Abstract
Hematopoietic stem cell transplantation is the oldest and successful form of stem cell therapy. High dose therapy (HDT) followed by hematopoietic stem cell transplantation allows physicians to administer increased amounts of chemotherapy and/or radiation while minimizing negative side effects such as damage to blood-producing bone marrow cells. Although HDT is successful in treating a wide range of cancers, it leads to lethal therapy-related myelodysplasia syndrome or acute myeloid leukemia (t-MDS/AML) in 5–10% of patients undergoing autologous hematopoietic cell transplantation for Hodgkin lymphoma and non-Hodgkin lymphoma. In this study, we carried out metabolomic analysis of peripheral blood stem cell samples collected in a cohort of patients before hematopoietic cell transplantation in order to gain insights into the molecular and cellular pathogenesis of t-MDS. Nonparametric tests and multivariate analyses were used to compare the metabolite concentrations in samples from patients that developed t-MDS within 5 years of transplantation and the patients that did not. The results suggest that the development of t-MDS is associated with dysfunctions in cellular metabolic pathways. The top canonical pathways suggested by the metabolomic analysis include alanine and aspartate metabolism, glyoxylate and dicarboxylate metabolism, phenylalanine metabolism, citrate acid cycle, and aminoacyl-t-RNA biosynthesis. Dysfunctions in these pathways indicate mitochondrial dysfunction that would result in decreased ability to detoxify reactive oxygen species generated by chemo and radiation therapy, therefore leading to cancer causing mutations. These observations suggest predisposing factors for the development of t-MDS.
Keywords: NMR, metabolomics, peripheral blood stem cells, leukemia
Introduction
Stem cell therapy is one of the most active new frontiers in biomedical research with the potential to provide treatment for a variety of diseases and injuries for which the current therapy is inadequate. Hematopoietic stem cell transplantation is the oldest and most successful form of stem cell therapy. High dose therapy (HDT) followed by hematopoietic stem cell transplantation allows physicians to administer increased amounts of chemotherapy and/or radiation with subsequent rescue by autologous blood-producing bone marrow or peripheral blood stem cells.1 Although HDT is successful in treating a wide range of cancers, it leads to therapy-related myelodysplasia or acute myeloid leukemia (t-MDS/AML) in 5–10% patients undergoing autologous hematopoietic cell transplantation (aHCT) for Hodgkin lymphoma (HL) and non-Hodgkin lymphoma (NHL).2–5 The molecular mechanisms that cause t-MDS/AML are unknown; epidemiologic studies suggest that pre-aHCT therapeutic exposures, transplant conditioning regimens, autograft collection and hematopoietic regeneration play a role in the development of t-MDS.2,6,7 The bone marrow of patients that do not develop t-MDS/AML renew and produce blood cells normally; however, this process becomes interrupted in response to chemotherapy or radiation in some patients and damage to hematopoietic stem cells (HSCs) can propagate through future differentiated cells ultimately leading to lethal disease.
Metabolomics is emerging as an important discipline in systems biology and directly reflects acute cellular status.8, 9 Metabolomics also has the potential to discover new biomarkers for disease diagnosis and prognosis.10–12 For example, a recent metabolomic study has identified the metabolite known as sarcosine as playing an important role in prostate cancer progression and as a biomarker for prostate cancer diagnosis.13 NMR or mass spectrometry-based metabolomics have analyzed metabolites of total cell extracts. The advantage of NMR-based analysis is that the signal intensity is directly proportional to the quantity of the molecules that produce the signals;14 the levels of various metabolites are important indicators of activities of different cellular pathways. In addition, NMR-based metabolomics do not require extensive fractionation of the samples by chromatography methods, because identification of individual metabolite from a mixture is achieved by examining multiple resonances from the same molecule simultaneously, and multi-dimensional NMR methods can be used to resolve resonance degeneracy occurring in a mixture of many metabolites.
In this study, we carried out metabolomic analysis in order to gain insights into the molecular and cellular pathogenesis of t-MDS/AML, and to identify biomarkers that allow the identification of patients that are predisposed to the development of t-MDS/AML. We have analyzed the metabolome of peripheral blood stem cell (PBSC) samples collected from twelve patients before aHCT. These included six patients who subsequently developed t-MDS/AML (“cases”) after undergoing aHCT for HL and NHL, and six patients who did not (“controls”). Nonparametric tests and multivariate analyses were used to compare the concentrations calculated from each sample. These analyses have identified metabolites that show statistically significant differences between the t-MDS/AML cases and controls. The top canonical pathways associated with these metabolites include alanine and aspartate metabolism, glyoxylate and dicarboxylate metabolism, phenylalanine metabolism, citrate acid cycle, and aminoacyl-t-RNA biosynthesis. These observations suggest predisposing factors for the development of t-MDS/AML.
Experimental Procedures
Patient Selection and Sample Collection
PBSC samples were obtained pre-HCT. For the current study a nested case-control design was used. Six patients who developed t-MDS/AML (“cases”) after undergoing aHCT for NHL or HL at the City of Hope National Medical Center were studied. Six controls were selected from the prospectively followed study cohort of patients who had undergone aHCT for HL or NHL, but did not develop t-MDS after aHCT. Information about the 6 patients and 6 controls are given in Table 1. These studies were approved by the institutional review board of City of Hope in accordance with an assurance filed with and approved by the Department of Health and Human Services and met all requirements of the Declaration of Helsinki. Informed consent was obtained from all subjects after the nature and possible consequences of the studies were explained.
Table 1.
Information about the 6 patients and 6 controls.
