Abstract
The mTOR pathway is the central regulator of cell size1. External signals from growth factors and nutrients converge on the mTORC1 multi-protein complex to modulate downstream targets, but how the different inputs are integrated and translated into specific cellular responses is incompletely understood2–4. Deregulation of the mTOR pathway occurs in polycystic kidney disease (PKD)5–7, where cilia (filiform sensory organelles) fail to sense urine flow because of inherited mutations in ciliary proteins8. We therefore investigated if cilia have a role in mTOR regulation. Here, we show that ablation of cilia in transgenic mice results in enlarged cells when compared with control animals. In vitro analysis demonstrated that bending of the cilia by flow is required for mTOR downregulation and cell-size control. Surprisingly, regulation of cell size by cilia is independent of flow-induced calcium transients, or Akt. However, the tumour-suppressor protein Lkb1 localises in the cilium, and flow results in increased AMPK phosphorylation at the basal body. Conversely, knockdown of Lkb1 prevents normal cell-size regulation under flow conditions. Our results demonstrate that the cilium regulates mTOR signalling and cell size, and identify the cilium-basal body compartment as a spatially restricted activation site for Lkb1 signalling.
The mammalian target of rapamycin (mTOR) pathway has a crucial role in metabolism and cell growth1. mTOR signalling is executed by two multi-protein complexes, mTORC1 and mTORC2. mTORC1 is activated by the GTPase Rheb to phosphorylate p70S6 kinase (S6K) and 4E-BP1 and thereby stimulate protein synthesis, but is effectively inhibited by treatment of cells with rapamycin. mTORC1 activity is regulated by diverse signals2–4. Growth factors and amino acids activate mTORC1, whereas energy stress and the tumour suppressor Lkb1 inhibit mTORC1-mediated signalling through the energy sensor, AMP-activated protein kinase (AMPK)9. Although knowledge of intracellular signal-transduction events is rapidly increasing, little information exists on where different external signals are processed to regulate mTOR signalling4.
We hypothesized that cilia have a role in mTOR signalling. Cilia are signalling platforms that protrude as filiform organelles from the plasma membrane, and rely on kinesin-driven intraflagellar transport (IFT) for their form and function10,11. They function as mechanosensors, which generates calcium currents12, have a pivotal role in the hedgehog pathway13,14 and are involved in Wnt signalling15. Mutations of ciliary proteins result in developmental defects, including situs-inversus and polydactyly, and postnatal diseases, such as retinal degeneration, obesity and polycystic kidney disease (PKD)8. In PKD the tubular geometry of kidneys is distorted and fluid-filled cysts replace renal parenchyma16. One hypothesis of why tubular cells are unable to maintain the tubular diameter is that there is a failure by the cilia to sense urine flow17; however, the downstream effects of flow sensing are unknown. Cyst epithelia have increased mTORC1 activity7. Interestingly, mTOR inhibitors markedly reduce cyst formation in PKD animal models5–7, and are currently being tested in clinical trials18,19. However, the mechanism of mTOR deregulation in PKD is not established. Polycystin-1, the most commonly mutated protein in autosomal dominant PKD (ADPKD), interacts with mTOR7 and reduces mTORC1 activity20, but the role of cilia in mTOR regulation has not been investigated.
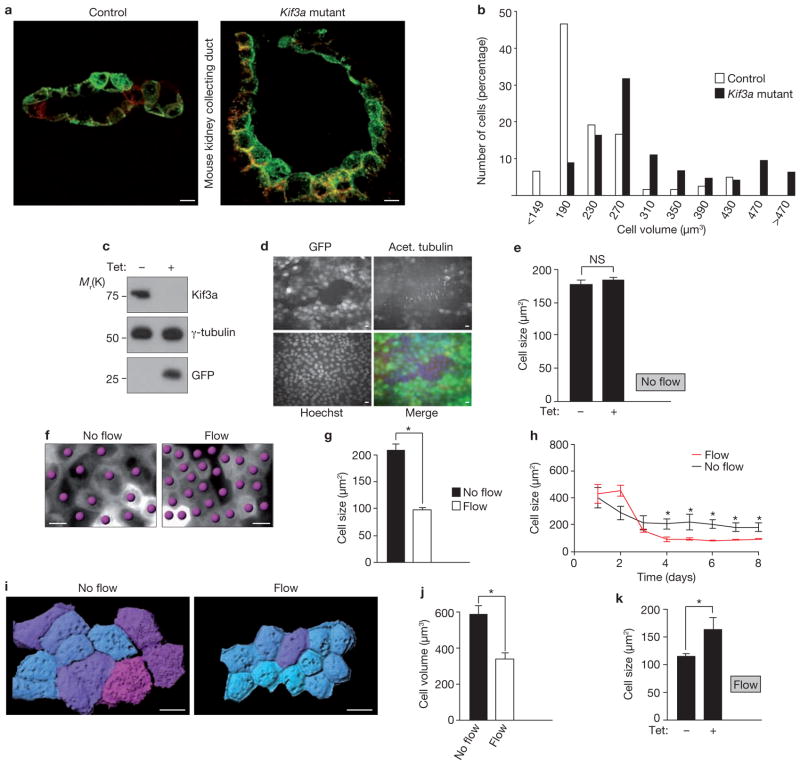
In polycystic kidneys, cells lining the cysts are larger than normal tubular cells21, raising the possibility that cilia have a role in cell-size control. To determine whether the loss of primary cilia affects cell size in vivo, we measured cell volumes in renal collecting ducts of mouse kidneys with a mutation in the kinesin subunit, Kif3a22. These mice lose cilia by postnatal day 10 (P10) and start to form cysts at P14 (ref. 22). The collecting duct cells of Kif3a mutants were larger than cells in control animals (Fig. 1a) and the size distribution of Kif3a-mutant cells was shifted to the right (Fig. 1b), suggesting that primary cilia have a role in the regulation of cell volume.
Figure 1.
Cilia regulate cell size under flow conditions. (a) 3D reconstruction of collecting ducts in kidney sections from control mice (left) and kidney-specific Kif3a-mutant-mice (right). Apical membrane stained with DBA; green, basolateral membrane stained with aquaporin-3; red. Mean cell volumes: controls, 233 ± 26 μm3 (n = 120); Kif3a mutants, 314 ± 24 μm3 (n = 189). (b) Cell volumes in control and Kif3a-mutant kidneys. In the Kif3a mutants, 22% of cells had volumes > 391 μm3 versus 6.3% in controls (P < 0.001, Mann-Whitney U test). (c) Western blot of lysates from Kif3a-i MDCK cells. Cells were established so that expression of shRNA against Kif3a, and a GFP reporter, were inducible with tetracycline. Cells were treated with or without tetracycline, as indicated. (d) Representative fluorescence microscopy images of Kif3a-i cells treated with tetracycline to express GFP (green in merged image) and knockdown Kif3a, and stained with anti-acetylated tubulin (red in merged image) and Hoechst (blue in merged image). GFP-positive cells: 85% (Supplementary Information Fig. S1a). (e) Size of Kif3a-i cells with and without tetracycline under non-flow conditions. n = 3. (f) Representative DIC microscopy images of MDCK cells grown under flow or non-flow conditions. Cells indicated by dots. (g) MDCK cell size after 6 days under indicated flow conditions (no flow, n = 12; flow, n = 21; asterisk indicates P < 0.0001). (h) Time course microscopy analysis of cell size. Data are from a single representative experiment for each condition. 5–10 visual fields per data point. Data are means ± s.d. Asterisks indicate P < 0.001. (i) 3D rendering of cells after in vivo membrane staining. Cells grown without flow, or after 5 days of flow. Cyan; small cells, magenta; large cells. (j) Mean cell volumes of wild-type MDCK cells under non-flow (n = 9, 153 cells), compared with flow (n = 6, 194 cells, P < 0.001). (k) Cell sizes of Kif3a-i cells grown under flow conditions with tetracycline (n = 21) and without tetracycline (n = 13). Asterisk indicates P < 0.001. Data in e, g, j and k are means ± s.e.m. Scale bars, 10 μm.
