Preface
In all eukaryotes, the target of rapamycin (TOR) signaling pathway couples energy and nutrient abundance to the execution of cell growth and division, owing to the ability of TOR protein kinase to simultaneously sense energy, nutrients and stress, and, in metazoan, growth factors. Mammalian TOR complexes 1 and 2 (mTORC1 and mTORC2) exert their actions by regulating other important kinases, such as S6K and Akt. In the last few years, a significant advance in our understanding of the regulation and functions of mTOR has revealed its critical involvement in the onset and progression of diabetes, cancer and ageing.
Introduction
‘Growth’ indicates the set of biochemical processes, intimately linked to the availability of nutrients and energy, by which organisms increase their size and cell number through the synthesis of new cellular components, including proteins, nucleic acids, and lipids. Cells also rely on a complex set of programs to cope with times of nutrient starvation and low energy. To avoid energy imbalance and death, cells quickly suppress biosynthetic programs, increase the recycling of ‘aged’ proteins and organelles to provide an internal source of metabolites, and slow or halt proliferation.
At the interface between growth and starvation stands a signaling pathway centered on a kinase known as the Target of Rapamycin (TOR). The appearance of TOR in early eukaryotes enabled these unicellular organisms to sense the availability of nutrients and to promote growth whenever encountering favorable environmental conditions. With the emergence of multicellularity, TOR acquired additional roles as a central controller of organism growth and homeostasis. As such, mammalian TOR (mTOR) is implicated in disease states where growth is deregulated and homeostasis is compromised, namely cancer, metabolic diseases, and ageing. Dysregulated mTOR signaling fuels the destructive growth of cancers. Over-stimulation of the mTOR pathway by excess food consumption may be a critical factor underlying the diabetes epidemics. Finally, recent findings suggest that mTOR signaling controls the rate at which cells and tissues age, and that inhibiting mTOR may represent a promising avenue to increase longevity.
In this Review we begin by summarizing our current understanding of the regulatory inputs and the cellular actions of mTOR. We then discuss how this integrated knowledge, together with the availability of new chemical agents, could pave the way towards the manipulation of human metabolism in order to maximize healthy lifespan and help to stave off ageing, cancer and metabolic diseases.
In the remainder of this review we will use the term mTOR when discussing TOR in mammalian organisms and TOR with species-specific prefixes when discussing non-mammalian organisms (that is, ceTOR in Caenorhabditis elegans, dTOR in Drosophila melanogaster, dyTOR in Dictyostelium discoideum and scTOR in Saccharomyces cerevisiae). It should be noted that while mTOR originally stood for ‘mammalian TOR’, it is now officially an abbreviation for ‘mechanistic TOR’.
Organization and actions of mTOR complexes
In yeast and mammalian organisms, genetic and biochemical approaches, respectively, led to the discovery of TOR as the target of the immunosuppressant rapamycin, a macrolide produced by a soil bacterium found on Easter island1–4. TOR is a protein kinase belonging to the phosphoinositide 3-kinase related protein kinases (PIKK) family, which comprises large proteins that enable organisms to cope with metabolic, environmental and genetic stress.
Molecular composition of mTORC1 and mTORC2
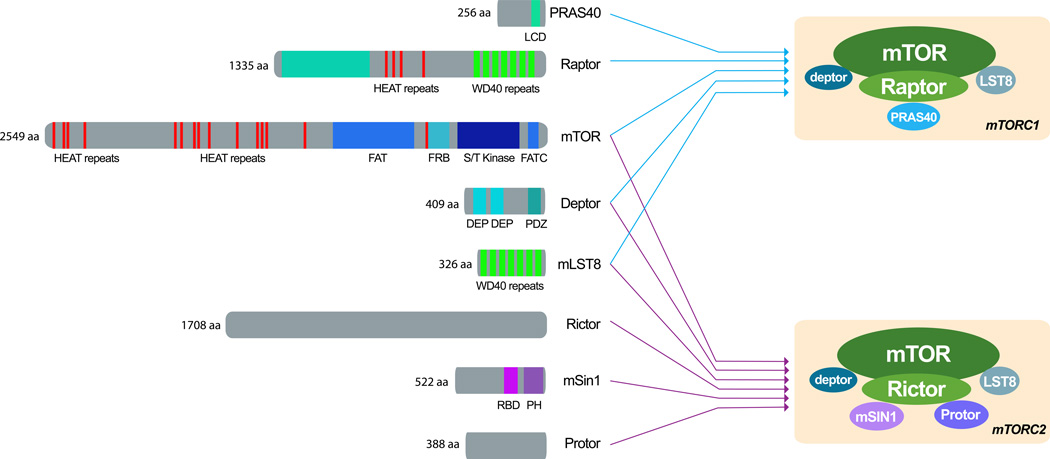
mTOR is the catalytic subunit of two distinct complexes, called mTOR Complex 1 (mTORC1) and 2 (mTORC2) (reviewed in5, 6) (FIG.1). Unique accessory proteins distinguish the two complexes: raptor and rictor define mTORC1 and mTORC2, respectively7–9. These companions function as scaffolds for assembling the complexes and for binding substrates as well as regulators7, 8, 10–15. Further unique components exist in both complexes: mTORC1 comprises a negative regulator, PRAS4014, 16, whereas mTORC2 contains Protor and mSin1, which likely help complex assembly and localization, respectively15, 17–19. mTORC1 and mTORC2 share mLST8 and the recently identified Deptor, which function as positive and negative regulators, respectively20 (FIG.1). Biochemical and structural evidence suggests that both mTORC1 and mTORC2 may exist as dimers12, 21.
Figure 1. Domain organization of mTOR and mTOR-interacting proteins.
A. mTORC1 and mTORC2 have shared (mTOR, mLST8 and deptor) and unique components. Raptor and PRAS40 are unique to mTORC1 and rictor, mSin1 and Protor are specific to mTORC2. The domain organization of mTOR resembles that of other PIKK family members. At the N-terminus there is a cluster of Huntingtin, Elongation factor 3, a subunit of protein phosphatase 2A and TOR1 (HEAT) repeats, which mediate protein-protein interactions. These are followed by, a FRAP, ATM and TRRAP (FAT) domain, the FKBP12-Rapamycin Binding (FRB) domain (which mediates the inhibitory action of rapamycin), the serine/threonine kinase catalytic domain and the C-terminal FATC domain. mLST8 is highly conserved; its seven WD40 domains form a beta propeller that mediates protein-protein interactions. Deptor consists of a tandem dishevelled, egl-10, pleckstrin (DEP) domains, followed by a single postsynaptic density 95, discs large, zonula occludens-1 (PDZ) domain. The scaffolding function of raptor is reflected by its composition of protein-binding domains; it consists of several HEAT repeats, followed by seven WD40 motifs, probably arranged in a beta propeller.Rictor has no clearly identifiable domains or motifs despite its key role in mTORC2 function; likewise, Protor lacks identifiable domains or motifs, and its function awaits clarification.
Both in yeast and mammals, rapamycin only inhibits the activities of mTORC1, but not mTORC29, 22– 25. Rapamycin binds to the small protein FKBP-12 and, in turn, rapamycin-FKBP-12 binds to and inhibits raptor-bound, but not rictor-bound, mTOR9, 22, 26, 27. Rapamycin might inhibit mTORC1 by dissociating raptor from mTOR, thus limiting the access of mTOR to some substrates8, 21. Complicating this picture, prolonged treatment with rapamycin can inhibit mTORC2 in a subset of tissues and cell lines28. This effect may involve a progressive sequestration of the cellular pool of mTOR in a complex with rapamycin-FKBP12, thus making it unavailable for assembly into mTORC2.
Substrates and actions of mTORC1
The complex in which it participates dictates the substrate specificity of mTOR. The mTORC1 substrates S6 Kinase 1 (S6K1) and eIF-4E binding proten 1 (4E-BP1) associate with mRNAs and regulate both mRNA translation initiation and progression, thus enhancing protein synthesis29–31 (reviewed in32) (FIG.2a). 4E-BP1 suppresses mRNA translation; when phosphorylated by mTORC1, it dissociates from eIF4E, allowing the latter to recruit the translation initiation factor eIF4G to the 5' end of most mRNAs30, 33. S6K1 is a positive regulator of translation initiation and elongation when it is phosphorylated by mTORC1, via its ability to phosphorylate multiple substrates including S634, eEF2K35, SKAR36, CBP8037 and eIF4B36, 38, 39.
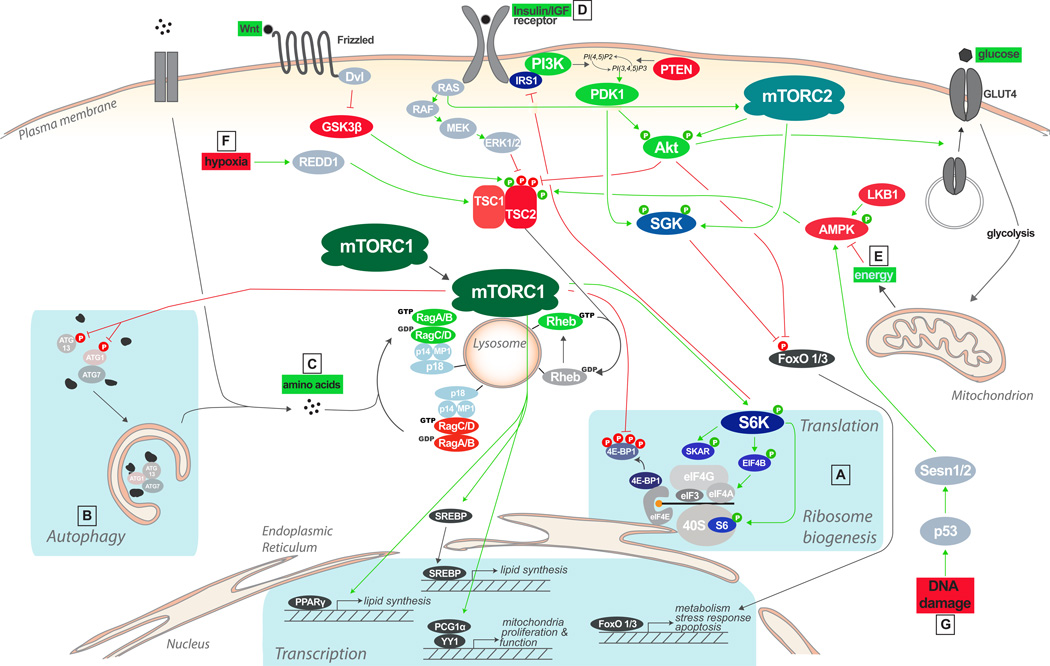
Figure 2. The mTOR signaling pathway.
mTORC1 promotes translation (A) and inhibits autophagy (B), by integrating nutrient signals generated by amino acids (C), growth factor signals relayed by the insulin and insulin-like growth factors (D), energy signals acting through AMP-activated kinase (AMPK) (E) and various stressors including hypoxia (F) and DNA damage (G). A first level of integration occurs at the level of the Tuberous Sclerosis Complex (TSC). Akt and extracellular regulated kinase (ERK) 1/2 phosphorylate and inhibit TSC, while AMPK, glycogen synthase kinase (GSK) 3-beta and hypoxic factor REDD1 activate it. A second level of integration occurs at the lysosome: the Rag GTPases (held in place by the p18/p14/MP1 ragulator) recruit mTORC1 to the lysosomal surface in response to amino acids; in turn, lysosomal recruitment enables mTORC1 to interact with GTP-bound Rheb, the end point of growth factor, energy and stress inputs. Growth factor receptors activate mTORC2, probably via Ras, near the plasma membrane, where mTORC2 may be recruited through binding of mSin1 to phospholipids. Because of its role in phosphorylating and activating Akt, mTORC2 forms a core component of the PI3K pathway.
