Abstract
Mitochondrial fission and fusion have been observed, and their importance revealed, in almost every tissue and cell type except adult cardiac myocytes. As each human heart is uniquely dependent upon mitochondria to generate massive amounts of ATP that fuel its approximately 38 million contractions per year, it seems odd that cardiac myocytes are the sole exception to the general rule that mitochondrial dynamism is important to function. Here, I briefly review the mechanisms for mitochondrial fusion and fission and examine current data that dispel the previous notion that mitochondrial fusion is dispensable in the heart. Rare and generally overlooked examples of cardiomyopathies linked either to naturally-occurring mutations or to experimentally-induced mutagenesis of mitochondrial fusion/fission genes are described. New findings from genetically targeted Drosophila and mouse models wherein mitochondrial fusion deficiency has specifically been induced in cardiac myocytes are discussed.
Keywords: Mitochondrial fusion, mitochondrial fission, cardiomyopathy, Drosophila, genetic mouse models, human mutations
Overview
Cardiologists like to point out that the heart is the hardest working organ in the human body. It is widely reported that the human heart consumes 30 kilograms of ATP per day fueling basal metabolism and normal contraction that is essential to sustaining systemic and pulmonary blood pressure [1]. Obviously, a ~300 gram heart that daily consumes 100 times its weight in ATP does not do so by relying upon stored reserves, but must constantly be generating it. Almost all ATP is produced in cardiomyocyte mitochondria, with trivial amounts created in the cytosol. Accordingly, ventricular myocardium is literally packed full of mitochondria, which account for ~35% of cardiomyocyte volume [2]. There are more mitochondria in mammalian myocardium than there are individual muscle sarcomeres (~2:1) (Figure 1).
Figure 1. Subcellular arrangement of mitochondria in normal adult cardiomyocytes.
Transmission electron micrograph of normal 8 week old mouse myocardium. Note lanes of interfibrillar mitochondria running between individual myofibrils and cluster of mitochondria surrounding nucleus (bisected on right margin). The “sardines in a can” arrangement appears to enforce inter-organelle contact and does not permit meaningful intra-cellular mitochondrial transport.
Mitochondrial ATP production is tightly regulated to meet varying metabolic demands resulting from minute-by-minute (or beta-to-beat) changes in cardiac work. While the components of oxidative phosphorylation and the electron transport chain are fully understood, the molecular details of their regulatory pathways remain controversial. Two major mechanisms have been proposed: Regulation by ADP and inorganic phosphate (Pi) that are the products of ATP hydrolysis, and regulation by cytosolic calcium ([Ca2+]c) that is released from the SR and sensed by the mitochondria in response to stimuli for increased work. The former classical hypothesis [3] has been disputed by Balaban and colleagues who found that large changes in cardiac work and ATP consumption are not associated with measurable changes in ADP/Pi levels [4, 5], while the latter hypothesis has gained credence from elegant studies of O’Rourke and coworkers [6, 7]. It seems likely that modulatory functions may exist for both ATP/ADP/Pi and [Ca2+]c, the former playing a greater role regulating mitochondrial oxidative phosphorylation while the latter mainly affects Krebs cycle dehydrogenases [8].
The unique bioenergetic requirements of the heart not only dictate that it generate and maintain the largest proportional density of mitochondria of any organ in the body, but that the mitochondria are organized throughout the cardiac myocyte in a highly structured and stable manner. As their names suggests, repeating ribbon-like clusters of interfibrillar mitochondria run between myofibrils arranged parallel to the long axis of the cardiomyocyte. This enforced proximity of mitochondria to sarcomeres throughout the cardiomyocyte provides for a ready and continuous supply of ATP to all components of actin-myosin contractile machinery. Similarly, deep T-tubular invaginations of the cardiomyocyte plasma membrane (sarcolemma) interdigitate at close intervals into the myofibrillar elements and mitochondrial clusters. Synchronization of calcium entry throughout an electrically depolarized cardiomyocyte is critical for harmoniously initiating and terminating normal excitation-contraction coupling [9]. The T-tubules facilitate coordinated calcium (Ca2+) import and export throughout all parts of the cardiomyocyte simultaneously.
A third cellular organelle structure, the sarcoplamsic reticulum, likewise forms a continuous network that surrounds and permeates myofibrils and is intimately associated with mitochondrial clusters. Sarcoplasmic reticulum (SR) is the primary calcium storage and release organelle for cardiomyocytes, and the source of the vast majority of the free cytosolic Ca2+ that drives sarcomeric contraction [9]. SR Ca2+ release is a passive process. In contrast, large amounts of ATP are required to fuel SR Ca2+ reuptake via the SR Ca2+ ATPase (SERCA) that, together with a smaller amount of cardiomyocyte calcium export through the Na/Ca exchanger (NCX; [10]) and plasmalemmal Ca2+ ATPase (PMCA; [11, 12]), terminates contraction [13]. Physical proximity between cardiomyocyte mitochondria and SR is enforced in the same way that proximity between individual sardines is assured in a can, they are confined in groups within a highly ordered compacted structure. This arrangement ensures an ample supply of mitochondrial ATP to power SR calcium reuptake pumps and may provide a means whereby mitochondria sense increased SR calcium release that presages increased cardiac contraction. Mitochondrial sensing of cytosolic Ca2+ released from the SR stimulates an anticipatory increase in ATP production to fuel the impending greater workload, thus preventing the energy deficit that would otherwise occur after acutely increased ATP consumption [14].
The paradigm described above is consistent with a static cardiomyocyte subcellular architecture in which organelle interactions are dictated by physical proximity, the “sardines in a can” model (Figure 1). Sarcomeric myofilaments run the length of cardiac myocytes with ribbons of mitochondria interspersed between, and the SR exists as an intercalated network surrounding myofilaments and mitochondria like fish net stockings. This model contrasts with that described for neurons, in which individual mitochondria are widely dispersed and are transported from the cell body along the axons to the synapse, and perhaps back again [15]. During the course of their axonal journeys, neuronal mitochondria transiently link to and fuse with other mitochondria in cycles of tethering, fusion, and fission that promote organelle regeneration through exchange of internal mitochondrial contents [16]. Indeed, the molecular and biophysical mechanisms for mitochondrial fusion and fission have been elucidated in detail in neurons, fibroblasts, and other cell types, as described in other articles within this compendium. Until recently however (when results of studies that manipulated mitochondrial fusion proteins in mouse and fruit fly hearts suggested otherwise, vide infra), it was generally assumed that the unique subcellular architecture and apparent absence of mitochondrial mobility in adult cardiac myocytes either precluded, or obviated the need for, mitochondrial tethering, fusion and fission [17]. Here, we review new findings that have altered the perception that cardiac mitochondria are static, exploring the evidence for and roles of mitochondrial dynamics in normal and diseased hearts.
The machinery of mitochondrial fusion/fission
The striking morphometric diversity of mitochondria has been recognized for over a century. Indeed the word “mitochondrion” is widely reported to refer to this morphological heterogeneity as it is derived from the Greek words “mitos” (thread) and “khondros” (grain or granule), which would seem to represent different observed organelle shapes [18]. However, in medical parlance “chondros” primarily refers to cartilage, and the term “mitochondria” was first used in 1898 by Carl Benda who postulated that the many microscopic intracellular structures he detected by light microscopy acted like posts or pillars to support and maintain the cell’s overall size and shape. Accordingly, he called these presumed cytoskeletal structures mitochondria, or “threads of cartilage”. Subsequent advances in optics and biochemistry and the development of live-cell microscopy led to the realization that mitochondria were cellular power supplies, correcting this functional misconception. In 1914 Lewis and Lewis discovered mitochondrial dynamism, describing cycles of mitochondrial fusion and fission in various cultured chick embryo tissues [19]: “We find in the living (cells) that (mitochondria) can be seen to fuse together into rods or chains, and these to elongate into threads, which in turn anastomose with each other and may unite into a complicated network, which in turn may again break down into threads, rods, loops and rings.” Mitochondrial mobility, fusion and fission have since been observed in almost every species (from yeast to human) and cell type except adult cardiac myocytes. Mitochondrial fission is seen in neonatal cardiac myocytes and various immortalized cardiomyocyte-like cells undergoing apoptosis [20–24]. However, cultured neonatal cardiomyocytes have a much less structured internal architecture than adult cardiomyocytes, entirely unlike the highly organized “sardine can” design described above. Indeed, there are no published direct observations of mitochondrial fusion or fission in normal adult cardiac myocytes. Specific attempts to detect mitochondrial fusion in adult cardiomyocytes using live-cell confocal examination have not born fruit: “Mitochondrial fusion or fission was seen only in NB HL-1 cells but not in adult cardiomyocytes” [23]. Nevertheless, the past year has seen publication of compelling genetic evidence indirectly supporting not only the occurrence of, but the necessity for, mitochondrial fusion/fission in adult invertebrate and mammalian hearts [25, 26]. In this context, here I will briefly review the molecular machinery of mitochondrial dynamics as elucidated in non-cardiac systems, so that the discussion of various genetic and pharmacological interrogations of cardiac mitochondrial fusion and fission factors that follows is readily understood.