| ID No. | Primary diagnosis | Age at diagnosis of Primary | Age at HCT | Sex | Race/ethnicity | Months to t-MDS from HCT | t-MDS morphology | t-MDS cytogenetics |
|---|---|---|---|---|---|---|---|---|
| 99 | NHL | 56 | 57 | M | W | N/A | N/A | N/A |
| 138 | NHL | 42 | 43 | F | W | N/A | N/A | N/A |
| 165 | NHL | 47 | 48 | M | H | N/A | N/A | N/A |
| 474 | HD | 24 | 25 | M | W | N/A | N/A | N/A |
| 559 | NHL | 44 | 44 | F | W | N/A | N/A | N/A |
| 745 | NHL | 45 | 46 | F | W | N/A | N/A | N/A |
| 125 | NHL | 50 | 51 | F | W | 49.7 | Normal | Del20q (7.5%) |
| 211 | NHL | 42 | 46 | M | W | 18.5 | AML | Multiple: del 6q, −7, del7q, add12, −9 |
| 650 | HD | 17 | 18 | M | H | 13 | RARS | −5, add17p |
| 15002 | NHL | 67 | 68 | M | W | 24.8 | AML | t(10;11)(p13;q13) |
| 15004 | HD | 54 | 57 | M | H | 46.9 | RARS | Multiple: 3, −5,del13q,del 20q,add15,a dd13 |
| 15007 | HD | 33 | 35 | M | H | 74.3 | RARS | Multiple: −5, −19, −20, del13q, −13 |
Sample Preparation for NMR Spectroscopy
For each sample, between 1–4 × 106 PBSCs were incubated in IMDM supplemented with 20% FBS and 2mM uniformly-labeled 13C-glutamine for 18 hours at 37°C. Cells were pelleted and washed with PBS to remove any residual medium prior to recording the wet mass. Cellular metabolites were extracted in a final ratio of methanol/chloroform/water of 2.0:2.0:1.8, as described previously.15 Briefly, the cell pellet was sonicated in 7.4 ml/g cold methanol and 2.22 ml/g cold water at 25% duty cycle for 1 minute on ice. The homogenate was transferred to a borosilicate glass culture tube and 3.7 ml/g cold chloroform was added. The suspension was vortexed and kept at 4°C overnight. To this monophasic solution, 3.7 ml/g cold chloroform and 3.7 ml/g cold water were added, and the solutions were vortexed and centrifuged for 5 minutes. The upper hydrophilic layer and the lower lipophilic layer were carefully transferred to clean tubes and dried in a speed-vac concentrator.
1H NMR Spectroscopy
Hydrophilic samples dried from the methanol-water fractions were each resuspended in 400 μl 100% D2O containing 6.4 μM DSS (4,4-dimethyl-4-silapentane-1-sulfonic acid; Cambridge Isotope Laboratories, MA). The internal standard DSS served as a chemical shift reference and a concentration standard. One-dimensional spectra of polar samples were acquired at 25°C on a Bruker Avance 600 spectrometer using a sequence with water suppression method WATERGATE W5; 1024 transients and 32 K data points were collected with a spectral width of 16 ppm.
Dried lipophilic samples were each resuspended in 400 μl deuterated chloroform containing 0.05% v/v tetramethylsilane (TMS; Cambridge Isotope Laboratories, MA) as a chemical shift reference. Standard one-dimensional 1H spectra were acquired at 25°C on a Bruker Avance 600 spectrometer with a 90° RF pulse, 10 K data points, and a spectral width of 13 ppm. Due to the small quantity of available sample, 8 K transients were recorded to achieve a better signal-to-noise ratio.
NMR Data and Statistical Analysis
Spectra of hydrophilic samples were imported into the Chenomx NMR Suite Processor (version 6.1, Chenomx Inc., Edmonton, Canada) for Fourier transformation, a cubic-spline-based baseline adjustment and phasing. Chemical shifts were referenced to the methyl protons of DSS. The integral of the DSS methyl proton peak was set within Chenomx to represent 6.4 μM DSS. The Chenomx NMR Suite Profiler was used to identify metabolites by fitting compound signatures from the provided NMR spectral library. Metabolite concentrations were calculated using Chenomx by determining heights of compound signatures that best fit the sample spectra. The table of identified and calculated concentrations were exported and saved in an Excel worksheet. The concentrations were converted from micromolar units to nanomoles per 3 × 106 cells by multiplying the sample volume and adjusting for the number of cells in the sample. Individual Mann-Whitney tests were performed using StatistiXL software (version 1.8; StatistiXL, Nedlands, Western Australia) for each of these metabolites to compare their quantities between patient groups. Metabolite quantities for each sample were used as an input for analysis using MetaboAnalyst;16 data were row-wise normalized to constant sum and column-wise normalized by log2 transformation prior to principal components analysis (PCA), partial least squares-discriminant analysis (PLS-DA), and Pearson’s r correlation.
Spectra from lipophilic samples were collected as discussed above, with a few modifications. Spectra of hydrophilic samples were imported into the Chenomx NMR Suite Processor for Fourier transformation, 1-Hz line broadening, cubic-spline-based baseline adjustment and phasing; chemical shifts were referenced to the methyl protons of TMS. The Chenomx NMR Suite Profiler was used for spectral binning; the spectral regions between 0.75–6 ppm were divided into 0.025 ppm bin windows and the values normalized to the total area of the spectrum. The regions above 6 ppm and less than 0.75 contained only noise and the chemical shift reference, respectively, and were therefore excluded from analyses. The bins were exported and used as input for the MetaboAnalyst software. Baseline noise cutoff was set to 7×10−5. Bins were row-wise normalized to constant sum and column-wise normalized by log2 transformation prior to principal components analysis (PCA), partial least squares-discriminant analysis (PLS-DA), and Pearson’s r correlation.