To facilitate the study of how cilia regulate cell size, we generated MDCK cells with inducible knockdown of Kif3a (Kif3a-i). On incubation of these cells with tetracycline, Kif3a is knocked down and GFP (green fluorescent protein) is expressed (Fig. 1c). Immunostaining for acetylated tubulin, a cilia marker, confirmed that GFP-positive knockdown cells lack cilia (Fig. 1d and Supplementary Information, Fig. S1a). However, in contrast to the in vivo findings, no size difference was seen in Kif3a-depleted cells, compared with non-induced control cells (Fig. 1e). This discrepancy suggested that physiological requirements for cell-size control were missing in the in vitro experiment. In renal tubules, cilia function as flow sensors12,23, so we hypothesized that bending of the cilia by fluid flow might be the physiological stimulus that regulates cell size. To test this hypothesis, we analysed ciliated MDCK cells in a flow chamber that allows cultivation of cells for several days under permanent fluid flow, mimicking the physiological conditions in renal tubules24. Interestingly, after 6 days under flow conditions the average cell size appeared smaller than in cells grown without flow (Fig. 1f, g). To further validate this finding, we performed a time-course analysis and found that cell size decreases between day 1 and 4, but with no further decrease from day 4 to 8 (Fig. 1h and Supplementary Information, Fig. S1b). Cells in stationary medium also reached a plateau after 4 days, but remained significantly larger, despite a similarly low mitotic index24 (Supplementary Information, Fig. S1c). Further analysis of cross-sections was performed to ensure that differences in the x–y plane are not offset by different cell heights, but no difference was found in z-axis measurements (Supplementary Information, Fig. S1d, e). Similarly, three-dimensional (3D) volume estimation of membrane-labelled cells demonstrated that cells under flow were significantly smaller than controls (Fig. 1i, j). These findings raised the possibility that cilia require flow to regulate cell size. Indeed, when Kif3a-i cells were analysed in the flow chamber the cell size of Kif3a-depleted cells was significantly larger than non-induced cells (Fig. 1k and Supplementary Information, Fig. S1f). Thus, our in vitro findings in the flow chamber mimic the deregulation of cell size in Kif3a-mutant mice and demonstrate that bending of cilia by flow is critical for cell size regulation.
We tested our hypothesis in a second cilia-deficient cell line, as Kif3a has known extra-ciliary functions25. We constructed MDCK cells for the inducible depletion of the IFT-molecule Ift88 (Supplementary Information, Fig. S1g–i). Mutation of Ift88 leads to loss of cilia and causes PKD in mice26,27. Knockdown of Ift88 effectively suppressed cilia formation (Supplementary Information, Fig. S1j). Similarly to Kif3a-depleted cells, unciliated Ift88-i cells under flow were significantly larger when compared with non-induced cells (Supplementary Information, Fig. S2a–d), but no size difference was observed in two independent control cell-lines (Supplementary Information, Fig. S2e–g). These results confirm that cell-size regulation under flow requires cilia.
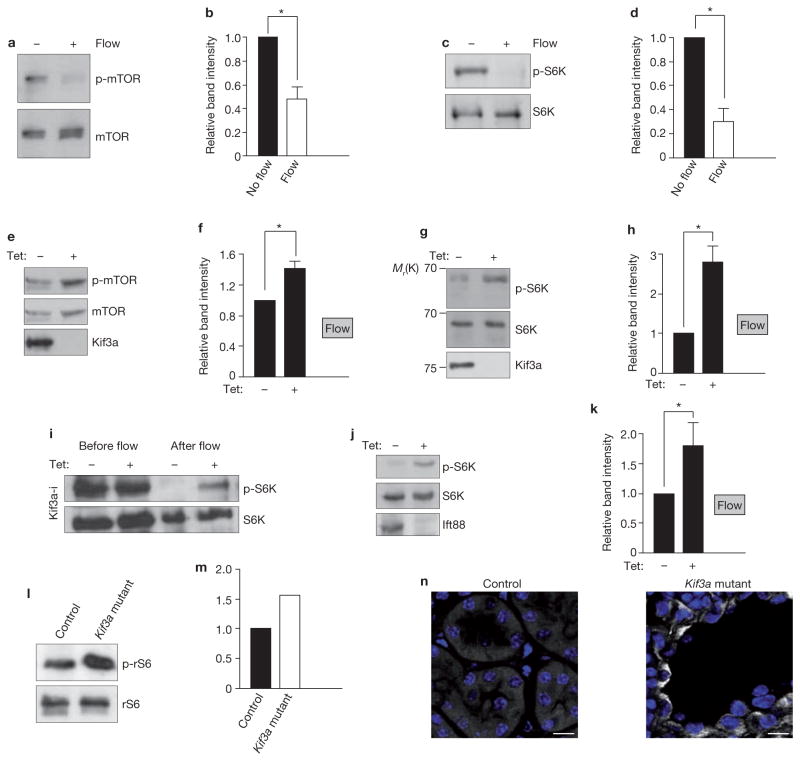
To test whether cilia regulate cell size through the mTOR pathway, we analysed cell lysates from ciliated MDCK cells for phosphorylation of mTOR and its target S6K, established markers of mTORC1 actvity28. We found that cells under flow had less phosphorylated mTOR and S6K when compared with cells in stationary medium (Fig. 2a–d), thus correlating with the decreased cell size. Conversely, the levels of phosphorylated mTOR and S6K increased after depletion of Kif3a in cells grown under flow (Fig. 2e–h), which corresponds to the larger size of these cells, compared with control cells. We hypothesized that flow represses mTORC1 in ciliated cells, and that cells without cilia fail to downregulate mTOR. Indeed, in ciliated Kif3a-i cells after 5 days under flow, phosphorylated S6K was decreased, compared with baseline before flow, whereas in Kif3a-i cells induced with tetracycline and therefore unciliated, the downregulation of phosphorylated S6K was less pronounced (Fig. 2i). Analogous results were obtained in Ift88-i cells (Fig. 2j, k), but not in control lines (Supplementary Information, Fig. S2h, i). Similarly to our in vitro data, Kif3a mutant kidneys contained more phosphorylated ribosomal-S6-protein (rS6), an S6K target, than kidneys from control animals (Fig. 2l, m) and epithelial phosphorylated rS6 staining was found in mutant mice but not in controls (Fig. 2n). These results confirm a positive correlation between cell size and mTORC1 activity and suggested that the cell size changes are mediated through cilia-dependent downregulation of the mTOR pathway.