What is the evidence that mitochondrial fusion/fission are necessary?
Essential roles for mitochondrial fusion and fission in basic cellular and organism function were revealed using genetic interruption of the fusion/fission cycle. Organism-wide suppression of the Drosophila outer mitochondrial membrane (OMM) fusion factor MARF (mitochondrial assembly regulatory factor, also called dMFN) induces lethality at the fly larval stage, approximately 2–3 days after hatching (2nd instar larvae) [25]. Likewise, genetic ablation of either of the mammalian OMM fusion factors mitofusin (Mfn) 1 or 2, or the mitochondrial fission factor dynamin-related protein 1 (Drp1, a.k.a. Dlp-1), induces embryonic lethality in mice [27, 28]. Our own experience with mitochondrial fusion-deficient murine embryonic fibroblasts derived from David Chan’s SV40 transformed Mfn1/Mfn2 double null mouse embryos [29] suggests that, while fusion-defective cells are viable with highly fragmented mitochondria, their doubling rate is 3–4 fold slower than either wild-type or single Mfn1 or Mfn2 knockout cells. Thus, normal cell viability and both invertebrate and vertebrate development require mitochondrial fusion/fission.
There are several consequences of normal mitochondrial fusion/fission that may be necessary to maintain cellular and organism homeostasis. Mitochondria are unique among cellular organelles in that they possess their own genomes (mtDNA), encoding proteins essential for mitochondrial biogenesis and respiratory function [30, 31]. Because a normal and pathological byproduct of mitochondrial respiration is reactive oxygen species (ROS) that can damage DNA, and DNA repair mechanisms in mitochondria are imperfect compared to those in the nucleus, the rate of accumulation of mitochondrial genomic mutations is higher than that of nuclear genes. The overall prevalence of mtDNA mutations therefore increases over time and with age [32]. One of the mechanisms for correcting mtDNA mutations is through complementation of genomes exchanged through the fusion of different organelles. Disruption of mitochondrial fusion in mouse skeletal muscle through combined tissue-specific ablation of Mfn1 and Mfn2 increased the number of mtDNA mutations, decreased mitochondrial mass, and caused cellular respiratory dysfunction [33]. Mitochondrial fusion/fission may also play a broader role in mitochondrial regeneration and quality control by promoting exchange of proteins and lipids in addition to mtDNA [34]. Thus, the functional, genetic, and structural integrity of a cell’s mitochondrial population requires cycles of organelle fusion and fission.
As fission/fusion coupled to selective elimination of damaged daughter organelles can protect the mitochondrial pool from cumulative effects of senescence and degeneration [35], it has been postulated that mitochondrial dynamics is mechanistically linked to the process of mitophagy. A detailed description of mitophagy, the term coined to describe selective degradation of mitochondria via autophagic mechanisms [36], is beyond the scope of this review and can be accessed elsewhere [37]. It is worth noting here that interfering with normal mitochondria-endopasmic reticulum tethering by suppressing Mfn2 (see below) prevented autophagy that is normally induced by starvation in cultured cancer cells [38], and that disturbances in mitochondrial fission/fusion and mitophagy contribute to the pathology of Alzheimer’s disease and other neurodegenerative syndromes [39]. Mitophagy has also been implicated in heart disease [40], and factors that mediate mitochondrial fusion and fission are ubiquitinated by the same events that target damaged mitochondria for mitophagic elimination [41–43]. However, delineation of causal mechanisms linking disturbances in cardiomyocyte mitochondrial dynamics to mitophagic dysfunction must await the results of ongoing experimentation.
What are the essential components of mitochondrial fission and fusion?
No matter their size, shape, or cell type of origin, mitochondria are double-membrane delimited organelles. The outer mitochondrial membrane (OMM) contains the organelle, separating its contents from the cytoplasm (think of an egg shell). The convoluted inner mitochondrial membrane (IMM) separates the internal organelle into a central core or matrix (egg yolk) and an inter-membranous space (egg white). This segregated structure is essential for compartmentalization of the many mitochondrial enzyme complexes necessary for oxidative phosphorylation and ATP generation, but vastly complicates mitochondrial fusion. Further extending the egg metaphor, combining the contents two eggs while maintaining the original egg white/yolk internal structure requires a sequential process in which the two whites are first merged, and then (within the double-yolk giant egg), the two yolks are combined. Mitochondria fuse in a similar manner. First, two mitochondria become physically connected while retaining their individuality (“tethered” is the preferred term for two organelles physically linked, but not exchanging contents). The OMMs of the two organelles then fuse, creating a common inter-membranous space with two distinct matrices, analogous to the double-yolk giant egg. There is direct physical evidence for this distinct and separable phase of mitochondrial fusion in the form of giant mitochondria in which a single (fused) OMM encompasses two distinct matrix structures contained within their own (not-yet fused) IMM. These bizarre organelles are produced by pharmacological inhibition of IMM fusion in yeast and mammalian cells [44, 45].
Given the sequential nature and functional distinctiveness of OMM and IMM fusion, it is not surprising that different sets of fusion proteins mediate these two phases. “Mitofusin” (Mfn) is the name given to the family of OMM tethering and fusing proteins. A single Mfn, called dMfn or MARF, mediates OMM fusion in somatic Drosophila cells, whereas mammals have two largely functionally redundant mitofusin proteins, Mfn1 and Mfn2. Vertebrate Mfns are highly conserved across evolution, as is (to a somewhat lesser degree) Drosophila MARF. Structurally, they share an extracellular amino terminal GTPase domain, a first heptad repeat, two closely approximated transmembrane (TM) domains, and a second heptad repeat. The first TM domain passes through the OMM, and the second TM does so again, giving the Mfn molecule a fishhook-like configuration having both the amino and carboxyl termini exposed to the cytosol. Because the second heptad repeat of one Mfn molecule will interact with that of another molecule in opposition [46], the inter-molecular interaction will tether two mitochondria in much the same way that Velcro loops and hooks attach to each other. These Mfn interactions can be either homotypic (i.e. Mfn1-Mfn1 or Mfn2-Mfn2) or heterotypic (i.e. Mfn1-Mfn2), thus providing functional overlap for fusion. Accordingly, elimination of either Mfn1 or Mfn2 from cultured cells or specific tissues does not prevent fusion and has a relatively minor impact on mitochondrial morphometry and function. By comparison, concomitant elimination of both mitofusins induces mitochondrial fragmentation due to unopposed organelle fission [26, 29, 33, 47]. Although neurological disease is not the focus of this essay, it must be noted that loss-of-function mutations of human Mfn2 cause the neurodegenerative disease Charcot Marie Tooth Syndrome type 2A [48–52]. Since these disease-causing mutations are typically heterozygous, the dysfunctional mutant Mfn2 proteins likely exert dominant negative effects on mitochondrial fusion.
IMM fusion is mediated by another large GTPase, optic atrophy 1 (Opa1; [53, 54], mutations of which cause dominant optic atrophy in humans [55]. As befits its function in IMM fusion, Opa1 is located on the IMM where it also regulates cristae structure independent of organelle fusion [56]. Like Mfn deficiency, suppression or genetic ablation of Opa1 prevents mitochondrial fusion. Unlike mitochondrial fragmentation induced by mitofusin deficiency, which is the consequence of preventing the initial step in the fusion process, Opa1 deficiency produces both mitochondrial fragmentation (presumably the products of repeated cycles of organelle fission without normal fusion) and unusually large organelles containing multiple unfused internal matrix compartments (the products of partial fusion [25, 57, 58]).