Results
NMR Profiling of Hydrophilic Metabolites in pre-HCT PBSC Samples
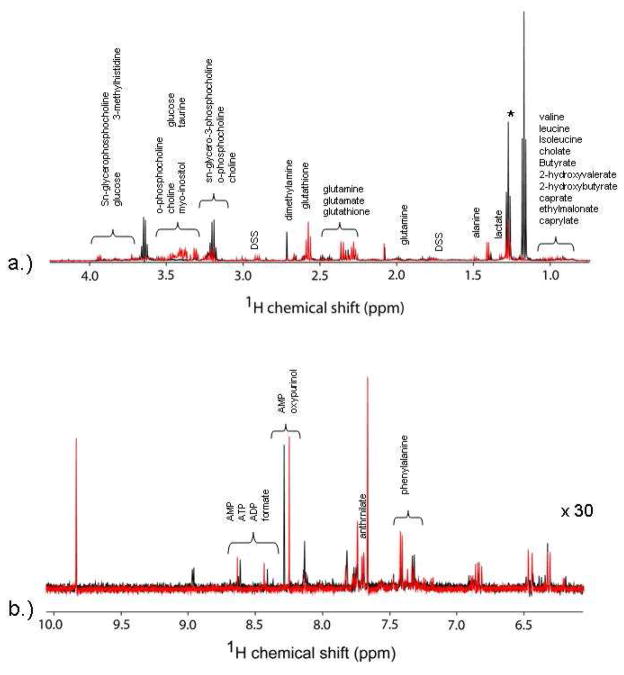
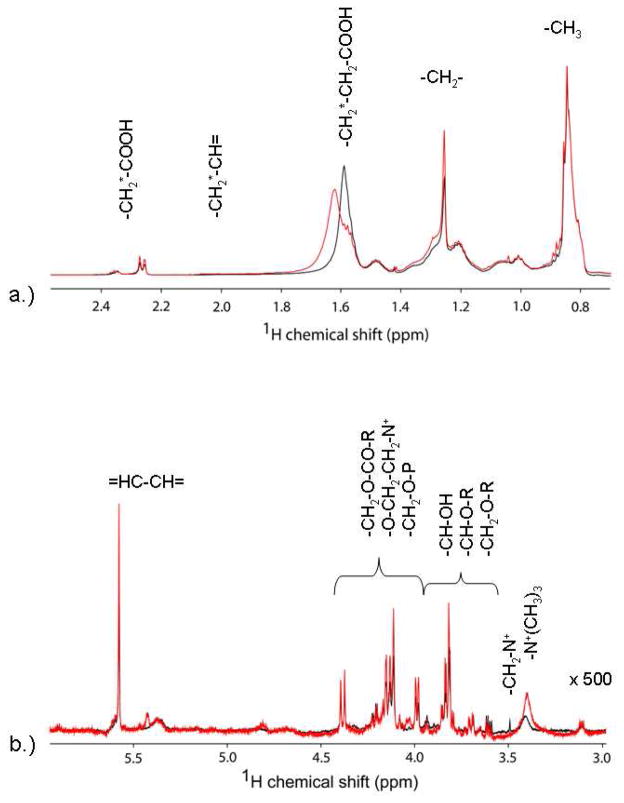
Figure 1 shows representative 1H NMR spectra of the control lymphoma patients and the t-MDS/AML case patients. Most of the information is located in the upfield region between 0 and 4 ppm, which corresponds to aliphatic groups of metabolites. There are clear differences in spectral intensity of some compounds such as dimethylamine, glutamine, glutamate, glutathione, and leucine, in these instances. The Chenomx Profiler was utilized to identify the compounds visible in the spectra; however, some resonances remained unidentified in the library. As noted previously,15 sample contamination from the communal speed-vacuum led to variant peaks 2.9, 3, and 8 ppm from N,N-dimethylformamide and were excluded from concentration analyses.
Figure 1.
Proton NMR spectra of the hydrophilic metabolites from PBSC samples. The intensity of the downfield region is increased to facilitate identification of resonances. Spectra in black represent typical results for a control patient, while those in red represent those for a t-MDS patient.
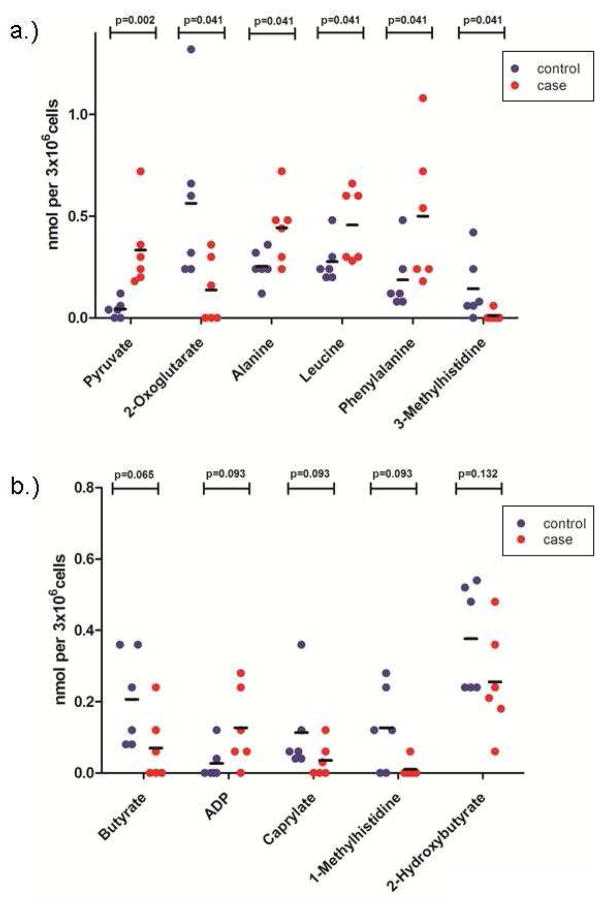
Nonparametric tests analyses were used to compare the concentrations calculated from each sample. From the approximately 50 metabolites identified, Mann-Whitney test classified six as statistically significant (p < 0.05; Figure 2). Pyruvate is on average lower in the samples from control patients than those from patients that develop t-MDS. However, 2-oxoglutarate (α-ketoglutarate) levels are higher on average in the control samples than t-MDS cases. These metabolites are directly linked to mitochondrial function. Additionally, PBSC cells from t-MDS/AML patients have increased levels of the amino acids alanine, leucine, and phenylalanine. Alanine is derived from pyruvate directly, and its increased level is correlated to the increased levels of pyruvate. Leucine and phenylalanine are synthesized in the mitochondria from acetyl-CoA, which is also derived from pyruvate. The increased levels of these metabolites in t-MDS/AML patients could be due to the reduced utilization of these compounds or due to their increased synthesis. Nonetheless, these results suggest different mitochondrial activity in t-MDS/AML patients relative to the controls.
Figure 2.
Nonparametric testing of PBSC metabolites. (A) Metabolites significantly altered in PBSC samples from cases (red) compared with controls (blue) are shown (MWU p<0.05; n=6 each). (B) Metabolites with possible alterations in cases (red) compared to controls (blue) (MWU 0.05 < p <0.15; n=6 each).