Figure 2.
Cilia and flow modulate mTORC1 activity. (a) Western blot to assess mTOR phosphorylation in ciliated wild-type MDCK cells grown under flow and non-flow conditions. (b) Intensity of phosphorylated mTOR (p-mTOR) bands, relative to intensity of mTOR bands, from four independent experiments performed as in a. Asterisk indicates P < 0.05. (c) Western blot to assess phosphorylation of S6K (p-S6K) in ciliated MDCK cells grown under flow and non-flow conditions. (d) Intensity of p-S6K bands, relative to intensity of S6K bands, from three independent experiments performed as in c. Asterisk indicates P < 0.05. (e) Western blot to assess how induction of Kif3a knockdown in Kif3a-i cells under flow affects mTOR phosphorylation. (f) Intensity of p-mTOR bands, relative to intensity of mTOR bands, from four independent experiments performed as in e. Asterisk indicates P < 0.05. (g) Western blot to assess how induction of Kif3a knockdown in Kif3a-i cells under flow affects S6K phosphorylation. (h) Intensity of p-S6K bands relative to intensity of S6K bands, from four independent experiments performed as in g. Asterisk indicates P < 0.05. (i) Western blot of lysates from Kif3a-i cells, treated with tetracycline as indicated, before the onset of flow and after 5 days under flow conditions, to assess S6K phosphorylation. (j) Western blot of lysates from cells with tetracycline-inducible expression of Ift88 shRNA, treated with tetracycline as indicated, and grown under flow conditions. (k) Intensity of p-S6K bands, relative to intensity of S6K bands, from five independent experiments, performed as in j. Asterisk indicates P < 0.01. (l) Western blot to assess levels of phosphorylated ribosomal S6 protein (p-rS6) in the kidneys of wild-type and Kif3a-mutant mice. (m) Intensity of the p-rS6 bands, relative to the intensity of rS6 bands, from two independent experiments performed as in l. (n) Representative images from immunofluorescence microscopy of wild-type and Kif3a-mutant mice kidney sections at postnatal day 21. Sections are stained with antibodies against p-rS6 and Hoechst (blue). Scale bars, 10 μm. Data in b, d, f, h and k are means ± s.e.m. Uncropped images of blots are shown in Supplementary Information, Fig. S6.
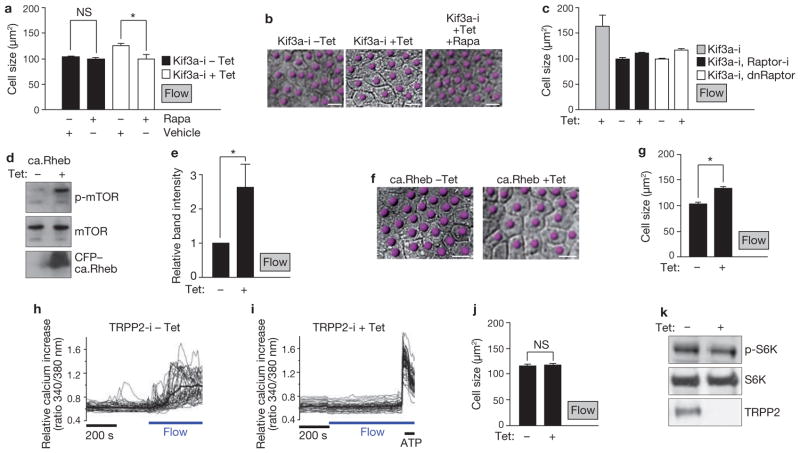
To further test if cell-size regulation in vitro occurs through mTOR we treated ciliated cells under flow with rapamycin29. Compared with vehicle-treated cells, rapamycin had no effect on the size of ciliated Kif3a-i cells (Kif3a-i cells without tetracycline; Fig. 3a) suggesting that flow maximally suppresses mTORC1 activity in ciliated cells. However, in unciliated cells, rapamycin compensated for the absence of cilia and resulted in smaller cells (Kif3a-i cells with tetracycline; Fig. 3a, b). In a complementary approach, co-expression of dominant-negative Raptor, a scaffold protein of mTORC129,30, or Raptor knockdown, in Kif3a-depleted cells, resulted in a similar size decrease under flow as in ciliated cells (Fig. 3c and Supplementary Information, Fig. S3a, b) and thus demonstrated that the size defect with absent cilia is because of ineffective downregulation of mTORC1.
Figure 3.
Flow-induced cell-size regulation is mTOR dependent and ablation of flow-induced calcium transients has no effect on cell size. (a) Size of Kif3a-i cells, in absence or presence of tetracycline and treated as indicated, under flow conditions. Uninduced Kif3a-i cells (filled bars), n = 6; with rapamycin, n = 6; P = 0.3. Kif3a-i knockdown cells (unfilled bars), n = 10; with rapamycin, n = 9. Asterisk indicates P < 0.001. (b) DIC microscopy images of Kif3a-i cells treated as indicated and grown under flow conditions for 5 days. Cells are indicated by dots. (c) Size of the indicated Kif3a-i cells, in absence or presence of tetracycline, under flow conditions. Raptor-i, Kif3a-i cells with tetracycline-inducible expression of Raptor shRNA; dnRaptor; cells with tetracycline-inducible expression of a dominant-negative Raptor mutant. Kif3a-i cells with tetracycline, n = 21. Kif3a-i, Raptor-i cells, n = 6. Kif3a-i, dnRaptor cells, n = 5. (d) Western blot to assess phosphorylation of mTOR after induction of a gain of function Rheb mutant in cells under flow conditions. (e) Intensity of p-mTOR bands, relative to intensity of mTOR bands, from five independent experiments performed as in d. Asterisk indicates P < 0.05. (f) Representative DIC microscopy images of MDCK cells expressing the tetracycline-inducible Rheb mutant, grown under flow conditions for 5 days. Cells are indicated by dots. (g) Size of MDCK cells expressing the tetracycline-inducible Rheb mutant, grown under flow conditions for 5 days. n = 14 (cells without tetracycline) and n = 16. Asterisk indicates P < 0.0001. (h, i) Intracellular calcium measurements of MDCK cells with tetracycline-inducible expression of TRPP2 shRNA (TRPP2-i cells), without tetracycline induction (h), and with tetracycline induction (i). Thin traces, individual cells; bold trace, mean calcium value. ATP stimulation demonstrates cell viability. Scale bar indicates length of 200 s on x axis. Flow indicates onset of flow. (j) Size of TRPP2-i cells under flow conditions. Without tetracycline; n = 7 and with; n = 10; P = 0.8. (k) Western blots of lysates from TRPP2-i cells under flow conditions and treated with tetracycline as indicated. Data in a, c, e, g and j are means ± s.e.m. Scale bars, 10 μm. Uncropped images of blots are shown in Supplementary Information, Fig. S6.