Compared to mitochondrial fusion, which requires sequential merging of OMM and then IMM by different families of GTPases, mitochondrial fission seems to be a less complex monophasic process. Returning to the egg analogy, slicing a boiled egg into two halves retains the overall egg white/egg yolk structure. This is pretty much what occurs at a molecular level for mitochondrial fission: Individual mitochondria are ligated and separated by a molecular garrote formed by self-assembled oligomers of another GTPase, Drp1/Dlp1 (dynamin-related or dynamin-like protein 1) [59]. The precise molecular events mediating mitochondrial fission are incompletely understood, but involve translocation of cytosolic monomeric Drp1/Dlp1 to the mitochondria, its oligomerization, and GTP-driven constriction. Activated Drp1/Dlp1 binds to the scaffolding protein Fis1 on the OMM [60]; Fis1 may direct sub-organelle localization of the forming Drp1/Dlp1 oligomer to the mitochondrial constriction site. GTP-binding to Drp1/Dlp1 (in exchange for GDP) alters its molecular configuration, creating tight spirals that, working in concert, constrict the organelle in the same way that prototypical dynamin works as a GTP-driven molecular motor [61, 62]. Consistent with this molecular model, genetic ablation of mouse Drp1/Dlp1 induces mitochondrial elongation as a result of unopposed mitochondrial fusion [28].
Experimental evidence that mitochondrial fusion/fission is essential to normal hearts
As noted above, mitochondrial fusion and fission have never been directly observed in adult cardiac myocytes. Indeed, the author is not aware that mitochondrial fusion and fission have been observed in normal skeletal muscle, which shares with cardiac muscle a highly ordered mitochondrial and myofilament subcellular architecture, high mitochondrial content, and high metabolic demands. Nevertheless, it is accepted that mitochondrial fusion and fission not only occur in skeletal muscle, but that these are regenerative processes critical to maintaining normal mitochondrial morphometry and function essential for muscle homeostasis. These conclusions derive largely from the results of tissue-specific manipulation of Drosophila and mouse mitochondrial fusion genes: The Drosophila mitochondrial fusion proteins MARF/dMfn and Opa1 can be efficiently suppressed in a tissue-specific manner using RNAi, inducing mitochondrial fragmentation in skeletal (wing) muscle [57]. A similar picture of mitochondrial fragmentation associated with myasthenia, muscle atrophy, and premature lethality was subsequently induced by combined Mfn1 and Mfn2 gene ablation in mouse skeletal muscle [33]. These results show that interrupting mitochondria fusion in skeletal muscle of both invertebrates and mammals is deleterious to muscle function.
We undertook an analogous approach to answer the question of whether mitochondrial fusion occurs in and is essential to the normal functioning of hearts. The first examination of mitochondrial fusion in working heart used the same Drosophila MARF RNAi and Opa1-RNAi pUAS transgenic lines described above for the mitochondrial studies in skeletal muscle [57], but we suppressed the mitochondrial fusion proteins by driving RNAi expression using a cardiomyocyte-specific tinman promoter (tincΔ4-gal4). The Drosophila tinman gene is analogous to mammalian Nkx2.5 in that it directs cardiomyocyte specification and differentiation; its promoter therefore drives transgene expression in a highly cardiomyocyte-specific manner [63]. By breeding fruit flies carrying the tincΔ4-gal4 driver with either a pUAS-MARF RNAi or a pUAS-OPA1 RNAi, we suppressed mitochondrial fusion specifically in cardiac myocytes of fly heart tubes.
Results from genetically-manipulated Drosophila heart models
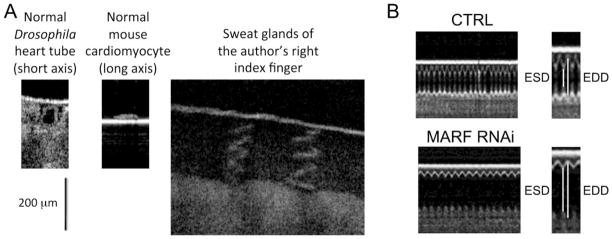
There was little precedent for our use of Drosophila to examine the need for mitochondrial fusion in functioning hearts. The Drosophila heart tube is a linear structure without the complex multi-chamber and valve morphology of vertebrates. Nevertheless, at the cellular and tissue levels Drosophila and mammalian myocardium are strikingly similar, and much has been learned about mammalian heart development and function from research performed in fruit flies [64]. Accordingly, we further miniaturized the same types of procedures routinely used to evaluate cardiac phenotypes in genetic mouse models (which were in turn adapted from clinical evaluation of human cardiomyopathy). Experimental endpoints in mouse cardiac studies typically include: 1. Non-invasive echocardiographic or pathological assessment of cardiac remodeling assessed as chamber dimension at end diastole (larger end diastolic dimensions show cardiac dilatation and indicate pathological remodeling); 2. An assessment of cardiac pump performance, typically as fractional ventricular shortening (the percent difference in chamber dimension between systole and diastole; a higher value is better and a lower value suggests cardiomyopathy); 3. Histological and/or ultrastructural assessment of myofibril and cardiomyocyte integrity. The Drosophila heart tube has a short axis dimension of ~150 microns, far too small to be resolved by ultrasound. However, real-time laser imaging of contracting Drosophila heart tubes had been described by Rolph Bodmer using optical coherence tomography (OCT) [65], and we found that a commercially available OCT microscope (Michelson Diagnostics, UK) provided excellent images of Drosophila heart tubes with a resolution of 4 microns (Figure 2a). Using OCT we observed that cardiomyocyte-specific suppression of either MARF/dMfn or Opa1 induced dilated cardiomyopathy, defined as increased diastolic chamber dimension and reduced fractional shortening [25] (Figure 2b). We took advantage of the ability to simultaneously drive multiple transgenes in Drosophila to interrogate cardiomyocyte organelle structure with genetically-encoded green fluorescence proteins (GFP) that specifically localized to and decorated mitochondria (mito-GFP) or sarcoplasmic reticulum (SR-GFP). Cardiomyocyte mitochondria in normal heart tubes were rounded, homogenous in size, and arranged in the typical inter-myofibrillar lanes. MARF/dMfn-deficient heart tubes showed evidence of unopposed mitochondrial fission, with proliferation of very small mitochondria and striking heterogeneity of mitochondrial size. Mitochondria of Opa1-deficient heart tubes also showed evidence of fragmentation, but these tiny organelles were accompanied by many abnormally large mitochondria as described in Opa1-null MEFS (likely representing organelles that have undergone outer membrane fusion without inner membrane fusion) [58]. The architecture of calcium-containing cardiomyocyte SR was not altered in the MARF/dMFN or Opa1 RNAi cardiomyocytes. Together, these results indicate that inhibition of mitochondrial fusion in cardiomyocytes by suppression of either the critical OMM or IMM fusion protein is sufficient to induce dilated cardiomyopathy.
Figure 2. Optical coherence tomography of normal and mitochondrial fusion-defective Drosophila hearts.
A. Representative images of a fruit fly heart tube in cross section (left), one isolated mouse cardiac myocyte (middle) and human epidermal sweat glands (right), each acquired under the same conditions. Scale bar is 200 microns. B. Line-scan images of representative Drosophila heart tube contractions, showing chamber diameter as a function of time. Top is control fly and bottom is MARF RNAi fly. End-systolic and end-diastolic dimensions (ESD and EDD) are indicated for one cardiac cycle each to the right.
We tested the translatability of the Drosophila heart tube results by investigating functional overlap between fruit fly MARF/dMfn and the human Mfn proteins. Transgenic Drosophila lines expressing human Mfn1 or Mfn2 were created and crossed onto the cardiomyopathic MARF-RNAi fly. Whereas simple human Mfn overexpression had no detectable effect on normal heart tube dimension or function, expression of either hMfn1 or hMfn2 on the MARF/dMfn RNAi background rescued both the characteristic dilated cardiomyopathy and mitochondrial fragmentation. The human mitofusins did not, however, rescue the cardiomyopathy induced by Opa1-RNAi (unpublished results). These results indicate that mammalian mitofusins perform the same essential function as Drosophila MARF/dMfn in the heart.
Finally, we tested whether the cardiomyopathy induced by unopposed mitochondrial fission might be a consequence of mitochondrial production of toxic ROS. As the mass of a Drosophila heart tube was not sufficient for biochemical analysis of cardiomyocyte mitochondrial ROS production, we again used a genetic approach by expressing superoxide dismutase (SOD1) in the MARF-RNAi fly. We hypothesized that SOD1 would rescue phenotypes induced by ROS, but would not affect phenotypes induced by other factors, such as ATP deficiency produced by loss of normal mitochondria. Indeed, the dilated cardiomyopathy conferred by MARF suppression was almost completely normalized by concomitant expression of SOD1, whereas mitochondrial fragmentation was incompletely rescued. This result suggests that the cardiomyopathy caused by disrupting mitochondrial fusion in cardiomyocytes results in part from mitochondrial production of toxic ROS, but that ROS are not the (only) cause of mitochondrial fragmentation.
