbrecht C, Ewanick S., and, MacLachlan I. A scalable, extrusion-free method for efficient liposomal encapsulation of plasmid DNA. Pharm Res. 2005;22:362–372. doi: 10.1007/s11095-004-1873-z. [DOI] [PubMed] [Google Scholar]
- Zimmermann TS, Lee AC, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN.et al. (2006RNAi-mediated gene silencing in non-human primates Nature 441111–114. [DOI] [PubMed] [Google Scholar]
- Heyes J, Palmer L, Bremner K., and, MacLachlan I. Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids. J Control Release. 2005;107:276–287. doi: 10.1016/j.jconrel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- Whitehead KA, Langer R., and, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., and, Rossi JJ. Cell-specific aptamer-mediated targeted drug delivery. Oligonucleotides. 2011;21:1–10. doi: 10.1089/oli.2010.0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dua P, Kim S., and, Lee DK. Nucleic acid aptamers targeting cell-surface proteins. Methods. 2011;54:215–225. doi: 10.1016/j.ymeth.2011.02.002. [DOI] [PubMed] [Google Scholar]
- Davis ME. The first targeted delivery of siRNA in humans via a self-assembling, cyclodextrin polymer-based nanoparticle: from concept to clinic. Mol Pharm. 2009;6:659–668. doi: 10.1021/mp900015y. [DOI] [PubMed] [Google Scholar]
- Davis ME, Zuckerman JE, Choi CH, Seligson D, Tolcher A, Alabi CA.et al. (2010Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles Nature 4641067–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calando Pharmaceuticals Safety Study of CALAA-01 to Treat Solid Tumor Cancers ClinicalTrialsgov [Internet] Bethesda (MD): National Library of Medicine (US) 20111006Available from: URL of the record NLM Identifier: NCT00689065.
- Yu YJ, Zhang Y, Kenrick M, Hoyte K, Luk W, Lu Y.et al. (2011Boosting brain uptake of a therapeutic antibody by reducing its affinity for a transcytosis target Sci Transl Med 384ra44. [DOI] [PubMed] [Google Scholar]
- Ni X, Zhang Y, Ribas J, Chowdhury WH, Castanares M, Zhang Z.et al. (2011Prostate-targeted radiosensitization via aptamer-shRNA chimeras in human tumor xenografts J Clin Invest 1212383–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Swiderski P, Li H, Zhang J, Neff CP, Akkina R.et al. (2009Selection, characterization and application of new RNA HIV gp 120 aptamers for facile delivery of Dicer substrate siRNAs into HIV infected cells Nucleic Acids Res 373094–3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Li H, Li S, Zaia J., and, Rossi JJ. Novel dual inhibitory function aptamer-siRNA delivery system for HIV-1 therapy. Mol Ther. 2008;16:1481–1489. doi: 10.1038/mt.2008.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler LA, Trifonova R, Vrbanac V, Basar E, McKernan S, Xu Z.et al. (2011Inhibition of HIV transmission in human cervicovaginal explants and humanized mice using CD4 aptamer-siRNA chimeras J Clin Invest 1212401–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara JO, 2nd, Andrechek ER, Wang Y, Viles KD, Rempel RE, Gilboa E.et al. (2006Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras Nat Biotechnol 241005–1015. [DOI] [PubMed] [Google Scholar]
- Dassie JP, Liu XY, Thomas GS, Whitaker RM, Thiel KW, Stockdale KR.et al. (2009Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors Nat Biotechnol 27839–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar S, Gu FX, Langer R, Farokhzad OC., and, Lippard SJ. Targeted delivery of cisplatin to prostate cancer cells by aptamer functionalized Pt(IV) prodrug-PLGA-PEG nanoparticles. Proc Natl Acad Sci USA. 2008;105:17356–17361. doi: 10.1073/pnas.0809154105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farokhzad OC, Cheng J, Teply BA, Sherifi I, Jon S, Kantoff PW.et al. (2006Targeted nanoparticle-aptamer bioconjugates for cancer chemotherapy in vivo Proc Natl Acad Sci USA 1036315–6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann AP, Bhavane RC, Somasunderam A, Liz Montalvo-Ortiz B, Ghaghada KB, Volk D.et al. (2011Thioaptamer conjugated liposomes for tumor vasculature targeting Oncotarget 2298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H, O'Donoghue MB, Liu H., and, Tan W. A liposome-based nanostructure for aptamer directed delivery . Chem Commun (Camb) 2010;46:249–251. doi: 10.1039/b916911c. [DOI] [PubMed] [Google Scholar]
- Tan L, Neoh KG, Kang ET, Choe WS., and, Su X. PEGylated anti-MUC1 aptamer-doxorubicin complex for targeted drug delivery to MCF7 breast cancer cells. Macromol Biosci. 2011;11:1331–1335. doi: 10.1002/mabi.201100173. [DOI] [PubMed] [Google Scholar]
- Judge AD, Robbins M, Tavakoli I, Levi J, Hu L, Fronda A.et al. (2009Confirming the RNAi-mediated mechanism of action of siRNA-based cancer therapeutics in mice J Clin Invest 119661–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekmira Pharmaceuticals Corporation Dose Escalation Study to Determine Safety, Pharmacokinetics, and Pharmacodynamics of Intravenous TKM-080301 ClinicalTrialsgov [Internet] Bethesda (MD): National Library of Medicine (US) 20111006Available from: URL of the record NLM Identifier: NCT01262235.