Ribosome biogenesis is highly energy-intensive and, as such, it is tightly coupled to the energetic status of the cell. Both the synthesis of ribosomal RNAs (rRNAs) and that of ribosomal proteins are positively regulated by mTORC1 (FIG.2a). mTORC1, via S6K1 kinase activity, upregulates the transcriptional activity of the rRNA polymerase, RNA PolI40, 41. In yeast, the transcription factors Rrn3 (TIF1 in mammalians), FHL1 and SFP1 mediate the transcription of ribosomal RNAs and proteins downstream of scTORC141–44.
Autophagy is the controlled self-degradation of cellular components, ranging from individual proteins (microautophagy) to entire organelles (macroautophagy), which mediates the recycling of damaged, redundant, or even dangerous cellular constituents. Autophagy provides an important source of substrates for energy production during periods of low extracellular nutrients (reviewed in45). mTORC1 actively suppresses autophagy, and conversely, inhibition of mTORC1 (by small molecules or by amino acid withdrawal) strongly induces it46, 47. In yeast, scTOR-dependent phosphorylation of Atg13 disrupts the Atg1-Atg13 complex that is critical for the formation of the autophagosome48. Similarly, mTORC1-mediated phosphorylation negatively regulates the mammalian Atg13 and Atg1 homologues, Atg13 and ULK1 and ULK249, 50 (FIG.2b). However, the mammalian-specific autophagic regulators FIP200 and Atg101 are also mTORC1 substrates51, 52, indicating additional mechanisms for the regulation of autophagy by mTOR.
Substrates and actions of mTORC2
scTORC2 was identified as a mediator of actin cytoskeletal organization and cell polarization23–25. scTORC2 controls a number of cytoskeletal regulators, including the chaperonin TCP-1, ROM2 (a GEF for Rho1 and Rho2) and the AGC kinase Ypk253, 54. The role of TORC2 in controlling cytoskeletal polarity has been confirmed in D. discoideum and mammalian cells.
Recent findings have revealed novel important roles for mTORC2 in the phosphorylation of AGC kinase family members. mTORC2 phosphorylates and activates Akt, SGK, and PKC, which regulate cell survival, cell cycle progression and anabolism55–58. Among AGC kinases, Akt is especially important because of its role in the pathogenesis of cancer and diabetes. Using RNAi in D. melanogaster S2 cells, it was found that dTORC2 mediates the phosphorylation of Akt at S505, located in the hydrophobic motif and homologous to S473 in mammals55, 57, 58. This is a key finding, because phosphorylation at S473 primes Akt for further phosphorylation at T308, located in the catalytic domain, by PDK1. Together, these two phosphorylation events cause full activation of Akt. Loss of Rictor in worm, fly, mouse and human cells results in complete loss of Akt phosphorylation at S473 but, interestingly, this affects only some substrates of Akt18, 58–60. Particularly, phosphorylation of the FOXO1/3 transcription factors was suppressed, but that of Tuberous Sclerosis Complex 2 (TSC2), a substrate of Akt that acts upstream of mTORC1, was not. Because phosphorylation of FOXO1/3 by Akt promotes avoidance of apoptosis, mTORC2 may regulate cell survival59, 61.
Collectively, these findings place mTORC2 upstream of key cellular processes such as cell-cycle progression, anabolism and cell survival.
Upstream regulators of mTOR complexes
mTORC1 acts as a signal integrator for four major regulatory inputs: nutrients, growth factors, energy, and stress (FIG.2). These inputs can cooperate and antagonize each other, enabling the cell to fine-tune the action of mTORC1. In contrast, the regulation of mTORC2 is only beginning to be discovered, but the available evidence seems to suggest that only growth factors directly regulate this complex.
Nutrients regulate mTORC1
Amino acids are the building blocks of proteins, and are also used in the synthesis of DNA, glucose and ATP. Early studies showed that amino acids are absolutely required for mTORC1 signaling in cultured cells, and cannot be compensated for by other activating stimuli13, 62, 63. However, the molecular components of amino acid signaling to mTORC1 have long remained mysterious. We now know that amino acids regulate mTORC1 via the Rag family of small GTPases13, 64 (FIG.2c). Rag GTPases are heterodimers of either Rag A or B with either Rag C or D; crucially, the two members of the heterodimer have opposite nucleotide loading states. In the absence of amino acids, RagA or RagB is GDP loaded and RagC or RagD contains GTP, and this is the inactive Rag conformation. Amino acids cause Rag GTPases to switch to the active conformation, where RagA or RagB is GTP loaded and RagC or RagD is GDP loaded. The active Rag heterodimer physically interacts with raptor, causing the clustering of mTORC1 onto the surface of late endosomes and lysosomes, where the Rags reside13, 64, 65. This relocalization may enable mTORC1 to interact with the small GTPase Rheb, an essential activator of mTORC1 that is controlled by growth factor inputs13, 66, 67. This model has been largely confirmed in S. cerevisiae, where the Rag GTPase homologs, GTR1 and GTR2, interact physically and genetically with scTORC168–71. However, the rheb homologue in S. Cerevisiae is not essential for life and does not appear to function upstream of scTORC172. Thus, some aspects of scTORC1 regulation may differ from metazoans and await clarification.
Rag proteins do not show any obvious membrane targeting signals. However, three small molecules p14, MP1 and p18, collectively known as the Ragulator, form a molecular scaffold that binds the Rag GTPases to the lysosomal surface. When the ragulator is genetically inactivated, the Rag GTPases become cytoplasmic, recruitment of mTORC1 to the lysosome fails, and amino acid signaling to mTORC1 is blocked65.
Collectively, these findings add to growing evidence for a key role of endomembranes in controlling the activity of mTOR (BOX 2); moreover, they suggest that subcellular localization of mTORC1 underlies its ability to integrate nutrient and growth factor signals (BOX 1).
BOX2: mTOR and membranes.
Increasing evidence points to a role for membrane traffic in the regulation of mTOR signaling. Amino acid signaling through the Rag GTPases causes mTORC1 to shuttle to lysosomes, which enables mTORC1 to respond to amino acids and to insulin signal-induced Rheb activation (Fig. 2). The lysosomal surface hosts the molecular machinery for mTORC1 activation that includes the Rag GTPases, the trimeric ragulator complex, and possibly GAPs and GEFs for the Rag GTPases13, 65. Vps39, a GEF for Rab7, has been implicated in regulating the nucleotide loading of GTR1, the RagA/B orthologue in yeast68. Moreover, work from the Thomas lab has implicated Vps34, the type III pPI3 Kinase that controls endosomal maturation and autophagy, in mediating amino acid signaling to mTORC1177.
The presence of mTORC1 at the lysosome also suggests a physical basis for the regulation of autophagy: because autophagic membranes fuse with lysosomes to promote degradation of their contents, mTORC1 may be optimally placed to phosphorylate and inhibit key autophagy-promoting proteins. Of notice, yeast GTR1 and GTR2 are part of the EGO complex, which are involved in the regulation of microautophagy69. It would be interesting to assess whether mammalian Rag GTPases play similar roles; in fact, recent evidence points to a role for mTORC1 in the reformation of primary lysosomes following autophagy178 In addition, nutrient storage and release by the lysosome may allow the rapid activation of nutrient signals upstream of mTORC1.
In contrast to mTORC1 the subcellular localization of mTORC2 is unclear, although recent studies suggest it may also associate with membranes. Experimental approaches based on genetic fluorescent tags revealed that yeast TORC2 localizes to discrete, spot-like domains on the plasma membrane. Notably, membrane localization of scTORC2 depends on Avo1, the mammalian orthologue of mSin1, which has a phospholipid-binding PH domain that may mediate the association of mTORC2 with membranes179. The localization of mTORC2 on peripheral membranes would be consistent with its activation by PI3K and the insulin receptor, and with its role in phosphorylating Akt. However, indirect evidence suggests that mTORC2 might localize, and be activated at, signaling endosomes near the cell surface.
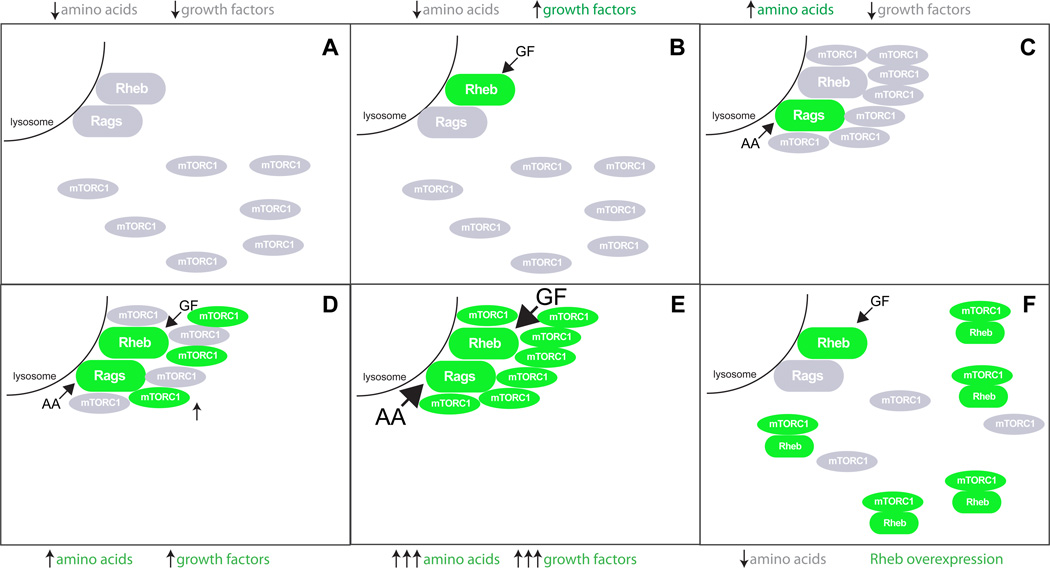
BOX1: mTORC1 as a signal integrator.
How are mTORC1 inputs integrated to generate a coherent signaling response? Signal integration by mTORC1 is likely based on convergent regulation of physical interactions and localization; growth factors induce GTP loading of Rheb, enabling it to physically interact with mTORC1, while amino acids cause the Rag GTPase-mediated shuttling of mTORC1 to the endomembrane system, where Rheb resides. This scenario provides an explanation for the requirement of both inputs for the activation of mTORC1. In the absence of growth factors and amino acids, the Rag and Rheb GTPases are inactive and mTORC1 is physically removed from Rheb (figure, part a). In the presence of abnormally high growth factor inputs, (caused, for instance, by activating mutations in the PI3K pathway), Rheb is hyperactive (green). However, because mTORC1 is not recruited to Rheb, it remains inactive (figure, part b). Amino acids activate the Rag GTPases, which recruit mTORC1 and allow it to bind to Rheb. However, in the absence of growth factors Rheb is inactive, and so is mTORC1 (figure, part c). When both amino acids and growth factors are present, mTORC1 is recruited to active Rheb and activated (figure, part d).