Results from genetically-manipulated mouse cardiac models
To extend our Drosophila studies to mammals and perform a more detailed mechanistic interrogation of cardiac mitochondrial fusion, we performed analogous genetic manipulations (i.e. cardiac-specific ablation of the genes encoding the OMM fusion proteins, Mfn1 and Mfn2) in mice [26]. These studies used tissue-specific Cre- recombination of the floxed allele Mfn1 and Mfn2 mice created in David Chan’s laboratory [47]. Ablating Mfn1 and Mfn2 genes with an Nkx2.5 Cre-driver [66], which has the transcriptional characteristics of the tinman Gal4 driver used in our Drosophila studies (e.g. cardiomyocyte-specific expression beginning very early in heart development), induced 100% lethality at approximately embryonic day 10.5. This result proves that cardiomyocyte mitochondrial fusion is essential to normal cardiac development, but unfortunately does not address the issue of whether mitochondrial fusion is equally important for normal day-to-day cardiac function in adult mammals. To answer this question, and to obtain sufficient Mfn1/Mfn2 double knockout mouse myocardium for functional, cellular, and ultrastructural analyses, we moved to a compound genetic and pharmacological approach. The two Mfn flox allele mice were crossed, and the double flox allele mice were combined with a cardiomyocyte-specific Cre transgene that expresses a modified estrogen receptor (MER)-Cre chimeric protein only after birth (Myh6-MER-Cre-MER; [67]), thereby avoiding any developmental effects. The MER moieties retain the transgenic Cre in the cytosol until activated by synthetic estrogen receptor ligands such as tamoxifen or Raloxifene that induce nuclear translocation. Recombination of floxed alleles is therefore specifically induced in cardiac myocytes after systemic administration of one or the other of these two drugs. We found that mice carrying four floxed Mfn alleles (two for Mfn1 and two for Mfn2) and expressing the MER-Cre-MER transgene were normal at least up to 20 weeks of age. Administration of tamoxifen or Raloxifene at 8 weeks of age induced complete cardiac-specific recombination of a ROSA-26 lacZ reporter within 7 days; loss of Mfn1 and Mfn2 immunoreactivity in myocardial samples followed the same time course. The mitochondrial fission protein Drp1/Dlp1 was not affected by Mfn1/Mfn2 ablation, although immunoreactivity of the IMM fusion protein Opa1 increased slightly. Thus, we created a population of healthy adult mice in which we could interrupt the expression both OMM fusion proteins at a time and under conditions optimal for our investigations.
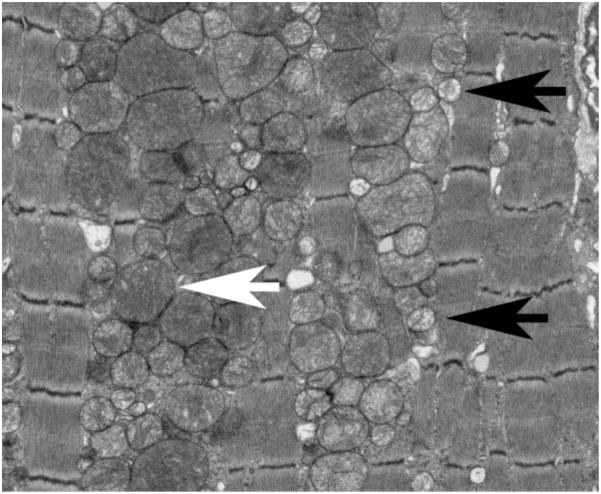
Achieving the conditional cardiac Mfn1/Mfn2 double knockout mice permitted us to ask “What are the consequences of disrupting mitochondrial fusion in situ on cardiomyocyte mitochondria, on isolated cardiac myocytes, and on functioning of the integrated organ/organism?” We started small, at the level of the mitochondria. Ultrastructural examination revealed severe mitochondrial fragmentation and apparent mitochondrial proliferation 3 weeks after induced combined Mfn gene ablation (Figure 3). Approximately 30% of the mitochondria appeared normal in size and crista structure (Figure 3, white arrow), but the majority of organelles were unusually small with loss of the typical matrix membrane structure, manifested as varying degrees of increased transparency (Figure 3, black arrow). The overall density of mitochondria was also increased. By comparison, sarcomeric and myofibrillar structures appeared healthy in the same cells, consistent with a primary defect in mitochondrial dynamics. The impressions obtained by transmission electron microscopic visualization of a few thousand mitochondria from representative hearts were confirmed by flow cytometric analysis of 30,000 isolated cardiac mitochondria/heart. Forward scatter (directly proportional to organelle size) was decreased ~40% and side scatter (inversely proportional to organelle sphericity) was decreased by ~70%. Mitochondrial content measured as mitochondrial protein/heart mass was increased ~60% in the fusion-defective hearts, indicating mitochondrial proliferation [26].
Figure 3. Mitochondrial fragmentation in mouse hearts with defective mitochondrial fusion.
Transmission electron micrograph of Mfn1/Mfn2 double null mouse myocardium 3 weeks after conditional gene ablation with tamoxifen. White arrow indicates a normal appearing mitochondrion. Black arrows point to representative small and translucent “fragmented” mitochondria.
The mitochondrial morphometric abnormalities observed three weeks after conditional combined Mfn1 and Mfn2 ablation did not obviously adversely impact the mice, which appeared outwardly normal. However, non-invasive echocardiographic assessment of ventricular size and contractility revealed chamber dilatation and decreased ejection performance, suggesting subclinical cardiac dysfunction. We therefore examined respiratory function of cardiomyocytes and mitochondria isolated from these hearts, compared to identically treated controls: Succinate-stimulated whole cell respiration was only modestly depressed, and both succinate and glutamate/malate-stimulated isolated mitochondrial respiration were normal (no difference in state3/state2 respiration between knockout and control mitochondria). However, uncoupling mitochondrial respiration from ATP synthesis with FCCP (think of pressing the gas pedal for respiration all the way to the floor) uncovered defective peak respiration in Mfn1/Mfn2 double knockout cardiac mitochondria. Taken together, these morphometric and functional studies suggested that mitochondrial fragmentation and degeneration induced by three weeks of combined Mfn1/Mfn2 deficiency in otherwise normal adult mice compromises peak respiration, despite mitochondrial proliferation that might be anticipated to compensate for individual organelle dysfunction.
MER-Cre-MER-mediated cardiomyocyte gene recombination induced by tamoxifen is both rapid and irreversible. Therefore, one would expect the consequences of conditional combined Mfn1 and Mfn2 ablation to progress. Indeed, all Mfn1/Mfn2 double cardiac knockout mice died of apparent heart failure approximately 9 weeks after tamoxifen treatment. Serial weekly echocardiographic studies showed progressive cardiac enlargement with a parallel decrease in ejection performance beginning the second week after gene ablation, reaching levels that are not compatible with life after 6 weeks in most mice. This disease was similar to the dilated cardiomyopathy induced by MAR/dMFN suppression in Drosophila [25]. Thus, we conclude that mitochondrial fusion is essential in mammals not only for cardiac development, but also for normal mitochondrial and cardiac function.
One of the collateral benefits of temporally-defined tissue-specific gene expression is the ability to interrupt cyclical phenomena such as mitochondrial fusion/fission at equilibrium and observe the time course of the resulting unidirectional function to calculate half-time and cycle length for the phenomenon at steady-state. This is like a pulse-chase study using in vivo gene ablation to start the clock. We observe that mean mitochondrial size decreased by ~40% two weeks after Mfn1 and Mfn2 were completely ablated from mouse hearts. If one assumes that the rate of mitochondrial fission was not affected by ablating the OMM fusion proteins, and that a fission event produces two equally sized daughter organelles from the parent, then the entire mitochondrial population had undergone (unopposed) fission just over two weeks after Mfn1/Mfn2 ablation. A two week fission/fusion cycle is sufficiently long that it explains why direct observation of mitochondrial fission and fusion in cultured cardiac myocytes over a period of hours fails to detect these events [23].