- (NCI) NCI TKM 080301 for Primary or Secondary Liver Cancer ClinicalTrialsgov [Internet] Bethesda (MD): National Library of Medicine (US) 20111006Available from: URL of the record NLM Identifier: NCT01437007.
- Akinc A, Querbes W, De S, Qin J, Frank-Kamenetsky M, Jayaprakash KN.et al. (2010Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms Mol Ther 181357–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffs LB, Palmer LR, Ambegia EG, Giesbrecht C, Ewanick S., and, MacLachlan I. A scalable, extrusion-free method for efficient liposomal encapsulation of plasmid DNA. Pharm Res. 2005;22:362–372. doi: 10.1007/s11095-004-1873-z. [DOI] [PubMed] [Google Scholar]
- Hall B, Micheletti JM, Satya P, Ogle K, Pollard J., and, Ellington AD. Design, synthesis, and amplification of DNA pools for in vitro selection. Curr Protoc Nucleic Acid Chem. 2009;Chapter 9:Unit 9.2. doi: 10.1002/0471142700.nc0902s39. [DOI] [PubMed] [Google Scholar]
- Wong I., and, Lohman TM. A double-filter method for nitrocellulose-filter binding: application to protein-nucleic acid interactions. Proc Natl Acad Sci USA. 1993;90::5428–5432. doi: 10.1073/pnas.90.12.5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw TE., and, Subtil A. Endocytosis: biochemical analyses. Curr Protoc Cell Biol Chapter 15. 2001. p. Unit 15.3. [DOI] [PubMed]
- Reulen SW, Brusselaars WW, Langereis S, Mulder WJ, Breurken M., and, Merkx M. Protein-liposome conjugates using cysteine-lipids and native chemical ligation. Bioconjug Chem. 2007;18:590–596. doi: 10.1021/bc0602782. [DOI] [PubMed] [Google Scholar]
- Collingwood MA, Rose SD, Huang L, Hillier C, Amarzguioui M, Wiiger MT.et al. (2008Chemical modification patterns compatible with high potency dicer-substrate small interfering RNAs Oligonucleotides 18187–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Analysis of minimized c2 constructs by flow cytometry.
Denaturing polyacrylamide gel analysis of liposomes showing nonencapsulated siRNA and unconjugated aptamer.
Denaturing polyacrylamide gel analysis of liposomes used to determine amount of unconjugated c2.
Denaturing polyacrylamide gel analysis of liposomes used to determine amount of unconjugated cntrl.36.
Effect of free transferrin on cell uptake and gene knockdown using nontargeted liposomes.
Effect of liposome concentration on cell uptake and gene knockdown using nontargeted liposomes.
Interferon response of HeLa-GFP cells treated with anti-TfR-targeted (c2), nontargeted (BME), or nonspecific aptamer (cntrl.36) functionalized SNALPs.
Influence of aptamer-conjugated SNALPs on interferon-β (IFNB) expression levels in HeLa-EGFP cells.
Transferrin competition binding assay using mouse transferrin (mTf).
SNALP stability at 4 °C.