Importantly, nutrients and energy can hardly be regarded as an on/off switch: their concentration varies smoothly in time as a function of the feeding cycle. Thus, mTORC1 likely senses fine variations in these parameters, continuously adjusting the rate of biosynthetic processes accordingly. Once activation of mTORC1 has been achieved, increasing concentrations of amino acids and growth factors can drive it further (figure, part e). Experimental overexpression of Rheb bypasses the requirement for Rag-mediated recruitment of mTORC1 and allows its activation in the absence of amino acids (figure, part f), supporting the signal integration model. Other regulatory inputs, such as energy-sensing AMPK, as well as feedback mechanisms can modulate or entirely suppress this signal integration mechanism, ensuring that nutrients, growth factors and energy do not generate conflicting signals. Thus, the regulation of mTORC1 is a multi-step decision process that takes into account multiple indicators of the energy status of the cell before making a commitment to grow and proliferate.
Growth factors regulate mTORC1
Multicellular organisms rely on long-range communication to coordinate the distribution of nutrients and the growth of cell populations throughout the body. With the emergence of metazoans, the mTOR pathway became wired to signaling pathways initiated by growth factors, such as insulin, which carry information on the nutritional state of the organism as a whole. One key end point of growth factor signaling to mTORC1 is the small GTPase Rheb that, when GTP-loaded, stimulates the kinase activity of mTORC114, 66, 67. In metazoans Rheb plays an essential role, because its loss abolishes the activation of mTORC1 by both growth factors and nutrients. Conversely, overexpression of Rheb can maintain mTORC1 activity even when nutrients and growth factors have been withdrawn (BOX 1).
How do growth factors regulate the GTP-loading of Rheb? Binding of insulin to its receptor activates the phosphatidylinositol 3-kinase (PI3K) pathway, which leads to phosphorylation and activation of Akt. In turn, Akt phosphorylates tuberin/TSC2, a large protein that, together with hamartin/TSC1, forms the Tuberous Sclerosis Complex (TSC)73–75 (FIG.2d). TSC acts as a GAP for Rheb76–79; because GDP-loaded Rheb is unable to activate mTORC1, TSC effectively shuts off mTORC1 signaling80, 81. Akt-mediated phoshorylation of TSC2 inhibits its GAP activity for Rheb82, 83 and thus promotes the activation of mTORC1. Akt also phosphorylates PRAS40, causing its binding to 14-3-3 proteins and preventing it from inhibiting mTORC114, 16, 84. Moreover, in a positive feedback loop, mTOR phosphorylates and inhibits PRAS4085, 86.
Growth factors can signal to mTORC1 via alternative pathways to the PI3K-Akt axis. For example, extracellular signal-regulated kinase (Erk) activates TSC1/2 downstream of the Ras-Raf-Erk axis87. Moreover, the Wnt pathway has been implicated in mTORC1 signaling88, 89. Glycogen synthase kinase 3 (GSK3) acts as a negative regulator of mTORC1 by phosphorylating TSC1/2; by inhibiting GSK, Wnt activates mTORC1.
The convergence of multiple growth factor-initiated pathways onto TSC2 likely allows mTORC1 to participate in many developmental and physiological processes. This is supported by the absolute requirement for mTORC1 in early embryonic development59, 90, 91.
Energy and stress regulate mTORC1
Chemical inhibitors of glycolysis and mitochondrial function suppress mTORC1 activity, indicating that mTORC1 senses cellular energy8, 92. Energy sensing by mTORC1 is critical, because mTORC1-driven growth processes consume a large fraction of cellular energy, and thus could be deleterious to starving cells.
Glycolysis and mitochondrial respiration convert nutrients into energy, stored in the form of ATP, and cellular ATP quickly drops upon nutrient deprivation. The mTORC1 pathway indirectly senses low ATP, via a mechanism centered on the AMP-activated protein kinase (AMPK) (reviewed in93) (FIG.2e). Both AMP and ATP are allosteric regulators of AMPK: when the AMP:ATP ratio increases, AMPK phosphorylates TSC1/2, possibly stimulating its GAP activity toward Rheb89, 94, 95. Moreover, AMPK phosphorylates raptor, causing its binding to 14-3-3, which leads to the inhibition of mTORC1 through allosteric mechanisms96.
Numerous stressors affect ATP levels, and thus may regulate mTOR via the AMP-AMPK axis. During hypoxia, mitochondrial respiration is impaired, leading to low ATP levels and activation of AMPK (reviewed in97). Hypoxia also affects mTORC1 in AMPK-independent ways, by inducing the expression of the REDD1/2 genes, which then suppresses mTORC1 through a mechanism that requires TSC1/298, 99 (FIG.2f). Conversely, other stressors that do not primarily impinge on cellular energy signal through AMPK. DNA damage results in inhibition of mTORC1 activity via p53-dependent upregulation of AMPK95, 100, 101 (FIG.2g). Sestrin 1 and 2 are two transcriptional targets of p53 implicated in the DNA damage response. Recently, it was shown that sestrins potently activate AMPK, thus mediating the p53-dependent suppression of mTOR activity upon DNA damage102.
Upstream regulation of mTORC2
Surprisingly little is known about the upstream activators of mTORC2. Given its role in regulating Akt, SGK and PKC, it is generally thought that growth factors control mTORC2, directly or indirectly. In fact, insulin stimulation of cultured cells promotes S473 phosphorylation of Akt by mTORC258.
Because Akt, SGK and PKC respond to different growth factors, the range of upstream regulators of mTORC2 may be quite wide. Thus, how can signaling specificity be achieved? One potential solution may come from the existence of multiple isoforms of mSin1. Three out of the five known splice variants of mSin1 participate in mTORC2, effectively defining three distinct complexes; of these, only two are regulated by insulin17. Thus, mSin1 may function as an adaptor between mTORC2 and specific growth factor receptors. The mSin1 orthologue in D. discoideum, RIP3, mediates the enhanced migratory behavior driven by activated RAS; thus, RAS may provide a link between growth factors and mTORC2103, 104.
mTOR regulates metabolism in mammals
Due to the intermittency of food intake and the necessity to keep nutrient levels in the bloodstream within a narrow physiological range, multicellular organisms have acquired mechanisms to store energy after feeding and to later mobilize this energy. Long-range signals, primarily insulin, provide whole-body coordination of energy consumption and storage. mTOR regulates all these responses by its ability to integrate insulin and nutrients, mediate energy storage and consumption and prevent the triggering of conflicting metabolic signals by suppressing catabolism (FIG.3).
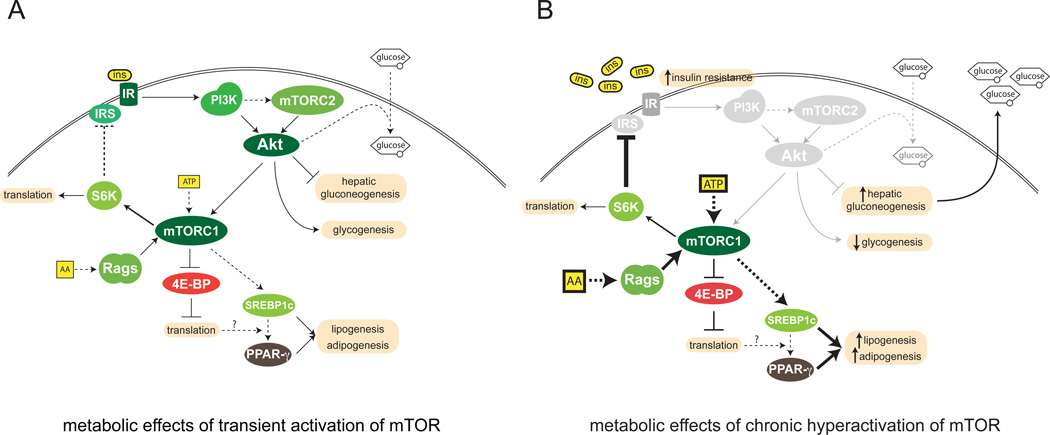
Figure 3. mTOR in metabolism and insulin resistance.
A. Effects of mTOR in metabolism. mTOR links nutrient abundance with growth and the accumulation of energy stores in anticipation of future nutrient shortage. Feeding triggers a raise in insulin and nutrient levels in the bloodstream; these converge as activating inputs to mTOR. In turn, mTORC1 activates protein synthesis, cell mass increase, and lipid accumulation, whereas mTORC2 promotes glucose usage by the cell by positively regulating glucose import, glycogen synthesis and by inhibiting gluconeogenesis. B. In the western diet, overabundance of nutrients leads to chronic mTOR activation, which disrupts energetic homeostasis. During chronic insulin stimulation, as occurs in over-feeding states, mTORC1 activity towards S6K inhibits insulin receptor signaling at the cellular membrane, contributing to the onset of the diabetic state. In an insulin-resistance state, phosphatidylinositol 3 kinase (PI3K) and Akt are not activated, leading to decreased glucose uptake and to hepatic gluconeogenesis, which in turn worsen hyperglycaemia. Despite decreased insulin signaling, mTORC1 remains active, maintaining the negative feedback loop to IR. The nutrient inputs to mTORC1 may explain sustained mTORC1 activity in the context of insulin insensitivity.
(AA: amino acids; ins: insulin; IR: insulin receptor; IRS; insulin receptor substrate)
mTOR in fasting and starvation
Having evolved in conditions of limited nutrient availability, mammals (including humans) have developed a striking ability to maximize the available resources of energy anticipating periods of shortage.
Early during fasting, glucose and amino acid blood levels decrease, causing a drop in circulating insulin. In addition, the imbalance between energy expenditure and food intake leads to an increased AMP:ATP ratio. These factors converge as inhibitory inputs on mTORC1, placing a brake on energyintensive biosynthetic processes and upregulating macroautophagy105. Mitophagy (the autophagic degradation of mitochondria) provides an immediate source of energy at the expense of nutrient-intensive, long-term ATP production (reviewed in106). The prosurvival role of autophagy was elegantly demonstrated in vivo in mice with ATG5 genetically deleted. Immediately after birth, interruption of the placental nutrient supply causes an energetic shortage that is compensated for by upregulation of autophagy in several organs. ATG5-deficient mice are unable to overcome this energetic challenge and die within one day after birth107.
In the liver, induction of autophagy causes simultaneous recycling of mitochondria, cytoplasmic proteins and stored glycogen. This effect is so dramatic that murine liver reduces to around 30% of its normal size in a 24h fasting period. Furthermore, during starvation, white adipose tissue (WAT) and liver mobilize lipid stores, converting them into free fatty acids that are utilized by the liver and muscle via β-oxidation. Recent evidence indicates that, during fasting, autophagosomes sequester lipid droplets and break them down into free fatty acids108. Autophagy also mediates massive protein breakdown in the muscle105, releasing amino acids to the bloodstream. The liver absorbs these amino acids, especially alanine, and converts them into glucose, which is then exported to the blood. The flow of nutrients released by autophagy feeds back onto mTORC1, causing its partial reactivation109.
