Abstract
We aimed to identify genetic variants associated with cortical bone thickness (CBT) and bone mineral density (BMD) by performing two separate genome-wide association study (GWAS) meta-analyses for CBT in 3 cohorts comprising 5,878 European subjects and for BMD in 5 cohorts comprising 5,672 individuals. We then assessed selected single-nucleotide polymorphisms (SNPs) for osteoporotic fracture in 2,023 cases and 3,740 controls. Association with CBT and forearm BMD was tested for ∼2.5 million SNPs in each cohort separately, and results were meta-analyzed using fixed effect meta-analysis. We identified a missense SNP (Thr>Ile; rs2707466) located in the WNT16 gene (7q31), associated with CBT (effect size of −0.11 standard deviations [SD] per C allele, P = 6.2×10−9). This SNP, as well as another nonsynonymous SNP rs2908004 (Gly>Arg), also had genome-wide significant association with forearm BMD (−0.14 SD per C allele, P = 2.3×10−12, and −0.16 SD per G allele, P = 1.2×10−15, respectively). Four genome-wide significant SNPs arising from BMD meta-analysis were tested for association with forearm fracture. SNP rs7776725 in FAM3C, a gene adjacent to WNT16, was associated with a genome-wide significant increased risk of forearm fracture (OR = 1.33, P = 7.3×10−9), with genome-wide suggestive signals from the two missense variants in WNT16 (rs2908004: OR = 1.22, P = 4.9×10−6 and rs2707466: OR = 1.22, P = 7.2×10−6). We next generated a homozygous mouse with targeted disruption of Wnt16. Female Wnt16−/− mice had 27% (P<0.001) thinner cortical bones at the femur midshaft, and bone strength measures were reduced between 43%–61% (6.5×10−13<P<5.9×10−4) at both femur and tibia, compared with their wild-type littermates. Natural variation in humans and targeted disruption in mice demonstrate that WNT16 is an important determinant of CBT, BMD, bone strength, and risk of fracture.
Author Summary
Bone traits are highly dependent on genetic factors. To date, numerous genetic loci for bone mineral density (BMD) and only one locus for osteoporotic fracture have been previously identified to be genome-wide significant. Cortical bone has been reported to be an important determinant of bone strength; so far, no genome-wide association studies (GWAS) have been performed for cortical bone thickness (CBT) of the tibial and radial diaphysis or BMD at forearm, a skeletal site rich in cortical bone. Therefore, we performed two separated meta-analyses of GWAS for cortical thickness of the tibia in 3 independent cohorts of 5,878 men and women, and for forearm BMD in 5 cohorts of 5,672 individuals. We identified the 7q31 locus, which contains WNT16, to be associated with CBT and BMD. Four SNPs from this locus were then tested in 2,023 osteoporotic fracture cases and 3,740 controls. One of these SNPs was genome-wide significant, and two were genome-wide suggestive, for forearm fracture. Generating a mouse with targeted disruption of Wnt16, we also demonstrated that mice lacking this protein had substantially thinner bone cortices and reduced bone strength than their wild-type littermates. These findings highlight WNT16 as a clinically relevant member of the Wnt signaling pathway and increase our understanding of the etiology of osteoporosis-related phenotypes and fracture.
Introduction
Osteoporosis is a common skeletal disease characterized by reduced areal bone mineral density (BMD) and defects in the microarchitecture of bone, resulting in an increased risk of fragility fracture [1]. Osteoporotic fractures affect between one third to one half of white women [2] and currently incur direct costs exceeding $19 billion per year in the United States alone [3]; and this socio-economic burden is increasing with the ageing of industrial societies [4].
Twin and family studies have revealed that genetic factors can explain up to 85% of the variation in peak BMD [5], [6]. Since 2007, we and others have published several genome-wide association studies (GWAS) for osteoporosis and related traits [7], [8], [9], [10], [11], [12], [13], [14] identifying multiple common variants associated with BMD and highlighting biologic pathways that influence BMD.
Most osteoporotic fractures occur at peripheral sites, mainly containing cortical bone, after the age of 65 [15]. As indicated by a recent study, bone loss at this age is mainly due to loss in cortical and not trabecular bone [16]. In human cadaver femurs, cortical bone has been reported to be the main determinant of the femoral neck bone strength, while trabecular bone only contributes marginally to bone strength at this site [17]. Evidence implicating cortical thinning as a risk factor for hip fracture has also been presented [18]. The heritability for cortical thickness, measured using computed tomography, has been reported to be as high as 51% [19].
BMD is a complex trait, obtained from a 2-dimensional projectional scan of the given bone with dual x-ray absorptiometry (DXA). Although BMD is the most clinical useful measure for diagnosing bone fragility (osteoporosis), it fails to provide a detailed skeletal phenotype necessary to discern traits such as bone geometry and volumetric BMD (vBMD) [20]. Most of the loci or genes identified have been associated with BMD at lumbar spine and/or femoral neck, sites rich in trabecular bone. Therefore, we hypothesized that investigating BMD at the forearm, a primarily cortical bone site, as well cortical bone thickness, a trait with high heritability, would serve as successful strategies to identify novel bone related genetic loci.
Forearm fractures are among the most common fractures, affecting 1.7 million individuals per year. In contrast to hip fractures [21], forearm fractures have been shown to be highly heritable, with estimates of 54% [22]. To our knowledge, no GWA studies for cortical bone thickness, forearm BMD or fractures have been published. Importantly, we are aware of only one previous locus [12] that has been associated with risk of fracture even in large-scale meta-analytic efforts at a genome-wide significant level (reviewed previously) [23], [24], [25].
In this study, we performed two separate GWA meta-analyses in order to identify loci for cortical bone thickness of tibial diaphysis and BMD at the distal radius. Firstly, we performed a GWA study of three large and well-characterized independent discovery cohorts of 5,878 samples with the aim of identifying genetic loci for cortical thickness. SNPs meeting GWAS significance in the discovery meta-analysis were also tested for association in a large replication cohort (N = 1032). In the second and separate GWA meta-analysis, we combined genome-wide association results of 5,672 samples with BMD measurement at the forearm site from five cohorts; we then sought evidence of association of selected genome-wide significant signals in three cohorts comprising 5,763 individuals for forearm fracture.
To determine the possible functional role of the identified genes on cortical bone thickness and bone strength, we generated mice with inactivated genes and investigated their skeletal phenotype. The resultant findings increase our understanding of the genetic basis of osteoporosis and osteoporotic fracture.
Results
GWAS Meta-Analysis of Cortical Thickness
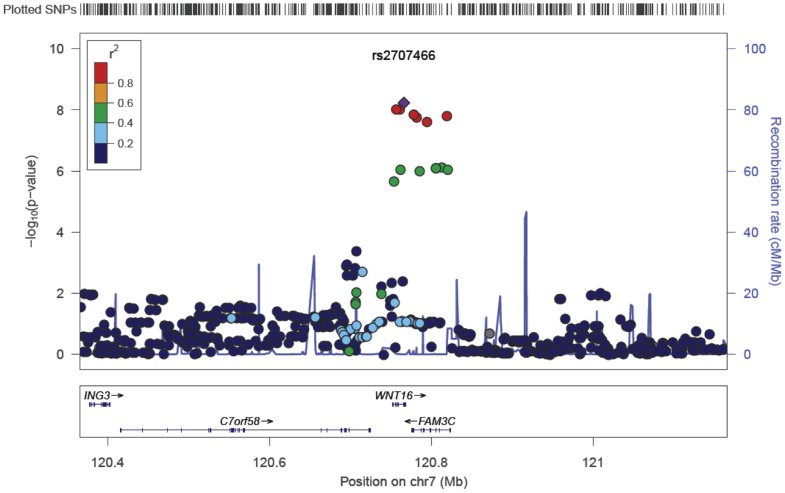
Anthropometrics, and bone variables for the three discovery GWAS cohorts and one replication cohort are presented in Table S1. Marked deviation from the null distribution amongst the lowest observed p-values were observed for the meta-analysis results (Figure S1). The results showed that the greatest evidence for association between genetic variation and tibial cortical thickness was seen for rs9525638 on chromosome 13, slightly upstream of the RANKL gene (−0.11 standard deviations [SD] per T allele, P = 3.3×10−10) (Table 1, Figure S2 and Figure S3). The second strongest genetic signal (rs2707466) for cortical thickness was located at the WNT16 locus (−0.10 SD per C allele, P = 5.9×10−9) (Table 1, Figure 1 and Figure S2). The SNP rs2707466 represents a missense polymorphism (Thr>Ile) located in the fourth exon of WNT16.
Table 1. Top cortical thickness GWA meta-analysis hits, with replication and meta-analysis of all four cohorts.
| Discovery | Meta-Analysis | Replication | Combined | ||||||||||||||||||
| Alspac discovery | GOOD discovery | YFS Discovery | MrOs Sweden | All cohorts | |||||||||||||||||
| Gene/Position | SNP | Effect allele | N | Beta (se) | p | N | Beta (se) | p | N | Beta (se) | p | Allele freq | N | Beta (se) | p | N | Beta (se) | p | N | Beta (se) | p |
| TNFSF11 | rs9525638 | T | 3382 | −0.11 | 2.1E−07 | 938 | −0.08 | 0.06 | 1558 | −0.11 | 2.1E−03 | 0.54 | 5878 | −0.11 | 3.3E−10 | 1021 | −0.02 | 0.74 | 6899 | −0.09 | 3.6E−9 |
| 13: 42026577 | (0.02) | (0.04) | (0.04) | (0.02) | (0.04) | (0.02) | |||||||||||||||
| WNT16 | rs2707466 | C | 3382 | −0.08 | 1.6E−04 | 938 | −0.14 | 2.2E−03 | 1558 | −0.14 | 1.7E−04 | 0.58 | 5878 | −0.10 | 5.9E−9 | 1032 | −0.11 | 8.0E−03 | 6910 | −0.11 | 1.5E−10 |
| 7: 120766325 | (0.02) | (0.05) | (0.04) | (0.02) | (0.04) | (0.02) | |||||||||||||||
Models adjusted for sex (ALSPAC and YFS), age, height, weight (ln). Betas in standard deviations and standard errors are presented.