Using an opposite approach, we reasoned that activation of mTOR would counteract the flow-mediated cell-size reduction. We transduced MDCK cells with a tetracycline-inducible gain-of-function mutant of the mTORC1 activator, Rheb31 (RhebS16H; Supplementary Information, Fig. S3c). Induction of this construct activated mTOR (Fig. 3d, e) and resulted in significantly larger cells under flow than the same cells without induction (Fig. 3f, g). These results demonstrate that mechanical stimulation of cilia downregulates the mTOR pathway and cell size.
After establishing a link between cilia, mTOR and cell size, we were interested in the underlying mechanism. Protein kinase B (Akt) is a major activator of mTOR. Akt activates mTORC1 directly, as well as indirectly by releasing Rheb from inhibition by the Tsc complex2. We hypothesized that if mTOR deregulation through lack of cilia involved Akt activation, loss of Akt function would rescue the phenotype. However, expression of a dominant-negative Akt mutant in Kif3a-depleted cells neither abrogated cell-size deregulation nor S6K phosphorylation under flow (Supplementary Information, Fig. S3d–f), suggesting that a role for Akt in ciliary mTOR regulation is unlikely.
The mechanosensory function of cilia in the kidney involves the cation channel polycystin-2 (TRPP2), which is mutated in a subset of patients with ADPKD. TRPP2 translates fluid flow into calcium transients17. However, it is unclear if deficient flow-mediated calcium transients have a role in cyst formation32. We hypothesized that inhibition of the mTOR pathway through ciliary bending might be mediated by a calcium signal. Ciliated MDCK cells are a well-established model to analyse calcium signalling under flow12,23. Consequently, we generated tetracycline-inducible TRPP2 knockdown cells. Tetracycline induction of these cells resulted in efficient depletion of TRPP2 (Supplementary Information, Fig. S3g) and inhibited flow-induced calcium transients (Fig. 3h, i and Supplementary Information, Fig. S3h). Surprisingly, in cells grown under flow conditions, no difference was found in cell size or levels of phosphorylated S6K after knockdown of TRPP2, compared with control cells (Fig. 3j, k and Supplementary Information, Fig. S3i, j). This finding indicated that cell size and mTORC1 control by cilia occurs independently of flow-induced calcium signalling.
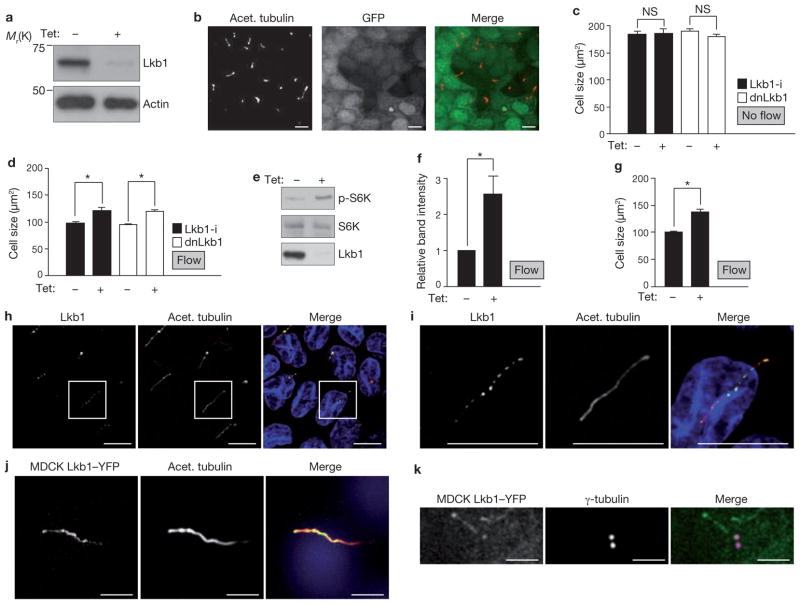
Next, we investigated the tumour suppressor, Lkb1, a negative regulator of mTOR9,33. In fact, loss of Lkb1 increases cell mass in pancreatic β-cells through activation of mTOR34. To test if Lkb1 has a role in the ciliary regulation of cell size, we generated MDCK cells with tetracycline-inducible knockdown of Lkb1 (Fig. 4a). Similarly to previous observations35, depletion of Lkb1 in MDCK cells did not affect markers of apico-basal cell polarity or cilia formation (Fig. 4b and Supplementary Information, Fig. S4). As expected, phosphorylation of AMPK, the target of Lkb1, was significantly decreased (Supplementary Information, Fig. S5a, b), thus validating the knockdown approach. Similarly to unciliated cells, Lkb1-depleted cells showed no difference in size when compared with non-induced cells, in the absence of flow (Fig. 4c). However, when subjected to flow, Lkb1-i cells were significantly larger than the controls without induction (Fig. 4d). Similar results were obtained in cells expressing dominant-negative mutant Lkb136 (Lkb1D194A; Fig. 4c, d and Supplementary Information, Fig. S5c), confirming that Lkb1 has a role in cell-size regulation under flow. The phosphorylation of S6K was increased in Lkb1 knockdown cells under flow (Fig. 4e, f) demonstrating that the size increase of Lkb1-deficient cells is accompanied by mTOR activation. Notably, expression of Lkb1 (Supplementary Information, Fig. S5d) in unciliated Kif3a-depleted cells did not rescue the absent-cilia phenotype (Fig. 4g and Supplementary Information, Fig. S5e) and suggested that cilia are required for Lkb1 activation.
Figure 4.
Lkb1 modulates flow-dependent mTOR and cell-size regulation and is localized in the basal body and the cilium. (a) Western blot of lysates from Lkb1-i MDCK cells. Cells were established so that expression of shRNA against Lkb1, and expression of GFP, were inducible with tetracycline. Cells were treated with or without tetracycline, as indicated. (b) Representative fluorescence microscopy images of Lkb1-i cells treated with tetracycline, which induces Lkb1 knockdown and GFP expression (green in merged image). Cilia are identified by acetylated tubulin (red). (c) Size of indicated cells, treated with tetracycline as indicated, under non-flow conditions. dnLkb1 cells; MDCK cells with tetracycline-inducible expression of a dominant-negative Lkb1 mutant. Lkb1-i, n = 3, dnLkb1, n = 3, 10 visual fields per n. (d) Size of indicated cells, treated with tetracycline as indicated, under flow (Lkb1-i cells, n = 8, P < 0.001, dnLkb1 cells, n = 12, P < 0.001). (e) Western blot to assess S6K phosphorylation in Lkb1-i cells treated with tetracycline as indicated under flow. (f) Intensity of p-S6K bands relative to intensity of S6K bands, from three independent experiments performed as in e. Asterisk indicates P < 0.05. (g) Kif3a-i cells overexpressing Lkb1 were treated with tetracycline, as indicated, under flow, and size of the cells was measured (n = 6, P < 0.001). (h) Immunofluorescence microscopy of cilia from wild-type MDCK cells immunostained with antibodies against the indicated proteins. (i) Higher-magnification images of h. (j) Fluorescence microscopy images of MDCK cells expressing Lkb1–YFP and immunostained with antibodies against acetylated tubulin. (k) Fluorescence microscopy images of MDCK cells expressing Lkb1–YFP and immunostained with antibodies against γ-tubulin. Scale bars, 2 μm. Data in (c, d, f and g are means ± s.e.m. An uncropped image of the blot in e is shown in Supplementary Information, Fig. S6. Scale bars: a, b, h–j, 10 μm; k, 2 μm.