Changes in mitochondrial function stemming from inhibition of mTORC1 may also contribute to fasting responses. Activation of 4E-BP1 under limiting nutrients in D. melanogaster led to the selective translation of mRNAs encoding the mitochondrial respiratory chain110, consistent with an attempt to increase the efficiency of ATP production. Moreover, mTORC1 promotes mitochondria biogenesis and enhances respiration through a ternary complex with the transcription factors PGC-1α and YY1111. Thus, inhibition of mTORC1 during starvation acts on mitochondrial function at 3 different levels: by placing a brake on the synthesis of new mitochondria, eliminating a subset of the existing mitochondria via mitophagy and by increasing the efficiency of existing mitochondria through the 4E-BP1 translational program.
mTOR, over-feeding, and insulin sensitivity
Growth control programs have evolved under conditions of scarce nutrients that were prevalent during mammalian evolution, and not in the western-life conditions of the last few decades. Overfeeding may be pathological because selection has favored organismal responses, partly mediated by mTOR, that accumulate and store energy in anticipation of periods of shortage. This translates into aberrant cellular responses when food and energy are plentiful and constantly available.
One of the most efficient forms of energy storage are triglycerides, because they provide a high energetic yield per unit of mass. mTORC1 mediates lipid accumulation in fat cells and favors formation of WAT (FIG.3). Adipocyte-specific deletion of Raptor in mice leads to reduced WAT tissue and enhanced fatty-acid oxidation112. Although the molecular mechanisms underlying these effects are not fully-understood, mTORC1 facilitates both differentiation of preadipocytes113 and lipid accumulation114 by indirectly upregulating Peroxysome proliferator-activated receptor (PPAR)-γ, a factor necessary and sufficient for adipocyte differentiation115, 116. Consistently, TSC2-deficient MEFs, which have increased mTORC1 activity as a result, show enhanced adipogenesis and PPAR-γlevels117.
mTORC1 may impact on PPAR-γ activity by increasing its translation118 and by activating the transcription factor SREBP-1c (FIG.3a). Active SREBP-1c enhances PPAR-γ activity119, 120, and transactivates a set of genes directly involved in lipid synthesis121. At present, the molecular links between mTORC1, SREBP-1c and PPAR-γ activity remain to be clarified.
Recent evidence also implicates mTORC2 in lipid biogenesis. Akt activation by mTORC2 leads to induction of PPAR-γ through parallel mechanisms that include mTORC1 activation as well as direct inhibition of FoxO1122, 123. In worms, ceTORC2 participates in lipid accumulation through both Akt and SGK60.
From a clinical perspective, chronic mTORC1 hyperactivation mediates the aberrant lipogenesis in WAT, liver and muscle that occurs in obesity, and which contributes to insulin resistance. Furthermore, mTORC1 hyperactivation during overfeeding may trigger an S6K-dependent negative feedback loop; activated S6K dampens the function of IRS1, an adaptor protein that recruits key downstream effectors to the insulin receptor, thus leading to insulin desensitization. The resulting dampened Akt activation leads to reduced glucose uptake and glycogen synthesis, and promotes gluconeogenesis in the liver. Collectively, these effects lead to a worsening of the hyperglycemic and hyperinsulinemic condition (FIG. 3b). Supporting the role of the S6K-IRS1 feedback loop in the pathogenesis of type 2 diabetes, S6K1-deficient mice displayed enhanced insulin sensitivity even when chronically maintained on a high fat diet124.
Of note, mTORC1 hyperactivation in the context of insulin insensitivity poses a paradox: how can mTORC1 be constitutively active in an insulin insensitivity state if insulin is responsible for the activation of mTORC1? This apparent paradox can be explained by mTORC1 being hyperactive as a consequence of the nutrient branch. Chronic high blood levels of amino acids, as occurs in obesity125, will keep mTORC1 at work together with an active S6K-IRS loop and, consequently, within a context of insulin insensitivity (FIG.3b).
Strikingly, the same molecular circuitry that controls metabolism in peripheral tissues also influences food intake in the central nervous system. Leucine locally applied to the hypothalamus induced satiety through activation of mTORC1; conversely, inhibition of mTORC1 in the hypothalamus by rapamycin injection increased food intake126.
Thus, mTORC1 coordinates food intake with energy storage at multiple levels, from central control of food seeking to energy storage and expenditure in peripheral tissues. This multi-level regulation explains the profound consequences that dysregulated mTOR signaling exerts on human metabolism.
mTOR in cancer etiology and therapy
The most direct evidence that mTOR can drive tumorigenesis comes from familial cancer syndromes arising from mutations of negative mTOR regulators such as TSC1/2, LKB1 and PTEN (see Supplementary FIG.1). mTOR-dependent mechanisms also contribute to the growth of many sporadic cancers. For example, upon mTORC1-mediated inhibition of 4E-BP1, activated eIF4E preferentially drives the translation of mRNAs for pro-tumorigenic genes, including cell cycle regulators (FIG.4a). Indeed, eIF4E promotes cell survival in in vivo mouse models of lymphoma by upregulating the translation of the anti-apoptotic protein Mcl-1127–129. mTOR also upregulates Fatty Acid Synthase (FAS), an enzyme involved in lipid synthesis that favors rapid proliferation of cancer cells (FIG.4a) (reviewed in130).
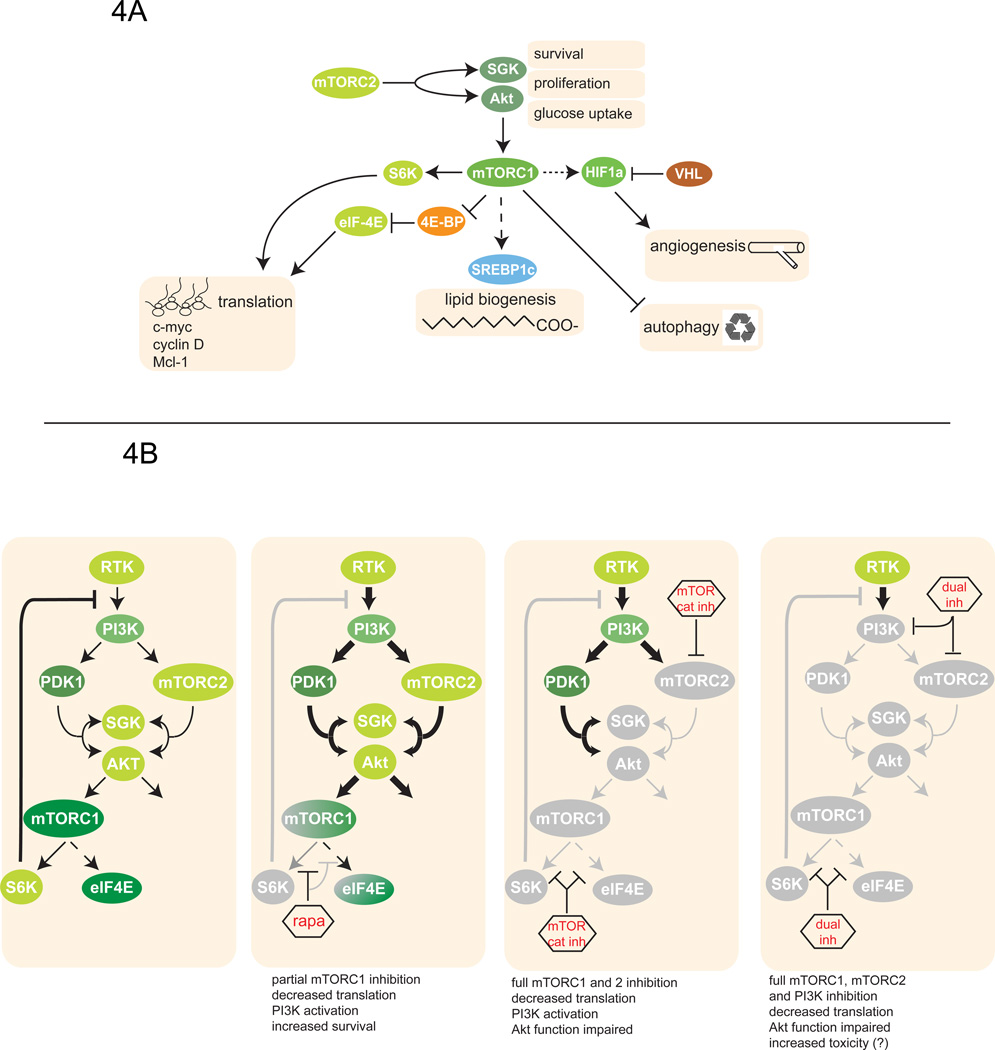
Figure 4. mTOR in cancer.
A. mTOR-regulated cellular processes that play a role in cancer. mTORC1 favors tumorigenesis by driving translation of oncogenes, by inhibiting autophagy, by upregulating angiogenesis, and by enhancing the accumulation of lipids. mTORC2 plays a role in tumorigenesis by activating Akt and other AGC family proteins which promote proliferation and survival. Moreover, by promoting Akt-mediated glucose uptake, mTORC2 fuels the metabolism of cancer cells.
B. Therapeutic inhibition of mTOR activity by rapamycin, mTOR catalytic inhibitors (mTOR cat inh) and dual PI3K-mTOR inhibitors (dual inh). Rapamycin only partially suppresses mTORC1 function, efficiently inhibiting S6K but not eIF4E; thus, it only partially inhibits translation. Moreover, due to the inhibition of the S6K-dependent feedback loops, rapamycin indirectly upregulates phosphatidylinositol 3 kinase (PI3K) activity and promotes cell survival. In contrast, mTOR ATP-competitive inhibitors target all known functions of mTORC1 as well as mTORC2; thus, they inhibit translation more potently. Although PI3K overactivation still occurs, Akt phosphorylation by mTORC2 is impaired. Dual PI3K/mTOR inhibitors block all functions of PI3K, including PDK1- and mTORC2-mediated activation of Akt. However, they might cause increased toxicity.
Increasing evidence suggests that autophagy plays a role in tumor suppression. The most direct data supporting the tumor suppressive roles of autophagy come from mice heterozygous for the autophagic protein Beclin131, 132 and from Atg4C-deficient mice133, both of which are tumor-prone. Thus, constitutive mTORC1 activation may indirectly favor tumorigenesis by suppressing autophagy (FIG.4a).
By activating Akt58 and SGK56 mTORC2 may directly drive tumorigenesis. Akt promotes proliferation, survival and nutrient uptake in cancer cells (reviewed in134). Tumors driven by mutations in the tumor suppressor PTEN (which usually inhibits Akt signalling) or by oncogenic mutations in PI3K (which promotes Akt signalling) may be especially dependent on the pro-survival activities of Akt, and this is where targeting mTORC2 may prove especially useful. In fact, Rictor is required for the growth of prostate tumors caused by PTEN deletion in mice61.
Rapamycin as a mTOR-centered cancer therapy
The existence of a potent, naturally occurring inhibitor of mTOR, rapamycin, appeared to be a lucky strike for cancer therapies. However, to date the limited success of rapamycin as an anti-cancer drug in clinical trials has generated some disappointment. Here we discuss the limitations of rapamycin and the current efforts to move beyond this drug towards an effective mTOR-centered therapy of cancer.
Rapamycin-based therapeutic approaches may have encountered a stumbling block in the S6Kmediated feedback loop, which leads to a severe upregulation of PI3K signaling, and provides important pro-survival and proliferative signals through Akt and other AGC kinases, when mTORC1 is inhibited and unable to activate S6K. Additionally, S6K inhibition activates the MEK-ERK signaling cascade135, as well as transcription of platelet-derived growth factor receptor (PDGFR)136. These loops counteract the effects of rapamycin, dampening its effectiveness in cancer models and in patients (FIG.4b)135, 137 (reviewed in138), and may explain why rapamycin is cytostatic but not cytotoxic in many tumors.