Figure 1. SNP rs2707466 regional association plot of the discovery genome-wide meta-analysis of cortical thickness.
Circles show GWA meta-analysis p-values, with different colors indicating varying linkage disequilibrium with rs2707466 (diamond).
We selected our top two regions, the RANKL and WNT16 loci, with SNPs with P<1×10−5 and carried out analyses conditional on the most associated SNPs in each region: rs9525638 and rs2707466, respectively. When conditioning on the most significant SNP in the WNT16 region (rs2707466) an additional suggestive signal (rs12706314 in C7orf58, P condition = 7.3×10−5) appeared, but did not achieve genome-wide significance. Using a similar conditional analysis (with rs9525638) for the RANKL locus, no additional SNPs with an independent signal appeared.
Cortical Thickness Replication Study
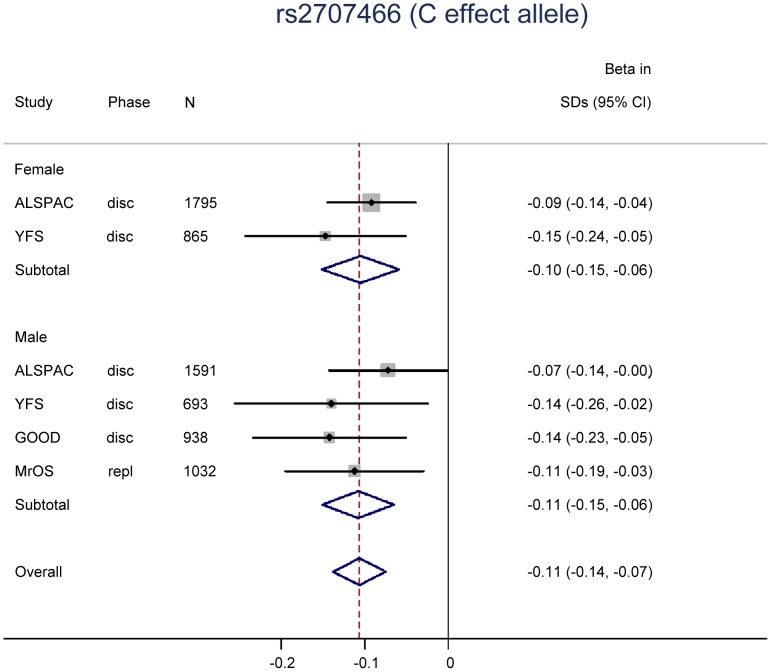
Two SNPs (rs9525638, rs2707466) were selected for replication in the MrOS Sweden cohort. In the replication stage, SNP rs2707466 at the WNT16 locus was significantly associated with tibial cortical thickness (−0.11 SD per C allele, P = 0.008), whilst no strong evidence of association was seen for rs9525638 near the RANKL locus although the estimated effect was in the same direction as in the discovery meta-analysis (Table 1). Thus, rs2707466 was the only SNP that was significantly associated with cortical thickness in both the discovery and replication cohorts (combined −0.11 SD per C allele, P = 1.5×10−10). Therefore, further analysis of associations with bone traits was constrained to rs2707466 at the WNT16 locus. Associations between rs2707466 and cortical thickness were highly similar when performed according to sex (Figure 2). No evidence of a significant impact of age for the association between rs2707466 and cortical thickness was found (ALSPAC (young) vs. GOOD, YFS and MrOS combined (adult and older): −0.09 SD vs. −0.13 SD per C allele, P = 0.135, for heterogeneity between the two groups). In the combined meta-analysis, rs2707466 was not associated with either cortical vBMD or periosteal circumference (Table S2). In the GOOD cohort, rs2707466 was associated with cortical bone thickness also at the radius (−0.12 SD per C allele, P = 0.008).
Figure 2. The genome-wide meta-analysis with cortical thickness according to sex.
GWAS Meta-Analysis of Forearm BMD
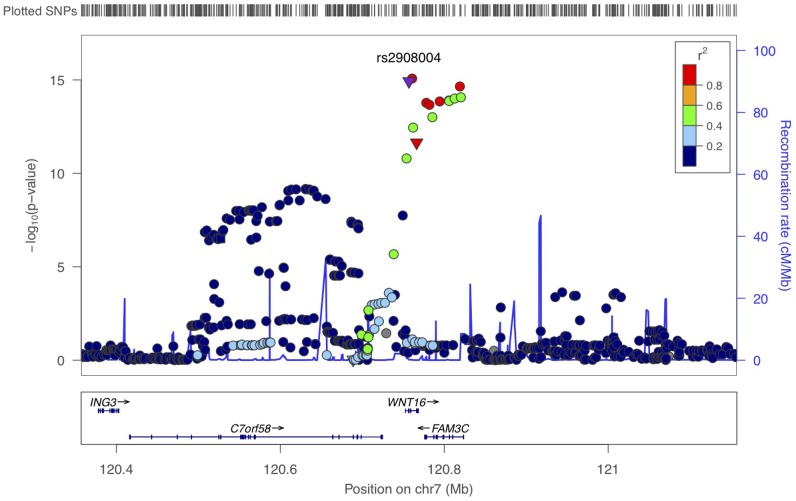
The information of the five forearm BMD cohorts is presented in Table S3. A quantile-quantile plot of the observed P values showed a clear deviation at the tail of the distribution from the null distribution (Figure S4). The meta-analysis revealed that 54 SNPs within the 7q31 locus had genome-wide significant associations (P<4.6×10−8) with forearm BMD (Table S4 and Figure S5). The most significant SNP was at rs2536189 (−0.16 SD per C allele, P = 8.5×10−16). Two common amino acid substitutions at WNT16, rs2908004 (Gly>Arg) (−0.16 SD per G allele, P = 1.2×10−15) and rs2707466 (Thr>Ile as described in cortical thickness study) (−0.14 SD per C allele, P = 2.3×10−12) also demonstrated genome-wide significance (Table 2). The highlighted locus at 7q31 locus included genome-wide significant SNPs at the WNT16 (wingless-type MMTV integration site family, member 16), FAM3C (family with sequence similarity 3, member C) and C7orf58 (chromosome 7 open reading frame 58) genes (Figure 3). To identify the possible secondary signals in this locus, we carried out a conditional analysis. When conditioning on rs2536189, the most significant SNP at WNT16 for BMD, an additional signal (rs1554634 in C7orf58, P = 7.8×10−8) was highlighted at a genome-wide suggestive level of association (Figure S6). This SNP is in LD with rs12706314 in C7orf58 (r2 = 0.35 and D' = 0.72 in HapMap CEU), which showed suggestive association with cortical thickness when conditioning on the top signal for cortical thickness.
Table 2. Association results of forearm BMD meta-analysis and fracture for selected SNPs.
| Meta Analysis of BMD GWAS | Meta Analysis of Fracture Results | ||||||||||||
| CHR | SNP | POSITION | EA | NEA | EAF | Beta | P-Value | RA | NEA | OR (95% CI) | P-Value | I2 | Gene Annotation |
| 7 | rs7776725 | 120820357 | T | C | 0.74 | −0.17 | 8.54E−15 | T | C | 1.33 (1.20–1.46) | 7.27E−09 | 11 | FAM3C |
| 7 | rs2908004 | 120757005 | G | A | 0.58 | −0.16 | 1.17E−15 | G | A | 1.22(1.12–1.33) | 4.90E−06 | 0 | WNT16 Missense |
| 7 | rs2707466 | 120766325 | C | T | 0.59 | −0.14 | 2.25E−12 | C | T | 1.22 (1.11–1.33) | 7.19E−06 | 0 | WNT16 Missense |
| 7 | rs10274324 | 120686577 | T | G | 0.94 | −0.21 | 3.82E−08 | T | G | 1.13 (0.92–1.35) | 1.50E−01 | 0 | C7orf58 |
EA: effect allele; NEA: non-effect allele; EAF: effect allele frequency; RA: risk allele.
See Table S4 for a list of all genome-wide significant SNPs.
Figure 3. Scatter plots of the observed association of 7q31 locus with forearm BMD.
The P values of SNPs (shown as −log10 values in y-axis, from the genome-wide single-marker association analysis using the linear regression model) are plotted against their map position (b36) (x-axis). The color of each SNP spot reflects its r2 with rs2908004. Missense SNPs are plotted as triangles, and other SNPs are plotted as circles.