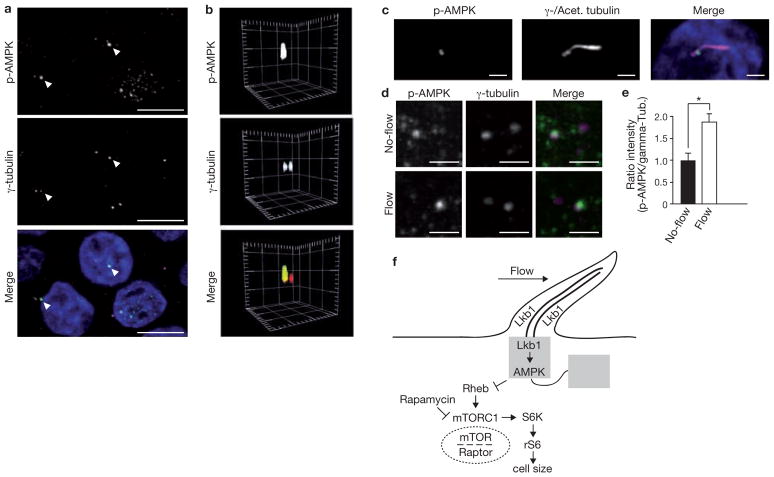
We then investigated the link between Lkb1 and the cilium. Lkb1 was present in cilia and the basal body, as visualized by immunostaining (Fig. 4h, i) and by overexpression of an Lkb1–YFP (yellow fluorescent protein) fusion protein (Fig. 4j, k and Supplementary Information, Fig. S5f). We hypothesized that the bending of cilia may activate the Lkb1 pathway, but levels of phosphorylated AMPK in whole cell lysates were similar in cells grown under flow and without (Supplementary Information, Fig. S5g, h). Strikingly, we found an intense accumulation of phosphorylated AMPK at the basal body (Fig. 5a–c). Furthermore, quantitative image analysis revealed that phosphorylated AMPK was significantly increased at the basal body of cells under flow, compared with cells grown in stationary medium (Fig. 5d, e). In summary, these findings demonstrate that bending of the cilium by flow activates the Lkb1 pathway in the cilia-basal body compartment to inhibit mTORC1 activity and cell size (Fig. 5f for schematic representation of a model).
Our data establish that cilia function as regulators in the mTOR signalling cascade and provide important insight into the mechanism of how extracellular signals are transduced to regulate mTOR activity. It is currently unclear how input–output specificity is achieved for a large number of extracellular signals that are translated into different mTOR effectors4. Our data suggest that the cilia-basal body compartment is where the Lkb1 pathway is activated on mechanical engagement of the cilium, and thus provide evidence of a spatially restricted regulatory site for mTOR signalling. Our data further demonstrate that, apart from calcium transients, mTOR regulation by Lkb1 in the cilium-basal body compartment represents an independent mechanosensory signal transduction mechanism that translates shear stress through bending of the cilium into cellular programmes. Our findings raise the intriguing question of whether cell size is important for tubular geometry of the kidney. Previous evidence links cell shape and surface tension with complex cellular programmes and morphogenesis37. Perfusion studies have shown that fluid flow and shear forces depend on tubular diameter38. It is conceivable that larger cells change the tubular diameter and thereby alter flow sensing. Whether cell-size-mediated changes in shear stress contribute to cyst formation remains to be shown in future work. Recent data indicate that polycystin 1 downregulates mTOR independently of flow20, suggesting that other pathways exist to modulate mTOR in renal epithelial cells.
It is possible that the cilium might have a role in mTOR signalling in contexts other than mechanotransduction. The cilium is a signalling organelle equipped with an intraflagellar transport system providing fast access for signalling molecules to a restricted compartment ideally suited for environmental sensing11, such as light in photoreceptors, smell in odorant receptors, and the regulation of Hedgehog and Wnt signalling11. Interestingly, the glucose transporter, Glut2, is localized in ependymal cilia in the brain39. Furthermore, cilia-deficient mice have impaired metabolic control after glucose challenge40, thus providing the basis for a functional link between glucose metabolism and cilia. Insulin signalling activates mTORC1 and glucose control is, at least in part, regulated by Lkb1 and AMPK in a complex mechanism that is not fully understood34. Together with these findings our results raise the possibility that, beyond mechanosensation, cilia may have a broader role in metabolic control through Lkb1, AMPK and mTOR signalling.
METHODS
Animals and in vivo cell volume measurements
Kidney-specific Kif3a-mutant mice were produced as described22. Briefly, Pkhd1–Cre transgenic mice expressing Cre recombinase under the control of the Pkhd1 promoter were crossed with Kif3aflox/− mice, and the resulting progeny were intercrossed to generate Pkhd1–Cre; Kif3aflox/− mice (the Kif3a mutant). Pkhd1–Cre; Kif3aflox/+ or Kif3a+/− littermates were used as controls. Mice were killed at postnatal day 16, and the kidneys were fixed and sectioned as described22. All experiments involving animals were performed under the auspices of the UT Southwestern Institutional Animal Care and Research Advisory Committee.
Tissue cryosections (6 μm thick) were co-stained with antibody against aquaporin-3 and FITC (fluorescein isothiocyanate)-coupled Dolichos biflorus agglutinin (DBA). Sections were prepared as previously described22. Images of the stained sections were acquired using an LSM 510 META laser scanning confocal microscope (Carl Zeiss MicroImaging GmbH). Mutant and control kidneys were imaged under identical magnification and exposure conditions. 3D images were reconstructed from z-stack images using Imaris software (Bitplane, Zurich, Switzerland). Single collecting duct cells stained with DBA and aquaporin-3 antibody were selected by cropping, and the cell volume was measured using Imaris software. Sections from mice for immunofluoresence microscopy staining of phosphorylated ribosomal S6 protein were obtained at postnatal day 21.
Transgenic cell lines
EB1–YFP has been previously described24. For inducible knockdown of Kif3a, Ift88, TRPP2, Lkb1 and Raptor, MDCK cells were first transduced with lentivirus encoding the tetracycline- sensitive tTR-KRAB repressor and a DsRed reporter41. Cells were then transfected with lentivirus encoding the specific shRNA, and a GFP reporter, (pLVTH vector), both under the control of tTR-KRAB. Target sequences: Kif3a (5′-AGGCTAGAGCTGAATTAGAG-3′), Ift88 (5′-GAAGGCAGCTGAATTCTAT-3′), TRPP2 (5′-GGAGG AGGCAAGTTA AACT-3′), Lkb1 (5′-GCTGGTGGACGTGTTATAC-3′) and Raptor (5′-GGCTAGTCTGTTTCGAAATTT-3′). Empty-vector control cells were obtained in the same way but lentivirus for the second transduction was prepared with pLVTH vector without shRNA, but still expressing GFP. Luci-i cells were obtained identically to Kif3a-i, with shRNA specific to luciferase (target sequence: 5′-CGTACGCGGAATACTTCGA-3′). To generate cells expressing the constitutively active Rheb mutant, mutant Rheb (RhebS16H; construct was a gift from R. Lamb, University of Alberta, Canada)31 was cloned into empty pLVTH vector in-frame with amino-terminal CFP (cyan fluorescent protein) after excision of GFP, and lentiviral transduction was performed as described above. Lkb1–YFP was cloned from canine cDNA into pLXSN in-frame with N-terminal Venus, a variant of the yellow fluorescent protein42, and stably expressed in MDCK cells after retroviral transduction. To generate Kif3a-i cells overexpressing Lkb1, canine Lkb1 was cloned into the GFP cassette of pLVTH and transduced by lentivirus into Kif3a-i cells. Dominant-negative Lkb1D194A was acquired through site-directed mutagenesis. Dominant-negative Raptor43 was generated by deleting the carboxy terminus of human Raptor (Addgene plasmid #1859) with primers (5′-ATGGAGTCCGAAATGCTGCAA-3′ and 5′-CGAGACTTGCCTTCTGGCCGGTGA-3′), and it was then cloned into pLVTH. The same was done for mouse dominant-negative AKT (construct was a gift from L. Cantley, Harvard Medical School, USA)44.