While high doses of rapamycin or its prolonged delivery can block mTORC2 in some cell lines28, 139, rapamycin is largely selective for mTORC1. Given the role of mTORC2 and especially Akt as drivers of tumorigenesis61, this poses a major concern. Furthermore, rapamycin does not inhibit all the functions of mTORC1; of notice, rapamycin only affects 4E-BP1 phosphorylation transiently and partially47, 139–142. Thus rapamycin, by suppressing the S6K1-IRS1 feedback loop and hyperactivating the PI3K-AKT pathway, may ultimately stimulate 4E-BP1 phosphorylation and perhaps other tumor-promoting functions of mTORC1. These two major drawbacks have motivated the search for second-generation inhibitors of mTOR function.
mTOR and mTOR-PI3K catalytic inhibitors
Recently, independent groups generated a series of synthetic small molecules47, 142–145, which function as ATP-competitive inhibitors and block all known mTORC1 and mTORC2 activities. For example, Torin 1 inhibits 4E-BP1 phosphorylation and triggers autophagy to a far greater extent than rapamycin47. Moreover, unlike rapamycin, Torin 1 and the other catalytic inhibitors also completely block mTORC2-mediated phosphorylation of Akt47, 142–145. Supporting the applicability of this drug in a clinical setting, pre-clinical data in genetically engineered mice argue that even full inhibition of mTORC1 and 2 could be well tolerated in adult tissues61, 146. Moreover, the mTOR-catalytic inhibitor PP-242 showed a better response than rapamycin in a mouse model of experimental leukemia, together with a surprising milder effect on normal lymphocytes147.
However, mTOR catalytic inhibitors are not immune to potential drawbacks. Loss of the S6Kmediated feedback loop resulting from mTORC1 inhibition enhances Akt phosphorylation at T308 by PDK1 (FIG.4b). Consequently, when suboptimal doses of mTOR catalytic inhibitors were used, the residual mTORC2 activity towards S473 potently activated Akt20. Furthermore, while acute inhibition of mTORC2 by one such inhibitor, Ku-0063794, effectively suppressed T308 phosphorylation in wild-type cells, it failed to do so in cells where mTORC2 is genetically inactivated144. This result suggests that under chronic inhibition of mTORC2, which may occur in a clinical setting, alternative pathways may ensure Akt phosphorylation at T308 even in the absence of the priming S473 phosphorylation.
The similarity between the catalytic domains of mTOR and class I PI3-Kinase has also enabled the design of ATP-competitive drugs that simultaneously block the activity of both kinases (FIG.4b). When two inhibitors of this kind, PI-103 and NVP-BEZ235, were delivered to tumor cells driven by PI3K, they strongly suppressed both S6K1 and Akt activation148, 149. More importantly, this type of compound suppressed the proliferation of cancer cells more efficiently than rapamycin or the PI3K inhibitor LY294002, and to a similar degree as a combination of the two.
Dual PI3K-mTOR inhibitors may be conceptually superior to catalytic mTOR inhibitors, because they disable both inputs to Akt, namely PI3K-PDK1 and mTORC2. However, this broad inhibition of signaling pathways may be toxic to normal cells. In fact, while both the ATP-competitive mTOR inhibitor PP-242 and the dual PI3K-mTOR inhibitor PI-103 showed an antileukemic effect in vivo, the latter also harmed normal lymphocytes, suggesting that the therapeutic window of dual inhibitors might be narrow147.
It still remains unclear to what extent mTOR inhibitors and mTOR/PI3K dual inhibitors are effective in inducing the death of cancer cells143, 148–150, although apoptotic responses were observed in cells from gliomas, breast and hematological tumors147, 151–153. Nevertheless, these compounds show a consistently good effect against tumors driven by PI3K/AKT, while they were ineffective against tumors driven by mutations of Ras, which has the ability to signal through multiple pathways, such as the MEKERK pathway148. In this case, a combination therapy of dual mTOR-PI3K inhibitors together with a MEK inhibitor was required to achieve antitumoral effects.
mTOR and ageing
Ageing can be defined as a time-dependent decline of the physiological functions of cells, tissues and organs. Ageing can favor the insurgence of sporadic diseases such as cancer, or can itself lead to death through organ failure. Because of this, ageing is increasingly viewed as a disease in its own right, for which molecular therapies can be designed.
In recent years, the manipulation of nutrient sensing and stress response pathways has resulted in an extended lifespan of organisms from yeast to mammals. The rationale behind this observation is that growth-promoting programs may accelerate ageing by generating metabolic by-products and by directly inhibiting homeostasis and repair. Conversely, suppression of growth programs through chemical and genetic manipulations, or by reducing food intake, results in the activation of salvage programs that preserve the functionality of cells and tissues for extended periods of time (reviewed in154).
Due to its role at the interface of growth and starvation, mTOR is a prime target in the genetic control of ageing, and evidence from genetic studies supports the view that mTOR may be a master determinant of lifespan and ageing in yeast155, 156, C. elegans157, 158, flies159, 160 and mice161.
The only ‘natural’ method available to counter ageing is dietary restriction (DR), where the caloric intake is decreased anywhere from 10% to 50%. DR appears to act mainly through the inhibition of mTORC1, and genetic inactivation of mTORC1 pathway components provides no additional benefit over DR156, 159, 160. In mice, DR causes lifespan extension and changes in gene expression profile similar to those resulting from loss of S6K1162, further supporting the view that DR acts through inhibition of mTORC1.
mTOR and cellular ageing
Cellular ageing may stem from the cumulative action of metabolic by-products, exogenous chemicals and ionizing radiation, and order-degrading stochastic processes. To cope with these sources of chaos and damage, the cell relies on rescue processes that either attempt to repair the damaged molecules (such as DNA repair), or mediate their clearance (such as autophagy and proteasomal degradation).
Inhibition of mTORC1 may both counter the sources of damage and enhance repair mechanisms (FIG.5a). Inhibiting mTORC1, and translation as a consequence, extended the lifespan of yeast, worms and flies163–166. Reduced translation may place smaller demands on the protein folding systems, thus decreasing the by-production of misfolded protein aggregates. Accordingly, prolonged rapamycin treatment in a mouse model of Huntington Disease decreased the formation of toxic huntingtin aggregates167.
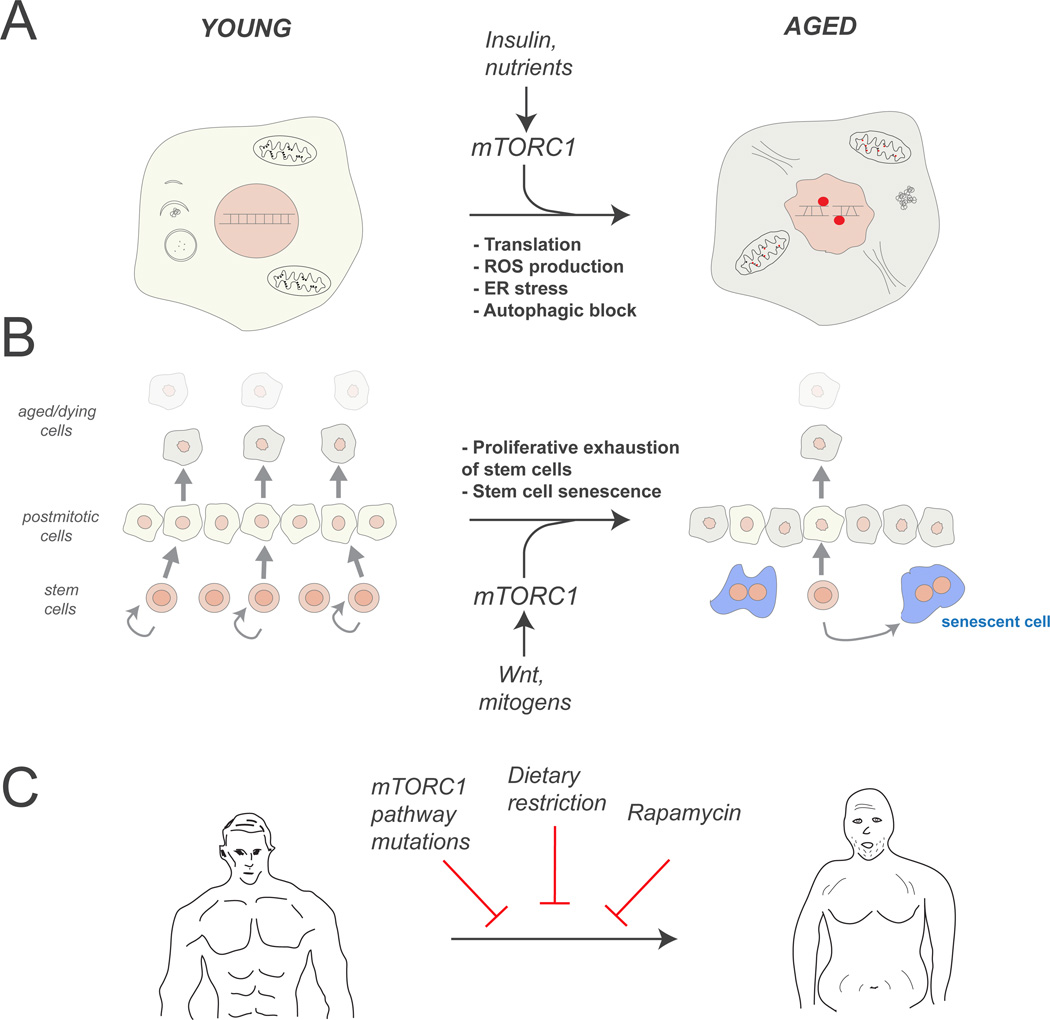
Figure 5. mTOR in ageing.
A. mTORC1-regulated processes that promote cellular ageing. mTORC1-dependent translation may overcome the protein folding capacity of the cell, resulting in accumulation of unfolded proteins and ER stress. Stimulation of mitochondrial function may increase ROS production, resulting in oxidative damage to DNA, proteins and membranes. Inhibition of autophagy by mTORC1 reduces the turnover of cellular components and promotes the accumulation of their damaged forms (red).
B. mTORC1 promotes stem cell exhaustion and tissue ageing. In young tissues, stem cells divide asymmetrically to generate new stem cells as well as postmitotic cell that replace those that have undergone turnover (left). Continued exposure to mitogens that signal through mTORC1 causes stem cell exhaustion through hyperproliferation or senescence (right); thus, in aged tissues post-mitotic cells are no longer replaced and the overall performance of the tissue is degraded.
C. Inhibiting mTORC1 activity by various means allows lifespan extension in multiple organisms, and may have beneficial effects on human ageing.
Lifespan extension can also be explained by an increase in autophagy following mTORC1 inhibition. Autophagy was required for lifespan extension by virtually all protocols in worms and flies159, 168, 169. Interestingly, an age-dependent drop in the expression of LAMP2A, which mediates chaperonmediated autophagy, contributed to the ageing of hepatocytes170. DR and rapamycin deliver a boost to the autophagic pathway that may compensate for its age-dependent decline.
Interestingly, inhibiting mTORC1 may lead to the increased translation of genes that exert a protective function. In a recent report, the activation of 4E-BP1 by DR in D. melanogaster resulted in the increased translation of several components of the mitochondrial electron transport chain110. This selective upregulation led to improved mitochondrial respiration. One may speculate that the resulting decrease in ROS production should result in less cellular damage.