Forearm Fracture Association Study
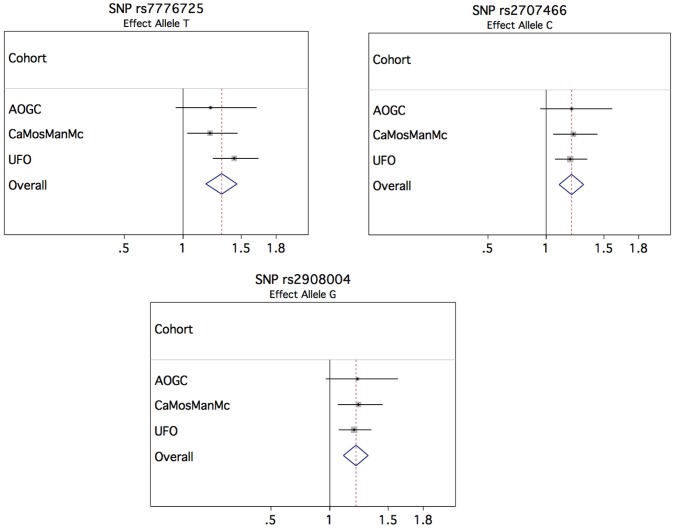
In order to investigate whether the variants showing association with forearm BMD also have an effect on the risk of forearm fracture, we selected 4 genome-wide significant SNPs from the BMD analysis for de novo genotyping in samples with forearm fracture and their controls (Table S5), including the two missense SNPs in WNT16 (rs2707466 and rs2908004), one from FAM3C (rs7776725) and one from C7orf58 (rs10274324). In the meta-analysis for osteoporotic fracture, comprising 2,023 forearm fracture cases and 3,740 controls, from 3 cohorts, we identified the rs7776725 SNP in FAM3C as being genome-wide significant for forearm fracture, with each C allele increasing the odds of fracture by 1.33 (95% confidence interval [CI]: 1.20–1.46, P-value = 7.3×10−9) (Table 2 and Figure 4). The two missense SNPs in WNT16 also demonstrated strong associations with risk of fracture (rs2908004, risk allele G, OR = 1.22 [95% CI: 1.12–1.33], P-value = 4.9×10−6 and rs2707466, risk allele C, OR = 1.22 [95% CI: 1.11–1.33], P-value 7.2×10−6). SNP rs10274324 from C7orf58 was not associated with fracture in this study (P = 0.15). These results were consistent across the three cohorts (Table S6).
Figure 4. Forest plots of association of top SNPs for forearm fracture.
Mouse Gene Deletion Studies
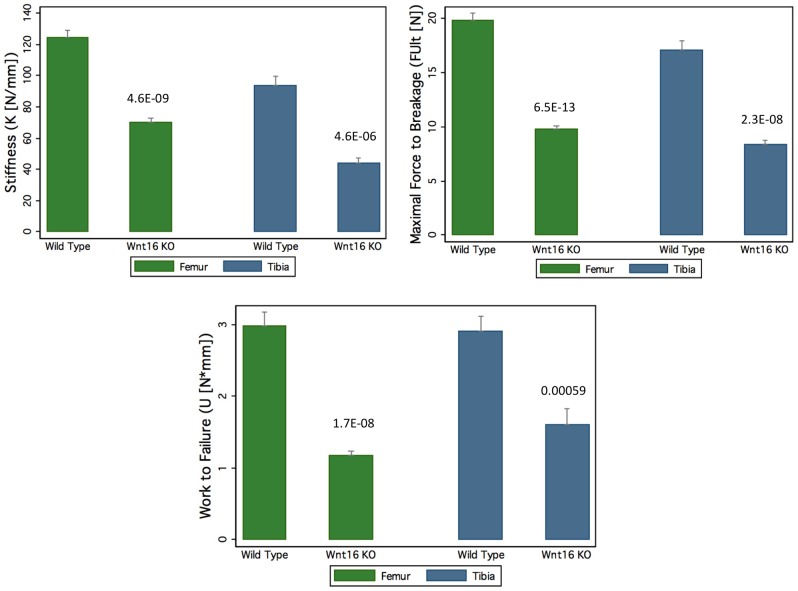
Mice with a gene deletion of Wnt16 (Wnt16−/−) appeared healthy with no discernible morphological or growth defects, and had normal body weight and femur length at 24 weeks. In microCT analyses of the femoral diaphysis, male Wnt16−/− mice had a trend suggestive of reduced cortical thickness (−7%, P = 0.14), and reduced cortical bone polar moment of inertia (−16%, P<0.001) (Table 3); female Wnt16−/− mice had substantially reduced cortical cross sectional area (−36%, P<0.001) and cortical thickness (−27%, P<0.001) and calculated bone strength (polar moment of inertia, −55%) (Table 3). Trabecular bone volume fraction (bone volume/total volume), as measured by microCT of 5th lumbar vertebrae (LV5), was similar in wild-type and Wnt16−/− mice (Table 3). In three-point bending tests, measures of bone strength (stiffness, maximal force to breakage and work to failure) were decreased between 43–61% (6.5×10−13<P<5.9×10−4) in Wnt16 −/− female mice at both femur and tibia (Figure 5). However, microCT parameters of the femoral shaft and LV5 in male wild type and Fam3c−/− mice did not reveal any consistent differences across the three targeting strategies (Table S7).
Table 3. Body weight, femoral length, and MicroCT data in Wnt16−/− mice, males (WT = 9; Wnt16−/− = 11) and females (WT = 24, Wnt16−/− = 16).
| Parameter | Male WT Mice | Male Wnt16−/− Mice | Statistics | Female WT Mice | Female Wnt16−/− Mice | Statistics |
| Body Weight (grams) | 35.4±1.8 | 37.6±1.3 | Δ = ↑10%, P = 0.35 | 27.3±0.8 | 25.9±1.3 | Δ = ↓5%, P = 0.34 |
| Femur Length (mm) | 16.3±0.2 | 16.2±0.2 | Δ = ↑1%, P = 0.58 | 16.2±0.1 | 16.1±0.1 | Δ = 0%, P = 0.86 |
| LV5 BV/TV (%) | 20.0±1.7 | 22.9±1.1 | Δ = ↑15%, P = 0.15 | 15.0±1.2 | 16.0±1.1 | Δ = ↑7%, P = 0.54 |
| Femur Shaft Total Area (mm2) | 1.83±0.09 | 1.60±0.08 | Δ = ↓13%, P = 0.07 | 1.57±0.04 | 1.07±0.03 | Δ = ↓32%, P<0.001 |
| Femur Shaft Bone Area (mm2) | 0.99±0.04 | 0.88±0.04 | Δ = ↓11%, P = 0.08 | 0.88±0.02 | 0.56±0.01 | Δ = ↓36%, P<0.001 |
| Femur Shaft Marrow Area (mm2) | 0.84±0.06 | 0.72±0.06 | Δ = ↓15%, P = 0.15 | 0.69±0.03 | 0.51±0.02 | Δ = ↓24%, P<0.001 |
| Femur Shaft Cortical Thickness (µm) | 251±9 | 233±8 | Δ = ↓7%, P = 0.14 | 246±4 | 186±4 | Δ = ↓27%, P<0.001 |
| Femur Shaft Polar MOI (mm4) | 0.507±0.047 | 0.428±0.49 | Δ = ↓16%, P<0.001 | 0.376±0.016 | 0.168±0.008 | Δ = ↓55%, P<0.001 |
Figure 5. Decrease of bone strength of Wnt16 knockout mice at femur and tibia.
In Femur group, the sample size are 23 wide type (WT) mice and 13 knock out (KO) mice, and in Tibia group, the sample size are 12 WT mice and 9 KO mice. The P values for each group are shown in the figure.
Discussion
Forearm fractures are a common and costly condition. In two separate GWASs for forearm BMD and cortical bone thickness we have identified variants that are genome-wide significant for these traits and, importantly, for forearm fracture at the 7q31 locus. Further, we have provided functional data from mice demonstrating that Wnt16 −/− mice have reduced cortical bone thickness and bone strength. These results are among the first to demonstrate a genome-wide significant locus for osteoporotic fracture suggesting that this locus is an important genomic determinant of cortical bone thickness and forearm BMD and fracture as well.
WNT16 is a member of the wingless-type MMTV integration site family, which has been reported to mediate signaling via canonical or non-canonical Wnt pathways. The canonical Wnt pathway has been shown to regulate bone mass. Specifically, loss of function mutations in the Wnt-co receptor LRP5, as seen in osteoporosis pseudoglioma syndrome, result in a dramatic loss in bone mass [26], while gain of function mutations give rise to extremely high BMD (5 SD above normal) [27]. Wnt16 has been proposed to signal via the non-canonical pathway [28], regulating haematopoetic stem cell specification in zebra fish, but whether this signaling system involves regulation of osteoblasts, which are of mesenchymal origin, is unclear. Little is known of the role of WNT16 in skeletal development and function, but Wnt16 has previously been implicated in synovial joint development in mice [29]. Several genes involved in the Wnt pathway have been previously identified to be associated with BMD by GWAS. These include known loci CTNNB [10], SOST [30], LRP4 [9], [10], LRP5 [8], [10], FOXC2 [10], GPR177 [10], and MEF2C [10]. Our study adds WNT16 to this list of bone-influencing Wnt factors. In addition to our GWA meta-analyses results for cortical bone thickness and forearm BMD, we present a functional study demonstrating that Wnt16 −/− mice have a substantial decrease in cortical bone thickness (27%) and bone strength (43–61%), but not bone length. Further, Medina et al (accompanying submission) provide data indicating that variation in WNT16 also influences BMD in children, suggesting that WNT16 may influence peak bone mass. Importantly, the clinical relevance of these findings at WNT16 is supported by our observation that WNT16 influences clinical fractures in humans and bone strength in mice.