Antibodies, reagents, western blots and band quantification
For immunofluorescence microscopy staining, the following antibodies were used: anti-aquaporin-3 (1:400; Chemicon International), FITC-coupled Dolichos biflorus agglutinin (DBA, 1:400; Vector Laboratories), anti-Lkb1 (1:400; 27D19, Cell Signalling), anti-Zo-1 (1:400; #40-2200, Invitrogen), anti-acetylated tubulin (1:3000; T7451, Sigma-Aldrich), anti-Scribble (1:200; C20, SantaCruz), anti-Par3 (1:200; 07-330, Millipore), anti-E-cadherin (1:400; 610181, BD Transduction Laboratories), anti-β-Catenin (1:200; 610154, BD Transduction Laboratories), anti-GP135 (1:20; G. Ojakian, Suny Downstate Medical center, USA), anti-phospho-AMPK (1:200; 2535, Cell Signalling), anti-Ift88 (1:200; B. Yoder, University of Alabama, USA) and anti-Ift88 (1:200, 13967-1 AP, ProteinTech Group). Antibodies were visualized using Cy5-, Cy3- or Alexa-488-labelled secondary antibodies (1:1000; Jackson Immunoresearch) and Fluor 594-coupled goat anti-rabbit IgG (immunoglobulin G; 1:400). Hoechst 33342 was used for staining nuclei. Rapamycin (553211, Calbiochem) was solubilized in vehicle (DMSO) and used at 100 nM.
For western blotting, the following antibodies were used (1:1,000 dilution and from Cell Signaling unless otherwise indicated): anti-phospho-mTOR (Ser 2448) (2971), anti-total mTOR (2972), anti-phospho-AMPK (2535), anti-total AMPK (2532), anti-p70S6 kinase (9202), anti-phospho-p70S6 kinase (9205), anti-phospho-S6 ribosomal protein (4857), anti-Raptor (4978), anti-Lkb1 (27D10), anti-V5 (MCA1360, Serotec), anti-GFP (SC9996, Santa Cruz), anti-CFP (S98, MBL), anti-tubulin (1:3,000; T6557), anti-actin (1:5,000; A1978, both Sigma-Aldrich) and anti-Kif3a (611508, BD Transduction Labs). Monoclonal anti-TRPP2 (1:800) was raised against amino-acid residues 698–799 of human TRPP2 in collaboration with Nanotools (www.nanotools.de). Cells were grown in 10 cm cell-culture dishes. Cell lysis and western blot were performed as previously described24. Preparation of lysates from the flow chambers was done with urea buffer (8 M Urea, 10 mM Tris-HCl at pH 8.0, 100 mM Na2HPO4/NaH2PO4 and 0.2% (v/v) Triton X-100). Before blotting, the membranes were fixed in glutaraldehyde. Quantification of non-saturated bands was performed using ImageJ (http://rsb.info.nih.gov/ij/).
Immunofluorescence microscopy staining
For staining of cilia, cells were grown on glass coverslips for 10 days. The cells were fixed and immunofluorescence microscopy staining was performed as previously described24. For improved rendering of images acquired by epifluorescence microscopy, gamma adjustments were performed in Photoshop for Figure 1d and Supplementary Information Figure S1j. For the tight junction (staining with anti-Zo-1 and anti-Par3) and adherence junction (staining with anti-β-Catenin, anti-E-cadherin and anti-Scribble) staining, cells were grown for 2 days.
Phosphorylated AMPK quantification at the basal body was performed using an inverted laser-scanning microscope. Within the cell monolayer (5 days old, grown under non-flow or flow conditions) random fields of view were selected and z-stacks (15 z-planes, 0.5 μm z-distance, pixel time 1.61 μs and area 44.91 × 44.91 μm) were recorded. 3D reconstruction and measurements of phosphorylated AMPK and γ-tubulin were conducted with Zeiss Zen 2010 software.
Flow-chamber and imaging
Transgenic MDCK cells were seeded into closed perfusion chambers (Microslide I or Microslide VI; channel dimensions 50 × 5 × 0.4 mm and 17 × 3.8 × 0.4 mm, respectively, both with iBiTreat and collagen IV; Ibidi) as previously described24, and allowed to adhere for 24 h. The chamber was then connected to a computer-controlled setup containing an air-pressure pump and a two-way switching valve (Ibidi pump system 10902), pumping 40 ml of phenol-red-free cell-culture medium unidirectionally between two reservoirs through the flow channel at a rate of 1.0 or 0.6 ml min−1 (Microslide I or VI), corresponding to a shear stress of approximately 0.75–1 dyn per cm2. Each day, single plane images were randomly recorded (Axiovert 200M, Zeiss) at ten locations with a C-Apochromat ×63, 1.2 W objective for differential interference contrast (DIC) microscopy, or YFP, images. YFP excitation was performed with 490–510 nm and emission was collected by using a filter combination of Q515LP and HQ535/30 (AHF).
The mean cell size was calculated by counting cells manually in a defined area of 2700 μm2 in the centre of full-frame images, using Imaris software. Fully visible cells were counted as one, cells at the top and right margins were counted when ≥ 50% was visible and at the bottom and left margins cells were counted when ≥ 90% was visible. The mean cell size in a visual field was obtained by dividing the area size by the number of counted cells in each visual field. Ten visual fields were analysed in each flow channel once, after day 5, during the plateau phase.
For cell-height measurements, EB1–YFP, Kif3a-i and Ift88-i cell lines were grown under flow and non-flow conditions. Cell height was analysed using an inverted laser scanning microscope (LSM 510 Meta Duo Live) equipped with a Plan-Apochromat ×40/1.2 N.A. water objective. Within the cell monolayer 5–6 random fields of view (225 × 225 μm) for each condition were selected and z-stacks (33–54 z-planes, 0.5 μm z-distance, pixel time 1.28 μs) were recorded. 3D reconstruction and measurements were performed using LSM 510 Software (Version 4.2, Zeiss).
Fura2 ratio imaging was performed as previously described23. Briefly, ciliated cells grown on coverslips for 7–10 days were incubated with fura-2 AM (10 μM) and Probenecid (1 mM) and inserted into the parallel-plate flow chamber. Ratio imaging was performed in temperature-controlled conditions under an inverted microscope (Axiovert 200M, Zeiss).