Finally, inhibition of mTORC1 may result in the activation of specific gene expression programs related with the regulation of lifespan; stress-response programs controlled by the transcription factor Gis-1 function downstream of scTOR in regulating S cerevisiae lifespan170.
mTOR and tissue ageing
Tissue-specific stem cells are key in maintaining organ function, by replacing differentiated cells undergoing turnover as well as those that have succumbed to damage (for comprehensive reviews, see171). There is evidence that the number of stem cells, as well as their propensity to undergo novel divisions for tissue turnover and repair purposes, declines over time, leading to an irreversible degradation of organ function and thus ageing. For instance, the cell cycle regulators p16Ink4a and p19Arf act as a brake to limit the proliferation of the stem cell pool, but their age-dependent accumulation ultimately causes stem cells to undergo senescence172, 173.
Moreover, aberrant growth signals or stress signals can accelerate stem cell senescence and tissue ageing (FIG.5B). In a recent report, persistent expression of Wnt proteins in mouse epidermis caused hyperproliferation of epithelial stem cells, ultimately causing them to undergo senescence and exhausting the stem cell niche. Importantly, these actions seemed to occur through Wnt-mediated activation of the mTOR pathway: rapamycin treatment prevented both the hyperproliferation and premature senescence of epidermal stem cell exposed to excess Wnt88. In mouse hematopoietic stem cells (HSCs), constitutive mTORC1 activation via deletion of TSC1 led to increased expression of p16Ink4a, p19Arf and p21CIP1, resulting in HSCs depletion. An age-dependent increase in the activity of mTORC1 was detected and, moreover, prolonged rapamycin treatment preserved the HSCs pool to levels similar to young animals174.
Finally, over-activation of the PI3K pathway via deletion of PTEN also led to HSCs hyperproliferation followed by their depletion, likely via mTORC1. In fact, treatment with rapamycin restored the capacity of PTEN-null HSCs to reconstitute the blood lineage of irradiated mice175.
Altogether, these findings point to mTORC1 as a key mediator of growth signals that drive exit of tissue stem cells from quiescence, and to mTORC1 inhibition as a viable approach to preserve the stem cell pool, and thus the functionality of tissues and organs over time.
Concluding remarks
The recent identification of novel regulators and their mode of action further strengthen the idea that the basic layout of the mTOR pathway is that of a signal integrator. The TSC node computes signals from growth factors, stressors and energy to regulate Rheb. A second node is mTORC1 itself, where raptor and PRAS40 directly relay energy and growth factors. Finally, the lysosomal membrane acts as a platform for integration of nutrient inputs with the Rheb axis. It remains to be determined whether signal integration also occurs at the level of mTORC2, and whether mTORC1 and mTORC2 are coordinated to a greater extent than is currently known. The identification of upstream regulators of mTORC2 will likely shed light on these questions.
Given its many cellular actions, it is puzzling that only few substrates of mTOR have been identified so far. This is partly due to the weak and transient nature of the mTOR-substrate interaction; the continuous improvement of mass spectrometry techniques, combined with the use of the novel mTOR catalytic inhibitors will likely bring important advances in this area. Moreover, the emerging concept that whole classes of genes may be co-regulated by mTORC1-mediated translational control further extends the range of its downstream effectors.
A more integrated understanding of the mTOR pathway will pave the way for novel approaches to old diseases; mTOR has evolved to accelerate growth, but it also speeds up cancer, metabolic derangement and ageing in adulthood. For these reasons, a chronic but well tolerated inhibition of mTOR starting in mid-life could bring significant improvements to human health.
This lifestyle improvement may come at a cost. For instance, it has been observed that lifespan extension by various manipulations comes at the expense of fertility and reproductive success, but recent findings indicate that there may be a way around it: in D. melanogaster, supply of a single amino acid, methionine, allows maintenance of reproductive potency in the context of CR-induced lifespan extension176. Another potential risk is that over-stimulation of autophagy by chronic mTOR inhibition may actually speed up ageing. Furthermore, the many feedback loops in which mTOR participates may actually result in harmful outcomes. Thus, the particular regimen of mTOR inhibition may have to be carefully chosen while considering the advantages of rapamycin versus catalytic inhibitors and chronic versus intermittent administration.
Finally, it remains to be seen whether limiting mTOR activity in adult humans would really enable a longer lifespan, or it would only bring an increase in the quality of life and the way we age, without necessarily affecting how long we live.
Supplementary Material
Box1 Figure.
References
- 1.Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- 2.Sabatini DM, Erdjument-Bromage H, Lui M, Tempst P, Snyder SH. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell. 1994;78:35–43. doi: 10.1016/0092-8674(94)90570-3. [DOI] [PubMed] [Google Scholar]
- 3.Brown EJ, et al. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994;369:756–758. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- 4.Sabers CJ, et al. Isolation of a protein target of the FKBP12-rapamycin complex in mammalian cells. J Biol Chem. 1995;270:815–822. doi: 10.1074/jbc.270.2.815. [DOI] [PubMed] [Google Scholar]
- 5.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 6.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 7.Hara K, et al. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110:177–189. doi: 10.1016/s0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- 8.Kim DH, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 9.Sarbassov DD, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 10.Nojima H, et al. The mammalian target of rapamycin (mTOR) partner, raptor, binds the mTOR substrates p70 S6 kinase and 4E-BP1 through their TOR signaling (TOS) motif. J Biol Chem. 2003;278:15461–15464. doi: 10.1074/jbc.C200665200. [DOI] [PubMed] [Google Scholar]
- 11.Schalm SS, Fingar DC, Sabatini DM, Blenis J. TOS motif-mediated raptor binding regulates 4E-BP1 multisite phosphorylation and function. Curr Biol. 2003;13:797–806. doi: 10.1016/s0960-9822(03)00329-4. [DOI] [PubMed] [Google Scholar]
- 12.Wullschleger S, Loewith R, Oppliger W, Hall MN. Molecular organization of target of rapamycin complex 2. J Biol Chem. 2005;280:30697–30704. doi: 10.1074/jbc.M505553200. [DOI] [PubMed] [Google Scholar]
- 13.Sancak Y, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sancak Y, et al. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell. 2007;25:903–915. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 15.Pearce LR, et al. Identification of Protor as a novel Rictor-binding component of mTOR complex-2. Biochem J. 2007;405:513–522. doi: 10.1042/BJ20070540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vander Haar E, Lee SI, Bandhakavi S, Griffin TJ, Kim DH. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat Cell Biol. 2007;9:316–323. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- 17.Frias MA, et al. mSin1 is necessary for Akt/PKB phosphorylation, and its isoforms define three distinct mTORC2s. Curr Biol. 2006;16:1865–1870. doi: 10.1016/j.cub.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 18.Yang Q, Inoki K, Ikenoue T, Guan KL. Identification of Sin1 as an essential TORC2 component required for complex formation and kinase activity. Genes Dev. 2006;20:2820–2832. doi: 10.1101/gad.1461206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woo SY, et al. PRR5, a novel component of mTOR complex 2, regulates platelet-derived growth factor receptor beta expression and signaling. J Biol Chem. 2007;282:25604–25612. doi: 10.1074/jbc.M704343200. [DOI] [PubMed] [Google Scholar]
- 20.Peterson TR, et al. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 2009;137:873–886. doi: 10.1016/j.cell.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yip CK, Murata K, Walz T, Sabatini DM, Kang SA. Structure of the human mTOR complex I and its implications for rapamycin inhibition. Mol Cell. 2010;38:768–774. doi: 10.1016/j.molcel.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacinto E, et al. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol. 2004;6:1122–1128. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- 23.Barbet NC, et al. TOR controls translation initiation and early G1 progression in yeast. Mol Biol Cell. 1996;7:25–42. doi: 10.1091/mbc.7.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Helliwell SB, Howald I, Barbet N, Hall MN. TOR2 is part of two related signaling pathways coordinating cell growth in Saccharomyces cerevisiae. Genetics. 1998;148:99–112. doi: 10.1093/genetics/148.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng XF, Florentino D, Chen J, Crabtree GR, Schreiber SL. TOR kinase domains are required for two distinct functions, only one of which is inhibited by rapamycin. Cell. 1995;82:121–130. doi: 10.1016/0092-8674(95)90058-6. [DOI] [PubMed] [Google Scholar]
- 26.Choi J, Chen J, Schreiber SL, Clardy J. Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science. 1996;273:239–242. doi: 10.1126/science.273.5272.239. [DOI] [PubMed] [Google Scholar]
- 27.Loewith R, et al. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell. 2002;10:457–468. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- 28.Sarbassov DD, et al. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 29.Brown EJ, et al. Control of p70 s6 kinase by kinase activity of FRAP in vivo. Nature. 1995;377:441–441. doi: 10.1038/377441a0. [DOI] [PubMed] [Google Scholar]
- 30.Hara K, et al. Regulation of eIF-4E BP1 phosphorylation by mTOR. J Biol Chem. 1997;272:26457–26463. doi: 10.1074/jbc.272.42.26457. [DOI] [PubMed] [Google Scholar]
- 31.von Manteuffel SR, Gingras AC, Ming XF, Sonenberg N, Thomas G. 4E-BP1 phosphorylation is mediated by the FRAP-p70s6k pathway and is independent of mitogen-activated protein kinase. Proc Natl Acad Sci U S A. 1996;93:4076–4080. doi: 10.1073/pnas.93.9.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 33.Haghighat A, Mader S, Pause A, Sonenberg N. Repression of cap-dependent translation by 4E-binding protein 1: competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J. 1995;14:5701–5709. doi: 10.1002/j.1460-2075.1995.tb00257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeno P, Ballou LM, Novak-Hofer I, Thomas G. Identification and characterization of a mitogen-activated S6 kinase. Proc Natl Acad Sci U S A. 1988;85:406–410. doi: 10.1073/pnas.85.2.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lenz G, Avruch J. Glutamatergic regulation of the p70S6 kinase in primary mouse neurons. J Biol Chem. 2005;280:38121–38124. doi: 10.1074/jbc.C500363200. [DOI] [PubMed] [Google Scholar]
- 36.Ma XM, Yoon SO, Richardson CJ, Julich K, Blenis J. SKAR links pre-mRNA splicing to mTOR/S6K1-mediated enhanced translation efficiency of spliced mRNAs. Cell. 2008;133:303–313. doi: 10.1016/j.cell.2008.02.031. [DOI] [PubMed] [Google Scholar]
- 37.Wilson KF, Wu WJ, Cerione RA. Cdc42 stimulates RNA splicing via the S6 kinase and a novel S6 kinase target, the nuclear cap-binding complex. J Biol Chem. 2000;275:37307–37310. doi: 10.1074/jbc.C000482200. [DOI] [PubMed] [Google Scholar]
- 38.Holz MK, Ballif BA, Gygi SP, Blenis J. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell. 2005;123:569–580. doi: 10.1016/j.cell.2005.10.024. [DOI] [PubMed] [Google Scholar]