In an experimental study using forearms from cadavers, cortical bone thickness was highly correlated (r = 0.93) to the 3-point bending failure load and could improve the prediction of this strength measure, in combination with bone mineral content derived from DXA [31]. Among individuals suffering a fracture at the radius, cortical bone thickness at the same site was 33% lower than in controls, which was the largest difference observed for the cortical bone traits [32]. The present study constitutes the first GWA study of cortical bone thickness, a trait with crucial importance for bone strength. The SNP rs2707466, which causes a missense amino acid substitution (Thr>Ile) at the WNT16 locus, was consistently associated with cortical thickness in a meta-analysis of three large discovery cohorts and in a replication cohort (combined P = 1.5×10−10). In the GOOD cohort, the rs2707466 was also associated with cortical bone thickness at the radius, with an effect size similar to what was seen for the tibia, indicating that the WNT16 locus affects cortical bone thickness at both the forearm and leg. Interestingly, the most significant 14 of the 54 genome-wide significant SNPs in the forearm BMD GWAS, were all in the WNT16 or FAM3C genes, and showed high correlation with the two WNT16 missense SNPs: rs2908004 and rs2707466 (0.58<r2<1, in HapMap CEU data). This finding might suggest that the association signals are driven by these coding SNPs in WNT16. These two missense variants were also strongly associated with risk of fracture in our study, but did not achieve genome-wide significance. All together, our results implicate the WNT16 locus as important for fracture risk, which would likely be mediated via an effect on cortical bone, particularly the thickness of the cortical shell.
FAM3C, which is predicted to be expressed in osteoblasts and encodes a newly identified cytokine necessary for epithelial to mesenchymal transition and retinal laminar formation in vertebrates [33]. We identified the SNP rs7776725 within the first intron of FAM3C to be genome-wide significant for forearm BMD (P = 8.5×10−15) and forearm fracture (P = 8.6×10−9). Since the fracture cohorts do not have available BMD data, except for the AOGC cohort, which comprised only 7% of the fracture case population, no meaningful conclusions could be drawn for the independence of the association between fracture and forearm BMD. While candidate gene studies have previously described relationships between genetic variants and fracture [34], [35], [36], we are aware of only one other variant that has been demonstrated to be genome-wide significant for any type of osteoporotic fracture, arising from the ALDH7A1 gene [12]. Interestingly, SNP rs7776725 in FAM3C was previously reported to be associated with speed of sound (SOS) as analyzed by quantitative ultrasound at the radius (P = 1.0×10−11) in an un-replicated GWAS carried out in Asian populations [13]. This SNP was also associated with BMD in a Caucasian population [37]. The high-throughput DEXA and microCT screen which initially identified reduced cortical bone thickness and bone strength in Wnt16 knockout mice failed to observe any skeletal phenotype changes in three independent knockouts of mouse Fam3c. Since the sample size of Fam3c−/− mice was small (N = 18), the possibility of a false negative result cannot be excluded. All together, our functional studies indicate that Wnt16 rather than Fam3c is responsible for the observed genetic signal arising from this locus. However, we provide no data as to whether or not gain of functions variants in Fam3c could have effects on the studied bone traits and fracture risk.
C7orf58 (FLJ21986), which codes for a hypothetical protein, has recently been identified to be associated with blood pressure in a study on Nigerians [38]. As an open reading frame, C7orf58 has no known function. In the forearm GWAS, the other 40 out of the 54 genome wide significant SNPs are from C7orf58, and show low LD with rs2908004 and rs2707466 (r2<0.2, in HapMap CEU data), when conditioning for the top SNP (rs2536189) in WNT16, resulted in an additional signal (rs1554634) in C7orf58. Similarly, conditioning for rs2707466 at the WNT16 locus, in the GWAS for cortical bone thickness, resulted in an additional, suggestive signal (rs12706314, which was in LD with rs1554634) located in C7orf58. Furthermore, Medina et al (accompanying submission) demonstrate in a conditional analysis that a separate signal, other than the signal derived from WNT16,, located in C7orf58 was associated with total body BMD. Thus, these studies reveal an independent genetic signal for several bone traits, arising from C7orf58, indicating a possible functional role of this protein. Even though our functional studies imply that Wnt16 determines the bone effects of the 7q31 locus, further studies are necessary to elucidate the role of C7orf58.
In summary, we provide the first evidence of association of common variants across the genome with cortical bone thickness, forearm BMD and forearm fracture. We also provide functional data implicating WNT16 at this locus. Importantly, our findings report one of two genome-wide significant variants for osteoporotic fracture. These results suggest a critical role of Wnt signaling pathway on cortical bone thickness and bone strength determination as well as fracture susceptibility.
Materials and Methods
Ethics Statement
All study participants provided informed written consent. Approval by local institutional review boards was obtained in all studies.
Study Samples of Bone Cortical Thickness GWAS
GOOD cohort
The Gothenburg Osteoporosis and Obesity Determinants (GOOD) study was initiated to determine both environmental and genetic factors involved in the regulation of bone and fat mass [39] [40]. Young men were randomly identified in the greater Gothenburg area in Sweden using national population registers, contacted by telephone, and invited to participate. Enrolled subjects were between 18 and 20 years of age. There were no other exclusion criteria, and 49% of the study candidates agreed to participate (n = 1068). Genotypes from 938 individuals passed the sample quality control criteria (Table S8). We carried out imputation to HapMap2 using Mach 1.0, Markov Chain Haplotyping [41], giving a total of 2,608,508 SNPs
YFS cohort
The Cardiovascular Risk in Young Finns Study (YFS) is an ongoing multi-centre follow-up of atherosclerosis risk factors in young Finns [42]. The first cross-sectional survey conducted in 1980 comprised a total of 3,596 subjects (83% of those invited) aged 3, 6, 9, 12, 15 and 18 years. The subjects were randomly selected from the national population register from five university cities in Finland (Helsinki, Turku, Tampere, Kuopio and Oulu) and the rural municipalities in their vicinity. In 2008, 1,884 subjects (1,058 women and 826 men) aged 31–46 years participated in pQCT measurements organized in five study centers (Turku, Helsinki, Tampere, Oulu and Kuopio) between February and December 2008. Trained technologists in each center performed the measurements. The same pQCT device was used in all study centers (Stratec XCT 2000R). Pregnant women were excluded from the pQCT measurements. Subjects gave written informed consent. Bone measures are described in detail elsewhere [43]. Both pQCT measurements and genotype information were available for 1558 study subjects. Genotype imputation was performed using MACH 1.0 [41] and HapMap II CEU samples as the reference set. See more detail in Table S8.
ALSPAC cohort
The Avon Longitudinal Study of Parents and their Children (ALSPAC) is a geographically based birth cohort study investigating factors influencing the health, growth, and development of children. All pregnant women resident within a defined part of the former county of Avon in South West England with an expected date of delivery between April 1991 and December 1992 were eligible for recruitment, of whom 14,541 were enrolled (http://www.alspac.bris.ac.uk) [44]. Both mothers and children have been extensively followed from the 8th gestational week onwards using a combination of self-reported questionnaires, medical records and physical examinations. Blood samples were taken and DNA extracted as previously described [45]. 3382 study subjects had both pQCT measurements and genotype information. We carried out imputation using MACH 1.0.16, Markov Chain Haplotyping [41], using CEPH individuals from phase 2 of the HapMap project as a reference set. See more detail in Table S8.
Study Samples of Bone Cortical Thickness Replication Study
MrOS Sweden cohort
The Osteoporotic Fractures in Men (MrOS) study is a prospective multicenter study including older men in Sweden (3014), Hong Kong (>2000), and the United States (>6000). In the present study, associations between candidate polymorphisms and skeletal parameters were investigated in the Swedish cohort (Table 1), which consists of three sub-cohorts from three different Swedish cities (n = 1005 in Malmö, n = 1010 in Gothenburg, and n = 999 in Uppsala). Study subjects were randomly identified using national population registers, contacted and asked to participate. To be eligible for the study, the subjects had to be able to walk without assistance, provide self reported data, and sign an informed consent; there were no other exclusion criteria [46]. See more detail in Table S8.
Study Samples of Forearm BMD GWAS
5,672 samples from five cohorts of European descent participated in this meta-analysis (Tables S3 and S9). BMD at forearm in all cohorts was measured by dual-energy X-ray absorptiometry following standard manufacturer protocols.
TwinUK1 and TwinUK23 cohorts
TwinsUK (http://www.twinsuk.ac.uk/) is a population-based registry of British Twins representative of the general British population [8], [36], [47]. Genotyping of the TwinUK1 was done by Illumina HumanHap300, and TwinUK23 by HumanHap610Q. Imputation was performed using the IMPUTE software package version 2 [48] based on HapMap2, release 22.
AFOS cohort
The Amish Family Osteoporosis Study (AFOS) study was designed to identify genetic determinants of osteoporosis in the Old Order Amish (OOA) population from Lancaster County, PA USA [49], [50]. Genotyping was done using either the Affymetrix 500K or 6.0 genotyping chip. The Birdseed genotype-calling algorithm was used. Imputation was performed using MACH on the HapMap2, rel 22 data.
GOOD cohort
The GOOD study subjects (study inclusion criteria described under the cortical bone thickness GWA study) were contacted and invited to participate in a five-year follow-up exam [51]. Forearm BMD measurements were available in 731 men (Table S3).
AOGC cohort
The Anglo-Australasian Osteoporosis Genetics Consortium (AOGC) study collected unrelated individuals with extreme BMD phenotypes as a powerful strategy for gene discovery in quantitative traits [11], [52]. Genotyping was performed using Illumina Infinium II HumHap370CNVQuad chips at the University of Queensland Diamantina Institute, Brisbane, Australia. Subsequent imputation was done based on the HapMap2 release 22 data using MACH program.