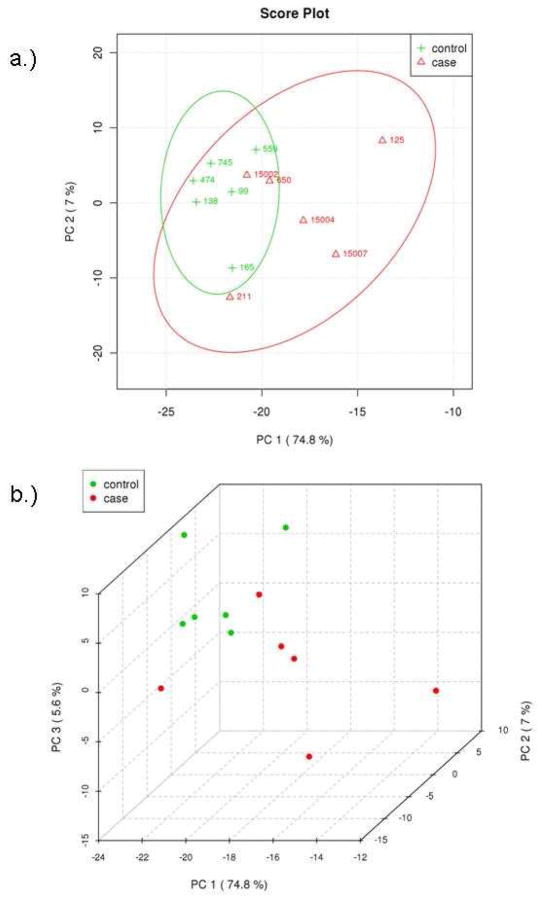
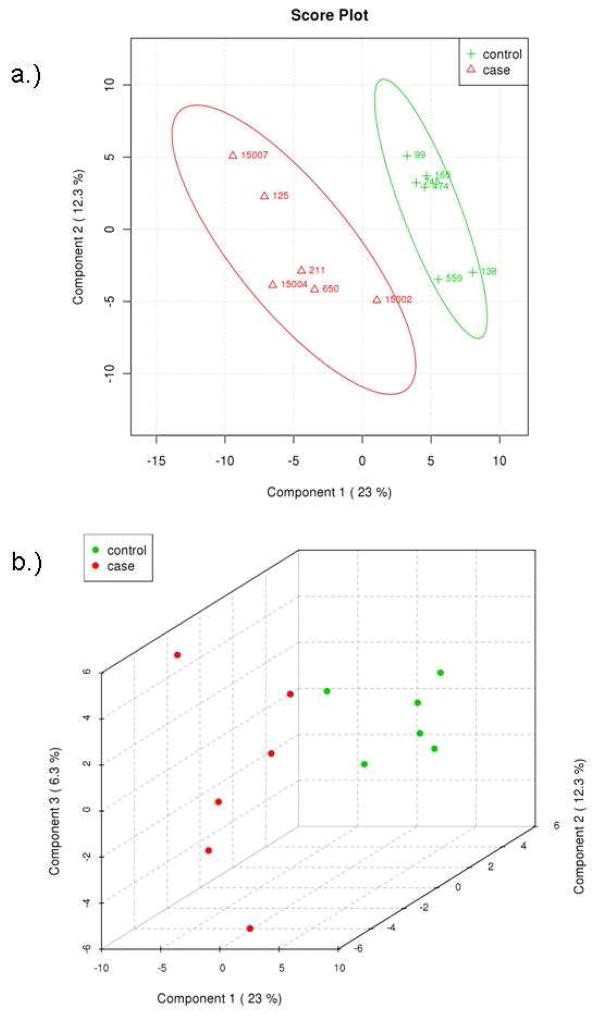
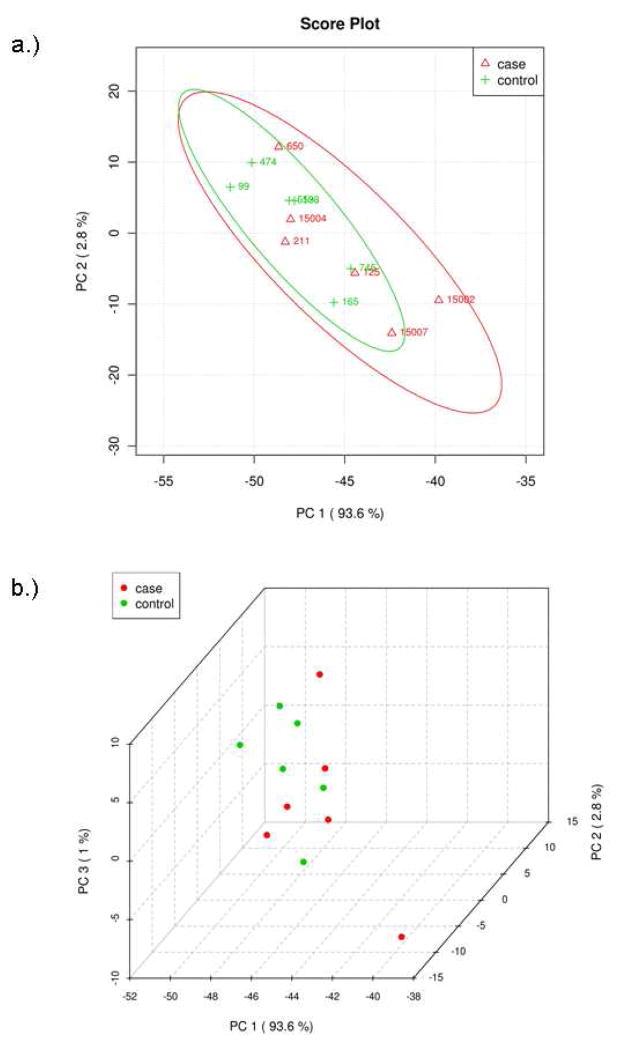
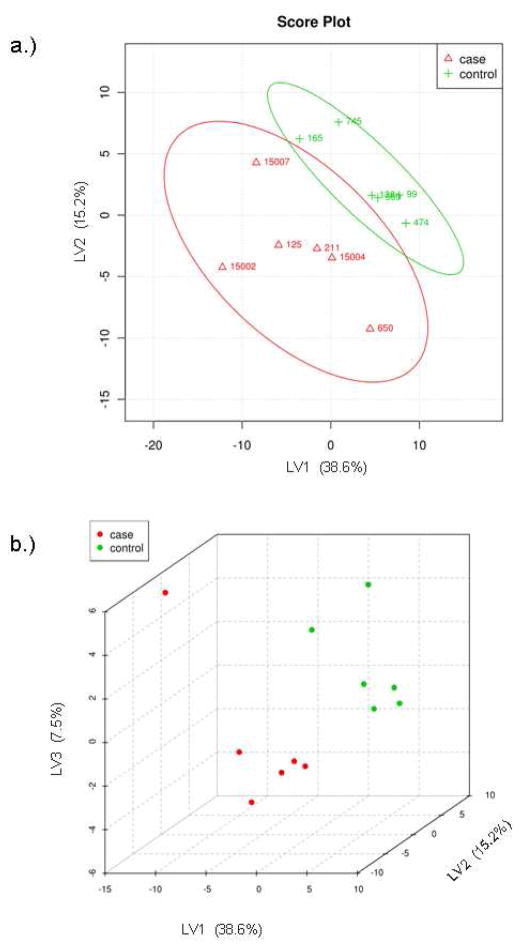
Multivariate analyses were also applied to identify differences between the two groups of patients. Principal components analysis (PCA) was used to analyze the concentrations of the identified metabolites. PCA shows that the two groups are not fully separated by the two largest principle components, PC1 and PC2 (Figure 3a). The two groups, however, can be better separated by the three largest principle components, PC1, PC2 and PC3 (Figure 3b). The difficulty in separating the two groups is likely due to the fact that their metabolomic differences that separate control and t-MDS/AML samples are overshadowed by the intrinsic variations found from patient to patient. Thus an alternative statistical method was applied to further examine the data. Analysis by partial least squares-discriminated analysis (PLS-DA) is a supervised method increasingly used in metabolomics when clusters are not distinctly separated by PCA. PLS-DA also provides identification of the metabolites that affect the separation. The same input used for PCA was used for PLS-DA and the results are shown in Figure 4. There is a clear distinction between control and t-MDS/AML case samples along the latent variable (LV) 1 axis, which correspond to the component with the highest differences (Figure 4a). The distinction is independent of the smaller components, LV2 and LV3 (Figure 4b).