- 39.Shahbazian D, et al. The mTOR/PI3K and MAPK pathways converge on eIF4B to control its phosphorylation and activity. EMBO J. 2006;25:2781–2791. doi: 10.1038/sj.emboj.7601166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hannan KM, et al. mTOR-dependent regulation of ribosomal gene transcription requires S6K1 and is mediated by phosphorylation of the carboxy-terminal activation domain of the nucleolar transcription factor UBF. Mol Cell Biol. 2003;23:8862–8877. doi: 10.1128/MCB.23.23.8862-8877.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mayer C, Zhao J, Yuan X, Grummt I. mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability. Genes Dev. 2004;18:423–434. doi: 10.1101/gad.285504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Claypool JA, et al. Tor pathway regulates Rrn3p-dependent recruitment of yeast RNA polymerase I to the promoter but does not participate in alteration of the number of active genes. Mol Biol Cell. 2004;15:946–956. doi: 10.1091/mbc.E03-08-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin DE, Soulard A, Hall MN. TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell. 2004;119:969–979. doi: 10.1016/j.cell.2004.11.047. [DOI] [PubMed] [Google Scholar]
- 44.Schawalder SB, et al. Growth-regulated recruitment of the essential yeast ribosomal protein gene activator Ifh1. Nature. 2004;432:1058–1061. doi: 10.1038/nature03200. [DOI] [PubMed] [Google Scholar]
- 45.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem. 1998;273:3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- 47.Thoreen CC, et al. An ATP-competitive Mammalian Target of Rapamycin Inhibitor Reveals Rapamycin-resistant Functions of mTORC1. J Biol Chem. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kamada Y, et al. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hosokawa N, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jung CH, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hara T, et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hosokawa N, et al. Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy. 2009;5:973–979. doi: 10.4161/auto.5.7.9296. [DOI] [PubMed] [Google Scholar]
- 53.Kamada Y, et al. Tor2 directly phosphorylates the AGC kinase Ypk2 to regulate actin polarization. Mol Cell Biol. 2005;25:7239–7248. doi: 10.1128/MCB.25.16.7239-7248.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmidt A, Bickle M, Beck T, Hall MN. The yeast phosphatidylinositol kinase homolog TOR2 activates RHO1 and RHO2 via the exchange factor ROM2. Cell. 1997;88:531–542. doi: 10.1016/s0092-8674(00)81893-0. [DOI] [PubMed] [Google Scholar]
- 55.Facchinetti V, et al. The mammalian target of rapamycin complex 2 controls folding and stability of Akt and protein kinase C. EMBO J. 2008;27:1932–1943. doi: 10.1038/emboj.2008.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Garcia-Martinez JM, Alessi DR. mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1) Biochem J. 2008;416:375–385. doi: 10.1042/BJ20081668. [DOI] [PubMed] [Google Scholar]
- 57.Ikenoue T, Inoki K, Yang Q, Zhou X, Guan KL. Essential function of TORC2 in PKC and Akt turn motif phosphorylation, maturation and signalling. EMBO J. 2008;27:1919–1931. doi: 10.1038/emboj.2008.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 59.Guertin DA, et al. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev Cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 60.Soukas AA, Kane EA, Carr CE, Melo JA, Ruvkun G. Rictor/TORC2 regulates fat metabolism, feeding, growth, and life span in Caenorhabditis elegans. Genes Dev. 2009;23:496–511. doi: 10.1101/gad.1775409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guertin DA, et al. mTOR complex 2 is required for the development of prostate cancer induced by Pten loss in mice. Cancer Cell. 2009;15:148–159. doi: 10.1016/j.ccr.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hara K, et al. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J Biol Chem. 1998;273:14484–14494. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- 63.Wang X, Campbell LE, Miller CM, Proud CG. Amino acid availability regulates p70 S6 kinase and multiple translation factors. Biochem J. 1998;334(Pt 1):261–267. doi: 10.1042/bj3340261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sancak Y, et al. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saucedo LJ, et al. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat Cell Biol. 2003;5:566–571. doi: 10.1038/ncb996. [DOI] [PubMed] [Google Scholar]
- 67.Stocker H, et al. Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat Cell Biol. 2003;5:559–565. doi: 10.1038/ncb995. [DOI] [PubMed] [Google Scholar]
- 68.Binda M, et al. The Vam6 GEF controls TORC1 by activating the EGO complex. Mol Cell. 2009;35:563–573. doi: 10.1016/j.molcel.2009.06.033. [DOI] [PubMed] [Google Scholar]
- 69.Dubouloz F, Deloche O, Wanke V, Cameroni E, De Virgilio C. The TOR and EGO protein complexes orchestrate microautophagy in yeast. Mol Cell. 2005;19:15–26. doi: 10.1016/j.molcel.2005.05.020. [DOI] [PubMed] [Google Scholar]
- 70.Gao M, Kaiser CA. A conserved GTPase-containing complex is required for intracellular sorting of the general amino-acid permease in yeast. Nat Cell Biol. 2006;8:657–667. doi: 10.1038/ncb1419. [DOI] [PubMed] [Google Scholar]
- 71.Zurita-Martinez SA, Puria R, Pan X, Boeke JD, Cardenas ME. Efficient Tor signaling requires a functional class C Vps protein complex in Saccharomyces cerevisiae. Genetics. 2007;176:2139–2150. doi: 10.1534/genetics.107.072835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Urano J, Tabancay AP, Yang W, Tamanoi F. The Saccharomyces cerevisiae Rheb G-protein is involved in regulating canavanine resistance and arginine uptake. J Biol Chem. 2000;275:11198–11206. doi: 10.1074/jbc.275.15.11198. [DOI] [PubMed] [Google Scholar]
- 73.Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 74.Manning BD, Tee AR, Logsdon MN, Blenis J, Cantley LC. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol Cell. 2002;10:151–162. doi: 10.1016/s1097-2765(02)00568-3. [DOI] [PubMed] [Google Scholar]
- 75.Potter CJ, Pedraza LG, Xu T. Akt regulates growth by directly phosphorylating Tsc2. Nat Cell Biol. 2002;4:658–665. doi: 10.1038/ncb840. [DOI] [PubMed] [Google Scholar]
- 76.Garami A, et al. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol Cell. 2003;11:1457–1466. doi: 10.1016/s1097-2765(03)00220-x. [DOI] [PubMed] [Google Scholar]
- 77.Inoki K, Li Y, Xu T, Guan KL. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang Y, et al. Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat Cell Biol. 2003;5:578–581. doi: 10.1038/ncb999. [DOI] [PubMed] [Google Scholar]
- 79.Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr Biol. 2003;13:1259–1268. doi: 10.1016/s0960-9822(03)00506-2. [DOI] [PubMed] [Google Scholar]
- 80.Gao X, Pan D. TSC1 and TSC2 tumor suppressors antagonize insulin signaling in cell growth. Genes Dev. 2001;15:1383–1392. doi: 10.1101/gad.901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tapon N, Ito N, Dickson BJ, Treisman JE, Hariharan IK. The Drosophila tuberous sclerosis complex gene homologs restrict cell growth and cell proliferation. Cell. 2001;105:345–355. doi: 10.1016/s0092-8674(01)00332-4. [DOI] [PubMed] [Google Scholar]
- 82.Cai SL, et al. Activity of TSC2 is inhibited by AKT-mediated phosphorylation and membrane partitioning. J Cell Biol. 2006;173:279–289. doi: 10.1083/jcb.200507119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li Y, Inoki K, Yeung R, Guan KL. Regulation of TSC2 by 14-3-3 binding. J Biol Chem. 2002;277:44593–44596. doi: 10.1074/jbc.C200510200. [DOI] [PubMed] [Google Scholar]
- 84.Kovacina KS, et al. Identification of a proline-rich Akt substrate as a 14-3-3 binding partner. J Biol Chem. 2003;278:10189–10194. doi: 10.1074/jbc.M210837200. [DOI] [PubMed] [Google Scholar]
- 85.Fonseca BD, Smith EM, Lee VH, MacKintosh C, Proud CG. PRAS40 is a target for mammalian target of rapamycin complex 1 and is required for signaling downstream of this complex. J Biol Chem. 2007;282:24514–24524. doi: 10.1074/jbc.M704406200. [DOI] [PubMed] [Google Scholar]
- 86.Oshiro N, et al. The proline-rich Akt substrate of 40 kDa (PRAS40) is a physiological substrate of mammalian target of rapamycin complex 1. J Biol Chem. 2007;282:20329–20339. doi: 10.1074/jbc.M702636200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ma L, Chen Z, Erdjument-Bromage H, Tempst P, Pandolfi PP. Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell. 2005;121:179–193. doi: 10.1016/j.cell.2005.02.031. [DOI] [PubMed] [Google Scholar]
- 88.Castilho RM, Squarize CH, Chodosh LA, Williams BO, Gutkind JS. mTOR mediates Wnt-induced epidermal stem cell exhaustion and aging. Cell Stem Cell. 2009;5:279–289. doi: 10.1016/j.stem.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Inoki K, et al. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell. 2006;126:955–968. doi: 10.1016/j.cell.2006.06.055. [DOI] [PubMed] [Google Scholar]
- 90.Gangloff YG, et al. Disruption of the mouse mTOR gene leads to early postimplantation lethality and prohibits embryonic stem cell development. Mol Cell Biol. 2004;24:9508–9516. doi: 10.1128/MCB.24.21.9508-9516.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Murakami M, et al. mTOR is essential for growth and proliferation in early mouse embryos and embryonic stem cells. Mol Cell Biol. 2004;24:6710–6718. doi: 10.1128/MCB.24.15.6710-6718.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dennis PB, et al. Mammalian TOR: a homeostatic ATP sensor. Science. 2001;294:1102–1105. doi: 10.1126/science.1063518. [DOI] [PubMed] [Google Scholar]
- 93.Hardie DG. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol. 2007;8:774–785. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- 94.Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 95.Corradetti MN, Inoki K, Bardeesy N, DePinho RA, Guan KL. Regulation of the TSC pathway by LKB1: evidence of a molecular link between tuberous sclerosis complex and Peutz-Jeghers syndrome. Genes Dev. 2004;18:1533–1538. doi: 10.1101/gad.1199104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gwinn DM, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Reiling JH, Sabatini DM. Stress and mTORture signaling. Oncogene. 2006;25:6373–6383. doi: 10.1038/sj.onc.1209889. [DOI] [PubMed] [Google Scholar]
- 98.Brugarolas J, et al. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 2004;18:2893–2904. doi: 10.1101/gad.1256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Reiling JH, Hafen E. The hypoxia-induced paralogs Scylla and Charybdis inhibit growth by down-regulating S6K activity upstream of TSC in Drosophila. Genes Dev. 2004;18:2879–2892. doi: 10.1101/gad.322704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Feng Z, et al. The regulation of AMPK beta1, TSC2, and PTEN expression by p53: stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1-AKT-mTOR pathways. Cancer Res. 2007;67:3043–3053. doi: 10.1158/0008-5472.CAN-06-4149. [DOI] [PubMed] [Google Scholar]
- 101.Shaw RJ, et al. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell. 2004;6:91–99. doi: 10.1016/j.ccr.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 102.Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134:451–460. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Charest PG, et al. A Ras signaling complex controls the RasC-TORC2 pathway and directed cell migration. Dev Cell. 2010;18:737–749. doi: 10.1016/j.devcel.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lee S, et al. TOR complex 2 integrates cell movement during chemotaxis and signal relay in Dictyostelium. Mol Biol Cell. 2005;16:4572–4583. doi: 10.1091/mbc.E05-04-0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kuma A, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 108.Singh R, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yu L, et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 2010;465:942–946. doi: 10.1038/nature09076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zid BM, et al. 4E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell. 2009;139:149–160. doi: 10.1016/j.cell.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cunningham JT, et al. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 2007;450:736–740. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- 112.Polak P, et al. Adipose-specific knockout of raptor results in lean mice with enhanced mitochondrial respiration. Cell Metab. 2008;8:399–410. doi: 10.1016/j.cmet.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 113.Yeh WC, Bierer BE, McKnight SL. Rapamycin inhibits clonal expansion and adipogenic differentiation of 3T3-L1 cells. Proc Natl Acad Sci U S A. 1995;92:11086–11090. doi: 10.1073/pnas.92.24.11086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gagnon A, Lau S, Sorisky A. Rapamycin-sensitive phase of 3T3-L1 preadipocyte differentiation after clonal expansion. J Cell Physiol. 2001;189:14–22. doi: 10.1002/jcp.1132. [DOI] [PubMed] [Google Scholar]