Defective mitochondrial fusion/fission in genetic heart disease
As introduced above, human disease is linked to two naturally occurring loss-of-function mutations in mitochondrial fusion proteins, Charcot Marie Tooth Syndrome type 2A (CMT) caused by Mfn2 mutations and Autosomal Dominant Optic Atrophy (DOA) caused by Opa1 mutations. Mitochondrial morphological abnormalities in the respective affected tissues are characteristic of both syndromes and are consistent with impaired mitochondrial fusion. Two features of these human diseases are notable in the current context: First, the mutations are typically heterozygous, suggesting that pathology is induced either through dominant inhibition (Mfn2) or haploinsufficiency (Opa1). And second, notwithstanding the ubiquity of mitochondria and these two mitochondrial proteins, the mutations selectively affect specific tissue types (neurons and the retina, respectively), apparently sparing all others. It is notable that there are no convincing data suggesting that the heart is compromised in either CMT or in DOA. The absence of cardiac involvement in two diseases caused by defects in mitochondrial fusion seems to support the idea that mitochondrial fusion and fission are dispensable for normal heart function. There may be other reasons, however, to explain why the heart seems to be spared in CMT and DOA, such as different fusion protein expression levels between neurological and cardiac tissue, variable expression of functionally overlapping proteins (i.e. Mfn1), and different roles of mitochondrial fusion in neurons versus striated muscle.
Mutations causing human degenerative neurological diseases have not suggested that defective mitochondrial fusion/fission is essential to cardiac health, but genetic studies of dilated cardiomyopathy in the animal kingdom tell a very different story: Bovine dilated cardiomyopathy (BDCMP) is a heritable (autosomal recessive) form of lethal heart failure that afflicts young Holstein-Friesian cattle [68–70]. Linkage analysis within affected bovine pedigrees mapped the disease locus to chromosome 18 [71]. Subsequent fine SNP mapping and deep resequencing identified a mutation that perfectly segregated with cardiomyopathy phenotype; the mutation encodes a premature termination codon (Q115X) in the bovine Opa3 mitochondrial fusion factor [72]. Functional studies uncovered decreased myocardial expression of the mutant Opa3 transcript as a result of nonsense-mediated decay, and immunoblot analysis of isolated mitochondria from affected cattle revealed loss of one of the two normal Opa3 isoforms. Serendipitously, human mutations in Opa3 had been implicated in rare Costeff optic atrophy syndrome [73] and so an ENU-induced mouse Opa3 mutation model has been carefully phenotyped. Mice homozygous for the Opa3 (L122P) mutation develop (in addition to the optic and metabolic features of Costeff syndrome) biventricular dilatation, cardiomyocyte hypertrophy, and myocardial fibrosis. These mice succumb at approximately 4 months of age with heart failure [74]. Thus, Opa3 mutations in cows and mice cause cardiomyopathy.
A second, purely experimental form of dilated cardiomyopathy further supports an essential role for mitochondrial fusion/fission in the mammalian heart. Here, mouse ENU mutagenesis induced autosomal dominant dilated cardiomyopathy in a mutant line called Python, and the responsible missense mutation (C452F) was identified in the mitochondrial fission protein gene, Dnm1l [75]. Yeast two-hybrid studies showed that the mutation impairs critical intra-molecular interactions; functional studies revealed decreased mitochondrial fission and altered myocardial metabolism. Taken together, these examples of inherited cardiomyopathy prove that mutational alterations affecting mitochondrial fusion/fission can adversely impact the heart
Abnormal mitochondrial fusion/fission in acquired heart disease
Mitochondrial fission occurs in, and facilitates programmed cell death from, apoptosis [76, 77]. Programmed cardiomyocyte loss plays a significant role in the progression from compensated hypertrophy to overt heart failure [78–81] and in delayed tissue loss after myocardial infarction [82]. Furthermore, failing (rat and human) hearts exhibit mitochondrial fragmentation associated with decreased expression of the IMM fusion protein, Opa1 [83]. Based on the links between mitochondrial fission and programmed cardiomyocyte death, it is not surprising that there has been great interest in targeting mitochondrial fission as a potential mediator of cardiac disease. Conceptually, there are two approaches to changing mitochondrial fusion/fission dynamics in the heart: Enhance fusion or inhibit fission.
The first attempt to protect cardiomyocytes from injury by enhancing mitochondrial fusion overexpressed Opa1 in cultured H9c2 cardiomyocytes, and then assessed apoptosis after simulated ischemia-reperfusion [83]. Although mitochondrial morphometry changed in a manner that indicated a shift in the balance toward more mitochondrial fusion, there was no associated protection from ischemia-induced apoptosis.
Two groups have tried to protect hearts from ischemia using the reciprocal approach of inhibiting mitochondrial fission. The strongest evidence to date supporting a causative or contributory role for increased mitochondrial fission in cardiac disease was provided by Ong, et al who used pharmacological antagonism of the mitochondrial fission protein Drp1 with a small molecule inhibitor, mdivi-1 (mitochondrial division inhibitor-1; [84]), to rescue mouse hearts from ischemic injury [85]. In vivo mdivi-1 treatment prior to ischemia increased mitochondrial length (i.e. it attenuated fission) and reduced myocardial infarction size by over half, compared to vehicle-treated controls. Direct in vivo measurements of cardiomyocyte apoptosis and programmed necrosis were not part of these experiments and it is possible that the large systemic intravenous doses of mdivi-1 administered (1.2 mg/kg) might have produced some effect in addition to inhibiting mitochondrial fission. However, these results strongly support a connection between mitochondrial dynamics and cardiac resistance to injury.
Another recent report described beneficial effects of mitochondrial fission inhibition induced (indirectly) by a cardiac microRNA (miR) [86]. Here the researchers were studying transgenic mice overexpressing a muscle-specific miR, miR-499, previously recognized primarily for regulating myosin isoforms in cardiac hypertrophy [87]. They found that increased miR-499 had few effects on normal hearts, but that it protected against cardiomyocyte apoptosis, myocardial infarction, and post-ischemic ventricular remodeling. They had observed Drp1 dephosphorylation and mitochondrial fission during cardiomyocyte apoptosis, and it was previously reported that calcineurin-modulated Drp1 phosphorylation can regulate mitochondrial fission [88]. The authors therefore used bioinformatics to identify isoforms of the phosphatase calcineurin A as putative miR-499 targets. They concluded that miR-499 downregulates calcineurin, which decreases dephosphorylated Drp1, thus preventing mitochondrial fission and protected the heart. This rather complex chain of causality is typical for microRNAs, which have multiple primary, secondary, and tertiary effects on multiple direct and indirect targets [89]. However, this complexity also offers the possibility that other factors may be influencing the outcome. Indeed, two other groups have expressed miR-499 in mouse hearts and found that, rather then being protected, they are predisposed to heart failure [90, 91]. Also, calcineurin A has not yet been confirmed as a valid in vitro target of miR-499 in myocardium despite attempts to do so using genome-wide deep resequencing approaches [91]. Thus, the possible collective impact of miR-499, calcineurin A, and phosphorylated/dephosphorylated Drp1 on mitochondrial fission and cardiomyocyte apoptosis is unclear at this time.
Summary
The cumulative results from studies described above prove that mitochondrial fusion/fission is essential to normal mitochondrial homeostasis and necessary for normal cardiac function. Recent reports have specifically implicated loss of mitochondrial fusion as the mechanism for the cardiomyopathies induced by mutation or ablation of mammalian mitofusins and Drosophila MARF/dMfn. These initial findings represent just the first steps in what will certainly be a much longer journey toward a more complete understanding of the roles of mitochondrial fusion and fission proteins play in the related functions of cardiac bioenergetics, calcium signaling, mitochondrial quality control, and biogenesis. The field appears well positioned to develop greater understanding of these specific areas, and I believe an information and insight explosion is just over the horizon.
Highlights.