Mitotic indices were calculated as previously described24. Briefly, MDCK cells stably expressing EB1–YFP were grown under flow and non-flow conditions. EB1 aggregates at mitotic spindles, which were counted daily together with the total number of cells (field of view size 14,477 μm2).
Automated volume measurements in non-fixed wild-type MDCK cells were performed using FM 1-43FX (F35355, Invitrogen, dilution 1:20, incubation for 10 min; FM 1-43FX exclusively stains membranes). Images were acquired with the LSM 510 Meta Duo Live microscope. Within the cell monolayer 4–5 random fields of view for each condition were selected and z-stacks (about 60 z-planes, 0.2 μm z-distance, pixel time 1.28 μs) were recorded. 3D reconstruction and measurements were performed using Imaris software.
Measurements were performed in the first 40 min after application of the membrane dye. To measure the cell volume, the acquired images were intensity-inverted: dark pixels ascribed with high intensity values and vice versa. Imaris ‘surface’ function was used without thresholding to auto-detect the cell volumes.
Epifluorescence images were obtained using an Axioplan2 with a Plan Neofluar ×63/1.2 N.A. oil objective (Zeiss). For confocal microscopy, fluorescence-imaging excitation of the fluorophores (Hoechst 33342, Alexa-488, Cy3/DsRed2 and Cy5) was performed at 405, 488, 561 and 633 nm, respectively. For detection of the emission signal at specified ranges, the spectral meta detector or normal photomultiplier channels were used with BP filter 420–480, BP 505–530, BP 575–615 and LP 650 nm. Confocal pinhole diameters were always adjusted to 1 μm sections.
Statistical analyses
Statistical analysis was carried out in Excel (Microsoft Cooperation). Because of variability between cell lines of different genetic background, only genetically identical cell lines were compared statistically. P values were calculated by unpaired t-test from the mean data of each flow channel (n), representing ten visual fields each. All values are given as mean of n ± s.e.m. The error bars in Figure 1h and Supplementary Information Figure 1f represent s.d.
The Mann-Whitney U test was used to compare the distributions of cell volumes in wild-type and Kif3a-mutant mice. For band intensity quantifications the paired t-test was performed. All statistic analyses were considered significant for P values of less than 0.05.
Supplementary Material
Figure 5.
Phosphorylated AMPK is localized at the basal body and increases under flow. (a) Immunofluorescence microscopy of MDCK cells under flow. Phosphorylated AMPK (p-AMPK, green) co-localizes with centrioles. The staining is more dominant at one centriole (arrowheads). Centrioles visualized by γ-tubulin (red). (b) 3D reconstruction of a centriole pair with phosphorylated AMPK at the apical pole of a single centriole. Spacing of grid marks 2 μm. (c) Immunofluorescence microscopy of wild-type MDCK cells. z-stack projection of the centrioles and the cilium (both magenta) reveals staining of p-AMPK (green) at the transition between basal body and the cilium. Scale bars, 2 μm. (d) Immunofluorescence microscopy of wild-type MDCK cells grown under the indicated flow conditions. Phosphorylated AMPK; green, centrioles; magenta. Scale bars, 2 μm. (e) Quantification of phosphorylated AMPK signal intensity, compared with γ-tubulin intensity, from immunofluorescence microscopy of wild-type MDCK cells grown under the indicated flow conditions. Asterisk indicates P < 0.05, n = 3, 33 cells per n. (f) Schematic representation depicting mTOR regulation through cilia. Data in e are means ± s.e.m.
Acknowledgments
This work was supported by grants DFG KFO 201 (to E.W.K.), WA597 and SFB 592 (to G.W.), SFB 592 Z2 and by the Excellence Initiative of the German Federal and State Governments (EXC 294 to R.N.), and NIH grants P30DK79328, R01DK67565 and T32DK07257 (to P.I.). We thank B. Yoder for anti-sera, R. Lamb, H. Clevers, Y. Mimori-Kiyosue, M. Sebbagh and A. Miyawaki for constructs, S. Arnold, M. Simons, D. Bennet and M. Bloech for critical appraisal of the manuscript and S. Lienkamp, F. Grahammer and members of the Walz lab for helpful discussions.
Footnotes
AUTHOR CONTRIBUTIONS
C.B., F.K., V.P., Si.B., H.V., Sa.B, T.B., H.J., C.H., K.M., Ma.H., Mi.H., M.D. and R.N. performed experiments. M.G. provided reagents. M.K., R.N., P.I., G.W. and E.W.K. conceived and planned the experiments and interpreted data. C.B., F.K. and E.W.K. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
Note: Supplementary Information is available on the Nature Cell Biology website
References
- 1.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 2.Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci. 2009;122:3589–3594. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Polak P, Hall MN. mTOR and the control of whole body metabolism. Curr Opin Cell Biol. 2009;21:209–218. doi: 10.1016/j.ceb.2009.01.024. [DOI] [PubMed] [Google Scholar]
- 4.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 5.Tao Y, Kim J, Schrier RW, Edelstein CL. Rapamycin markedly slows disease progression in a rat model of polycystic kidney disease. J Am Soc Nephrol. 2005;16:46–51. doi: 10.1681/ASN.2004080660. [DOI] [PubMed] [Google Scholar]
- 6.Wahl PR, et al. Inhibition of mTOR with sirolimus slows disease progression in Han:SPRD rats with autosomal dominant polycystic kidney disease (ADPKD) Nephrol Dial Transplant. 2006;21:598–604. doi: 10.1093/ndt/gfi181. [DOI] [PubMed] [Google Scholar]
- 7.Shillingford JM, et al. The mTOR pathway is regulated by polycystin-1, and its inhibition reverses renal cystogenesis in polycystic kidney disease. Proc Natl Acad Sci USA. 2006;103:5466–5471. doi: 10.1073/pnas.0509694103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fliegauf M, Benzing T, Omran H. When cilia go bad: cilia defects and ciliopathies. Nat Rev Mol Cell Biol. 2007;8:880–893. doi: 10.1038/nrm2278. [DOI] [PubMed] [Google Scholar]
- 9.Jansen M, Ten Klooster JP, Offerhaus GJ, Clevers H. LKB1 and AMPK family signaling: the intimate link between cell polarity and energy metabolism. Physiol Rev. 2009;89:777–798. doi: 10.1152/physrev.00026.2008. [DOI] [PubMed] [Google Scholar]
- 10.Pedersen LB, Rosenbaum JL. Intraflagellar transport (IFT) role in ciliary assembly, resorption and signalling. Curr Top Dev Biol. 2008;85:23–61. doi: 10.1016/S0070-2153(08)00802-8. [DOI] [PubMed] [Google Scholar]
- 11.Berbari NF, O’Connor AK, Haycraft CJ, Yoder BK. The primary cilium as a complex signaling center. Curr Biol. 2009;19:R526–535. doi: 10.1016/j.cub.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Praetorius HA, Spring KR. Bending the MDCK cell primary cilium increases intracellular calcium. J Membr Biol. 2001;184:71–79. doi: 10.1007/s00232-001-0075-4. [DOI] [PubMed] [Google Scholar]