Study Samples of Forearm Fracture Association Study
Four genome-wide significant SNPs for forearm BMD were selected to test the association with forearm fracture in 2,142 cases and 3,697 controls from three cohorts. Forearm fracture was defined as fractures resulting from low trauma (such as a fall from standing height) occurring at the wrist, ulna, radius, forearm, as well as Colles' fractures.
UFO cohort
The Umeå Fracture and Osteoporosis (UFO) study is a nested case-cohort, population-based study from Sweden designed to identify the genetic and gene-by-environmental determinants of osteoporotic fracture. This cohort is sampled from a population-based cohort study from Northern Sweden initiated to identify the risk factors for diabetes and cardiovascular disease [53], [54]. In total, 1,068 cases and 1,218 age-matched controls were included from this cohort. All fractures were confirmed by radiographic or surgical report. De-novo genotyping in the UFO study was undertaken at Kbiosciences (England).
CaMos and ManMc cohort
The Canadian MultiCentre Osteoporosis study (CaMos) is a population-based prospective study of 9,423 men and women from across Canada, followed for fourteen years for osteoporotic outcomes and risk factors [55]. All fractures in the CaMos study at forearm were confirmed with radiographic or surgical report. The Manitoba-McGill (ManMc) fracture study is a population-based cohort of women undergoing surgical repair of wrist and hip fractures within the Province of Manitoba, Canada [56]. All individuals had no prior history of concomitant disease or use of bone-altering drugs. All fractures occurred at forearm and were confirmed with surgical report. ManMc was designed to complement the CaMos cohort by recruiting individuals suffering osteoporotic fractures in the most heritable age range (prior to age 70) and compare them to controls from the CaMos cohort that had not suffered an osteoporotic fracture after up to fourteen years of follow-up and subjects reaching at least 70 years of age. Weight and height at time of fracture are not collected for these study participants. 800 cases with forearm fracture and 855 controls were included from the combined CaMos/ManMc study. De novo genotyping for the CaMos and ManMc cohorts at the 4 SNPs assessed for fracture was undertaken at Kbiosciences (England).
AOGC cohort
This in silico association study included 155 forearm fracture cases and 1,672 controls from the AOGC GWAS [11]. In some samples the level of trauma was not known. In these cases, if the fracture occurred after age 60 years, it was considered to be an osteoporotic low trauma fracture, fractures occurring prior to this age with unknown trauma level were conservatively excluded. Fracture cases were identified by self-report.
Statistical Analysis
Genome-wide meta-analysis and replication method for bone cortical thickness study
The ALSPAC (n = 3382), YFS (n = 1558) and GOOD (n = 938) discovery cohorts contributed to the genome-wide meta-analysis. We analyzed only those imputed SNPs which had a minor allele frequency of >0.01 and an r2 imputation quality score of >0.3 in all 3 sets (n = 2,401,124). We carried out genome-wide association analyses for cortical thickness using additive linear regression in Mach2QTL for ALSPAC, ProbABEL [57] for YFS and using GRIMP [58] for the GOOD analyses. We included age, sex, height and weight(ln) as covariates. We carried out meta-analyses of the results from the three cohorts using the inverse variance method. Standardized betas and standard errors from each study are combined using a fixed effect model which weights the studies using the inverse variance and applying genomic control to individual studies and the combined results. Genome-wide significance was taken to be p<5×10−8. We selected one SNP from each independent region that had a p<5×10−8 for replication in the MrOS Sweden cohort. We also repeated the analyses in each of the three discovery cohorts, conditional on these top SNPs, to identify any additional independent associations in the regions. Additive linear regression analyses were carried out for the associations between these SNPs and cortical thickness in SPSS Statistics 17.0 for MrOS Sweden, using age, sex, height and weight(ln) as covariates. The results of all four cohorts were combined using a fixed effects inverse-variance meta-analysis in Stata (version 11.2). Correlations between bone traits in the MrOS cohort were tested and presented as Spearman's rank correlation coefficients (rho).
Genome-wide meta-analysis method for forearm BMD study
All cohorts independently conducted the association analysis of SNP allele dosage with standardized BMD residuals, while adjusting for age, age2, gender, weight and population substructure where applicable, for centre of recruitment (AOGC), and for family structure in cohorts with family members. The analyses were performed for men and women combined. Details of each study's GWAS are found on Table S9. A meta-analysis of the GWAS results was conducted using the GWAMA software (Genome-Wide Association Meta Analysis) (http://www.well.ox.ac.uk/gwama/) [59], with which meta-analyses are performed for both directly genotyped and imputed SNPs using estimates of the allelic effect size and standard error for BMD, and estimates of the allelic odds ratio and 95% confidence interval for fracture. Poorly imputed SNPs (r2 in MACH<0.3 or proper_info in IMPUTE2<0.40) and SNPs with low MAF (<0.01) were excluded. The summary effect estimates for BMD and fracture risk were computed using fixed-effects inverse variance meta-analysis [60]. Cochran's Q statistic and I2 estimates were used to evaluate the heterogeneity. To control for possible inflation of statistics due to population stratification and family relations, genomic control was applied to each study as well as the overall meta-analysis [61]. Statistical significance for genome-wide BMD association study was set at P<5×10−8.
Fracture association analysis
Four of the 54 genome-wide significant SNPs, in the forearm BMD GWAS, were selected to test the association with fracture. The rationale for picking these SNPs included: 1) Given that the Wnt pathway is central to osteoporosis etiology, two missense SNPs (rs2908004 and rs2707466) from WNT16 were selected, considering that missense SNPs may have more functional consequence than synonymous or non-coding SNPs, and that these two SNPs can fully tag the top SNP rs2536189 (r2>0.98); 2) The SNP rs7776725, which is in FAM3C, was selected because it was previously reported to be associated with speed of sound through bone, as analyzed by quantitative ultrasound at the radius [13] and was in only moderate LD with the rs2908004 (r2 = 0.58); 3) The SNP rs10274324 was selected based on its genome wide significance (−0.21 SD per T allele, P = 3.8×10−8 for forearm BMD), LD information (r2 = 0.04 with the top SNP rs2536189) and location (in C7orf58 gene). Therefore, the SNP selection provided assessment across all three genes at this locus (WNT16, FAM3C and C7orf58). SNPs were assessed for association with fracture risk using logistic regression models adjusted for sex, height and weight. Age was included as an additional covariate where this was not controlled for through the study design. Again, a fixed effect meta-analysis was undertaken assessing the effect of allelic dose on risk of fracture.
Generation of Knockout Mice
Wnt16−/− mice
Mice with a gene deletion of Wnt16 were generated using homologous recombination techniques. The first three exons were disrupted, with confirmation by Southern hybridization analyses (Figure S7). F2 hybrid littermates, derived from C57BL/6J and 129 SvEv parental strains, were examined at 24 weeks of age.
Fam3c−/− mice
Three separate knockout strategies were employed to inactive mouse Fam3c: 1) gene trap disrupting the intron between the first two exons with confirmation by lack of gene expression by RT-PCR in kidney and spleen (Figure S8), 2) homologous recombination removing the first two coding exons with confirmation by Southern hybridization analysis (Figure S9), 3) homologous recombination involving replacement of the mouse gene by the human gene resulting in loss of function (Figure S10). F2 hybrid littermates, derived from C57BL/6J and 129 SvEv parental strains, were examined at 16 weeks of age. All studies were performed in accordance with institutional and regulatory guidelines for animal care.
Imaging
Male and female mice were scanned using a microCT (Scanco μCT40, Switzerland). The fifth lumbar vertebrae (LV5) were scanned with a voxel size of 16 µm. Midshaft femurs were scanned with a voxel size of 20 µm. All scans used a threshold of 240, an X-ray tube voltage of 55 keV, a current of 145 microamperes and an integration time of 200 microseconds. Three-point bending tests were performed using Mach-1TM Micromechanical System A300.100 (Bio Syntech Canada inc., Laval, Quebec, Canada). The extrinsic parameters (ultimate force [Fult], stiffness [K or S], and work to failure [W or U]) were determined from a force-displacement curve. The span of two support points was 7 mm. The de-formation rate was 50 µm/s.
Statistical analysis
Two-sided student's t-test was employed to determine statistical significance of the effect of gene inactivation for each gender. Results are shown as mean+SEM.
Supporting Information
Quantile-quantile plots of the observed P values versus the expected P values for association for GWAS Meta-Analysis of cortical thickness. The scatters in black showed a clear deviation at the tail of the distribution from the null distribution (the red line).
(DOCX)
Manhattan plot for GWAS Meta-Analysis of cortical thickness. Genome-wide P values (−log10 P) of the linear regression analysis plotted against position on each chromosome.
(DOCX)
SNP rs9525638 regional association plot of the discovery genome-wide meta-analysis of cortical thickness. Circles show GWA meta-analysis p-values, with different colors indicating varying linkage disequilibrium with rs9525638 (diamond).
(DOCX)
Quantile-quantile plots of the observed P values versus the expected P values for association of Forearm BMD. The scatters in blue were based on the entire set of SNPs, whereas the scatters in black were obtained after removing WNT16 region SNPs (+/−400KB either side of rs2908004). The black line was the distribution expected if there were no association.
(DOCX)
Manhattan plot for GWAS Meta-Analysis of Forearm BMD. Genome-wide P values (−log10 P) of the linear regression analysis plotted against position on each chromosome.