Mitochondrial fusion/fission has not been observed in normal adult cardiomyocytes (84)
Genetic inhibition of mitochondrial fusion causes fragmentation and heart failure (84)
Thus, mitochondrial fusion/fission is essential to normal cardiac health (75)
Mitochondrial fission is associated with programmed cardiomyocyte death (74)
Inhibiting mitochondrial fission may protect against cardiac injury (71)
Footnotes
The author declares that he has no conflicts of interest relating to this manuscript
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ferrari R, Censi S, Mastrorilli F, Boraso A. Prognostic benefits of heart rate reduction in cardiovascular disease. Eur Heart J Suppl. 2003;5:G10–G14. [Google Scholar]
- 2.Page E, McCallister LP. Quantitative electron microscopic description of heart muscle cells. Application to normal, hypertrophied and thyroxin-stimulated hearts. Am J Cardiol. 1973;31:172–181. doi: 10.1016/0002-9149(73)91030-8. [DOI] [PubMed] [Google Scholar]
- 3.Lardy HA, Wellman H. Oxidative phosphorylations; role of inorganic phosphate and acceptor systems in control of metabolic rates. J Biol Chem. 1952;195:215–224. [PubMed] [Google Scholar]
- 4.Balaban RS, Kantor HL, Katz LA, Briggs RW. Relation between work and phosphate metabolite in the in vivo paced mammalian heart. Science. 1986;232:1121–1123. doi: 10.1126/science.3704638. [DOI] [PubMed] [Google Scholar]
- 5.Katz LA, Swain JA, Portman MA, Balaban RS. Relation between phosphate metabolites and oxygen consumption of heart in vivo. Am J Physiol. 1989;256:H265–274. doi: 10.1152/ajpheart.1989.256.1.H265. [DOI] [PubMed] [Google Scholar]
- 6.Cortassa S, Aon MA, Marban E, Winslow RL, O'Rourke B. An integrated model of cardiac mitochondrial energy metabolism and calcium dynamics. Biophys J. 2003;84:2734–2755. doi: 10.1016/S0006-3495(03)75079-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maack C, Cortassa S, Aon MA, Ganesan AN, Liu T, O'Rourke B. Elevated cytosolic Na+ decreases mitochondrial Ca2+ uptake during excitation-contraction coupling and impairs energetic adaptation in cardiac myocytes. Circ Res. 2006;99:172–182. doi: 10.1161/01.RES.0000232546.92777.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cortassa S, Aon MA, O'Rourke B, Jacques R, Tseng HJ, Marban E, Winslow RL. A computational model integrating electrophysiology, contraction, and mitochondrial bioenergetics in the ventricular myocyte. Biophys J. 2006;91:1564–1589. doi: 10.1529/biophysj.105.076174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bers DM. Calcium cycling and signaling in cardiac myocytes. Annu Rev Physiol. 2008;70:23–49. doi: 10.1146/annurev.physiol.70.113006.100455. [DOI] [PubMed] [Google Scholar]
- 10.Bers DM, Despa S. Cardiac myocytes Ca2+ and Na+ regulation in normal and failing hearts. J Pharmacol Sci. 2006;100:315–322. doi: 10.1254/jphs.cpj06001x. [DOI] [PubMed] [Google Scholar]
- 11.Carafoli E. Calcium pump of the plasma membrane. Physiol Rev. 1991;71:129–153. doi: 10.1152/physrev.1991.71.1.129. [DOI] [PubMed] [Google Scholar]
- 12.Ruknudin AM, Lakatta EG. The regulation of the Na/Ca exchanger and plasmalemmal Ca2+ ATPase by other proteins. Ann N Y Acad Sci. 2007;1099:86–102. doi: 10.1196/annals.1387.045. [DOI] [PubMed] [Google Scholar]
- 13.Shull GE, Okunade G, Liu LH, Kozel P, Periasamy M, Lorenz JN, Prasad V. Physiological functions of plasma membrane and intracellular Ca2+ pumps revealed by analysis of null mutants. Ann N Y Acad Sci. 2003;986:453–460. doi: 10.1111/j.1749-6632.2003.tb07229.x. [DOI] [PubMed] [Google Scholar]
- 14.Maack C, O'Rourke B. Excitation-contraction coupling and mitochondrial energetics. Basic Res Cardiol. 2007;102:369–392. doi: 10.1007/s00395-007-0666-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reynolds IJ, Rintoul GL. Mitochondrial stop and go: signals that regulate organelle movement. Sci STKE. 2004;2004:PE46. doi: 10.1126/stke.2512004pe46. [DOI] [PubMed] [Google Scholar]
- 16.Westermann B. Mitochondrial fusion and fission in cell life and death. Nat Rev Mol Cell Biol. 2010;11:872–884. doi: 10.1038/nrm3013. [DOI] [PubMed] [Google Scholar]
- 17.Archer SL. The mitochondrion as a Swiss army knife: implications for cardiovascular disease. J Mol Med. 2010;88:963–965. doi: 10.1007/s00109-010-0665-7. [DOI] [PubMed] [Google Scholar]
- 18.Goodpasture EW. Observations on Mitochondria of Tumors. J Med Res. 1918;38:213–224. 211. [PMC free article] [PubMed] [Google Scholar]
- 19.Lewis MR, Lewis WH. Mitochondria in Tissue Culture. Science. 1914;39:330–333. doi: 10.1126/science.39.1000.330. [DOI] [PubMed] [Google Scholar]
- 20.Brady NR, Hamacher-Brady A, Gottlieb RA. Proapoptotic BCL-2 family members and mitochondrial dysfunction during ischemia/reperfusion injury, a study employing cardiac HL-1 cells and GFP biosensors. Biochim Biophys Acta. 2006;1757:667–678. doi: 10.1016/j.bbabio.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 21.Yu T, Sheu SS, Robotham JL, Yoon Y. Mitochondrial fission mediates high glucose-induced cell death through elevated production of reactive oxygen species. Cardiovasc Res. 2008;79:341–351. doi: 10.1093/cvr/cvn104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parra V, Eisner V, Chiong M, Criollo A, Moraga F, Garcia A, Hartel S, Jaimovich E, Zorzano A, Hidalgo C, Lavandero S. Changes in mitochondrial dynamics during ceramide-induced cardiomyocyte early apoptosis. Cardiovasc Res. 2008;77:387–397. doi: 10.1093/cvr/cvm029. [DOI] [PubMed] [Google Scholar]
- 23.Beraud N, Pelloux S, Usson Y, Kuznetsov AV, Ronot X, Tourneur Y, Saks V. Mitochondrial dynamics in heart cells: very low amplitude high frequency fluctuations in adult cardiomyocytes and flow motion in non beating Hl-1 cells. J Bioenerg Biomembr. 2009;41:195–214. doi: 10.1007/s10863-009-9214-x. [DOI] [PubMed] [Google Scholar]
- 24.Hom J, Yu T, Yoon Y, Porter G, Sheu SS. Regulation of mitochondrial fission by intracellular Ca2+ in rat ventricular myocytes. Biochim Biophys Acta. 2010;1797:913–921. doi: 10.1016/j.bbabio.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dorn GW, 2nd, Clark CF, Eschenbacher WH, Kang MY, Engelhard JT, Warner SJ, Matkovich SJ, Jowdy CC. MARF and Opa1 control mitochondrial and cardiac function in Drosophila. Circ Res. 2011;108:12–17. doi: 10.1161/CIRCRESAHA.110.236745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y, Liu Y, Dorn GW., 2nd Mitochondrial Fusion is Essential for Organelle Function and Cardiac Homeostasis. Circ Res. 2011;109:1327–1331. doi: 10.1161/CIRCRESAHA.111.258723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishihara N, Nomura M, Jofuku A, Kato H, Suzuki SO, Masuda K, Otera H, Nakanishi Y, Nonaka I, Goto Y, Taguchi N, Morinaga H, Maeda M, Takayanagi R, Yokota S, Mihara K. Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat Cell Biol. 2009;11:958–966. doi: 10.1038/ncb1907. [DOI] [PubMed] [Google Scholar]
- 29.Chen H, Chomyn A, Chan DC. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J Biol Chem. 2005;280:26185–26192. doi: 10.1074/jbc.M503062200. [DOI] [PubMed] [Google Scholar]
- 30.Bibb MJ, Van Etten RA, Wright CT, Walberg MW, Clayton DA. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981;26:167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- 31.Clayton DA. Transcription and replication of mitochondrial DNA. Hum Reprod. 2000;15:11–17. doi: 10.1093/humrep/15.suppl_2.11. [DOI] [PubMed] [Google Scholar]