- 13.Corbit KC, et al. Vertebrate Smoothened functions at the primary cilium. Nature. 2005;437:1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- 14.Haycraft CJ, et al. Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet. 2005;1:e53. doi: 10.1371/journal.pgen.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corbit KC, et al. Kif3a constrains β-catenin-dependent Wnt signalling through dual ciliary and non-ciliary mechanisms. Nat Cell Biol. 2008;10:70–76. doi: 10.1038/ncb1670. [DOI] [PubMed] [Google Scholar]
- 16.Torres VE, Harris PC. Autosomal dominant polycystic kidney disease: the last 3 years. Kidney Int. 2009;76:149–168. doi: 10.1038/ki.2009.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nauli SM, et al. Polycystins 1 and 2 mediate mechanosensation in the primary cilium of kidney cells. Nat Genet. 2003;33:129–137. doi: 10.1038/ng1076. [DOI] [PubMed] [Google Scholar]
- 18.Serra AL, et al. Sirolimus and kidney growth in autosomal dominant polycystic kidney disease. N Engl J Med. 2010;363:820–829. doi: 10.1056/NEJMoa0907419. [DOI] [PubMed] [Google Scholar]
- 19.Walz G, et al. Everolimus in patients with autosomal dominant polycystic kidney disease. N Engl J Med. 2010;363:830–840. doi: 10.1056/NEJMoa1003491. [DOI] [PubMed] [Google Scholar]
- 20.Distefano G, et al. Polycystin-1 regulates extracellular signal-regulated kinase-dependent phosphorylation of tuberin to control cell size through mTOR and its downstream effectors S6K and 4EBP1. Mol Cell Biol. 2009;29:2359–2371. doi: 10.1128/MCB.01259-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grantham JJ, Geiser JL, Evan AP. Cyst formation and growth in autosomal dominant polycystic kidney disease. Kidney Int. 1987;31:1145–1152. doi: 10.1038/ki.1987.121. [DOI] [PubMed] [Google Scholar]
- 22.Patel V, et al. Acute kidney injury and aberrant planar cell polarity induce cyst formation in mice lacking renal cilia. Hum Mol Genet. 2008;17:1578–1590. doi: 10.1093/hmg/ddn045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kotsis F, et al. Ciliary calcium signaling is modulated by kidney injury molecule-1 (Kim1) Pflugers Arch. 2007;453:819–829. doi: 10.1007/s00424-006-0168-0. [DOI] [PubMed] [Google Scholar]
- 24.Kotsis F, Nitschke R, Doerken M, Walz G, Kuehn EW. Flow modulates centriole movements in tubular epithelial cells. Pflugers Arch. 2008;456:1025–1035. doi: 10.1007/s00424-008-0475-8. [DOI] [PubMed] [Google Scholar]
- 25.Nishimura T, et al. Role of the PAR-3–KIF3 complex in the establishment of neuronal polarity. Nat Cell Biol. 2004;6:328–334. doi: 10.1038/ncb1118. [DOI] [PubMed] [Google Scholar]
- 26.Pazour GJ, et al. Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene Tg737, are required for assembly of cilia and flagella. J Cell Biol. 2000;151:709–718. doi: 10.1083/jcb.151.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murcia NS, et al. The oak ridge polycystic kidney (orpk) disease gene is required for left-right axis determination. Development. 2000;127:2347–2355. doi: 10.1242/dev.127.11.2347. [DOI] [PubMed] [Google Scholar]
- 28.Chiang GG, Abraham RT. Phosphorylation of mammalian target of rapamycin (mTOR) at Ser 2448 is mediated by p70S6 kinase. J Biol Chem. 2005;280:25485–25490. doi: 10.1074/jbc.M501707200. [DOI] [PubMed] [Google Scholar]
- 29.Kim DH, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 30.Polak P, et al. Adipose-specific knockout of raptor results in lean mice with enhanced mitochondrial respiration. Cell Metab. 2008;8:399–410. doi: 10.1016/j.cmet.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 31.Yan L, et al. Hyperactivation of mammalian target of rapamycin (mTOR) signaling by a gain-of-function mutant of the Rheb GTPase. J Biol Chem. 2006;281:19793–19797. doi: 10.1074/jbc.C600028200. [DOI] [PubMed] [Google Scholar]
- 32.Kottgen M, et al. TRPP2 and TRPV4 form a polymodal sensory channel complex. J Cell Biol. 2008;182:437–447. doi: 10.1083/jcb.200805124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alessi DR, Sakamoto K, Bayascas JR. LKB1-dependent signaling pathways. Annu Rev Biochem. 2006;75:137–163. doi: 10.1146/annurev.biochem.75.103004.142702. [DOI] [PubMed] [Google Scholar]
- 34.Granot Z, et al. LKB1 regulates pancreatic β cell size, polarity and function. Cell Metab. 2009;10:296–308. doi: 10.1016/j.cmet.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sebbagh M, Santoni MJ, Hall B, Borg JP, Schwartz MA. Regulation of LKB1/STRAD localization and function by E-cadherin. Curr Biol. 2009;19:37–42. doi: 10.1016/j.cub.2008.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie Z, et al. Identification of the serine 307 of LKB1 as a novel phosphorylation site essential for its nucleocytoplasmic transport and endothelial cell angiogenesis. Mol Cell Biol. 2009;29:3582–3596. doi: 10.1128/MCB.01417-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mammoto T, Ingber DE. Mechanical control of tissue and organ development. Development. 2010;137:1407–1420. doi: 10.1242/dev.024166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu W, et al. Effect of flow and stretch on the [Ca2+]i response of principal and intercalated cells in cortical collecting duct. Am J Physiol Renal Physiol. 2003;285:F998–F1012. doi: 10.1152/ajprenal.00067.2003. [DOI] [PubMed] [Google Scholar]
- 39.Maekawa F, et al. Localization of glucokinase-like immunoreactivity in the rat lower brain stem: for possible location of brain glucose-sensing mechanisms. Endocrinology. 2000;141:375–384. doi: 10.1210/endo.141.1.7234. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Q, Davenport JR, Croyle MJ, Haycraft CJ, Yoder BK. Disruption of IFT results in both exocrine and endocrine abnormalities in the pancreas of Tg737(orpk) mutant mice. Lab Invest. 2005;85:45–64. doi: 10.1038/labinvest.3700207. [DOI] [PubMed] [Google Scholar]
- 41.Wiznerowicz M, Trono D. Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. J Virol. 2003;77:8957–8961. doi: 10.1128/JVI.77.16.8957-8961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rekas A, Alattia JR, Nagai T, Miyawaki A, Ikura M. Crystal structure of venus, a yellow fluorescent protein with improved maturation and reduced environmental sensitivity. J Biol Chem. 2002;277:50573–50578. doi: 10.1074/jbc.M209524200. [DOI] [PubMed] [Google Scholar]
- 43.Hara K, et al. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110:177–189. doi: 10.1016/s0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- 44.Manning BD, Tee AR, Logsdon MN, Blenis J, Cantley LC. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol Cell. 2002;10:151–162. doi: 10.1016/s1097-2765(02)00568-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.