(DOCX)
Scatter plots of the observed association of 7q31 locus with forearm BMD after condition on the top SNP rs2536189. The P values of SNPs (shown as −log10 values in y-axis, from the genome-wide single-marker association analysis using the linear regression model) are plotted against their map position (b36) (x-axis).
(PDF)
A: Restriction map of the Wnt16 gene and construction of the neomycin-resistance (neo) vector. Wnt16 exons are shown as filled boxes. Sequence information (deletion, insertion site, flanking sequence) is provided on the Taconic Farms website (http://www.taconic.com/wmspage.cfm?parm1=16 catalogue number TF3785). B: confirmation by Southern blots.
(DOCX)
A: Retroviral insertion disrupted Fam3c gene prior to the exon encoding amino acid 19 in a protein of 227 amino acids. Sequence information (deletion, insertion site, flanking sequence) is provided on the Taconic Farms website (http://www.taconic.com/wmspage.cfm?parm1=16 catalogue number TF3786). B: RT-PCR analysis revealed that the wild-type transcript was absent in the (−/−) mouse analyzed. Larger transcripts were detected at low levels in both tissues of the (−/−) mouse due to the splicing of fragments from the retroviral vector into the target transcript as determined by nucleotide sequence analysis. However, the in-frame stop codon in the retroviral vector sequence was predicted to disrupt translation of this transcript.
(DOCX)
A: homologous recombination removing the first two coding exons of Fam3c. Sequence information (deletion, insertion site, flanking sequence) is provided on the Taconic Farms website (http://www.taconic.com/wmspage.cfm?parm1=16 catalogue number TF3787). B: confirmation by Southern hybridization analysis.
(DOCX)
A: homologous recombination involving replacement of the mouse gene by the human gene resulting in loss of function of Fam3c. Sequence information (deletion, insertion site, flanking sequence) is provided on the Taconic Farms website (http://www.taconic.com/wmspage.cfm?parm1=16 catalogue number TF3788). B: confirmation by Southern hybridization analysis.
(DOCX)
Characteristics of the included cohorts for GWAS meta-analysis of cortical bone thickness.
(DOCX)
SNP rs2707466 associations with pQCT derived bone parameters at different ages and meta-analyses results for cortical bone thickness study.
(DOCX)
Characteristics of the included cohorts for GWAS meta-analysis of forearm BMD.
(DOCX)
54 genome-wide significant SNPs in 7q31 for forearm BMD GWAS meta-analysis.
(XLSX)
Characteristics of the included cohorts for fracture study.
(DOCX)
Association results for the 3 fracture cohorts.
(DOCX)
Micro CT parameters of the femoral shaft and fifth lumbar vertebra (LV5) in male wild type and Fam3c−/− mice. Data are presented for each of the three KO strategies and also for the combined cohorts (WT = 2; Fam3c−/− = 4 for each of the individual cohorts).
(DOCX)
Bone measurement, genotyping, quality control, imputation by study for cortical bone thickness meta-analysis.
(XLSX)
Genome-wide genotyping, imputation and genotype-phenotype analysis by study for BMD meta-analysis.
(XLSX)
Acknowledgments
We are extremely grateful to all the individuals who took part in this study; the midwives (ALSPAC) for their help in recruiting them; and the whole TwinsUK, AOFS, AOGC, CaMos/ManMc, UFO, ALSPAC, YFS, and GOOD teams, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists, and nurses. We thank the Wellcome Trust Sanger Institute, 23andMe, and the Laboratory Corporation of America for generating the TwinsUK and ALSPAC GWA data. This publication is the work of the authors and they will serve as guarantors for the contents of this paper.
Footnotes
R Brommage and J Liu are full-time employees of Lexicon Pharmaceuticals. All other authors have declared that no competing interests exist.
The UK Medical Research Council (Grant ref: 74882), the Wellcome Trust (Grant ref: 076467), and the University of Bristol provide core support for the Avon Longitudinal Study of Parents and their Children (ALSPAC). The national German MediGRID and Services@MediGRID part of the German D-Grid (who provided resources for the GOOD analysis) are both funded by the German Bundesministerium fuer Forschung und Technology under grants #01 AK 803 A-H and #01 IG 07015 G. L Paternoster, DM Evans, and this work were supported by a Medical Research Council New Investigator Award (MRC G0800582 to DM Evans). JP Kemp is funded by a Wellcome Trust four-year PhD studentship in molecular, genetic, and life course epidemiology (WT083431MA). The GOOD and MrOS Sweden investigators were supported by the Swedish Research Council, the Swedish Foundation for Strategic Research, the ALF/LUA research grant in Gothenburg, the Lundberg Foundation, the Torsten and Ragnar Söderberg's Foundation, the Novo Nordisk Foundation, and the European Commission grant HEALTH-F2-2008-201865-GEFOS. The Young Finns Study has been financially supported by the Academy of Finland: grants 126925, 121584, 124282, 129378, 117797, 216310, and 41071; the Social Insurance Institution of Finland; Kuopio, Tampere, and Turku University Hospital Medical Funds; Juho Vainio Foundation; Paavo Nurmi Foundation; Finnish Foundation of Cardiovascular Research and Finnish Cultural Foundation; Sigrid Juselius Foundation; Tampere Tuberculosis Foundation; and Emil Aaltonen Foundation. AOFS: NIH research grants R01 AG18728, R01HL088119, R01AR046838, U01 HL084756, P30DK072488, and F32AR059469. AOGC: The Anglo-Australasian Osteoporosis Genetics Consortium was funded by a project grant from the Australian National Health and Medical Research Council (Australia) (grant reference 511132). Funding was also received from the Australian Cancer Research Foundation and Rebecca Cooper Foundation (NHMRC, Australia). MA Brown was funded by an NHMRC Senior Principal Research Fellowship (grant reference APP1024879). CaMos/ManMc: This work was supported by grants from the Canadian Foundation for Innovation, the Canadian Institutes of Health Research (CIHR), Fonds de la recherche en santé du Quebec and the Jewish General Hospital, Ministere Développement Économique, Innovation et Exportation du Québec, Fonds de la recherche en santé du Québec, Lady Davis Institute of Medical Research, and the Dairy Farmers of Canada. TwinsUK: NIHR Biomedical Research Centre (grant to Guys' and St. Thomas' Hospitals and King's College London); the Chronic Disease Research Foundation, Wellcome Trust; and National Institutes of Health Research, National Health and Medical Research Council (Australia) grant 1010494. UFO: The Umeå Fracture and Osteoporosis Study (UFO) is supported by BBMRI.se, the Swedish Research Council (K20006-72X-20155013), the Swedish Sports Research Council (87/06), the Swedish Society of Medicine, and the Kempe-Foundation (JCK-1021), and by grants from the Medical Faculty of Umeå Unviersity (ALFVLL:968:22-2005, ALFVLL-937-2006, ALFVLL 223:11-2007, ALFVLL-78151-2009) and from the county council of Västerbotten (Spjutspetsanslag VLL:159:33-2007). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Consensus development conference: diagnosis, prophylaxis, and treatment of osteoporosis. The American journal of medicine. 1993;94:646–650. doi: 10.1016/0002-9343(93)90218-e. [DOI] [PubMed] [Google Scholar]
- 2.Kanis JA, Johnell O, Oden A, Sembo I, Redlund-Johnell I, et al. Long-term risk of osteoporotic fracture in Malmo. Osteoporos Int. 2000;11:669–674. doi: 10.1007/s001980070064. [DOI] [PubMed] [Google Scholar]
- 3.Organization WH. WHO scientific group on the assessment of osteoporosis at primary health care level; 2007 May 5; Brussels, Belgium.