- 32.Wallace DC, Fan W. The pathophysiology of mitochondrial disease as modeled in the mouse. Genes Dev. 2009;23:1714–1736. doi: 10.1101/gad.1784909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen H, Vermulst M, Wang YE, Chomyn A, Prolla TA, McCaffery JM, Chan DC. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell. 2010;141:280–289. doi: 10.1016/j.cell.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tatsuta T, Langer T. Quality control of mitochondria: protection against neurodegeneration and ageing. EMBO J. 2008;27:306–314. doi: 10.1038/sj.emboj.7601972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, Alroy J, Wu M, Py BF, Yuan J, Deeney JT, Corkey BE, Shirihai OS. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lemasters JJ. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res. 2005;8:3–5. doi: 10.1089/rej.2005.8.3. [DOI] [PubMed] [Google Scholar]
- 37.Green DR, Galluzzi L, Kroemer G. Mitochondria and the autophagy-inflammation-cell death axis in organismal aging. Science. 2011;333:1109–1112. doi: 10.1126/science.1201940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, Lippincott-Schwartz J. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X, Su B, Siedlak SL, Moreira PI, Fujioka H, Wang Y, Casadesus G, Zhu X. Amyloid-beta overproduction causes abnormal mitochondrial dynamics via differential modulation of mitochondrial fission/fusion proteins. Proc Natl Acad Sci USA. 2008;105:19318–19323. doi: 10.1073/pnas.0804871105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gottlieb RA, Gustafsson AB. Mitochondrial turnover in the heart. Biochim Biophys Acta. 2011;1813:1295–1301. doi: 10.1016/j.bbamcr.2010.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gegg ME, Cooper JM, Chau KY, Rojo M, Schapira AH, Taanman JW. Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Hum Mol Genet. 2010;19:4861–4870. doi: 10.1093/hmg/ddq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rakovic A, Grunewald A, Kottwitz J, Bruggemann N, Pramstaller PP, Lohmann K, Klein C. Mutations in PINK1 and Parkin impair ubiquitination of Mitofusins in human fibroblasts. PLos One. 2011;6:e16746. doi: 10.1371/journal.pone.0016746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zungu M, Schisler J, Willis MS. All the little pieces. -Regulation of mitochondrial fusion and fission by ubiquitin and small ubiquitin-like modifer and their potential relevance in the heart. Circ J. 2011;75:2513–2521. doi: 10.1253/circj.cj-11-0967. [DOI] [PubMed] [Google Scholar]
- 44.Meeusen S, McCaffery JM, Nunnari J. Mitochondrial fusion intermediates revealed in vitro. Science. 2004;305:1747–1752. doi: 10.1126/science.1100612. [DOI] [PubMed] [Google Scholar]
- 45.Malka F, Guillery O, Cifuentes-Diaz C, Guillou E, Belenguer P, Lombes A, Rojo M. Separate fusion of outer and inner mitochondrial membranes. EMBO Rep. 2005;6:853–859. doi: 10.1038/sj.embor.7400488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koshiba T, Detmer SA, Kaiser JT, Chen H, McCaffery JM, Chan DC. Structural basis of mitochondrial tethering by mitofusin complexes. Science. 2004;305:858–862. doi: 10.1126/science.1099793. [DOI] [PubMed] [Google Scholar]
- 47.Chen H, McCaffery JM, Chan DC. Mitochondrial fusion protects against neurodegeneration in the cerebellum. Cell. 2007;130:548–562. doi: 10.1016/j.cell.2007.06.026. [DOI] [PubMed] [Google Scholar]
- 48.Zuchner S, Mersiyanova IV, Muglia M, Bissar-Tadmouri N, Rochelle J, Dadali EL, Zappia M, Nelis E, Patitucci A, Senderek J, Parman Y, Evgrafov O, Jonghe PD, Takahashi Y, Tsuji S, Pericak-Vance MA, Quattrone A, Battaloglu E, Polyakov AV, Timmerman V, Schroder JM, Vance JM. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat Genet. 2004;36:449–451. doi: 10.1038/ng1341. [DOI] [PubMed] [Google Scholar]
- 49.Kijima K, Numakura C, Izumino H, Umetsu K, Nezu A, Shiiki T, Ogawa M, Ishizaki Y, Kitamura T, Shozawa Y, Hayasaka K. Mitochondrial GTPase mitofusin 2 mutation in Charcot-Marie-Tooth neuropathy type 2A. Hum Genet. 2005;116:23–27. doi: 10.1007/s00439-004-1199-2. [DOI] [PubMed] [Google Scholar]
- 50.Verhoeven K, Claeys KG, Zuchner S, Schroder JM, Weis J, Ceuterick C, Jordanova A, Nelis E, De Vriendt E, Van Hul M, Seeman P, Mazanec R, Saifi GM, Szigeti K, Mancias P, Butler IJ, Kochanski A, Ryniewicz B, De Bleecker J, Van den Bergh P, Verellen C, Van Coster R, Goemans N, Auer-Grumbach M, Robberecht W, Milic Rasic V, Nevo Y, Tournev I, Guergueltcheva V, Roelens F, Vieregge P, Vinci P, Moreno MT, Christen HJ, Shy ME, Lupski JR, Vance JM, De Jonghe P, Timmerman V. MFN2 mutation distribution and genotype/phenotype correlation in Charcot-Marie-Tooth type 2. Brain. 2006;129:2093–2102. doi: 10.1093/brain/awl126. [DOI] [PubMed] [Google Scholar]
- 51.Engelfried K, Vorgerd M, Hagedorn M, Haas G, Gilles J, Epplen JT, Meins M. Charcot-Marie-Tooth neuropathy type 2A: novel mutations in the mitofusin 2 gene (MFN2) BMC Med Genet. 2006;7:53. doi: 10.1186/1471-2350-7-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McCorquodale DS, 3rd, Montenegro G, Peguero A, Carlson N, Speziani F, Price J, Taylor SW, Melanson M, Vance JM, Zuchner S. Mutation screening of mitofusin 2 in Charcot-Marie-Tooth disease type 2. J Neurol. 2011;258:1234–1239. doi: 10.1007/s00415-011-5910-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Griparic L, van der Wel NN, Orozco IJ, Peters PJ, van der Bliek AM. Loss of the intermembrane space protein Mgm1/OPA1 induces swelling and localized constrictions along the lengths of mitochondria. J Biol Chem. 2004;279:18792–18798. doi: 10.1074/jbc.M400920200. [DOI] [PubMed] [Google Scholar]
- 54.Cipolat S, Martins de Brito O, Dal Zilio B, Scorrano L. OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc Natl Acad Sci USA. 2004;101:15927–15932. doi: 10.1073/pnas.0407043101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Delettre C, Lenaers G, Griffoin JM, Gigarel N, Lorenzo C, Belenguer P, Pelloquin L, Grosgeorge J, Turc-Carel C, Perret E, Astarie-Dequeker C, Lasquellec L, Arnaud B, Ducommun B, Kaplan J, Hamel CP. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat Genet. 2000;26:207–210. doi: 10.1038/79936. [DOI] [PubMed] [Google Scholar]
- 56.Frezza C, Cipolat S, Martins de Brito O, Micaroni M, Beznoussenko GV, Rudka T, Bartoli D, Polishuck RS, Danial NN, De Strooper B, Scorrano L. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell. 2006;126:177–189. doi: 10.1016/j.cell.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 57.Deng H, Dodson MW, Huang H, Guo M. The Parkinson's disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proc Natl Acad Sci USA. 2008;105:14503–14508. doi: 10.1073/pnas.0803998105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Song Z, Ghochani M, McCaffery JM, Frey TG, Chan DC. Mitofusins and OPA1 mediate sequential steps in mitochondrial membrane fusion. Mol Biol Cell. 2009;20:3525–3532. doi: 10.1091/mbc.E09-03-0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smirnova E, Shurland DL, Ryazantsev SN, van der Bliek AM. A human dynamin-related protein controls the distribution of mitochondria. J Cell Biol. 1998;143:351–358. doi: 10.1083/jcb.143.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mozdy AD, McCaffery JM, Shaw JM. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J Cell Biol. 2000;151:367–380. doi: 10.1083/jcb.151.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Urrutia R, Henley JR, Cook T, McNiven MA. The dynamins: redundant or distinct functions for an expanding family of related GTPases? Proc Natl Acad Sci USA. 1997;94:377–384. doi: 10.1073/pnas.94.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lackner LL, Horner JS, Nunnari J. Mechanistic analysis of a dynamin effector. Science. 2009;325:874–877. doi: 10.1126/science.1176921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lo PC, Frasch M. A role for the COUP-TF-related gene seven-up in the diversification of cardioblast identities in the dorsal vessel of Drosophila. Mech Dev. 2001;104:49–60. doi: 10.1016/s0925-4773(01)00361-6. [DOI] [PubMed] [Google Scholar]