Figure 3.
Principal components analysis of metabolic profile differences between patient groups. (A) PCA scores of components 1 and 2 of calculated hydrophilic metabolite levels from control PBSC samples (green) and t-MDS patient PBSC (red). (B) PCA scores of components 1, 2, and 3 for the control PBSC (green) and t-MDS/AML PBSC (red). The explained variances are shown in parentheses.
Figure 4.
Partial least squares discriminant analysis of metabolite concentrations between patient groups. (A) PLS-DA scores of latent variables 1 and 2 of calculated hydrophilic metabolite levels from control PBSC samples (green) and t-MDS/AML patient samples (red). (B) PLS-DA scores of latent variables 1, 2, and 3 for the control PBSC (green) and t-MDS/AML PBSC (red). The explained variances are shown in parentheses.
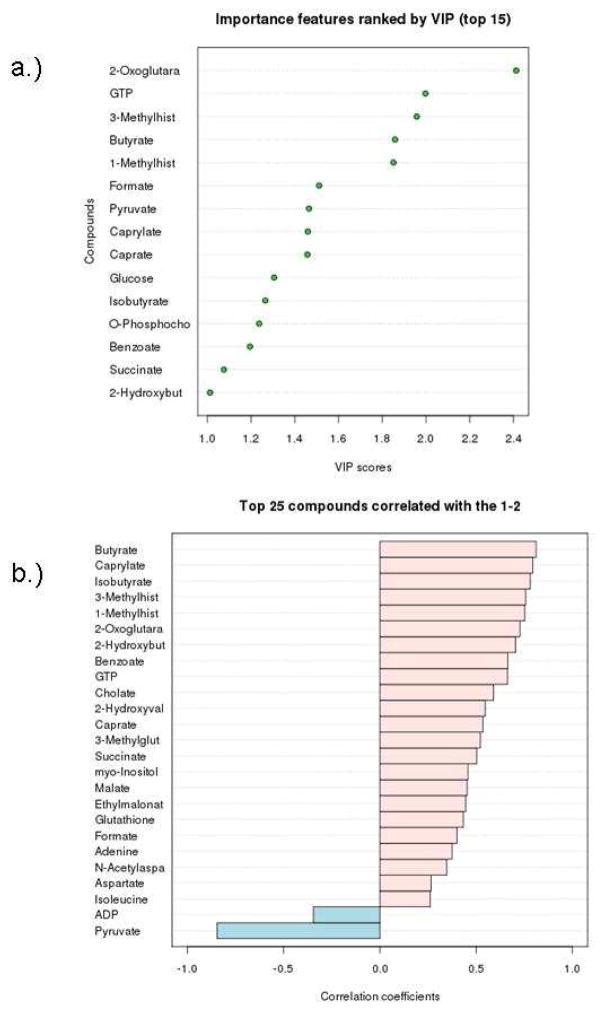
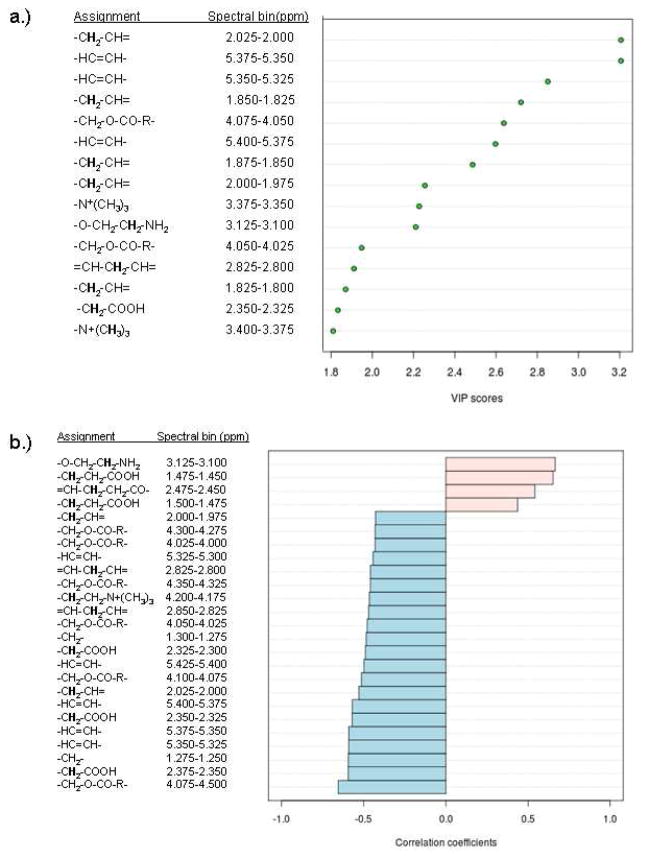
Metabolites contributing to the distinction of the t-MDS/AML and control groups were identified. In addition to those found to be significant by nonparametric testing, more compounds have been identified that separate the two groups by PLS-DA (Fig. 5). Seven compounds are commonly found as discriminators between the t-MDS/AML cases and controls by both nonparametric testing and multivariate analysis. They are pyruvate, 2-oxoglutarate, 3-methylhistidine, butyrate, 1-methylhistidine, caprylate, and 2-hydroxybutyrate. Variable Importance in Projection (VIP) implicated additional eight metabolites, whose concentrations are important features that separate the two groups (Fig. 5a). Correlation analysis also recognized the trends in the concentration levels of various metabolites associated with the two groups (Fig. 5b); correlation coefficients close to +1 or −1 are deemed strong correlations. A total of 30 metabolites were identified by the different statistical analyses as discriminators between the two groups of patients.