- 4.Burge R, Dawson-Hughes B, Solomon DH, Wong JB, King A, et al. Incidence and economic burden of osteoporosis-related fractures in the United States, 2005–2025. Journal of bone and mineral research. 2007;22:465–475. doi: 10.1359/jbmr.061113. [DOI] [PubMed] [Google Scholar]
- 5.Gueguen R, Jouanny P, Guillemin F, Kuntz C, Pourel J, et al. Segregation analysis and variance components analysis of bone mineral density in healthy families. J Bone Miner Res. 1995;10:2017–2022. doi: 10.1002/jbmr.5650101223. [DOI] [PubMed] [Google Scholar]
- 6.Smith DM, Nance WE, Kang KW, Christian JC, Johnston CC., Jr Genetic factors in determining bone mass. J Clin Invest. 1973;52:2800–2808. doi: 10.1172/JCI107476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kiel DP, Demissie S, Dupuis J, Lunetta KL, Murabito JM, et al. Genome-wide association with bone mass and geometry in the Framingham Heart Study. BMC medical genetics. 2007;8(Suppl 1):S14. doi: 10.1186/1471-2350-8-S1-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richards JB, Rivadeneira F, Inouye M, Pastinen TM, Soranzo N, et al. Bone mineral density, osteoporosis, and osteoporotic fractures: a genome-wide association study. Lancet. 2008;371:1505–1512. doi: 10.1016/S0140-6736(08)60599-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Styrkarsdottir U, Halldorsson BV, Gretarsdottir S, Gudbjartsson DF, Walters GB, et al. Multiple genetic loci for bone mineral density and fractures. N Engl J Med. 2008;358:2355–2365. doi: 10.1056/NEJMoa0801197. [DOI] [PubMed] [Google Scholar]
- 10.Rivadeneira F, Styrkarsdottir U, Estrada K, Halldorsson BV, Hsu YH, et al. Twenty bone-mineral-density loci identified by large-scale meta-analysis of genome-wide association studies. Nat Genet. 2009;41:1199–1206. doi: 10.1038/ng.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duncan EL, Danoy P, Kemp JP, Leo PJ, McCloskey E, et al. Genome-wide association study using extreme truncate selection identifies novel genes affecting bone mineral density and fracture risk. PLoS Genet. 2011;7:e1001372. doi: 10.1371/journal.pgen.1001372. doi: 10.1371/journal.pgen.1001372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo Y, Tan LJ, Lei SF, Yang TL, Chen XD, et al. Genome-wide association study identifies ALDH7A1 as a novel susceptibility gene for osteoporosis. PLoS Genet. 2010;6:e1000806. doi: 10.1371/journal.pgen.1000806. doi: 10.1371/journal.pgen.1000806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho YS, Go MJ, Kim YJ, Heo JY, Oh JH, et al. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat Genet. 2009;41:527–534. doi: 10.1038/ng.357. [DOI] [PubMed] [Google Scholar]
- 14.Paternoster L, Lorentzon M, Vandenput L, Karlsson MK, Ljunggren O, et al. Genome-wide association meta-analysis of cortical bone mineral density unravels allelic heterogeneity at the RANKL locus and potential pleiotropic effects on bone. PLoS Genet. 2010;6:e1001217. doi: 10.1371/journal.pgen.1001217. doi: 10.1371/journal.pgen.1001217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnell O, Kanis J. Epidemiology of osteoporotic fractures. Osteoporos Int. 2005;16(Suppl 2):S3–7. doi: 10.1007/s00198-004-1702-6. [DOI] [PubMed] [Google Scholar]
- 16.Zebaze RM, Ghasem-Zadeh A, Bohte A, Iuliano-Burns S, Mirams M, et al. Intracortical remodelling and porosity in the distal radius and post-mortem femurs of women: a cross-sectional study. Lancet. 2010;375:1729–1736. doi: 10.1016/S0140-6736(10)60320-0. [DOI] [PubMed] [Google Scholar]
- 17.Holzer G, von Skrbensky G, Holzer LA, Pichl W. Hip fractures and the contribution of cortical versus trabecular bone to femoral neck strength. J Bone Miner Res. 2009;24:468–474. doi: 10.1359/jbmr.081108. [DOI] [PubMed] [Google Scholar]
- 18.Johannesdottir F, Poole KE, Reeve J, Siggeirsdottir K, Aspelund T, et al. Distribution of cortical bone in the femoral neck and hip fracture: a prospective case-control analysis of 143 incident hip fractures; the AGES-REYKJAVIK Study. Bone. 2011;48:1268–1276. doi: 10.1016/j.bone.2011.03.776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Havill LM, Mahaney MC, T LB, Specker BL. Effects of genes, sex, age, and activity on BMC, bone size, and areal and volumetric BMD. J Bone Miner Res. 2007;22:737–746. doi: 10.1359/jbmr.070213. [DOI] [PubMed] [Google Scholar]
- 20.Pearson TA, Manolio TA. How to interpret a genome-wide association study. JAMA. 2008;299:1335–1344. doi: 10.1001/jama.299.11.1335. [DOI] [PubMed] [Google Scholar]
- 21.Michaelsson K, Melhus H, Ferm H, Ahlbom A, Pedersen NL. Genetic liability to fractures in the elderly. Archives of internal medicine. 2005;165:1825–1830. doi: 10.1001/archinte.165.16.1825. [DOI] [PubMed] [Google Scholar]
- 22.Andrew T, Antioniades L, Scurrah KJ, Macgregor AJ, Spector TD. Risk of wrist fracture in women is heritable and is influenced by genes that are largely independent of those influencing BMD. Journal of bone and mineral research. 2005;20:67–74. doi: 10.1359/JBMR.041015. [DOI] [PubMed] [Google Scholar]
- 23.Zheng HF, Spector TD, Richards JB. Insights into the genetics of osteoporosis from recent genome-wide association studies. Expert reviews in molecular medicine. 2011;13:e28. doi: 10.1017/S1462399411001980. [DOI] [PubMed] [Google Scholar]
- 24.Duncan EL, Brown MA. Genetic determinants of bone density and fracture risk–state of the art and future directions. The Journal of clinical endocrinology and metabolism. 2010;95:2576–2587. doi: 10.1210/jc.2009-2406. [DOI] [PubMed] [Google Scholar]
- 25.Qiu C, Papasian CJ, Deng HW, Shen H. Genetics of osteoporotic fracture. Orthop Res Rev. 2011;3:11–21. [Google Scholar]
- 26.Gong Y, Slee RB, Fukai N, Rawadi G, Roman-Roman S, et al. LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell. 2001;107:513–523. doi: 10.1016/s0092-8674(01)00571-2. [DOI] [PubMed] [Google Scholar]
- 27.Boyden LM, Mao J, Belsky J, Mitzner L, Farhi A, et al. High bone density due to a mutation in LDL-receptor-related protein 5. N Engl J Med. 2002;346:1513–1521. doi: 10.1056/NEJMoa013444. [DOI] [PubMed] [Google Scholar]
- 28.Clements WK, Kim AD, Ong KG, Moore JC, Lawson ND, et al. A somitic Wnt16/Notch pathway specifies haematopoietic stem cells. Nature. 2011;474:220–224. doi: 10.1038/nature10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo X, Day TF, Jiang X, Garrett-Beal L, Topol L, et al. Wnt/beta-catenin signaling is sufficient and necessary for synovial joint formation. Genes Dev. 2004;18:2404–2417. doi: 10.1101/gad.1230704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Styrkarsdottir U, Halldorsson BV, Gretarsdottir S, Gudbjartsson DF, Walters GB, et al. New sequence variants associated with bone mineral density. Nature genetics. 2009;41:15–17. doi: 10.1038/ng.284. [DOI] [PubMed] [Google Scholar]
- 31.Hudelmaier M, Kuhn V, Lochmuller EM, Well H, Priemel M, et al. Can geometry-based parameters from pQCT and material parameters from quantitative ultrasound (QUS) improve the prediction of radial bone strength over that by bone mass (DXA)? Osteoporos Int. 2004;15:375–381. doi: 10.1007/s00198-003-1551-8. [DOI] [PubMed] [Google Scholar]
- 32.Melton LJ, 3rd, Riggs BL, van Lenthe GH, Achenbach SJ, Muller R, et al. Contribution of in vivo structural measurements and load/strength ratios to the determination of forearm fracture risk in postmenopausal women. J Bone Miner Res. 2007;22:1442–1448. doi: 10.1359/jbmr.070514. [DOI] [PubMed] [Google Scholar]
- 33.Katahira T, Nakagiri S, Terada K, Furukawa T. Secreted factor FAM3C (ILEI) is involved in retinal laminar formation. Biochemical and biophysical research communications. 2010;392:301–306. doi: 10.1016/j.bbrc.2009.12.180. [DOI] [PubMed] [Google Scholar]
- 34.Ralston SH, Uitterlinden AG, Brandi ML, Balcells S, Langdahl BL, et al. Large-scale evidence for the effect of the COLIA1 Sp1 polymorphism on osteoporosis outcomes: the GENOMOS study. PLoS Med. 2006;3:e90. doi: 10.1371/journal.pmed.0030090. doi: 10.1371/journal.pmed.0030090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ioannidis JP, Ralston SH, Bennett ST, Brandi ML, Grinberg D, et al. Differential genetic effects of ESR1 gene polymorphisms on osteoporosis outcomes. JAMA. 2004;292:2105–2114. doi: 10.1001/jama.292.17.2105. [DOI] [PubMed] [Google Scholar]
- 36.Richards JB, Kavvoura FK, Rivadeneira F, Styrkarsdottir U, Estrada K, et al. Collaborative meta-analysis: associations of 150 candidate genes with osteoporosis and osteoporotic fracture. Annals of internal medicine. 2009;151:528–537. doi: 10.7326/0003-4819-151-8-200910200-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang LS, Hu HG, Liu YJ, Li J, Yu P, et al. A follow-up association study of two genetic variants for bone mineral density variation in Caucasians. Osteoporos Int. 2011 doi: 10.1007/s00198-011-1863-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tayo BO, Luke A, Zhu X, Adeyemo A, Cooper RS. Association of regions on chromosomes 6 and 7 with blood pressure in Nigerian families. Circulation Cardiovascular genetics. 2009;2:38–45. doi: 10.1161/CIRCGENETICS.108.817064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lorentzon M, Swanson C, Andersson N, Mellstrom D, Ohlsson C. Free testosterone is a positive, whereas free estradiol is a negative, predictor of cortical bone size in young Swedish men: the GOOD study. J Bone Miner Res. 2005;20:1334–1341. doi: 10.1359/JBMR.050404. [DOI] [PubMed] [Google Scholar]
- 40.Lorentzon M, Mellstrom D, Ohlsson C. Age of attainment of peak bone mass is site specific in Swedish men―The GOOD Study. J Bone Miner Res. 2005;20:1223–1227. doi: 10.1359/JBMR.050306. [DOI] [PubMed] [Google Scholar]