- 64.Bier E, Bodmer R. Drosophila, an emerging model for cardiac disease. Gene. 2004;342:1–11. doi: 10.1016/j.gene.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 65.Choma MA, Izatt SD, Wessells RJ, Bodmer R, Izatt JA. Images in cardiovascular medicine: in vivo imaging of the adult Drosophila melanogaster heart with real-time optical coherence tomography. Circulation. 2006;114:e35–36. doi: 10.1161/CIRCULATIONAHA.105.593541. [DOI] [PubMed] [Google Scholar]
- 66.Moses KA, DeMayo F, Braun RM, Reecy JL, Schwartz RJ. Embryonic expression of an Nkx2-5/Cre gene using ROSA26 reporter mice. Genesis. 2001;31:176–180. doi: 10.1002/gene.10022. [DOI] [PubMed] [Google Scholar]
- 67.Sohal DS, Nghiem M, Crackower MA, Witt SA, Kimball TR, Tymitz KM, Penninger JM, Molkentin JD. Temporally regulated and tissue-specific gene manipulations in the adult and embryonic heart using a tamoxifen-inducible Cre protein. Circ Res. 2001;89:20–25. doi: 10.1161/hh1301.092687. [DOI] [PubMed] [Google Scholar]
- 68.Baird JD, Maxie MG, Kennedy BW, Harris DJ. Dilated (congestive) cardiomyopathy in Holstein cattle in Canada: genetic analysis of 25 cases. Proceedings of the 14th World Congress on Diseases of Cattle; Dublin. 1986. pp. 89–94. [Google Scholar]
- 69.Dolf G, Stricker C, Tontis A, Martig J, Gaillard C. Evidence for autosomal recessive inheritance of a major gene for bovine dilated cardiomyopathy. J Anim Sci. 1998;76:1824–1829. doi: 10.2527/1998.7671824x. [DOI] [PubMed] [Google Scholar]
- 70.Konig F, Zwahlen R, Schaller J, Kampfer U, Roth D, Tontis A, Luginbuhl H. Bovine cardiomyopathy, pathomorphogenic and biochemical studies in yearling steers. Schweiz Arch Tierheilkd. 1990;132:439–440. [Google Scholar]
- 71.Owczarek-Lipska M, Denis C, Eggen A, Leeb T, Posthaus H, Dolf G, Braunschweig MH. The bovine dilated cardiomyopathy locus maps to a 1.0-Mb interval on chromosome 18. Mamm Genome. 2009;20:187–192. doi: 10.1007/s00335-009-9171-z. [DOI] [PubMed] [Google Scholar]
- 72.Owczarek-Lipska M, Plattet P, Zipperle L, Drogemuller C, Posthaus H, Dolf G, Braunschweig MH. A nonsense mutation in the optic atrophy 3 gene (OPA3) causes dilated cardiomyopathy in Red Holstein cattle. Genomics. 2011;97:51–57. doi: 10.1016/j.ygeno.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 73.Anikster Y, Kleta R, Shaag A, Gahl WA, Elpeleg O. Type III 3-methylglutaconic aciduria (optic atrophy plus syndrome, or Costeff optic atrophy syndrome): identification of the OPA3 gene and its founder mutation in Iraqi Jews. Am J Hum Genet. 2001;69:1218–1224. doi: 10.1086/324651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Davies VJ, Powell KA, White KE, Yip W, Hogan V, Hollins AJ, Davies JR, Piechota M, Brownstein DG, Moat SJ, Nichols PP, Wride MA, Boulton ME, Votruba M. A missense mutation in the murine Opa3 gene models human Costeff syndrome. Brain. 2008;131:368–380. doi: 10.1093/brain/awm333. [DOI] [PubMed] [Google Scholar]
- 75.Ashrafian H, Docherty L, Leo V, Towlson C, Neilan M, Steeples V, Lygate CA, Hough T, Townsend S, Williams D, Wells S, Norris D, Glyn-Jones S, Land J, Barbaric I, Lalanne Z, Denny P, Szumska D, Bhattacharya S, Griffin JL, Hargreaves I, Fernandez-Fuentes N, Cheeseman M, Watkins H, Dear TN. A mutation in the mitochondrial fission gene Dnm1l leads to cardiomyopathy. PLoS genetics. 2010;6:e1001000. doi: 10.1371/journal.pgen.1001000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Braschi E, McBride HM. Mitochondria and the culture of the Borg: understanding the integration of mitochondrial function within the reticulum, the cell, and the organism. BioEssays. 2010;32:958–966. doi: 10.1002/bies.201000073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Martinou JC, Youle RJ. Mitochondria in apoptosis: Bcl-2 family members and mitochondrial dynamics. Dev Cell. 2011;21:92–101. doi: 10.1016/j.devcel.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Adams JW, Sakata Y, Davis MG, Sah VP, Wang Y, Liggett SB, Chien KR, Brown JH, Dorn GW., 2nd Enhanced Galphaq signaling: a common pathway mediates cardiac hypertrophy and apoptotic heart failure. Proc Natl Acad Sci USA. 1998;95:10140–10145. doi: 10.1073/pnas.95.17.10140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yussman MG, Toyokawa T, Odley A, Lynch RA, Wu G, Colbert MC, Aronow BJ, Lorenz JN, Dorn GW., 2nd Mitochondrial death protein Nix is induced in cardiac hypertrophy and triggers apoptotic cardiomyopathy. Nat Med. 2002;8:725–730. doi: 10.1038/nm719. [DOI] [PubMed] [Google Scholar]
- 80.Diwan A, Wansapura J, Syed FM, Matkovich SJ, Lorenz JN, Dorn GW., 2nd Nix-mediated apoptosis links myocardial fibrosis, cardiac remodeling, and hypertrophy decompensation. Circulation. 2008;117:396–404. doi: 10.1161/CIRCULATIONAHA.107.727073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen Y, Lewis W, Diwan A, Cheng EH, Matkovich SJ, Dorn GW., 2nd Dual autonomous mitochondrial cell death pathways are activated by Nix/BNip3L and induce cardiomyopathy. Proc Natl Acad Sci USA. 2010;107:9035–9042. doi: 10.1073/pnas.0914013107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Diwan A, Krenz M, Syed FM, Wansapura J, Ren X, Koesters AG, Li H, Kirshenbaum LA, Hahn HS, Robbins J, Jones WK, Dorn GW., 2nd Inhibition of ischemic cardiomyocyte apoptosis through targeted ablation of Bnip3 restrains postinfarction remodeling in mice. J Clin Invest. 2007;117:2825–2833. doi: 10.1172/JCI32490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen L, Gong Q, Stice JP, Knowlton AA. Mitochondrial OPA1, apoptosis, and heart failure. Cardiovasc Res. 2009;84:91–99. doi: 10.1093/cvr/cvp181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cassidy-Stone A, Chipuk JE, Ingerman E, Song C, Yoo C, Kuwana T, Kurth MJ, Shaw JT, Hinshaw JE, Green DR, Nunnari J. Chemical inhibition of the mitochondrial division dynamin reveals its role in Bax/Bak-dependent mitochondrial outer membrane permeabilization. Dev Cell. 2008;14:193–204. doi: 10.1016/j.devcel.2007.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ong SB, Subrayan S, Lim SY, Yellon DM, Davidson SM, Hausenloy DJ. Inhibiting mitochondrial fission protects the heart against ischemia/reperfusion injury. Circulation. 2010;121:2012–2022. doi: 10.1161/CIRCULATIONAHA.109.906610. [DOI] [PubMed] [Google Scholar]
- 86.Wang JX, Jiao JQ, Li Q, Long B, Wang K, Liu JP, Li YR, Li PF. miR-499 regulates mitochondrial dynamics by targeting calcineurin and dynamin-related protein-1. Nat Med. 2011;17:71–78. doi: 10.1038/nm.2282. [DOI] [PubMed] [Google Scholar]
- 87.van Rooij E, Quiat D, Johnson BA, Sutherland LB, Qi X, Richardson JA, Kelm RJ, Jr, Olson EN. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell. 2009;17:662–673. doi: 10.1016/j.devcel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cribbs JT, Strack S. Reversible phosphorylation of Drp1 by cyclic AMP-dependent protein kinase and calcineurin regulates mitochondrial fission and cell death. EMBO Rep. 2007;8:939–944. doi: 10.1038/sj.embor.7401062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dorn GW., 2nd Decoding the Cardiac Message. Circ Res. 2012 doi: 10.1161/CIRCRESAHA.111.256768. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shieh JT, Huang Y, Gilmore J, Srivastava D. Elevated miR-499 levels blunt the cardiac stress response. PLos One. 2011;6:e19481. doi: 10.1371/journal.pone.0019481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dorn GW, 2nd, Matkovich SJ, Eschenbacher WH, Zhang Y. A human 3' miR-499 mutation alters cardiac mRNA targeting and function. Circ Res. 2012 doi: 10.1161/CIRCRESAHA.111.260752. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]