Figure 5.
(A) Top 15 important features identified by PLS-DA. (B) Top 25 important features selected by correlation analysis with positive correlation coefficients indicate metabolites increased in control PBSC, while negative correlations are associated with increased levels in tMDS/AML cases.
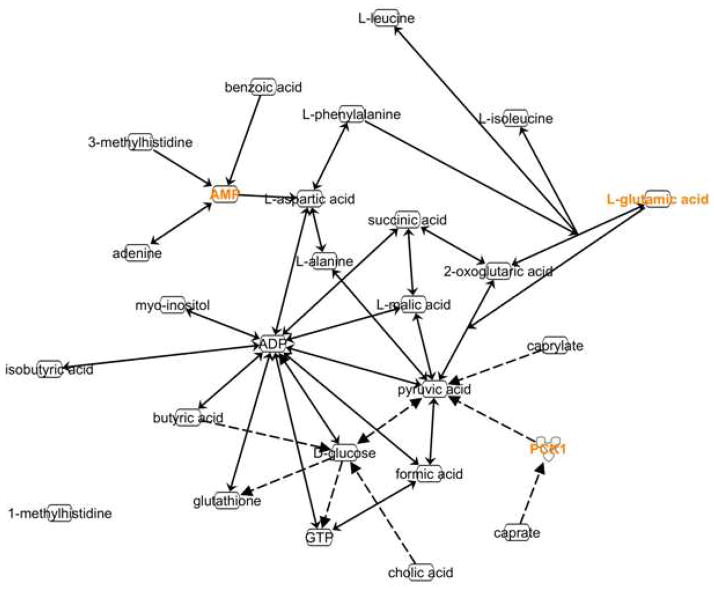
To gain insights into dysfunctions of the biological pathways associated with t-MDS/AML, the connections between the metabolites that discriminate the case group from the control group were established through the use of the Metabolomics module of Ingenuity Pathways Analysis (IPA, Ingenuity® Systems, www.ingenuity.com) (Fig. 6). Most of these metabolites are connected with each other through a direct chemical reaction (indicated by solid lines). Only three metabolites, caprate, caprylate, and cholate, are connected to the other metabolites indirectly (dashed lines). Three metabolites are connected to AMP, whose level was not significantly different according to the statistical analyses. An enzyme, phosphoenolpyruvate carboxykinase 1 (PCK1), was needed to connect pyruvate to caprate, suggesting that the activity of the enzyme is different between the two groups. Canonical pathways analysis identified the pathways from the IPA library of canonical pathways that were most significantly different between patients that develop and did not develop t-MDS (Table 2). The top canonical pathways are alanine and aspartate metabolism, glyoxylate and decarboxylate metabolism, phenyalanine metabolism, citrate cycle, aminoacyl-tRNA biosynthesis, and butanoate metabolism. All of these pathways are implicated by four or more molecules.
Figure 6.
Schematic representation of the interaction between significantly relevant hydrophilic metabolites. Ingenuity Pathway software was used to search connections between selected metabolites. Direct biochemical reactions are indicated by solid arrows; indirect effects are represented by dashed arrows.
Table 2.
Top Canonical Pathways identified through Ingenuity Pathway Analysis
| Ingenuity Canonical Pathway | p-value | Significant Metabolites |
|---|---|---|
| Alanine and Aspartate Metabolism | 1.25E-06 | L-alanine, pyruvic acid, L-malic acid, L-aspartic acid, 2-oxoglutaric acid, succinic acid |
| Glyoxylate and Dicarboxylate Metabolism | 1.03E-04 | pyruvic acid, L-malic acid, formic acid, 2-oxoglutaric acid, succinic acid |
| Phenylalanine Metabolism | 2.1E-04 | pyruvic acid, L-phenylalanine, benzoic acid, succinic acid |
| Citrate Cycle | 2.57E-04 | pyruvic acid, L-malic acid, 2-oxoglutaric acid, succinic acid |
| Aminoacyl-tRNA Biosynthesis | 3.92E-04 | L-alanine, L-isoleucine, L-leucine, L-phenylalanine, L-aspartic acid |
| Butanoate Metabolism | 5.19E-04 | pyruvic acid, 2-oxoglutaric acid, butyric acid, succinic acid |
| Valine, Leucine and Isoleucine Biosynthesis | 7.35E-04 | L-isoleucine, L-leucine, pyruvic acid |
| Glutamate Metabolism | 8.14E-04 | glutathione, L-malic acid, 2-oxoglutaric acid, succinic acid |
| Pyruvate Metabolism | 0.0035 | pyruvic acid, L-malic acid, formic acid |
| Cysteine Metabolism | 0.0035 | L-alanine, glutathione, pyruvic acid |
| Histidine Metabolism | 0.00527 | L-aspartic acid, 2-oxoglutaric acid,3-methylhistidine |
| Nitrogen Metabolism | 0.0075 | L-phenylalanine, formic acid, L-aspartic acid |
| Arginine and Proline Metabolism | 0.0509 | pyruvic acid, L-aspartic acid, adenine |
Fisher’s exact test was used to calculate a p-value determining the probability that the association between the genes in the dataset and the canonical pathway is explained by chance alone.
NMR Profiling of Lipophilic Metabolites in pre-aHCT PBSC samples
Spectra from the chloroform extracted fractions also showed significant differences between the two outcome groups (Figure 7). The Chenomx library does not currently include lipid groups, therefore, assignments were made with comparisons to published lipid NMR data.17–21 The most prominent resonances observed were those from lipid/cholesterol methyl (0.8–1.1 ppm) and methylene (1.1–1.4 ppm) protons, and lipid propionyl (1.4–1.7 ppm) protons, originating from different lipid molecules that contain such functional groups. Because of resonance degeneracy of lipid molecules in general, NMR spectra do not identify individual lipid molecules. Therefore, statistical analysis was carried out by comparing signal intensity (given as integration) of defined spectral regions. PCA and PLS-DA analyses were applied to identify the spectral features distinguishing the two groups of patients. Again, the largest principle components of PCA analysis did not allow separation of the two groups (Fig. 8). However, PLS-DA analysis provided good separation using the largest three latent variables (LV1–LV3) (Fig. 9).
Figure 7.