- 41.Li Y, Abecasis GR. Mach 1.0: Rapid Haplotype Reconstruction and Missing Genotype Inference. Am J Hum Genet. 2006;S79:2290. [Google Scholar]
- 42.Raitakari OT, Juonala M, Ronnemaa T, Keltikangas-Jarvinen L, Rasanen L, et al. Cohort profile: the cardiovascular risk in Young Finns Study. Int J Epidemiol. 2008;37:1220–1226. doi: 10.1093/ije/dym225. [DOI] [PubMed] [Google Scholar]
- 43.Laaksonen M, Sievanen H, Tolonen S, Mikkila V, Rasanen L, et al. Determinants of bone strength and fracture incidence in adult Finns: Cardiovascular Risk in Young Finns Study (the GENDI pQCT study). Arch Osteoporosis. 2010;5:119–130. [Google Scholar]
- 44.Golding J, Pembrey M, Jones R. ALSPAC–the Avon Longitudinal Study of Parents and Children. I. Study methodology. Paediatr Perinat Epidemiol. 2001;15:74–87. doi: 10.1046/j.1365-3016.2001.00325.x. [DOI] [PubMed] [Google Scholar]
- 45.Jones RW, Ring S, Tyfield L, Hamvas R, Simmons H, et al. A new human genetic resource: a DNA bank established as part of the Avon longitudinal study of pregnancy and childhood (ALSPAC). Eur J Hum Genet. 2000;8:653–660. doi: 10.1038/sj.ejhg.5200502. [DOI] [PubMed] [Google Scholar]
- 46.Mellstrom D, Johnell O, Ljunggren O, Eriksson AL, Lorentzon M, et al. Free testosterone is an independent predictor of BMD and prevalent fractures in elderly men: MrOS Sweden. J Bone Miner Res. 2006;21:529–535. doi: 10.1359/jbmr.060110. [DOI] [PubMed] [Google Scholar]
- 47.Andrew T, Hart DJ, Snieder H, de Lange M, Spector TD, et al. Are twins and singletons comparable? A study of disease-related and lifestyle characteristics in adult women. Twin research. 2001;4:464–477. doi: 10.1375/1369052012803. [DOI] [PubMed] [Google Scholar]
- 48.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Streeten EA, McBride DJ, Pollin TI, Ryan K, Shapiro J, et al. Quantitative trait loci for BMD identified by autosome-wide linkage scan to chromosomes 7q and 21q in men from the Amish Family Osteoporosis Study. Journal of bone and mineral research. 2006;21:1433–1442. doi: 10.1359/jbmr.060602. [DOI] [PubMed] [Google Scholar]
- 50.Streeten EA, McBride DJ, Lodge AL, Pollin TI, Stinchcomb DG, et al. Reduced incidence of hip fracture in the Old Order Amish. Journal of bone and mineral research. 2004;19:308–313. doi: 10.1359/JBMR.0301223. [DOI] [PubMed] [Google Scholar]
- 51.Ohlsson C, Darelid A, Nilsson M, Melin J, Mellstrom D, et al. Cortical consolidation due to increased mineralization and endosteal contraction in young adult men: a five-year longitudinal study. J Clin Endocrinol Metab. 2011;96:2262–2269. doi: 10.1210/jc.2010-2751. [DOI] [PubMed] [Google Scholar]
- 52.Sims AM, Shephard N, Carter K, Doan T, Dowling A, et al. Genetic analyses in a sample of individuals with high or low BMD shows association with multiple Wnt pathway genes. Journal of bone and mineral research. 2008;23:499–506. doi: 10.1359/jbmr.071113. [DOI] [PubMed] [Google Scholar]
- 53.Englund U, Nordstrom P, Nilsson J, Bucht G, Bjornstig U, et al. Physical activity in middle-aged women and hip fracture risk: the UFO study. Osteoporosis international. 2011;22:499–505. doi: 10.1007/s00198-010-1234-1. [DOI] [PubMed] [Google Scholar]
- 54.Hallmans G, Agren A, Johansson G, Johansson A, Stegmayr B, et al. Cardiovascular disease and diabetes in the Northern Sweden Health and Disease Study Cohort - evaluation of risk factors and their interactions. Scandinavian journal of public health. 2003;(Supplement 61):18–24. doi: 10.1080/14034950310001432. [DOI] [PubMed] [Google Scholar]
- 55.Richards JB, Papaioannou A, Adachi JD, Joseph L, Whitson HE, et al. Effect of selective serotonin reuptake inhibitors on the risk of fracture. Archives of internal medicine. 2007;167:188–194. doi: 10.1001/archinte.167.2.188. [DOI] [PubMed] [Google Scholar]
- 56.Ladouceur M, Leslie WD, Dastani Z, Goltzman D, Richards JB. An efficient paradigm for genetic epidemiology cohort creation. PLoS ONE. 2010;5:e14045. doi: 10.1371/journal.pone.0014045. doi: 10.1371/journal.pone.0014045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aulchenko YS, Struchalin MV, van Duijn CM. ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics. 2010;11:134. doi: 10.1186/1471-2105-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Estrada K, Abuseiris A, Grosveld FG, Uitterlinden AG, Knoch TA, et al. GRIMP: a web- and grid-based tool for high-speed analysis of large-scale genome-wide association using imputed data. Bioinformatics. 2009;25:2750–2752. doi: 10.1093/bioinformatics/btp497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Magi R, Morris AP. GWAMA: software for genome-wide association meta-analysis. BMC bioinformatics. 2010;11:288. doi: 10.1186/1471-2105-11-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pereira TV, Patsopoulos NA, Salanti G, Ioannidis JP. Discovery properties of genome-wide association signals from cumulatively combined data sets. American journal of epidemiology. 2009;170:1197–1206. doi: 10.1093/aje/kwp262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Devlin B, Roeder K, Wasserman L. Genomic control, a new approach to genetic-based association studies. Theoretical population biology. 2001;60:155–166. doi: 10.1006/tpbi.2001.1542. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Quantile-quantile plots of the observed P values versus the expected P values for association for GWAS Meta-Analysis of cortical thickness. The scatters in black showed a clear deviation at the tail of the distribution from the null distribution (the red line).
(DOCX)
Manhattan plot for GWAS Meta-Analysis of cortical thickness. Genome-wide P values (−log10 P) of the linear regression analysis plotted against position on each chromosome.
(DOCX)
SNP rs9525638 regional association plot of the discovery genome-wide meta-analysis of cortical thickness. Circles show GWA meta-analysis p-values, with different colors indicating varying linkage disequilibrium with rs9525638 (diamond).
(DOCX)
Quantile-quantile plots of the observed P values versus the expected P values for association of Forearm BMD. The scatters in blue were based on the entire set of SNPs, whereas the scatters in black were obtained after removing WNT16 region SNPs (+/−400KB either side of rs2908004). The black line was the distribution expected if there were no association.
(DOCX)
Manhattan plot for GWAS Meta-Analysis of Forearm BMD. Genome-wide P values (−log10 P) of the linear regression analysis plotted against position on each chromosome.
(DOCX)
Scatter plots of the observed association of 7q31 locus with forearm BMD after condition on the top SNP rs2536189. The P values of SNPs (shown as −log10 values in y-axis, from the genome-wide single-marker association analysis using the linear regression model) are plotted against their map position (b36) (x-axis).
(PDF)
A: Restriction map of the Wnt16 gene and construction of the neomycin-resistance (neo) vector. Wnt16 exons are shown as filled boxes. Sequence information (deletion, insertion site, flanking sequence) is provided on the Taconic Farms website (http://www.taconic.com/wmspage.cfm?parm1=16 catalogue number TF3785). B: confirmation by Southern blots.
(DOCX)
A: Retroviral insertion disrupted Fam3c gene prior to the exon encoding amino acid 19 in a protein of 227 amino acids. Sequence information (deletion, insertion site, flanking sequence) is provided on the Taconic Farms website (http://www.taconic.com/wmspage.cfm?parm1=16 catalogue number TF3786). B: RT-PCR analysis revealed that the wild-type transcript was absent in the (−/−) mouse analyzed. Larger transcripts were detected at low levels in both tissues of the (−/−) mouse due to the splicing of fragments from the retroviral vector into the target transcript as determined by nucleotide sequence analysis. However, the in-frame stop codon in the retroviral vector sequence was predicted to disrupt translation of this transcript.
(DOCX)
A: homologous recombination removing the first two coding exons of Fam3c. Sequence information (deletion, insertion site, flanking sequence) is provided on the Taconic Farms website (http://www.taconic.com/wmspage.cfm?parm1=16 catalogue number TF3787). B: confirmation by Southern hybridization analysis.
(DOCX)
A: homologous recombination involving replacement of the mouse gene by the human gene resulting in loss of function of Fam3c. Sequence information (deletion, insertion site, flanking sequence) is provided on the Taconic Farms website (http://www.taconic.com/wmspage.cfm?parm1=16 catalogue number TF3788). B: confirmation by Southern hybridization analysis.
(DOCX)
Characteristics of the included cohorts for GWAS meta-analysis of cortical bone thickness.
(DOCX)
SNP rs2707466 associations with pQCT derived bone parameters at different ages and meta-analyses results for cortical bone thickness study.
(DOCX)
Characteristics of the included cohorts for GWAS meta-analysis of forearm BMD.
(DOCX)
54 genome-wide significant SNPs in 7q31 for forearm BMD GWAS meta-analysis.
(XLSX)
Characteristics of the included cohorts for fracture study.
(DOCX)
Association results for the 3 fracture cohorts.
(DOCX)
Micro CT parameters of the femoral shaft and fifth lumbar vertebra (LV5) in male wild type and Fam3c−/− mice. Data are presented for each of the three KO strategies and also for the combined cohorts (WT = 2; Fam3c−/− = 4 for each of the individual cohorts).
(DOCX)
Bone measurement, genotyping, quality control, imputation by study for cortical bone thickness meta-analysis.
(XLSX)
Genome-wide genotyping, imputation and genotype-phenotype analysis by study for BMD meta-analysis.
(XLSX)