- 115.Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 116.Kim JE, Chen J. regulation of peroxisome proliferator-activated receptor-gamma activity by mammalian target of rapamycin and amino acids in adipogenesis. Diabetes. 2004;53:2748–2756. doi: 10.2337/diabetes.53.11.2748. [DOI] [PubMed] [Google Scholar]
- 117.Zhang HH, et al. Insulin stimulates adipogenesis through the Akt-TSC2-mTORC1 pathway. PLoS One. 2009;4:e6189. doi: 10.1371/journal.pone.0006189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Le Bacquer O, et al. Elevated sensitivity to diet-induced obesity and insulin resistance in mice lacking 4E-BP1 and 4E-BP2. J Clin Invest. 2007;117:387–396. doi: 10.1172/JCI29528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kim JB, Spiegelman BM. ADD1/SREBP1 promotes adipocyte differentiation and gene expression linked to fatty acid metabolism. Genes Dev. 1996;10:1096–1107. doi: 10.1101/gad.10.9.1096. [DOI] [PubMed] [Google Scholar]
- 120.Kim JB, Wright HM, Wright M, Spiegelman BM. ADD1/SREBP1 activates PPARgamma through the production of endogenous ligand. Proc Natl Acad Sci U S A. 1998;95:4333–4337. doi: 10.1073/pnas.95.8.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Porstmann T, et al. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 2008;8:224–236. doi: 10.1016/j.cmet.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dowell P, Otto TC, Adi S, Lane MD. Convergence of peroxisome proliferator-activated receptor gamma and Foxo1 signaling pathways. J Biol Chem. 2003;278:45485–45491. doi: 10.1074/jbc.M309069200. [DOI] [PubMed] [Google Scholar]
- 123.Nakae J, et al. Regulation of insulin action and pancreatic beta-cell function by mutated alleles of the gene encoding forkhead transcription factor Foxo1. Nat Genet. 2002;32:245–253. doi: 10.1038/ng890. [DOI] [PubMed] [Google Scholar]
- 124.Um SH, et al. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature. 2004;431:200–205. doi: 10.1038/nature02866. [DOI] [PubMed] [Google Scholar]
- 125.Newgard CB, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009;9:311–326. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Cota D, et al. Hypothalamic mTOR signaling regulates food intake. Science. 2006;312:927–930. doi: 10.1126/science.1124147. [DOI] [PubMed] [Google Scholar]
- 127.Hsieh AC, et al. Genetic dissection of the oncogenic mTOR pathway reveals druggable addiction to translational control via 4EBP-eIF4E. Cancer Cell. 2010;17:249–261. doi: 10.1016/j.ccr.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wendel HG, et al. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature. 2004;428:332–337. doi: 10.1038/nature02369. [DOI] [PubMed] [Google Scholar]
- 129.Wendel HG, et al. Dissecting eIF4E action in tumorigenesis. Genes Dev. 2007;21:3232–3237. doi: 10.1101/gad.1604407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer. 2007;7:763–777. doi: 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- 131.Qu X, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Marino G, et al. Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in Atg4C/autophagin-3. J Biol Chem. 2007;282:18573–18583. doi: 10.1074/jbc.M701194200. [DOI] [PubMed] [Google Scholar]
- 134.Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134:703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 135.Carracedo A, et al. Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K-dependent feedback loop in human cancer. J Clin Invest. 2008;118:3065–3074. doi: 10.1172/JCI34739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhang H, et al. PDGFRs are critical for PI3K/Akt activation and negatively regulated by mTOR. J Clin Invest. 2007;117:730–738. doi: 10.1172/JCI28984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.O'Reilly KE, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66:1500–1508. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Efeyan A, Sabatini DM. mTOR and cancer: many loops in one pathway. Curr Opin Cell Biol. 2010;22:169–176. doi: 10.1016/j.ceb.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Shor B, et al. A new pharmacologic action of CCI-779 involves FKBP12-independent inhibition of mTOR kinase activity and profound repression of global protein synthesis. Cancer Res. 2008;68:2934–2943. doi: 10.1158/0008-5472.CAN-07-6487. [DOI] [PubMed] [Google Scholar]
- 140.Choo AY, Yoon SO, Kim SG, Roux PP, Blenis J. Rapamycin differentially inhibits S6Ks and 4E-BP1 to mediate cell-type-specific repression of mRNA translation. Proc Natl Acad Sci U S A. 2008;105:17414–17419. doi: 10.1073/pnas.0809136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.McMahon LP, Choi KM, Lin TA, Abraham RT, Lawrence JC., Jr The rapamycin-binding domain governs substrate selectivity by the mammalian target of rapamycin. Mol Cell Biol. 2002;22:7428–7438. doi: 10.1128/MCB.22.21.7428-7438.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Feldman ME, et al. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 2009;7:e38. doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chresta CM, et al. AZD8055 is a potent, selective, and orally bioavailable ATP-competitive mammalian target of rapamycin kinase inhibitor with in vitro and in vivo antitumor activity. Cancer Res. 2010;70:288–298. doi: 10.1158/0008-5472.CAN-09-1751. [DOI] [PubMed] [Google Scholar]
- 144.Garcia-Martinez JM, et al. Ku-0063794 is a specific inhibitor of the mammalian target of rapamycin (mTOR) Biochem J. 2009;421:29–42. doi: 10.1042/BJ20090489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yu K, et al. Biochemical, cellular, and in vivo activity of novel ATP-competitive and selective inhibitors of the mammalian target of rapamycin. Cancer Res. 2009;69:6232–6240. doi: 10.1158/0008-5472.CAN-09-0299. [DOI] [PubMed] [Google Scholar]
- 146.Nardella C, et al. Differential requirement of mTOR in postmitotic tissues and tumorigenesis. Sci Signal. 2009;2:ra2. doi: 10.1126/scisignal.2000189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Janes MR, et al. Effective and selective targeting of leukemia cells using a TORC1/2 kinase inhibitor. Nat Med. 2010;16:205–213. doi: 10.1038/nm.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Engelman JA, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008;14:1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Fan QW, et al. A dual PI3 kinase/mTOR inhibitor reveals emergent efficacy in glioma. Cancer Cell. 2006;9:341–349. doi: 10.1016/j.ccr.2006.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Liu TJ, et al. NVP-BEZ235, a novel dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor, elicits multifaceted antitumor activities in human gliomas. Mol Cancer Ther. 2009;8:2204–2210. doi: 10.1158/1535-7163.MCT-09-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Brachmann SM, et al. Specific apoptosis induction by the dual PI3K/mTor inhibitor NVP-BEZ235 in HER2 amplified and PIK3CA mutant breast cancer cells. Proc Natl Acad Sci U S A. 2009;106:22299–22304. doi: 10.1073/pnas.0905152106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Chiarini F, et al. Dual inhibition of class IA phosphatidylinositol 3-kinase and mammalian target of rapamycin as a new therapeutic option for T-cell acute lymphoblastic leukemia. Cancer Res. 2009;69:3520–3528. doi: 10.1158/0008-5472.CAN-08-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.McMillin DW, et al. Antimyeloma activity of the orally bioavailable dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor NVP-BEZ235. Cancer Res. 2009;69:5835–5842. doi: 10.1158/0008-5472.CAN-08-4285. [DOI] [PubMed] [Google Scholar]
- 154.Fontana L, Partridge L, Longo VD. Extending healthy life span--from yeast to humans. Science. 2010;328:321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Fabrizio P, Pozza F, Pletcher SD, Gendron CM, Longo VD. Regulation of longevity and stress resistance by Sch9 in yeast. Science. 2001;292:288–290. doi: 10.1126/science.1059497. [DOI] [PubMed] [Google Scholar]
- 156.Kaeberlein M, et al. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- 157.Jia K, Chen D, Riddle DL. The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development. 2004;131:3897–3906. doi: 10.1242/dev.01255. [DOI] [PubMed] [Google Scholar]
- 158.Vellai T, et al. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426:620. doi: 10.1038/426620a. [DOI] [PubMed] [Google Scholar]
- 159.Bjedov I, et al. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 2010;11:35–46. doi: 10.1016/j.cmet.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kapahi P, et al. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr Biol. 2004;14:885–890. doi: 10.1016/j.cub.2004.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Harrison DE, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Selman C, et al. Ribosomal protein S6 kinase 1 signaling regulates mammalian life span. Science. 2009;326:140–144. doi: 10.1126/science.1177221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hansen M, et al. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6:95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- 164.Pan KZ, et al. Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging Cell. 2007;6:111–119. doi: 10.1111/j.1474-9726.2006.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Steffen KK, et al. Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell. 2008;133:292–302. doi: 10.1016/j.cell.2008.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Syntichaki P, Troulinaki K, Tavernarakis N. eIF4E function in somatic cells modulates ageing in Caenorhabditis elegans. Nature. 2007;445:922–926. doi: 10.1038/nature05603. [DOI] [PubMed] [Google Scholar]
- 167.Murphy CT, et al. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- 168.Hansen M, et al. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet. 2008;4:e24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Toth ML, et al. Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans. Autophagy. 2008;4:330–338. doi: 10.4161/auto.5618. [DOI] [PubMed] [Google Scholar]
- 170.Zhang C, Cuervo AM. Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nat Med. 2008;14:959–965. doi: 10.1038/nm.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.He S, Nakada D, Morrison SJ. Mechanisms of stem cell self-renewal. Annu Rev Cell Dev Biol. 2009;25:377–406. doi: 10.1146/annurev.cellbio.042308.113248. [DOI] [PubMed] [Google Scholar]
- 172.Janzen V, et al. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature. 2006;443:421–426. doi: 10.1038/nature05159. [DOI] [PubMed] [Google Scholar]
- 173.Molofsky AV, et al. Increasing p16INK4a expression decreases forebrain progenitors and neurogenesis during ageing. Nature. 2006;443:448–452. doi: 10.1038/nature05091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Chen C, Liu Y, Zheng P. mTOR regulation and therapeutic rejuvenation of aging hematopoietic stem cells. Sci Signal. 2009;2:ra75. doi: 10.1126/scisignal.2000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Yilmaz OH, et al. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 2006;441:475–482. doi: 10.1038/nature04703. [DOI] [PubMed] [Google Scholar]
- 176.Grandison RC, Piper MD, Partridge L. Amino-acid imbalance explains extension of lifespan by dietary restriction in Drosophila. Nature. 2009;462:1061–1064. doi: 10.1038/nature08619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Nobukuni T, et al. Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc Natl Acad Sci U S A. 2005;102:14238–14243. doi: 10.1073/pnas.0506925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Chang YY, Neufeld TP. An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol Biol Cell. 2009;20:2004–2014. doi: 10.1091/mbc.E08-12-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Berchtold D, Walther TC. TORC2 plasma membrane localization is essential for cell viability and restricted to a distinct domain. Mol Biol Cell. 2009;20:1565–1575. doi: 10.1091/mbc.E08-10-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.