Proton NMR of lipophilic fractions from PBSC samples. The intensity of the downfield region is increased to facilitate identification of resonances. Spectra in black represent typical results for a control patient, while those in red present those for a t-MDS/AML patient.
Figure 8.
Principal components analysis of lipophilic profile differences between patient groups. (A) PCA scores of components 1 and 2 from spectral binning of control PBSC samples (green) and t-MDS/AML patient PBSC (red) spectra. (B) PCA scores of components 1, 2, and 3 for the control PBSC (green) and t-MDS/AML PBSC (red). The explained variances are shown in parentheses.
Figure 9.
Partial least squares discriminant analysis of metabolite concentrations between patient groups. (A) PLS-DA scores of latent variables 1 and 2 of lipophilic spectral bins from control PBSC samples (green) and t-MDS/AML patient samples (red). (B) PLS-DA scores of latent variables 1, 2, and 3 for the control PBSC (green) and t-MDS/AML PBSC (red). The explained variances are shown in parentheses.
In similar fashion to the analysis of the hydrophilic metabolites, VIP and correlation analyses were used to determine spectral features that contribute to the successful separation of the patient groups by PLS-DA. The regions common to these results are 1.8–2.0 ppm (-CH2-CH=), 2.30–2.325 ppm (-CH2-COOH), 4.05–4.075 ppm (-CH2-O-P-), 4.175–4.2 ppm (-CH2-O-CO-R-, -O-CH2-CH2-N+-), and 5.35–5.40 ppm (-HC=CH-). Correlation analysis also identified increased intensities within the 1.45–1.50 ppm area, corresponding to the resonances from –CH2-CH2-COOH, from the control patient samples. These results indicate differences in the lipid synthesis and/or modifications between the two groups of samples. The cellular pathways identified from hydrophilic samples discussed above can lead to the differences in the lipophilic molecules observed here. For example, increase in signal intensity at 5.35–5.40 ppm (-HC=CH-) was shown to correlate to mitochondria damage from chemotherapeutic drugs.22,23 Taken together, the lipophilic fractions have also indicated intrinsic cellular dysfunctions in patients that develop t-MDS in comparison to the controls.
Discussion
The NMR-based metabolomic analyses have found significant differences between PBSC obtained from patients who develop t-MDS/AML and those from patients who do not develop t-MDS/AML. These differences have suggested cellular pathways whose dysfunctions are associated with the development of t-MDS/AML in patients undergoing aHCT for HL and NHL. These dysfunctions could be caused by pre-aHCT therapeutic exposures, transplant conditioning regimens, autograft collection and hematopoietic regeneration, as found to associate with t-MDS/AML development by epidemiology studies.2,6,7 The top canonical pathways include alanine and aspartate metabolism, glyoxylate and dicarboxylate metabolism, phenylalanine metabolism, citrate acid cycle, and aminoacyl-t-RNA biosynthesis. All of these pathways are indicated by at least four molecules and related to mitochondria function.
Dysfunctions in the above mentioned cellular pathways will result in decreased ability of the cells to tolerate chemotherapy and radiation, leading to DNA mutations. For example, decreased glutathione production is associated with t-MDS/AML cases (Fig. 5b), suggesting decreased ability of these patients to detoxify reactive oxygen species generated by chemo and radiation therapy. We attempted to use 13C-labeled glutamine to follow its mitochondrial metabolism to further confirm differences in mitochondria functions within the PBSC samples as was previously shown in cultured breast cancer cells;15 however, these primary cells grew too slowly to utilize glutamine, and as such, measurable levels of the labeled glutamine and its metabolites were not detected from cellular extracts by 1H-13C-HSQC.
Although the number of samples in each group is limited, due to the limited amount of PBSC samples available, the conclusion described here is very consistent with cellular pathway analysis carried using microarray studies of gene expression profiles for the same set of patient samples.24 The microarray analysis was carried out specifically for the CD34+ hematopoietic stem cells isolated from PBSC. More than thirty-five samples from t-MDS patients and fifty control samples were analyzed and have identified similar cellular pathways as identified by NMR-based metabolomics of the entire PBSC. Because other cells in PBSC are all differentiated from CD34+ stem cells, they likely carry similar cellular dysfunctions due to similar genetic backgrounds, consistent with the similar cellular dysfunctions found by NMR-based metabolomics of PBSC and microarray analysis of gene expression profiles of CD34+ stem cells. Thus, the conclusions obtained from NMR-based metabolomics are further supported by microarray analysis using a larger sample size. All of these results indicate that differences in patients, representing predisposing factors contributing to risk of t-MDS/AML, can be identified months to years prior to the development of t-MDS/AML. These results support the efforts to discover biomarkers for identification of patients at risk prior to exposure to further stem cell damaging therapy. Because PBSC is blood producing stem cells, similar metabolic signatures may also occur in plasma or serum samples for discovery of feasible biomarkers. Such biomarkers will allow physicians to properly allocate resources to follow selected patients more closely than all patients, and will have a major impact in enhanced overall outcome.
Figure 10.
(A) Top 15 important spectral bins identified by PLS-DA. (B) Top 25 important spectral bins selected by correlation analysis, with positive correlation coefficients correlating with control PBSC (increased levels compared to cases) and negative correlations indicating correlation with t-MDS/AML cases (increased levels compared to controls.
Acknowledgments
This work was supported by NIH grants R01 HL083050 to RB, R01 GM074748 and R01 086171 to YC, P50 CA107399, and General Clinical Research Center Grant #5M01 RR00043.
References
- 1.Hauke RJ, Armitage JO. Treatment of non-Hodgkin lymphoma. Current Opinion in Oncology. 2000;12(5):412–418. doi: 10.1097/00001622-200009000-00005. [DOI] [PubMed] [Google Scholar]
- 2.Bhatia S, Ramsay NK, Steinbuch M, Dusenbery KE, Shapiro RS, Weisdorf DJ, Robison LL, Miller JS, Neglia JP. Malignant neoplasms following bone marrow transplantation. Blood. 1996;87(9):3633–3639. [PubMed] [Google Scholar]
- 3.Miller JS, Arthur DC, Litz CE, Neglia JP, Miller WJ, Weisdorf DJ. Myelodysplastic syndrome after autologous bone marrow transplantation: an additional late complication of curative cancer therapy [see comments] Blood. 1994;83(12):3780–3786. [PubMed] [Google Scholar]
- 4.Pedersen-Bjergaard J, Andersen MK, Christiansen DH. Therapy-related acute myeloid leukemia and myelodysplasia after high-dose chemotherapy and autologous stem cell transplantation. Blood. 2000;95(11):3273–3279. [PubMed] [Google Scholar]
- 5.Stone RM, Neuberg D, Soiffer R, Takvorian T, Whelan M, Rabinowe SN, Aster JC, Leavitt P, Mauch P, Freedman AS. Myelodysplastic syndrome as a late complication following autologous bone marrow transplantation for non-Hodgkin’s lymphoma. J Clin Oncol. 1994;12(12):2535–2542. doi: 10.1200/JCO.1994.12.12.2535. [DOI] [PubMed] [Google Scholar]
- 6.Kalaycio M, Rybicki L, Pohlman B, Sobecks R, Andresen S, Kuczkowski E, Bolwell B. Risk Factors Before Autologous Stem-Cell Transplantation for Lymphoma Predict for Secondary Myelodysplasia and Acute Myelogenous Leukemia. J Clin Oncol. 2006;24(22):3604–3610. doi: 10.1200/JCO.2006.06.0673. [DOI] [PubMed] [Google Scholar]
- 7.Krishnan A, Bhatia S, Slovak ML, Arber DA, Niland JC, Nademanee A, Fung H, Bhatia R, Kashyap A, Molina A, O’Donnell MR, Parker PA, Sniecinski I, Snyder DS, Spielberger R, Stein A, Forman SJ. Predictors of therapy-related leukemia and myelodysplasia following autologous transplantation for lymphoma: an assessment of risk factors. Blood. 2000;95(5):1588–1593. [PubMed] [Google Scholar]
- 8.Daviss B. Growing Pains for Metabolomics. The Scientist. 2005;19(8):25–28. [Google Scholar]
- 9.Nicholson JK, Lindon JC, Holmes E. ‘Metabonomics’: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999;29(11):1181–1189. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- 10.Odunsi K. Cancer diagnostics using 1H-NMR-based metabonomics. Ernst Schering Foundation symposium proceedings; 2007. pp. 205–226. [DOI] [PubMed] [Google Scholar]
- 11.Vinayavekhin N, Homan EA, Saghatelian A. Exploring Disease through Metabolomics. ACS Chemical Biology. 2009;5(1):91–103. doi: 10.1021/cb900271r. [DOI] [PubMed] [Google Scholar]
- 12.Fiehn O. Metabolomics – the link between genotypes and phenotypes. Plant Molecular Biology. 2002;48(1):155–171. [PubMed] [Google Scholar]
- 13.Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y, Nyati MK, Ahsan A, Kalyana-Sundaram S, Han B, Cao X, Byun J, Omenn GS, Ghosh D, Pennathur S, Alexander DC, Berger A, Shuster JR, Wei JT, Varambally S, Beecher C, Chinnaiyan AM. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457(7231):910–4. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 14.Wishart DS. Quantitative metabolomics using NMR. TrAC Trends in Analytical Chemistry. 2008;27(3):228–237. [Google Scholar]
- 15.Cano KE, Li Y-J, Chen Y. NMR Metabolomic Profiling Reveals New Roles of SUMOylation in DNA Damage Response. Journal of Proteome Research. 2010;9(10):5382–5388. doi: 10.1021/pr100614a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xia J, Psychogios N, Young N, Wishart DS. MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucl Acids Res. 2009;37(suppl_2):W652–660. doi: 10.1093/nar/gkp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kriat M, Vion-Dury J, Confort-Gouny S, Favre R, Viout P, Sciaky M, Sari H, Cozzone PJ. Analysis of plasma lipids by NMR spectroscopy: application to modifications induced by malignant tumors. Journal of Lipid Research. 1993;34(6):1009–1019. [PubMed] [Google Scholar]
- 18.Rezzi S, Giani I, Héberger K, Axelson DE, Moretti VM, Reniero F, Guillou C. Classification of Gilthead Sea Bream (Sparus aurata) from 1H NMR Lipid Profiling Combined with Principal Component and Linear Discriminant Analysis. Journal of Agricultural and Food Chemistry. 2007;55(24):9963–9968. doi: 10.1021/jf070736g. [DOI] [PubMed] [Google Scholar]
- 19.Gottschalk M, Ivanova G, Collins DM, Eustace A, O’Connor R, Brougham DF. Metabolomic studies of human lung carcinoma cell lines using in vitro 1H NMR of whole cells and cellular extracts. NMR in Biomedicine. 2008;21(8):809–819. doi: 10.1002/nbm.1258. [DOI] [PubMed] [Google Scholar]
- 20.Bonzom PMA, Nicolaou A, Zloh M, Baldeo W, Gibbons WA. NMR lipid profile of Agaricus bisporus. Phytochemistry. 1999;50(8):1311–1321. [Google Scholar]
- 21.Adosraku RK, Choi GT, Constantinou-Kokotos V, Anderson MM, Gibbons WA. NMR lipid profiles of cells, tissues, and body fluids: proton NMR analysis of human erythrocyte lipids. J Lipid Research. 1994;35(11):1925–1931. [PubMed] [Google Scholar]
- 22.Griffin JL, Lehtimäki KK, Valonen PK, Grohn OHJ, Kettunen MI, Yla-Herttuala S, Pitkanen A, Nicholson JK, Kauppinen RA. Assignment of 1H Nuclear Magnetic Resonance Visible Polyunsaturated Fatty Acids in BT4C Gliomas Undergoing Ganciclovir-Thymidine Kinase Gene Therapy-induced Programmed Cell Death. Cancer Research. 2003;63(12):3195–3201. [PubMed] [Google Scholar]
- 23.Delikatny EJ, Cooper WA, Brammah S, Sathasivam N, Rideout DC. Nuclear Magnetic Resonance-visible Lipids Induced by Cationic Lipophilic Chemotherapeutic Agents Are Accompanied by Increased Lipid Droplet Formation and Damaged Mitochondria. Cancer Research. 2002;62(5):1394–1400. [PubMed] [Google Scholar]
- 24.Li L, Li M, Sun C-L, Sabado MD, Francisco L, McDonald T, Chang KL, Wang S, Radich J, Zhao LP, Forman SJ, Bhatia S, Bhatia R. Gene Expression Changes in CD34+ Cells Precede Development of Therapy-Related Leukemia (t-MDS/AML) After Autologous Hematopoietic Cell Transplantation (aHCT) for Hodgkin (HL) or Non-Hodgkin Lymphoma (NHL) ASH Annual Meeting Abstracts. 2009;114(22):677. [Google Scholar]